Abstract
A comparison between the BD MAX MRSA and Xpert MRSA assays was performed using 239 nares samples. A 97.9% overall agreement between the two molecular assays was observed. The BD MAX MRSA assay proved to be a reliable alternative for a highly automated system to detect methicillin-resistant Staphylococcus aureus (MRSA) in patient nares samples.
TEXT
Methicillin-resistant Staphylococcus aureus (MRSA) is an important cause of community and hospital-acquired infections (1, 2). Screening for MRSA colonization to help identify patients at greater risk for MRSA infection and to allow for initiation of isolation precautions has become standard practice in many institutions (3–7). Molecular amplification methods for MRSA screening of hospitalized patients provides rapid results and have been demonstrated to be more sensitive than culture-based methods (8–15). The BD MAX MRSA assay (BD Diagnostics, Québec, Canada) is a molecular test performed on the BD MAX system (BD Diagnostics, Sparks, MD). The BD MAX is an automated sample-in and answer-out instrument that combines sample extraction, PCR setup, and real-time PCR on a walkaway platform. The objective of this study was to compare the BD MAX MRSA assay to the only other fully automated walkaway MRSA PCR assay, the Xpert MRSA (Cepheid, Sunnyvale, CA), performed on the GeneXpert instrument (Cepheid).
A total of 239 nares samples were collected from August to September 2012 from all patients admitted to any intensive care unit (ICU) at Tampa General Hospital (TGH), using two individually collected traditional swabs (LQ Stuart; BD BBL). One of the swabs was used for the Xpert MRSA assay, and the other was tested by the BD MAX MRSA assay; both assays were performed in parallel on both instruments according to the manufacturer's instructions without the technician having knowledge of the result from either instrument. For the Xpert MRSA assay, samples were placed in the Xpert assay buffer and transferred to the Xpert MRSA cartridge for PCR on the GeneXpert 16 system (Cepheid). For the BD MAX MRSA assay, samples were placed in the BD MAX MRSA sample buffer tube, which was subjected to vortex mixing for 60 s on a multiposition vortex. Sample tubes were placed in the BD MAX system along with the extraction reagent strip and PCR cartridges. The BD MAX MRSA sample buffer supports MRSA viability, which allowed a limited set of samples to be tested by an in-house-validated mecA and femA PCR, using previously published primers/probes (1), to address discrepant samples.
The limit of detection (LOD) of each assay was assessed using a positive-control strain (MRSA, ATCC 33591). Initially, a 0.5-McFarland-standard suspension of the strain was prepared in saline using fresh colonies from a blood agar plate. The suspension was followed by a series of 10-fold dilutions in saline. Five dilutions (1 × 103 to 1 × 107 CFU/ml) were tested in triplicate by both the Xpert MRSA and the BD MAX MRSA assay. The highest dilution (i.e., lowest concentration) at which all three replicates were positive was considered the LOD of the assay. A challenge panel (Microbiologics Inc., MN, USA) comprising 10 lyophilized control strains was evaluated by both assays. The 10-member challenge panel included seven MRSA mec right extremity junction (MREJ) genotypes, a methicillin-susceptible S. aureus (MSSA) strain, a methicillin-susceptible S. aureus mecA dropout strain (MSSA), and a methicillin-susceptible S. epidermidis (MSSE) strain. Lyophilized bacterial organisms were reconstituted according to the manufacturer's instructions, and all were tested by both MRSA assays.
A workflow analysis was performed by four time and motion studies, focusing on the collection of the following metrics: total cycle time (amount of time required to perform testing across the batch of specimens; includes all manual and automated activities), total processing steps (number of steps required to perform testing across the batch of specimens), hands-on time (amount of time that lab personnel must devote to physically performing testing), and walkaway time (amount of time that testing was being performed but during which the operator was not actively engaged in hands-on testing activities). Four separate batches of specimens (2 batches of 16 specimens and 2 batches of 24 specimens) were analyzed with the two methods (BD MAX and GeneXpert), and a regression analysis was performed to ensure consistency in the measurements and processes. Time and process measurements were collected and analyzed from the time the specimens were acquired from the bins on the processing counter to the time that actionable results were generated. Times collected were then compared to the manufacturer's package insert to ensure that processing adhered to manufacturer guidelines.
Identical results were obtained for 234 of the 239 paired samples tested by both tests (Table 1). A total of 30 (12.5%) samples sets were positive and 204 samples sets were negative by both assays. Five sample sets were positive only by the BD MAX assay. BD MAX residual samples from these five sets were tested using the in-house-validated PCR test, and all of them were positive for both mecA and femA, suggesting that MRSA was present in these samples. Both assays demonstrated equivalent LODs by detecting up to a 1 × 105 dilution of the original suspension.
TABLE 1.
Results from 239 nares swab samples tested by the BD MAX and Xpert MRSA assays
| Xpert MRSA assay result | No. of samples with indicated BD MAX MRSA assay result |
|
|---|---|---|
| Positive | Negative | |
| Positive | 30 | 0 |
| Negative | 5 | 204 |
Strains from the challenge panel representing six out of seven MREJ types were correctly identified by both the Xpert and the BD MAX MRSA assay. Negative results were reported for MSSE, and one was reported for MSSA. The MSSA mecA dropout strain and the MRSA MREJ type xxi (mecC) were incorrectly identified by both tests (Table 2). The current BD MAX MRSA XT and StaphSR assays, however, can correctly identify both mecA dropout and mecC strains (16, 17). Workflow analysis results of hands-on times and walkaway times for the 16-sample and 24-sample batch studies, respectively, are summarized in Fig. 1 and 2. The total times to a result were as follow: (i) with 16 samples, 90 min for the GeneXpert and 125 min for the BD MAX assay, and (ii) with 24 samples, 169 min for the GeneXpert and 146 min for the BD MAX assay.
TABLE 2.
Challenge panel results for 10 strains tested by the Xpert MRSA and BD MAX MRSA assays
| Sample identification | Description | MREJ typea | Xpert MRSA assay result | BD MAX MRSA assay result |
|---|---|---|---|---|
| 1 | MRSA | MRSA i | Positive | Positive |
| 2800 | MRSA | MRSA ii | Positive | Positive |
| 9 | MRSA | MRSA iii | Positive | Positive |
| 11 | MRSA | MRSA iv | Positive | Positive |
| 16 | MRSA | MRSA v | Positive | Positive |
| 19 | MRSA | MRSA vii | Positive | Positive |
| ATCC 2312 | MRSA (mecC) | MRSA xxi | Negative | Negative |
| 3097 | MSSA (empty cassette) | NA | Positive | Positive |
| ATCC 29213 | MSSA | NA | Negative | Negative |
| ATCC 14990 | MSSE | NA | Negative | Negative |
NA, not applicable.
FIG 1.
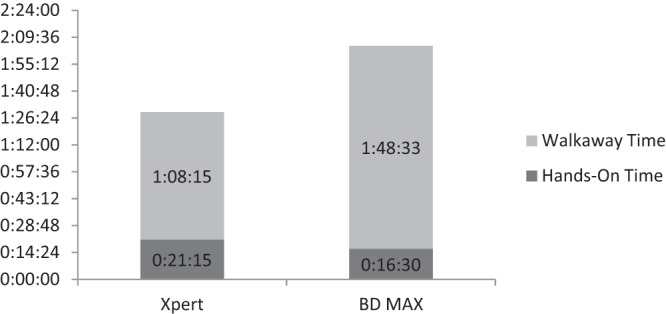
Comparison of hands-on times and walkaway times for the Xpert and BD MAX MRSA assays for 16 samples, displayed as hours:minutes:seconds.
FIG 2.
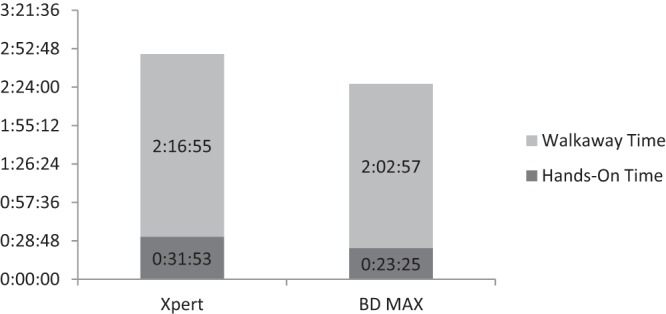
Comparison of hands-on times and walkaway times for the Xpert and BD MAX MRSA assays for 24 samples, displayed as hours:minutes:seconds.
Rapid detection of MRSA in nares samples using molecular amplification methods has been recognized as an effective method to address the problem of MRSA infection and the nosocomial transmission of MRSA (3, 9–12, 14, 15, 18–26). This study compared the two automated, sample-in, answer-out, walkaway PCR assays for MRSA, the Xpert MRSA assay and the BD MAX MRSA assay. Overall, there was 97.9% (234/239 samples) agreement between the two assays. In spite of exhibiting a small number of discrepant results, the two methods displayed an excellent concordance of results. Moreover, the two methods had identical LODs and were able to identify 8 out of 10 strains comprising the challenge panel. Both assays, however, produced a false-positive MRSA result with the mecA dropouts in the MSSA strain.
Workflow analysis showed that the Xpert MRSA assay provided a faster time to a result than did the BD MAX assay when 16 samples were tested. With 24 samples, the time to a result was faster with the BD MAX assay. The Xpert assay required slightly more total hands-on time than the BD MAX assay to process 16 and 24 samples.
There are several limitations to this study. The study was limited to a single site, with the primary service area being central Florida. We identified only 30 positives in the study, which is consistent with our historical positivity rate of just over 10%. A final but important limitation is the fact that two separate swabs were collected, leading to the possibility of errors due to unequal collection quality. In summary, both assays tested presented consistent and comparable results provided with a fast turn-around time. The BD MAX MRSA assay represents a reliable alternative for a highly automated system to detect MRSA in patient nares samples.
ACKNOWLEDGMENTS
This research was performed under University of South Florida Institutional Review Board number 101457 for use of residual pathology samples for assay development research.
The study was supported in part by a grant from BD Diagnostics (Sparks, MD).
Footnotes
Published ahead of print 14 May 2014
REFERENCES
- 1.Hardy K, Price C, Szczepura A, Gossain S, Davies R, Stallard N, Shabir S, McMurray C, Bradbury A, Hawkey PM. 2010. Reduction in the rate of methicillin-resistant Staphylococcus aureus acquisition in surgical wards by rapid screening for colonization: a prospective, cross-over study. Clin. Microbiol. Infection. 16:333–339. 10.1111/j.1469-0691.2009.02899.x [DOI] [PubMed] [Google Scholar]
- 2.Drews SJ, Willey BM, Kreiswirth N, Wang M, Ianes T, Mitchell J, Latchford M, McGeer AJ, Katz KC. 2006. Verification of the IDI-MRSA assay for detecting methicillin-resistant Staphylococcus aureus in diverse specimen types in a core laboratory setting. J. Clin. Microbiol. 44:3794–3796. 10.1128/JCM.01509-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donnio PY, Fevrier F, Bifani P, Dehem M, Kervegant C, Wilhelm N, Lerestif ALG, Lafforgue N, Cormier M, the MR-MSSA Study Group of the College de Bacteriologie-Virologie-Hygeine des Hopitaux de France. Le Coustumier A. 2007. Molecular and epidemiologic evidence for spread of multiresistant methicillin-susceptible Staphylococcus aureus strains in hospitals. Antimicrob. Agents Chemother. 51:4342–4350. 10.1128/AAC.01414-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein E, Smith DL, Laxminarayan R. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg. Infect. Dis. 13:1840–1846. 10.3201/eid1312.070629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peterson LR, Hacek DM, Robiscek A. 2007. Case study: an MRSA intervention at Evanston Northwestern Healthcare. Jt. Comm. J. Qual. Patient Saf. 33:732–738 [DOI] [PubMed] [Google Scholar]
- 6.Khoury J, Jones M, Grim A, Dunne WM, Fraser V. 2005. Eradication of methicillin-resistant Staphylococcus aureus from a neonatal intensive care unit by active surveillance and aggressive infection control measures. Infect. Control Hosp. Epidemiol. 26:616–621. 10.1086/502590 [DOI] [PubMed] [Google Scholar]
- 7.Snyder JW, Munier GK, Johnson CL. 2010. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR assay to culture by use of BBL Chromagar MRSA for detection of MRSA in nasal surveillance cultures from intensive care unit patients. J. Clin. Microbiol. 48:1305–1309. 10.1128/JCM.01326-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francois P, Pittet D, Bento M, Pepey B, Vaudaux P, Lew D, Schrenzel J. 2003. Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J. Clin. Microbiol. 41:254–260. 10.1128/JCM.41.1.254-260.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Struelens MJ, Denis O. 2006. Rapid molecular detection of methicillin-resistant Staphylococcus aureus: a cost effective tool for infection control in critical care? Crit. Care 10:128–130. 10.1186/cc4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jog S, Cunningham R, Cooper S, Wallis M, Marchbank A, Vasco-Knight Jenks PJ. 2008. Impact of prospective screening for methicillin-resistant Staphylococcus aureus by real-time polymerase chain reaction in patients undergoing cardiac surgery. J. Hosp. Infect. 69:124–130. 10.1016/j.jhin.2008.02.008 [DOI] [PubMed] [Google Scholar]
- 11.Robicsek A, Beaumont JL, Paule SM, Hacek DM, Thomson RB, Kaul KL, King P, Peterson LR. 2008. Universal surveillance for methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Ann. Intern. Med. 148:409–418. 10.7326/0003-4819-148-6-200803180-00003 [DOI] [PubMed] [Google Scholar]
- 12.van Loo L, vanDijk S, Verbakel-Schelle I, Buiting AG. 2007. Evaluation of a chromogenic agar (MRSASelect) for the detection of methicillin-resistant Staphylococcus aureus with clinical samples in The Netherlands. J. Med. Microbiol. 56:491–494. 10.1099/jmm.0.47016-0 [DOI] [PubMed] [Google Scholar]
- 13.Hombach H, Pfyffer GE, Roos M, Lucke K. 2010. Detection of methicillin-resistant Staphylococcus aureus (MRSA) in specimens from various body sites: performance characteristics of BD GeneOhm MRSA assay, the Xpert MRSA assay, and broth-enriched culture in an area with a low prevalence of MRSA infections. J. Clin. Microbiol. 48:3882–3887. 10.1128/JCM.00670-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Hal SJ, Stark D, Longwood B, Marriott D, Harkness J. 2007. Methicillin-resistant Staphylococcus aureus (MRSA) detection: comparison of two molecular methods (IDI MRSA PCR assay and Genotype MRSA Direct PCR assay) with three selective MRSA agars (MRSA ID, MRSAselect, and CHROMagar MRSA) for use with infection-control swabs. J. Clin. Microbiol. 45:2486–2490. 10.1128/JCM.00139-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stamper PD, Louie L, Wong H, Simor AE, Farley JE, Carroll KC. 2011. Genotypic and phenotypic characterization of methicillin-susceptible Staphylococcus aureus isolates misidentified as methicillin-resistant Staphylococcus aureus by the BD GeneOhm MRSA assay. J. Clin. Microbiol. 40:1240–1244. 10.1128/JCM.02220-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubasek C, Galambo F, Vendrone E, Silbert S, Widen R. 2014. Evaluation of the BD MAX™ MRSA XT assay in a clinical laboratory, abstr 1239. Abstr. 30th Annu. Clin. Virol. Symp [Google Scholar]
- 17.Vendrone E, Silbert S, Kubasek C, Galambo F, Widen R. 2014. Evaluation of the BD MAX™ STAPHSR assay in a clinical laboratory, abstr 1234. 30th Annu. Clin. Virol. Symp [Google Scholar]
- 18.Muto CA, Jernigan JA, Ostrowsky BE, Richel HM, Jarvis WR, Boyce JM, Farr BM. 2003. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus. Infect. Control Hosp. Epidemiol. 24:362–386. 10.1086/502213 [DOI] [PubMed] [Google Scholar]
- 19.Wolk DM, Picton E, Johnson D, Davis T, Pancholi P, Ginocchio CC, Finegold S, Welch DF, de Boer D, Fuller D, Solomon MC, Rogers B, Mehta MS, Peterson LR. 2009. Multicenter evaluation of the Cepheid Xpert methicillin-resistant Staphylococcus aureus (MRSA) test as a rapid screening method for detection of MRSA in nares. J. Clin. Microbiol. 47:758–764. 10.1128/JCM.01714-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conterno LO, Shymanski J, Ramotar K, Toye B, vanWalraven C, Coyle D, Roth VR. 2007. Real-time polymerase chain reaction detection of methicillin-resistant Staphylococcus aureus: impact on nosocomial transmission and costs. Infect. Control Hosp. Epidemiol. 28:1134–1141. 10.1086/520099 [DOI] [PubMed] [Google Scholar]
- 21.Harris AD, Furuno JP, Roghmann MC, Johnson JK, Conway LJ, Venezia RA, Standiford HC, Schweizer ML, Hebden JN, Moore AC, Perencevich EN. 2010. Targeted surveillance of methicillin-resistant Staphylococcus aureus and its potential use to guide empiric antibiotic therapy. Antimicrob. Agents Chemother. 54:3143–3148. 10.1128/AAC.01590-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Trijp MJCA, Melles DC, Hendriks DH, Parlevliet GA, Gommans M, Ott A. 2007. Successful control of widespread methicillin-resistant Staphylococcus aureus colonization and infection in a large teaching hospital in Ths Netherlands. Infect. Control Hosp. Epidemiol. 28:970–975. 10.1086/519210 [DOI] [PubMed] [Google Scholar]
- 23.Boyce JM, Havill NL. 2008. Comparison of BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR versus the CHROMagar MRSA assay for screening patients for the presence of MRSA strains. J. Clin. Microbiol. 46:350–351. 10.1128/JCM.02130-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossney AS, Herra CM, Brennan DD, Morgan PM, O'Connell B. 2008. Evaluation of the Xpert methicillin-resistant Staphylococcus aureus (MRSA) assay using the Cepheid GeneXpert real-time PCR platform for rapid detection of MRSA from screening specimens. J. Clin. Microbiol. 46:3285–3290. 10.1128/JCM.02487-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farley JE, Stamper PD, Ross T, Cal M, Speser S, Carroll KC. 2008. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) to culture by use of BBL CHROMagar MRSA for detection of MRSA in nasal surveillance cultures from an at-risk community population. J. Clin. Microbiol. 46:743–746. 10.1128/JCM.02071-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherkaoui A, Renzi G, Francois P, Schrenzel J. 2007. Comparison of four chromogenic media for culture-based screening of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 56:500–503. 10.1099/jmm.0.46981-0 [DOI] [PubMed] [Google Scholar]


