ABSTRACT
The interferon antiviral system is a primary barrier to virus replication triggered upon recognition of nonself RNAs by the cytoplasmic sensors encoded by retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology gene 2 (LGP2). Paramyxovirus V proteins are interferon antagonists that can selectively interact with MDA5 and LGP2 through contact with a discrete helicase domain region. Interaction with MDA5, an activator of antiviral signaling, disrupts interferon gene expression and antiviral responses. LGP2 has more diverse reported roles as both a coactivator of MDA5 and a negative regulator of both RIG-I and MDA5. This functional dichotomy, along with the concurrent interference with both cellular targets, has made it difficult to assess the unique consequences of V protein interaction with LGP2. To directly evaluate the impact of LGP2 interference, MDA5 and LGP2 variants unable to be recognized by measles virus and parainfluenza virus 5 (PIV5) V proteins were tested in signaling assays. Results indicate that interaction with LGP2 specifically prevents coactivation of MDA5 signaling and that LGP2's negative regulatory capacity was not affected. V proteins only partially antagonize RIG-I at high concentrations, and their expression had no additive effects on LGP2-mediated negative regulation. However, conversion of RIG-I to a direct V protein target was accomplished by only two amino acid substitutions that allowed both V protein interaction and efficient interference. These results clarify the unique consequences of MDA5 and LGP2 interference by paramyxovirus V proteins and help resolve the distinct roles of LGP2 in both activation and inhibition of antiviral signal transduction.
IMPORTANCE Paramyxovirus V proteins interact with two innate immune receptors, MDA5 and LGP2, but not RIG-I. V proteins prevent MDA5 from signaling to the beta interferon promoter, but the consequences of LGP2 targeting are poorly understood. As the V protein targets MDA5 and LGP2 simultaneously, and LGP2 is both a positive and negative regulator of both MDA5 and RIG-I, it has been difficult to evaluate the specific advantages conferred by LGP2 targeting. Experiments with V-insensitive proteins revealed that the primary outcome of LGP2 interference is suppression of its ability to synergize with MDA5. LGP2's negative regulation of MDA5 and RIG-I remains intact irrespective of V protein interaction. Complementary experiments demonstrate that RIG-I can be converted to V protein sensitivity by two amino acid substitutions. These findings clarify the functions of LGP2 as a positive regulator of MDA5 signaling, demonstrate the basis for V-mediated LGP2 targeting, and broaden our understanding of paramyxovirus-host interactions.
INTRODUCTION
The production of type 1 interferon (IFN) initiates a primary antiviral response in higher eukaryotes that activates innate immunity and primes long-term adaptive immunity. Diverse examples of virus-designed countermeasures, evasion strategies, and antagonists of antiviral signaling responses highlight the importance of the IFN antiviral system as an early barrier to virus replication.
Cytoplasmic RNA viruses can be detected by a group of sentry proteins known collectively as RIG-I-like receptor (RLR) proteins. The RLR proteins are cytoplasmic DECH box proteins that can specifically recognize virus-derived RNA species as a molecular feature discriminating the pathogen from the host (1). The RLR family is composed of three homologous proteins encoded by the retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology gene 2 (LGP2). All three RLR proteins share homologous DEXD box helicase regions that have intrinsic double-stranded RNA (dsRNA) binding and ATP hydrolysis activities and a C-terminal domain that has been implicated in recognizing RNA termini and autoregulation. Despite their similarities, the RLR proteins differ greatly in their properties with respect to virus recognition and biological functions. RIG-I and MDA5 both have tandem N-terminal caspase activation and recruitment domain (CARD) motifs, protein interaction domains that mediate interactions with upstream and downstream regulatory proteins. The CARD motifs of activated RLRs interact with the CARD of the mitochondrial antiviral signaling protein, IPS-1/MAVS, to initiate activation of serine kinases and other signaling proteins responsible for IFN-β gene activation and antiviral responses. In contrast, LGP2 lacks CARD regions and uses alternate means to participate in both antiviral signaling and signal attenuation (2).
The enveloped, nonsegmented negative-strand RNA viruses in the large paramyxovirus family are recognized to have evolved specific mechanisms to overcome host innate antiviral responses (3). Many viruses in this family encode an immune evasion protein known as the V protein, characterized by a signature highly conserved C-terminal zinc-binding domain. This conserved domain is required for many of the V protein's biological activities, including selective inactivation of cytosolic RNA recognition that leads to immediate antiviral responses, including production of IFN and other antiviral effectors and disruption of IFN-stimulated antiviral gene expression. Paramyxovirus V proteins are well known for their diverse, virus-specific abilities to degrade or interfere with STAT proteins to prevent IFN-stimulated gene expression. In addition, V proteins selectively interact with and antagonize a subset of RLRs that are required for IFN production (4, 5). The conserved C-terminal domains of diverse V proteins have the ability to bind to MDA5, inactivating its ATP hydrolysis activity and downstream signal-transducing ability (6–9). In addition, V proteins interact with LGP2, interfering with its ATP hydrolysis activity (7). The V proteins target MDA5 and LGP2 helicase domains through a shared interface referred to as the minimal V protein binding region (MVBR) (7). The MVBR encompasses helicase domain 2, a region that harbors essential helicase motifs IV, V, and VI (10) (see Fig. 1A). The MVBR is highly conserved between MDA5 and LGP2 but more divergent in RIG-I (7), and both structural (11) and experimental (12) evidence indicates that a single arginine residue of MDA5 (R806) is a critical mediator of this association. Substitution with the analogous leucine of RIG-I (L714) results in the V-insensitive MDA5 R806L mutant. Complementary substitution creates the RIG-I L714R mutant that acquired the ability to bind to V proteins (11, 12). For LGP2, the analogous arginine (R455) is required for recognition by measles virus V protein, but substitution of this single residue was not sufficient to abrogate LGP2 binding to parainfluenza virus 5 (PIV5), mumps virus, or Nipah virus V proteins (12). These findings indicated that paramyxoviruses have evolved related but distinct interactions with MDA5 and LGP2 but not RIG-I.
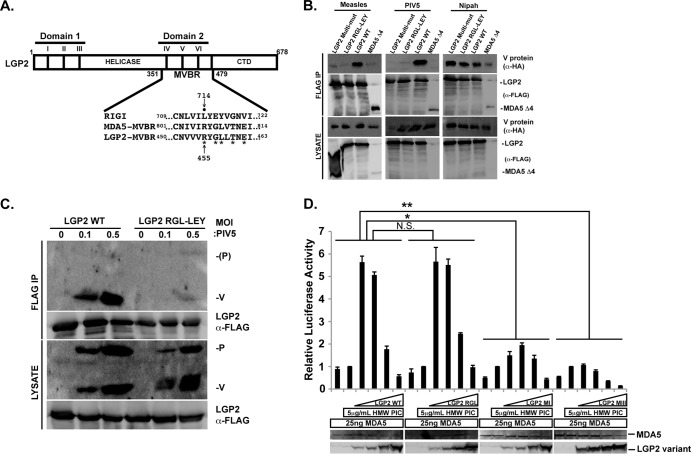
FIG 1.
Generation of a biologically active, V protein-insensitive LGP2. (A) Illustration of the key features of LGP2. LGP2 is shown as a box and positions of the helicase region, including conserved helicase motifs I to III of domain 1, motifs IV to VI of domain 2, and the C-terminal regulatory domain (CTD). Domain 2 coincides with the minimal V protein-binding region (MVBR) that is shared between MDA5 and LGP2 (7). Expanded sequence alignment illustrates relevant region of all three RLR proteins within the MVBR, and residues targeted are depicted with an asterisk. Arrow points to LGP2 R455, which is R806 of MDA5 and L714 of RIG-I; closed circle indicates RIG-I L714. (B) LGP2 interactions with V proteins. FLAG-tagged LGP2 or variants were coexpressed with HA-tagged V proteins from measles, PIV5, and Nipah virus in HEK293T cells. The cell lysates were subjected to FLAG immunoaffinity purification, and detection of coprecipitation was carried out by anti-HA immunoblotting. (C) LGP2 RGL-LEY is defective for interaction with PIV5 V protein in infected cells. HEK293T cells were transfected with FLAG-tagged LGP2 or mutant and infected with PIV5 at the indicated multiplicity of infection (MOI) for 24 h. The cell lysates were subjected to FLAG immunoaffinity purification, and detection of coprecipitation was carried out by immunoblotting with antiserum that recognizes P and V proteins. (D) LGP2 RGL-LEY retains biological activity. A total of 25 ng MDA5 was expressed with WT or mutant LGP2 titrated at 4, 20, 100, or 500 ng of vector. Cells were stimulated by transfection of high-molecular-weight (HMW) poly(I·C) for 6 h prior to luciferase assays. Student's t test indicated as follows: N.S. (not significant), P > 0.015; *, P < 0.015; **, P < 0.0015. LGP2 MI = motif I mutant (K30A); LGP2 MIII = motif III mutant (T167A S169A). Immunoblots below correspond to expression levels of representative lysates.
V protein inhibition of MDA5 is thought to provide an advantage to the virus by preventing downstream signaling leading to antiviral gene expression (9). The reasons for LGP2 interference are less apparent, primarily due to a lack of clarity surrounding LGP2's apparently antithetical functions in antiviral signaling (13). Expression of LGP2 in cells from plasmid vectors results in general inhibition of RIG-I and MDA5 signaling (14–17), and several mechanisms were proposed to explain LGP2 feedback inhibition (15–18). The fact that LGP2 binds with high affinity to dsRNA was interpreted as evidence for inhibition by RNA sequestration (15, 17). Characterization of a C-terminal regulatory domain in RIG-I that mediates autoinhibition in cis and trans via CARD interaction was proposed as another source of LGP2-mediated interference. By analogy, it was proposed that LGP2's C-terminal domain could also mediate RIG-I interference (16). Another mechanism for LGP2-negative regulation was revealed by experiments that demonstrated LGP2 can inhibit antiviral signaling independent of dsRNA or virus infection by engaging in a protein complex with IPS-1/MAVS (14). In this case, LGP2 is thought to accumulate during infection to result in competition for downstream kinase activation.
In addition to these negative effects of LGP2 expression, growing evidence implicates LGP2 as a positive regulator of IFN-β and antiviral responses. Mice with a targeted disruption in the LGP2 locus are more susceptible to specific virus infections, and LGP2 deficiency reduces IFN-β production and other host responses to several RNA viruses, notably the picornaviruses, including encephalomyocarditis virus (EMCV) and poliovirus. These viruses had been previously linked to detection by MDA5, supporting a positive role for LGP2 in IFN-β expression and antiviral signaling in connection with MDA5 (19). Experiments in LGP2- and MDA5-deficient cells revealed synergistic signal transduction resulting from coexpression of LGP2 with MDA5 (19, 20), suggesting that LGP2 may promote efficient MDA5 RNA detection or facilitate interactions with signaling machinery. This notion is supported by the characterization of an EMCV-derived MDA5 agonist RNA that was identified not on the basis of interaction with MDA5 but through its association with LGP2 (21). Replacing LGP2 with the enzymatically inactive K30A mutant by knock-in does not reconstitute defective positive signaling responses, indicating the importance of ATP hydrolysis in LGP2-positive regulation (19), but this same LGP2 mutant retains negative regulatory functions (14, 22). Biochemical analysis and single-molecule RNA binding studies demonstrate that LGP2 uses ATP hydrolysis to enhance its ability to associate with diverse dsRNA species, enabling it to act in concert with MDA5 to maximize antiviral signal transduction (22, 23). These results indicate an ATP-dependent function of LGP2 in promoting signal transduction in response to intracellular virus infections. The apparent synergy between MDA5 and LGP2 provides a plausible basis for their specific antagonism by paramyxovirus V proteins. A positive antiviral role for LGP2, and a connection to MDA5, is highlighted by the fact that V proteins can target both LGP2 and MDA5 helicase domains, disrupting ATP hydrolysis (7). Together, these findings are consistent with LGP2 acting as an upstream mediator of RNA recognition and signaling.
While evolution has produced paramyxovirus V proteins that interact well with MDA5 and LGP2, the benefits of selective LGP2 antagonism are poorly understood. Moreover, despite the fact that paramyxoviruses replicate better in RIG-I-deficient cells, direct V protein interference with RIG-I has not been observed. A potential indirect mechanism for RIG-I interference was proposed in which V protein inhibits RIG-I signaling by forming a complex with LGP2 (24). This proposed model contrasts with earlier studies of V protein-RLR interaction specificity, as well as the finding that LGP2–RIG-I coimmunoprecipitation is bridged by independent interactions with distinct regions of IPS-1/MAVS (14).
To more definitively investigate the consequences of MDA5 and LGP2 suppression and directly test the impact of V protein interference with LGP2 on both MDA5 and RIG-I signaling, an LGP2 protein was created that retains biological activity but is insensitive to measles virus or PIV5 V proteins. Data indicate that LGP2 is a negative regulator of RIG-I signaling irrespective of V protein sensitivity, and we demonstrate that as few as two amino acid substitutions can convert RIG-I into a bona fide V protein target. These findings support the conclusion that the primary consequence of V protein interaction with LGP2 is suppression of MDA5 signaling synergy and is not relevant to negative regulation of either RIG-I or MDA5.
MATERIALS AND METHODS
Cells and viruses.
HEK293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% cosmic calf serum (HyClone) and 1% penicillin-streptomycin (Gibco-BRL). PIV5 was propagated and titers were determined in CV1 cells.
Plasmids, immunoprecipitations, and immunoblotting.
MDA5, LGP2, and RIG-I were cloned into the mammalian expression plasmid p3XFLAG-CMV-10 to provide an amino-terminal FLAG epitope tag, and these plasmids were used as templates to generate point mutations with Agilent's QuikChange mutagenesis lightning kit. All the mutations were confirmed by DNA sequencing. V proteins expressed from plasmid pEF-hemagglutinin (HA) have been described (12).
For immunoprecipitation experiments, 7.5 μg of FLAG-helicase vector and 2.5 μg of HA-V vector were transfected by the CaPO4 method. Twenty-four hours later, cells were harvested by first being washed with cold phosphate-buffered saline and then lysed with whole-cell extract buffer (WCEB) (12). Cell lysates were then precleared with Sepharose beads, and 5% of the cleared lysates was retained for direct analysis. The helicase and V protein complexes were FLAG immunoaffinity purified overnight and washed extensively with WCEB, eluted with SDS sample buffer, separated by SDS-PAGE, and processed for immunoblotting.
For transfection/infection, HEK293T cells were transfected with 7.5 μg of FLAG-LGP2 or variant and infected with PIV5 at the indicated multiplicity of infection (MOI). Cells were harvested 24 h later and lysates prepared as described above.
For immunoblotting, the separated proteins were transferred to nitrocellulose and probed with commercial primary antibodies recognizing FLAG or HA (Sigma) or rabbit antiserum recognizing the PIV5 P and V proteins (7, 12). Proteins were visualized by enhanced chemiluminescence (NEN Life Sciences).
Luciferase reporter gene assays.
HEK293T cells were transfected with vectors for FLAG-tagged RLRs, HA-tagged V proteins, the −110 IFN-β luciferase reporter genes, and a Renilla luciferase control by Lipofectamine 2000. Twenty-four hours posttransfection, cells were stimulated for 6 h with poly(I·C) and assayed for luciferase activities using the dual Luciferase reporter assay system (Promega). Relative luciferase activity was calculated by dividing the firefly luciferase values by those of the Renilla luciferase. Data are plotted as average values (n ≥ 3), with error bars representing standard deviations.
RESULTS
Generation of a biologically active LGP2 that is insensitive to V proteins.
Our previous studies revealed that LGP2 arginine 455 (R455) is necessary for measles virus V protein interaction (12) (Fig. 1A). Conversion of R455 to the leucine of RIG-I (LGP2 R455L mutant) created an LGP2 protein unable to be recognized by the measles virus V protein. However, PIV5 and Nipah virus V proteins continued to coprecipitate with the LGP2 R455L mutant, indicating additional requirements for PIV5 disengagement (12). As the PIV5 V protein has served as a prototype V protein for several studies of MDA5 and LGP2 recognition, a PIV5 V protein-insensitive variant of LGP2 was sought. Site-directed mutagenesis was used to substitute additional LGP2 amino acids for their RIG-I analog. Substitution of R455, G457, and L458 created LGP2 RGL-LEY, and further substitution of T460 and E462 created LGP2 multi-Mut. To test if these amino acid changes disrupted PIV5 V protein interactions, a coimmunoprecipitation assay was carried out (Fig. 1B). The FLAG epitope-tagged LGP2 proteins were expressed in HEK293T cells along with HA epitope-tagged V proteins from measles virus, PIV5, or Nipah virus. The cell lysates were subjected to FLAG immunoaffinity purification, and FLAG peptide-eluted proteins were separated by SDS-PAGE and subjected to HA immunoblotting to determine if the mutations disrupted the LGP2-V protein interaction. The wild-type (WT) LGP2 coprecipitated with all three V proteins. In contrast, both of the LGP2 mutants were unable to coprecipitate with either measles virus or PIV5 V protein, similar to a previously characterized negative control, MDA5Δ4, that encompasses residues 747 to 1025 (7). All of the mutants retained coprecipitation with Nipah virus V protein, indicating a unique recognition mechanism. Importantly, this analysis identifies three amino acids that are required for LGP2 interactions with PIV5 V protein.
To verify the coimmunoprecipitation results, the ability of LGP2 RGL-LEY to disengage from physiologically expressed V protein was tested in the context of virus infection (Fig. 1C). Cells expressing WT LGP2 or LGP2 RGL-LEY proteins were infected with PIV5, and the whole-cell lysates were subjected to FLAG immunoprecipitation. Eluates were probed with antiserum that recognizes PIV5 P and V proteins. The PIV5 V protein, but not the P protein, coprecipitated with the WT LGP2. In contrast, the LGP2 RGL-LEY mutant was defective for binding to the PIV5 V protein, though a trace amount of V protein was detected, suggesting that the mutations greatly impair, but do not completely eliminate, interaction with PIV5 V protein.
To test if the mutagenesis had any effects on the biological activity of LGP2, an MDA5-dependent IFN-β-promoter luciferase reporter gene assay was conducted (Fig. 1D). We previously used a similar functional assay to demonstrate that LGP2 ATP hydrolysis and RNA recognition are required to synergistically enhance MDA5-mediated signaling (22). Expression of MDA5 activates IFN-β-luciferase regardless of poly(I·C) stimulation (7, 12, 22), and MDA5 signaling can be further enhanced by titration of LGP2. Increasing the amount of LGP2 expression by transfection with more plasmid DNA ultimately results in suppression of reporter gene activity. Titration of LGP2 RGL-LEY also enabled increased MDA5 signaling, resulting in a characteristic biphasic enhancement and interference profile nearly identical to WT LGP2. LGP2 mutants that target helicase motif I (MI; K30A; defective for ATP hydrolysis but able to bind RNA [10]) or motif III (MIII; T167A S169A; defective for both ATP hydrolysis and RNA binding [10]) served as negative controls. These mutants fail to augment MDA5 signaling but retain negative regulation (22) (Fig. 1D). Together, these experiments demonstrate that LGP2 RGL-LEY is a biologically active LGP2 protein that is insensitive to measles virus and PIV5 V proteins.
V proteins suppress MDA5 signaling enhancement by LGP2.
Biochemical studies established that paramyxovirus V proteins can directly bind to both MDA5 and LGP2 to interfere with their ATP hydrolysis and signal transduction activities (7, 9, 10, 23). The ability of LGP2 to synergize with MDA5 in the presence of a V protein was tested with a reporter gene assay (Fig. 2). In control experiments, MDA5-dependent activation of IFN-β-luciferase gene expression was enhanced by low concentrations of LGP2 and inhibited at higher concentrations of LGP2. Expression of either measles virus or PIV5 V protein antagonized MDA5 signaling and also prevented LGP2-mediated MDA5 enhancement (Fig. 2A), consistent with V protein targeting of both MDA5 and LGP2. Interestingly, negative effects of LGP2 expression remained intact irrespective of V proteins.
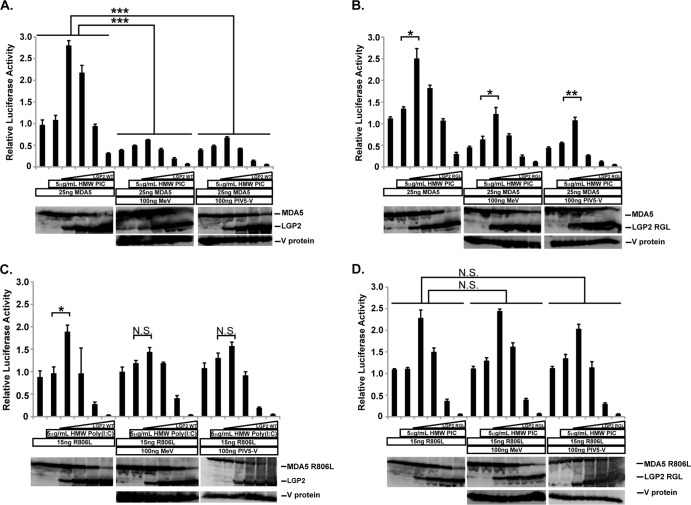
FIG 2.
V proteins suppress MDA5 signaling enhancement by LGP2. (A) Similar to that described for Fig. 1D, WT MDA5 was expressed with various amounts of the indicated LGP2 vector (4, 20, 100, 500 ng) and stimulated with high-molecular-weight (HMW) poly(I·C) for 6 h prior to luciferase assays. Parallel experiments included measles virus V protein (MeV) or PIV5 V protein (PIV5-V) as indicated. (B) Similar to that described for panel A, but using WT MDA5 and V protein-insensitive LGP2 RGL-LEY mutant. (C) Similar to that described for panel A, but using the V protein-insensitive MDA5 R806L mutant and WT LGP2. (D) Similar to that described for panel A, but using the V protein-insensitive MDA5 R806L mutant and V protein-insensitive LGP2 RGL-LEY. For all panels, Student's t test indicated as follows: N.S. (not significant), P > 0.015; *, P < 0.015; **, P < 0.0015; ***, P < 0.00015.
To more directly test the effects of V protein on either MDA5 or LGP2, V-insensitive mutants were used in signaling assays. First, a similar experiment was carried out with WT MDA5 and the V-insensitive LGP2 RGL-LEY mutant (Fig. 2B). The mutant LGP2 mediated a characteristic biphasic MDA5 signaling profile in the absence of V proteins; similar to the outcome with WT LGP2, signaling was activated at low expression levels but inhibited at high expression levels. V protein expression induced a dramatic reduction of MDA5 signaling, but the V-resistant LGP2 RGL-LEY mutant enhanced the small amount of residual signaling activity by MDA5. Again, it was observed that LGP2 RGL-LEY negative regulation remained intact in the presence of V proteins. This result indicates that V protein suppression of LGP2 prevents its positive effects on MDA5 signal transduction.
We previously described the MDA5 R806L mutant, which is insensitive to measles virus and PIV5 V proteins. In addition to V protein insensitivity, the MDA5 R806L mutant not only retains signaling activity but exhibits 3- to 5-fold-higher signaling activity than WT MDA5 (12). In the absence of V proteins, low expression of LGP2 enhanced MDA5 R806L and higher expression of LGP2 inhibited signaling (Fig. 2C). As expected, V proteins did not alter MDA5 R806L mutant activity, but LGP2 enhancement of MDA5 signaling was blunted by V protein expression. Again, negative regulation by LGP2 remained intact and unaffected by V proteins.
Biphasic MDA5 R806L signaling was also observed with LGP2 RGL-LEY (Fig. 2D). Low concentrations of LGP2 RGL-LEY enhanced MDA5 R806L signaling, and higher expression suppressed signaling. Consistent with their inability to engage the LGP2 mutant, V proteins did not alter this profile of biphasic LGP2 action. Again, negative regulation was induced by a high concentration of LGP2 RGL-LEY, and this was not influenced by V protein expression.
Together, these results indicate that when examined in the absence of V-mediated MDA5 suppression, V protein interaction with LGP2 disables its ability to augment MDA5 signal transduction, and that LGP2-mediated negative regulation remains unaltered by V protein associations.
RIG-I is inhibited by LGP2 independent of V protein interaction.
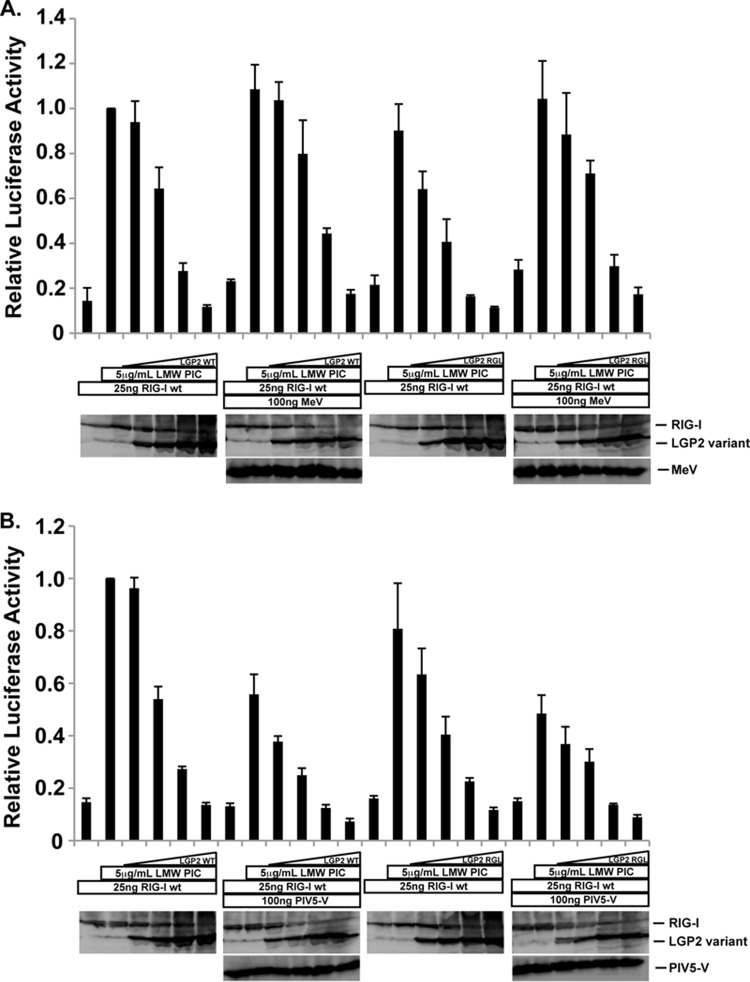
Unlike MDA5 and LGP2, RIG-I does not directly interact with paramyxovirus V proteins. Nonetheless, it was proposed that V protein interaction with LGP2 might enable negative regulation of RIG-I signaling (24). To test this V-dependent, LGP2-mediated mode of suppressing RIG-I signaling, and to distinguish it from the documented ability of LGP2 to suppress RIG-I signaling in the absence of V proteins, RIG-I was used to activate the IFN-β reporter gene in the presence and absence of LGP2 or LGP2 RGL-LEY (Fig. 3). Unlike MDA5, RIG-I expression alone does not activate the reporter gene unless stimulated with dsRNA ligands. Transfection of low-molecular-weight poly(I·C) was able to efficiently activate RIG-I. Titration of LGP2 resulted in concentration-dependent interference with RIG-I signaling (Fig. 3A). LGP2 did not enhance RIG-I signaling at any concentration tested, confirming the prior conclusion that LGP2 can negatively regulate RIG-I signaling (2, 14–17, 24, 25). A similar profile of RIG-I suppression by LGP2 was observed in the presence of measles virus V protein (Fig. 3A). The V protein-insensitive LGP2 RGL-LEY mutant was fully capable of RIG-I suppression, which was again observed irrespective of measles virus V protein expression. In this experiment, the addition of PIV5 V protein differed from measles virus V protein by inducing an overall decrease in RIG-I signaling, reducing the level of RIG-I activity by ∼40% in this experiment (Fig. 3B). Despite this difference, titration of LGP2 expression was able to reduce the signal to baseline levels. Importantly, as found with measles virus V protein, similar levels of RIG-I signaling were obtained using LGP2 RGL-LEY (Fig. 3B). Both WT LGP2 and mutant LGP2 suppress RIG-I signaling in the presence or absence of PIV5 V protein. These results indicate that LGP2 interferes with RIG-I signaling and that V protein interaction with LGP2 has little influence on RIG-I signal transduction or its attenuation.
FIG 3.
RIG-I inhibition by LGP2 is independent of V proteins. (A) Similar to that described for Fig. 2, but using WT RIG-I coexpressed with various amounts of either WT LGP2 or LGP2 RGL-LEY in the absence or presence of MeV. (B) Similar to that described for panel A, but using PIV5 V. Student's t test indicated no significant differences for all conditions (P > 0.015).
Mutations to RIG-I enable direct V protein interactions.
To try to clarify the potential for generalizable RIG-I suppression by V proteins, signaling assays were carried out into which V protein expression was titrated (Fig. 4A). Signaling from RIG-I remained largely intact in the presence of V proteins in this experiment, with measles virus inducing only a minor loss of activity at the highest V protein concentration, reducing luciferase expression by 15 to 20%. PIV5 V protein also reduced RLR signaling only at the highest concentration tested, reducing luciferase activity by 40 to 50% in this experiment.
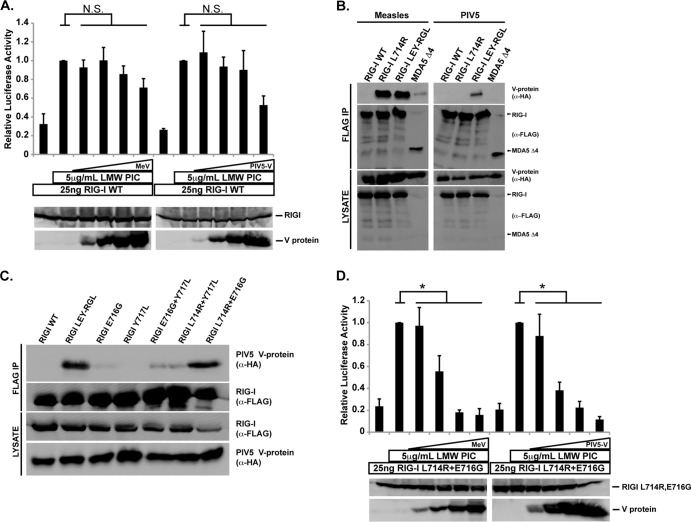
FIG 4.
Mutations to RIG-I enable direct V protein targeting. (A) RIG-I signaling assay similar to that described for Fig. 3A, but using a broader range of measles virus or PIV5 V protein expression (4, 20, 100, 500 ng). Immunoblots below demonstrate protein expression levels in representative lysates. (B) FLAG-tagged RIG-I or variants were coexpressed in HEK293T cells with HA-tagged V proteins from PIV5 and measles virus. Cell lysates were prepared and analyzed as described for Fig. 1B. (C) Combinatorial analysis of RIG-I mutations required for PIV5 V interaction. RIG-I mutants and PIV5 V were coexpressed and prepared as described above. (D) The RIG-I L714R E716G mutant is biologically active and suppressed by V proteins. Luciferase assays were carried out similar to those described for Fig. 4A, but using the RIG-I L714R E716G mutant in the presence and absence of measles virus or PIV5 V protein titration (4 ng, 20 ng, 100 ng, and 500 ng). Student's t test indicated as follows: N.S. (not significant), P > 0.05; *, P < 0.05.
Substitution of RIG-I L714 with the arginine of MDA5 or LGP2 was previously reported to create a RIG-I mutant (L714R) susceptible to measles virus V protein, but this single change did not cause generalized V protein recognition (11, 12). Another RIG-I variant, with five total changes, including L714 (multi-Mut), expanded V protein recognition to include PIV5 and Nipah virus and confirmed the importance of MDA5 R806, LGP2 R455, and several neighboring residues in mediating V protein associations (12). Unfortunately this version of RIG-I was found to be incompetent for signal transduction. To narrow down the minimum requirements for V protein recognition, RIG-I residues L714, E716, and Y717 were substituted to the MDA5/LGP2 correlate (creating RIG-I LEY-RGL; see Fig. 1A and 4B). Coimmunoprecipitation assays reveal that RIG-I LEY-RGL confers PIV5 and measles V protein recognition (Fig. 4B). The contributions of the three RIG-I mutations were further screened for PIV5 V protein interaction by introducing single and combinatorial substitutions (Fig. 4C). The RIG-I L714R E716G double mutant was sufficient to support interaction with the PIV5 V protein.
Despite the two amino acid modifications, the RIG-I L714R E716G mutant retained inducible signaling activity, resulting in ligand-induced IFN-β promoter activation (Fig. 4D). The RIG-I mutant was highly sensitive to both measles virus and PIV5 V proteins, which were both capable of reducing signaling activity to baseline, even at lower expression levels (Fig. 4D). These findings highlight the fact that while RIG-I is not a direct target for V protein antagonism, it can be converted to complete V protein sensitivity by a small number of amino acid alterations.
DISCUSSION
As a primary mediator of the cellular antiviral IFN response, the RLR signaling pathway provides many targets for virus-encoded host evasion and antagonism. The paramyxovirus V proteins share the ability to specifically disrupt MDA5 and LGP2, but not RIG-I, signaling. This interference is mediated by a discrete binding site within the helicase domains of MDA5 and LGP2 (6–9, 11), and this shared but specific recognition capacity allows V proteins to interfere with both MDA5 and LGP2 simultaneously. As a result, it has been impossible to discern the consequences of V protein antagonism of each RLR in an intact system, obscuring their nonredundant and redundant functions.
By altering the amino acid sequence surrounding the key contact region within their respective MVBRs, V-resistant variants of both MDA5 and LGP2 were created that disengage measles virus and PIV5 targeting ability but retain RLR signaling activity. These reagents allowed evaluation of the effects of V protein targeting on both positive and negative regulation mediated by LGP2 while maintaining MDA5 activity, addressing for the first time the unique consequences of LGP2-specific interference. Titration of wild-type LGP2 yields a biphasic effect on MDA5 signaling, and results indicate that a primary effect of LGP2 targeting by V protein is interference with its ability to enhance MDA5-mediated IFN-β production. The negative regulatory effects of LGP2 on MDA5 signaling were not altered by V protein interactions. The mutant LGP2 protein was insensitive to V protein antagonism and retained the ability to both enhance and inhibit MDA5 signaling.
The effects of V protein-sensitive and -insensitive LGP2 were also examined in the context of RIG-I signaling. No RIG-I enhancement was observed in the presence of WT or V-resistant LGP2, but both were able to suppress RIG-I signaling, in agreement with prior reports (14–17). WT and mutant LGP2 were similarly effective at RIG-I inhibition. In the context of RIG-I signaling, V protein expression exhibited only a small degree of signaling interference that was observed only at the highest expression levels tested, primarily for PIV5 V protein. Substitution of only two amino acids, L714 and E716, converted RIG-I into a bona fide V protein target, leading to dramatically heightened sensitivity to V protein expression and complete suppression. These findings demonstrate that a generalizable mechanism for RIG-I suppression is not an intrinsic property of the viral V proteins tested. Measles virus V protein is an ineffective RIG-I inhibitor at best, and this is not altered by LGP2 expression. While it was observed that high expression of PIV5 V protein could partially decrease RIG-I signaling in some experiments, no differences were found upon expression of LGP2, irrespective of its V protein sensitivity. Further research will be required to determine if the partial repression of RIG-I by PIV5 is biologically significant. As V proteins are well known to exhibit strain-specific specialization in their precise modes of IFN evasion, it is possible that both specific and nonspecific cellular targets are affected by high PIV5 expression.
A prior report published in The Journal of Virology (24) concluded that V proteins interact with LGP2 to inhibit RIG-I-dependent IFN induction. The present data indicate that V-resistant LGP2 has the same impact on RIG-I signaling as WT LGP2, even in the presence of V proteins. The prior report also suggested that LGP2 expression alone is inefficient at RIG-I inhibition in the absence of V proteins, a conclusion at odds with several descriptions of LGP2-mediated negative regulation (14–17, 22, 25) as well as the conclusions reported here. The reasons for this prior misapprehension are unclear, but because V proteins are multifunctional host antagonists that target both MDA5 and LGP2, the ability to dissociate the positive effects of LGP2 on MDA5 signaling from its negative impacts on both MDA5 and RIG-I signaling was essential to enable a more direct analysis.
These findings evoke an apparent conundrum surrounding paramyxovirus RLR inhibition. For example, paramyxoviruses like Sendai virus have been shown to have a replication advantage in RIG-I-deficient mice but not in the absence of MDA5 or LGP2, a finding broadly interpreted to indicate their detection specifically by RIG-I (19, 26–31). Despite this clear indication, RIG-I is not a target for V protein antagonism. The idea that MDA5 and LGP2 might be important for cellular antiparamyxovirus responses might seem inconsistent with the strict interpretation that RIG-I is the only relevant sensor of paramyxovirus RNA in vivo. Several considerations may help put this controversy into perspective. Foremost, the V protein interference with MDA5 and LGP2 already creates a virtual null environment for these RLRs. As such, the genetic absence of these factors could have only minor impacts on the intracellular milieu for virus replication.
In addition, several paramyxoviruses have been shown to use alternate methods in addition to V proteins to suppress RIG-I activity or downstream transcription factor activation (e.g., see references 32 and 33). An alternate explanation for the lack of RIG-I antagonism may be that it is simply not able to detect these viruses in the natural context. The observed RIG-I sensitivity may reflect the propensity of some paramyxoviruses to harbor defective interfering (DI) RNA genomes (34) that activate RIG-I (35–37). Many studies of RIG-I activation employ Sendai viruses that are known to accumulate defective genomes during replication. In many of these cases, the RIG-I activation observed is due primarily to the presence of defective genomes, which have been demonstrated to be the primary RIG-I ligand during Sendai virus infections (38). It is noteworthy that preparations of wild-type PIV5 do not induce appreciable amounts of IFN-β, but PIV5 with an inactivated V protein hyperactivates IFN-β and antiviral gene expression (39). This may suggest that RIG-I recognition of DI genomes actually supports virus fitness, controlling genome quality by using the cellular antiviral response to prevent further propagation of truncated genomes. RIG-I activation by these corrupted viruses would provide a means to eliminate unfit participants from the population by exploiting the host antiviral response system. Even in this context, LGP2 would be able to act as an endogenous negative regulator that could attenuate prolonged RLR signaling.
ACKNOWLEDGMENTS
We are grateful to Annie M. Bruns and Jean-Patrick Parisien for their guidance and helpful comments on this work and the manuscript.
Research on RLRs and V proteins in the Horvath lab was supported by NIH grants AI073919 and AI50707 to C.M.H.
Footnotes
Published ahead of print 15 May 2014
REFERENCES
- 1.Ramos HJ, Gale MJ. 2011. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr. Opin. Virol. 1:167–176. 10.1016/j.coviro.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruns AM, Horvath CM. 2012. Activation of RIG-I-like receptor signal transduction. Crit. Rev. Biochem. Mol. Biol. 47:194–206. 10.3109/10409238.2011.630974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlier D, Lyles DS. 2011. Interplay between innate immunity and negative-strand RNA viruses: towards a rational model. Microbiol. Mol. Biol. Rev. 75:468–490. 10.1128/MMBR.00007-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramachandran A, Horvath CM. 2009. Paramyxovirus disruption of interferon signal transduction: STATus report. J. Interferon Cytokine Res. 29:531–537. 10.1089/jir.2009.0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodbourn S, Randall RE. 2009. The regulation of type I interferon production by paramyxoviruses. J. Interferon Cytokine Res. 29:539–547. 10.1089/jir.2009.0071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall RE, Goodbourn S. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190–200. 10.1016/j.virol.2006.09.023 [DOI] [PubMed] [Google Scholar]
- 7.Parisien J-P, Bamming D, Komuro A, Ramachandran A, Rodriguez JJ, Barber G, Wojahn RD, Horvath CM. 2009. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 83:7252–7260. 10.1128/JVI.00153-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramachandran A, Horvath CM. 2010. Dissociation of paramyxovirus interferon evasion activities: universal and virus-specific requirements for conserved V protein amino acids in MDA5 interference. J. Virol. 84:11152–11163. 10.1128/JVI.01375-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrejeva J, Childs K, Young D, Carlos T, Stock N, Goodbourn S, Randall RE. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264–17269. 10.1073/pnas.0407639101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bamming D, Horvath CM. 2009. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 284:9700–9712. 10.1074/jbc.M807365200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, Conzelmann K-K, Hopfner K-P. 2013. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science 339:690–693. 10.1126/science.1230949 [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez KR, Horvath CM. 2013. Amino acid requirements for MDA5 and LGP2 recognition by paramyxovirus V proteins: a single arginine distinguishes MDA5 from RIG-I. J. Virol. 87:2974–2978. 10.1128/JVI.02843-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, Zhang X, Wang G, Zheng H. 2014. The laboratory of genetics and physiology 2: emerging insights into the controversial functions of this RIG-I-like receptor. Biomed. Res. Int. 2014:960190. 10.1155/2014/960190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komuro A, Horvath CM. 2006. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 80:12332–12342. 10.1128/JVI.01325-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175:5260–5268. 10.4049/jimmunol.175.8.5260 [DOI] [PubMed] [Google Scholar]
- 16.Saito T, Hirai R, Loo Y-M, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale MJ. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U. S. A. 104:582–587. 10.1073/pnas.0606699104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo Y-M, Gale MJ, Akira S, Yonehara S, Kato A, Fujita T. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851–2858. 10.4049/jimmunol.175.5.2851 [DOI] [PubMed] [Google Scholar]
- 18.Komuro A, Bamming D, Horvath CM. 2008. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine 43:350–358. 10.1016/j.cyto.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U. S. A. 107:1512–1517. 10.1073/pnas.0912986107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178:6444–6455. 10.4049/jimmunol.178.10.6444 [DOI] [PubMed] [Google Scholar]
- 21.Deddouche S, Goubau D, Rehwinkel J, Chakravarty P, Begum S, Maillard PV, Borg A, Matthews N, Feng Q, van Kuppeveld FJM, Reis e Sousa C. 2014. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. eLife 3:e01535. 10.7554/eLife.01535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruns AM, Pollpeter D, Hadizadeh N, Myong S, Marko JF, Horvath CM. 2013. ATP hydrolysis enhances RNA recognition and antiviral signal transduction by the innate immune sensor, laboratory of genetics and physiology 2 (LGP2). J. Biol. Chem. 288:938–946. 10.1074/jbc.M112.424416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Childs KS, Randall RE, Goodbourn S. 2013. LGP2 plays a critical role in sensitizing mda-5 to activation by double-stranded RNA. PLoS One 8:e64202–e64202. 10.1371/journal.pone.0064202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Childs KS, Randall RE, Goodbourn S. 2012. Paramyxovirus V proteins interact with the RNA helicase LGP2 to inhibit RIG-I-dependent interferon induction. J. Virol. 86:3411–3421. 10.1128/JVI.06405-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, Schmidt A, Rothenfusser S, Hopfner K-P. 2009. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 37:2014–2025. 10.1093/nar/gkp059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gitlin L, Benoit L, Song C, Cella M, Gilfillan S, Holtzman MJ, Colonna M. 2010. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6:e1000734. 10.1371/journal.ppat.1000734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale MJ. 2008. Establishment and maintenance of the innate antiviral response to West Nile virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 82:609–616. 10.1128/JVI.01305-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610. 10.1084/jem.20080091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh C-S, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- 30.McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Virgin HW, Colonna M. 2008. MDA-5 recognition of a murine norovirus. PLoS Pathog. 4:e1000108. 10.1371/journal.ppat.1000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 103:8459–8464. 10.1073/pnas.0603082103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strähle L, Marq J-B, Brini A, Hausmann S, Kolakofsky D, Garcin D. 2007. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J. Virol. 81:12227–12237. 10.1128/JVI.01300-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw ML, Cardenas WB, Zamarin D, Palese P, Basler CF. 2005. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and Toll-like receptor 3-triggered signaling pathways. J. Virol. 79:6078–6088. 10.1128/JVI.79.10.6078-6088.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnston MD. 1981. The characteristics required for a Sendai virus preparation to induce high levels of interferon in human lymphoblastoid cells. J. Gen. Virol. 56:175–184. 10.1099/0022-1317-56-1-175 [DOI] [PubMed] [Google Scholar]
- 35.Strähle L, Garcin D, Kolakofsky D. 2006. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 351:101–111. 10.1016/j.virol.2006.03.022 [DOI] [PubMed] [Google Scholar]
- 36.Yount JS, Kraus TA, Horvath CM, Moran TM, López CB. 2006. A novel role for viral-defective interfering particles in enhancing dendritic cell maturation. J. Immunol. 177:4503–4513. 10.4049/jimmunol.177.7.4503 [DOI] [PubMed] [Google Scholar]
- 37.Yount JS, Gitlin L, Moran TM, López CB. 2008. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai virus defective interfering particles. J. Immunol. 180:4910–4918. 10.4049/jimmunol.180.7.4910 [DOI] [PubMed] [Google Scholar]
- 38.Baum A, Sachidanandam R, García-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:16303–16308. 10.1073/pnas.1005077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole E, He B, Lamb RA, Randall RE, Goodbourn S. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33–46. 10.1006/viro.2002.1737 [DOI] [PubMed] [Google Scholar]