ABSTRACT
Influenza A viruses recruit components of the nuclear import pathway to enter the host cell nucleus and promote viral replication. Here, we analyzed the role of the nuclear import factor importin-α7 in H1N1 influenza virus pulmonary tropism by using various ex vivo imaging techniques (magnetic resonance imaging, confocal laser scanning microscopy, and correlative light-electron microscopy). We infected importin-α7 gene-deficient (α7−/−) mice with a recombinant H1N1 influenza virus and compared the in vivo viral kinetics with those in wild-type (WT) mice. In WT mice, influenza virus replication in the bronchial and alveolar epithelium already occurred a few days after infection. Accordingly, extensive mononuclear infiltration and alveolar destruction were present in the lungs of infected WT mice, followed by 100% lethality. Conversely, in α7−/− mice, virus replication was restricted mostly to the bronchial epithelium with marginal alveolar infection, resulting in significantly reduced lung damage and enhanced animal survival. To investigate the host immune response during alveolar virus replication, we studied the role of primary macrophages in virus propagation and clearance. The ability of macrophages to support or clear the virus infection, as well as the host cellular immune responses, did not significantly differ between WT and α7−/− mice. However, cytokine and chemokine responses were generally elevated in WT mice, likely reflective of increased viral replication in the lung. In summary, these data show that a cellular factor, importin-α7, is required for enhanced virus replication in the alveolar epithelium, resulting in elevated cytokine and chemokine levels, extensive mononuclear infiltration, and thus, severe pneumonia and enhanced virulence in mice.
IMPORTANCE Influenza A viruses are respiratory pathogens that may cause pneumonia in humans. Viral infection and replication in the alveoli of the respiratory tract are believed to be crucial for the development of the acute respiratory distress syndrome associated with fatal outcomes in influenza virus-infected patients. Here, we report the requirement of a cellular factor, importin-α7, for efficient virus replication in the alveolar epithelium of mice. Using complementary ex vivo imaging approaches, we show that influenza virus replication is restricted to the bronchial epithelium, followed by enhanced survival in importin-α7-deficient mice. In contrast, the presence of this gene results in enhanced virus replication in the alveoli, elevated cytokine and chemokine responses, mononuclear infiltration, alveolar destruction, and 100% lethality in wild-type mice. Taken together, our results show that importin-α7 is particularly required for virus replication in the alveolar epithelium in association with severe pneumonia and death in mice.
INTRODUCTION
Influenza A viruses cause yearly epidemics and recurrent pandemics. The emergence of new influenza virus strains from animal reservoirs further poses a continuous threat to human health (1). Influenza virus transcription and replication occur in the nuclei of respective host cells. Since cellular components that transport the viral ribonucleoprotein complex into the nucleus are divergent between species, viral polymerase subunit PB2 and the nucleoprotein (NP) require adaptation to the nuclear import machinery upon interspecies transmission (2). The classical nuclear import pathway consists of an adaptor protein, importin-α, that binds to cargo proteins containing a nuclear localization signal and the transport receptor protein, importin-β1, which then promotes nuclear entry (3). Influenza viruses with host adaptive signatures in their ribonucleoprotein complexes, such as PB2 627K or 701N, bind more efficiently to human importin-α7 proteins. It was shown that importin-α7 is a positive cellular factor required for efficient virus polymerase activity and replication in human cells (4, 5). Furthermore, deletion of the importin-α7 gene results in reduced pathogenicity of mammalian influenza viruses in mice (5, 6). These findings highlight the importance of importin-α7 for influenza virus replication and pathogenicity in the mammalian host.
The primary target cells for influenza virus replication are epithelial cells of the respiratory tract. Virus replication in respiratory cells and subsequent modulation of the host immune response may lead to compromised respiratory functions (7). Hereby, influenza virus infection of the alveolar epithelium is particularly crucial for the development of an acute respiratory distress syndrome (ARDS) that often results in fatal outcomes for humans (8). Viral replication in the alveolar epithelium can damage the epithelial-endothelial barrier, resulting in leakage of proteinaceous fluid into the alveolar lumen and thus respiratory insufficiency and death (8). Moreover, infected epithelial cells produce cytokines that then attract leukocytes, such as macrophages and neutrophils, stimulating further mononuclear infiltration and respiratory failure (7, 8).
It was previously reported that lung macrophages of wild-type (WT) mice were more abundantly positive for viral RNA than those of α7−/− mice (6). This observation suggests that a potential modulation of the host immune response might be a reason for enhanced pathogenicity in the mammalian host. To shed light on the role of the importin-α7 gene in lung pathology, we used a recombinant influenza virus that expresses a green fluorescent protein (GFP) fused to the viral NS1 gene of the A/Puerto Rico/8/34 PR8 (H1N1) virus (PR8:NS1-GFP), which replicates efficiently in the murine lung and causes severe pathogenicity in mice (9, 10). In both WT and α7−/− mice, we analyzed in vivo virus kinetics and pulmonary tropism. These studies were complemented by ex vivo imaging techniques (magnetic resonance imaging [MRI], confocal laser scanning microscopy, and correlative light-electron microscopy) to assess the role of importin-α7 in viral lung tropism and its potential effect on the host immune response.
MATERIALS AND METHODS
Ethical statement.
All of our animal experiments were approved by the relevant German authority (Behörde für Stadtentwicklung und Umwelt Hamburg) and performed according to the national guidelines of the animal protection law (Tierschutzgesetz, project 97/11) in Germany.
Cells and viruses.
Madin-Darby canine kidney (MDCK) cells were grown in minimal essential medium (PAA, Linz, Austria) supplemented with 10% fetal calf serum (PAA), 1% glutamine (PAA), and 1% penicillin-streptomycin (PAA).
Generation of the PR8:NS1-GFP (H1N1) virus was described previously (9). Briefly, the GFP used in this recombinant virus was derived from the pMAXGFP plasmid (Amaxa), which contains the enhanced GFP gene from the copepod Pontellina plumata.
Animal experiments.
Importin-α7−/− (α7−/−) mice (6, 11) and WT littermates thereof in the C57BL/6J genetic background were maintained at the animal facility of the Heinrich Pette Institute, Leibniz Institute for Experimental Virology, Hamburg, Germany.
Mice were anesthetized with ketamine-xylazine (70 and 7 mg/kg, respectively). Fifty percent minimum lethal dose (MLD50) studies were performed with serial virus dilutions of PR8:NS1-GFP (104, 105, and 106 PFU) in WT and α7−/− mice. In all other studies, WT and α7−/− mice were intranasally infected with 106 PFU of the PR8:NS1-GFP virus. Survival and weight loss were monitored for 14 days. Mice were humanely killed upon >25% weight loss according to the guidelines of the German animal protection law.
For detection of virus replication kinetics and lung tropism, whole lungs of WT and α7−/− mice infected with PR8:NS1-GFP influenza virus were harvested on days 1, 3, and 6 postinfection (p.i.). The lung tissue of each animal was divided into two parts; one was processed for immunohistochemical analysis, and the other was used for virus titration and cytokine measurements. The anatomical areas within the lung were chosen randomly to diminish detection bias. For cytokine and virus titer detection, lungs were further homogenized in 1 ml of phosphate-buffered saline (PBS) and centrifuged at 956 × g for 5 min at 4°C, and aliquots of supernatants were stored at −80°C. Virus titers were determined by plaque assays on MDCK cells.
Cytokine assay.
A mouse cytokine antibody array (RayBiotech, Inc.) was used according to the manufacturer's instructions to detect cytokines and chemokines in lung homogenates of influenza virus-infected mice. Samples were not thawed more than once for this assay. The lung homogenates of three to five mice were pooled and measured with one array. Intensity of signals was quantified with the ImageQuant LAS 4000 bioimager at nonsaturated levels. The array tests for the cytokines and chemokines granulocyte colony-stimulating factor (GCS-F), granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-2 (IL-2), IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 (p40p70), IL-12(p70), IL-13, IL-17, gamma interferon (IFN-γ), monocyte chemoattractant protein 1 (MCP-1), MCP-5, RANTES, stem cell factor, soluble tumor necrosis factor (TNF) receptor type I, TNF-α, thrombopoietin, and vascular endothelial growth factor.
Histological analysis.
Lungs were harvested and fixed with 3.7% paraformaldehyde (PFA) and embedded in paraffin as described before (12). Briefly, tissue sections were directly visualized by fluorescence microscopy or further processed by immunohistochemistry with anti-fowl plague virus serum and the ZytoChem Plus (HRP) Broad Spectrum (DAB) kit (Zytomed) according to the manufacturer's instructions.
Correlative imaging.
Mice were intranasally infected with 106 PFU of PR8:NS1-GFP or treated with PBS as a control. On day 3 p.i., they were sacrificed and subjected to intracardiac perfusion with 2% PFA. Lungs were removed en bloc, degassed in a 2% PFA solution overnight, and embedded in PBS-buffered low-melting-point agarose (1%). MRI examinations were performed with a dedicated small-animal MRI scanner at 7 T (ClinScan; Bruker BioSpin, Ettlingen, Germany) and a circularly polarized bird cage send-receive coil with an inner diameter of 40 mm (Bruker BioSpin, Ettlingen Germany). MRI is a noninvasive tomographic imaging technique that uses the sensitivity of the nuclear magnetic resonance of hydrogen to its chemical environment to generate contrast in gray-scale images of biological samples. The digital images generated are discrete, and the volume elements of the sample corresponding to the image pixels are called voxels. As the signal intensity, i.e., the gray value of each voxel, is given in arbitrary units, it is appropriate to use the signal-to-noise-ratio (SNR) to compare images acquired from different samples.
High-resolution images with an isotopic voxel dimensions of 90 μm were acquired with a three-dimensional fast-recovery, fast-spin echo sequence. The imaging parameters chosen were as follows: repetition time, 1,500 ms; echo time, 44 ms; turbo factor, 8; number of signal averages, 2; field of view, 35 mm; acquisition matrix size, 384 by 384 by 96. The total acquisition time was 6:43 h. This special sequence generates T2-weighted images in which a high SNR corresponds to high hydrogen density and motility, e.g., free watery fluid. In contrast, a low SNR indicates tissues with low hydrogen density and structural heterogeneities caused by, e.g., fibrotic tissue alterations, the presence of high protein density, or paramagnetic substances such as deoxyhemogoblin. In the animal models, low lung parenchyma SNRs represent regions of inflammatory lung tissue alterations.
Quantitative MR analyses were performed with ImageJ 1.37 (National Institutes of Health, Bethesda, MD). Therefore, the lung parenchyma was automatically segmented from the agarose gel background (lower and upper thresholds of signal intensity, 400 and 1,500, respectively). The distributions of the signal intensities within the lungs of all specimens were then determined. The SNR of each lung voxel was calculated by measuring the standard deviation of the background noise of the agar surrounding the lungs. By using these data, the mean SNR and normal standard deviation of all lung voxels were calculated for each lung separately.
After MRI imaging, the lungs were sectioned into 200-μm sections with a Vibratome (Leica VT1200 S). Several sections were preselected by fluorescence microscopy at low magnifications (Nikon AZ100) and imaged by confocal microscopy (Zeiss LSM 510 Meta and Nikon C2 Plus) after staining with the CellMask Orange Plasma membrane Stain (Life Technologies, Carlsbad, CA). Additionally, selected sections were stained with 1% osmium tetroxide in PBS and investigated fully hydrated by environmental scanning electron microscopy (ESEM; Philips XL30ESEM) with the backscattered electron detector. Alternative selected sections were postfixed with glutaraldehyde (2.5% in PBS) and processed for transmission electron microscopy (TEM) as described before (13).
Primary macrophage isolation and in vitro activity assays.
Primary macrophages were isolated either by bronchoalveolar lavage (BAL) to obtain alveolar macrophages or by peritoneal lavage to obtain naive tissue-resident macrophages, as described previously (14). Briefly, cells obtained from peritoneal exudates were harvested from euthanized naive mice by administration of 5 ml PBS–0.5% EDTA into the peritoneal cavity. BAL was performed with the same animal. Therefore, the chest cavity was opened and the trachea was cannulated with a needle and anchored by suturing. The lung was gently infused 10 times with 0.5 ml PBS–0.5% EDTA. Cells from mice (n = 4 to 6) were pooled and pelleted by centrifugation at 400 × g and 4°C for 10 min. Cells were resuspended in Macrophage-SFM (Gibco), and the phenotype was assessed by flow cytometry (see Table 2).
TABLE 2.
Phenotypic analysis of isolated primary alveolar and interstitial macrophages by flow cytometrya
| Cell type | % of total cells in: |
|||
|---|---|---|---|---|
| BAL fluid |
Peritoneal lavage fluid |
|||
| Before | After | Before | After | |
| B cells | 4.7 | 0.08 | 17.6 | 0.07 |
| T cells | 0.9 | 0.1 | 9.7 | 0.15 |
| NK cells | 0.6 | 0.14 | 2.9 | 0.04 |
| AM | 82.6 | 80 | ||
| DCs | 7.58 | 11 | 0.95 | |
| CD11c− macrophages | 0.08 | 50.8 | 85 | |
| CD11cmid macrophages | 0.38 | 2.89 | ||
| Granulocytes | 0.57 | 0.0 | 0.12 | 0.00 |
The distribution of cell components in BAL and peritoneal lavage fluid samples isolated from mice was assessed by flow cytometry (for the gating strategy used, see Fig. 10) before and after culturing of cells in Macrophage-SFM (Gibco) for 1 day.
For in vitro infection of macrophages, 105 cells/well were plated into the wells of a 24-well cell culture plate (Nunc, China). Sixteen hours after plating, more than 80% of the remaining cells were macrophages and were directly infected at a multiplicity of infection (MOI) of 1 in the presence of 0.25 μg/μl Na-p-tosyl-l-lysine chloromethyl ketone (TPCK)-trypsin (see Table 2). Viral titers were determined by plaque assay on MDCK cells at several time points after infection (0, 6, and 24 h p.i.).
Alternatively, isolated macrophages were directly used in virus clearance experiments as previously described (15). Briefly, MDCK cells (2 × 105 cells/well) were seeded into the wells of a 24-well cell culture plate (Nunc, China) and infected with PR8:NS1-GFP (MOI of 0.01) 24 h later. After 4 h, cells from BAL fluid or peritoneal exudates were added to the infected MDCK cell cultures. The ratio of effector to MDCK cells was 1:1 in cells from BAL fluid and 10:1 for cells obtained from peritoneal lavage fluid. Cell culture medium was supplemented with 0.25 μg/μl TPCK-trypsin. Supernatants were collected 7 h later, and virus titers (11 h p.i.) were determined on MDCK cells by plaque assay.
Flow cytometry.
Mice were euthanized and exsanguinated at the p.i. times indicated. BAL was performed as described above with 500 μl ice-cold PBS (2 mM EDTA). Lungs were removed after PBS perfusion of the right ventricle of the heart and digested with 30 IU DNase I (Sigma-Aldrich, Taufkirchen, Germany) and 2 mg/ml collagenase D (Roche, Mannheim, Germany) at 37°C for 30 min. Cells were then passed through a 40-μm cell strainer, and red blood cells were lysed with RCL buffer (eBiosience, Frankfurt am Main, Germany). Unspecific antibody binding was blocked by CD16/CD32 block (TruStain fcX BioLegend, San Diego, CA) and normal rat serum (Jackson Immuno Research, Suffolk, United Kingdom). Multicolor analyses were performed with an LSR II flow cytometer (BD, Heidelberg, Germany) and analyzed with FlowJo software (Tree Star, Ashland, OR). The gating strategy used (see Fig. 10) was adapted from previous studies especially to analyze the effect of viral infection on different macrophage populations that were distinguished as either resident in the lung (CD11c−) or as those recruited from the bloodstream and infiltrating the lung (CD11cmid) (16). Monoclonal antibodies specific for CD19 (6D5), NKp46 (29A1.4), CD8 (53-6.7), CD4 (RM4-5), CD11b (M1/70), Gr-1 (RB6-8C5), CD11c (N418), CD3e (145-2C11), F4/80 (CI:A3-1), and CD45 (30-F11) were purchased from BioLegend or eBioscience. Alexa Fluor 594 carboxylic acid, succinimidyl ester, was used to stain and exclude dead cells before fixation with 1% PFA for 15 min. Cells were reconstituted in 0.5% bovine serum albumin–2 mM EDTA–PBS before acquisition.
FIG 10.
Gating strategy used to define immune cell populations in mice infected with influenza virus. Cell suspensions were analyzed by flow cytometry with an antibody panel that allowed the following subpopulations to be distinguished: CD11b+, CD11c−, F4/80−, and Gr-1+ granulocytes; CD11b+, CD11c−, F4/80+, and Gr-1+ macrophages (Mϕ); CD11b+, CD11c−, F4/80+, and Gr-1− macrophages; CD11b+ and CD11cmid macrophages; CD11b− and CD11chigh alveolar macrophages; CD11b+ and CD11chigh DCs; CD11b− and CD11cmid DCs; NKp46+ and CD3e− NK cells; and NKp46− and CD3e+ T cells. Here, we illustrate the gating strategy used for the lung homogenates and BAL fluid samples from an uninfected (PBS-treated) mouse and an infected mouse at 3 days p.i.
Statistical analysis.
All of the data shown are presented as the mean ± the standard error of the mean. Student's t test (unpaired, two tailed), analysis of variance, and the Mantel-Cox test were calculated with GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA). A P value of <0.05 was considered significant.
RESULTS
Influenza virus replication and pathogenesis are reduced in α7−/− mice.
First, we have assessed PR8:NS1-GFP (H1N1) influenza virus pathogenesis in WT and α7−/− mice. Infected WT animals showed severe weight loss and pathogenicity, resulting in approximately three times more virulence than α7−/− mice (Fig. 1A and B; Table 1). Analysis of viral lung titers showed similar virus replication properties in WT and α7−/− mice on day 3 p.i. but reduced virus titers in α7−/− mice on day 6 p.i. (Fig. 1C).
FIG 1.
Pathogenicity of PR8:NS1-GFP (H1N1) influenza virus in WT and importin-α7−/− mice. WT (black squares; n = 10) or α7−/− (red open squares; n = 10) mice were intranasally inoculated with 106 PFU of PR8:NS1-GFP virus. Weight loss (A) and survival (B) were monitored for 14 days. Mice receiving PBS were used as controls (gray circles; n = 6). The data shown represent means ± the standard errors of the means. (C) Lungs were removed from WT or α7−/− mice at 1, 3, and 6 h p.i. (n = 2) and on days 1, 3, and 6 p.i. (n = 5), and virus titers were determined by plaque assay.
TABLE 1.
MLD50 of PR8:NS1-GFP virus for WT and α7−/− micea
| Virus dose (PFU) | % Survival |
P value | MLD50 (PFU) |
||
|---|---|---|---|---|---|
| WT | α7−/− | WT | α7−/− | ||
| 106 | 10 | 50 | 0.1018 | 105.5 | 106.0 |
| 105 | 90 | 100 | 0.3173 | ||
| 104 | 100 | 100 | 1 | ||
The MLD50 was calculated as described in reference 29. The significance of differences in survival between WT and α7−/− mice was calculated by the Mantel-Cox test.
These findings confirm and extend previous reports (5, 6) showing that importin-α7 is required for enhanced H1N1 influenza virus replication and pathogenicity in mice.
Influenza virus replication is reduced in the alveoli of α7−/− mice.
First, we correlated the NS1-GFP expression profile with the viral-NP-positive cells in the lungs of PR8:NS1-GFP (H1N1) influenza virus-infected WT and α7−/− mice by histochemical and immunohistochemical analyses (Fig. 2).
FIG 2.
In vivo kinetics of influenza virus replication in lungs of WT and importin-α7−/− mice. WT or α7−/− mice were intranasally inoculated with 106 PFU of PR8:NS1-GFP virus. Control mice received PBS. On days 1, 3, and 6 p.i., lungs were harvested, formalin fixed, paraffin embedded, and visualized in a wide-field fluorescence microscope. (A) Direct visualization of GFP expression in histological tissue sections. (B) Viral NP detection in tissue sections by immunohistochemical staining. Virus antigen-positive cells are red-brown. Representative infected cells are indicated by arrows.
NS1-GFP-positive pulmonary cells were already detected in bronchial epithelium and alveolar tissue on day 1 p.i. in WT mice (Fig. 2A). At 3 days p.i., viral replication manifested in bronchial epithelial cells with increasing levels of GFP-positive cells in the alveoli. Virus replication further increased in both bronchial epithelium, and alveolar tissue on day 6 p.i. in WT mice. In contrast, virus replication was strongly reduced in the lungs of α7−/− mice. GFP-positive epithelial cells were not detectable in the bronchi until day 3 p.i. and only marginally present in the alveoli on day 6 p.i. (Fig. 2A). Consistently, NP-positive cells were most abundant in bronchial and alveolar cells of WT lungs, following the kinetics of GFP expression. Again, in α7−/− mice, virus replication was largely restricted to epithelial cells of the bronchi with single NP-positive alveolar cells on day 6 p.i. (Fig. 2B). Furthermore, enhanced virus replication in the alveoli correlated with mononuclear infiltration as early as day 3 p.i. and severe alveolar destruction on day 6 p.i. in WT mice. In contrast, alveolar damage was strongly reduced in the lungs of α7−/− mice, where the first single mononuclear infiltrations were observed on day 6 p.i. (Fig. 2).
These findings show that importin-α7 is required for enhanced virus replication in alveolar cells of the respiratory tract.
Lung damage upon influenza virus replication is strongly reduced in α7−/− mice.
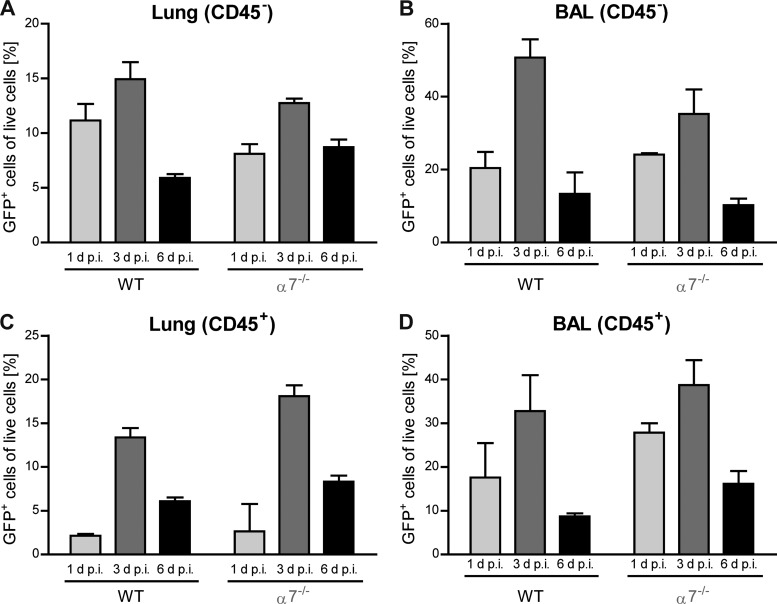
Comparable expression patterns of GFP- and NP-positive cells in the lungs of PR8:NS1-GFP (H1N1) influenza virus-infected mice confirmed the feasibility of using GFP expression as a marker of active virus replication in subsequent ex vivo imaging. Since the highest number of GFP-positive stromal and immune cells, as well as the highest virus titers, were detected on day 3 p.i. (Fig. 1C and 3), we used this time point of productive virus replication for further lung imaging.
FIG 3.
Kinetics of influenza virus infection in live cells from infected WT and importin-α7−/− mice. WT or α7−/− mice were intranasally infected with 106 PFU of PR8:NS1-GFP virus. Percentages of GFP+ stromal cells (CD45−) (A and B) or immune cells (CD45+) (C and D) from homogenized lungs (A and C) or BAL fluid samples (B and D) were assessed by flow cytometry at days 1, 3, and 6 p.i. (n = 2 to 6). For a description of the gating strategy used, see Fig. 10.
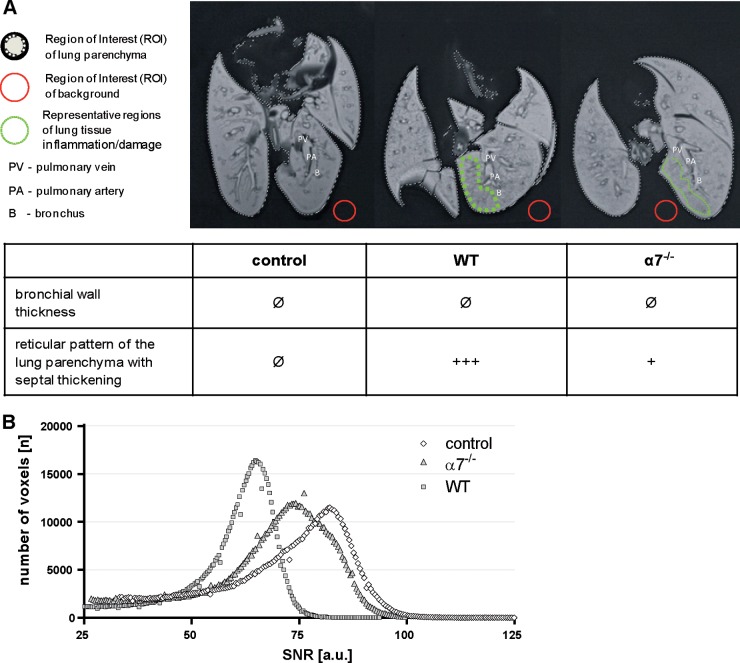
Therefore, we developed an ex vivo method to analyze the macroscopic damage of infected lungs by MRI (Fig. 4; see Videos S1, S2, and S3 in the supplemental material). The major advantages of this nondestructive method are the high soft tissue contrast and the relatively rapid examination of the whole-lung pathology, in contrast to histological analysis, which is restricted to certain pulmonary areas.
FIG 4.
Total-lung imaging of WT and importin-α7−/− mice upon influenza virus infection. Macroscopic lung tissue damage was analyzed ex vivo by MRI. Control mice treated with PBS were compared with WT and α7−/− mice infected with 106 PFU of PR8:NS1-GFP virus. Lungs were processed at day 3 p.i. (A) T2-weighted MRI presents virus-induced lung inflammation and damage with the formation of a hypointense reticular pattern within the lung parenchyma, which was most severe in WT mice, followed by α7−/− mice. In contrast, uninfected control mice showed homogeneous lung tissues. (B) According to this qualitative rating, the quantitative distribution analysis of the SNR of each voxel of the lungs demonstrated the highest values in control mice (white diamonds) and reduced SNRs in α7−/− (light gray triangles) and WT mice (dark gray squares). a.u., arbitrary units. See also Videos S1, S2, and S3 in the supplemental material.
Ex vivo MRI of the lungs of uninfected control mice differed from that of the lungs of WT and α7−/− mice (Fig. 4). Bronchi, bronchioles, alveoli, and all of the pulmonary vessels (pulmonary arteries and veins) were fluid filled, and therefore, the lung tissue appears artificially hyperintense in T2-weighted MRI. Control mice showed a regular anatomical structure and texture in T2w MRI with a homogeneous tissue pattern. In contrast, WT and α7−/− mice presented virus-induced lung damage with typical inflammatory changes, such as septal thickening leading to reticular consolidations and more tissue heterogeneity (Fig. 4A). In consequence, these changes result in a reduced SNR of the lung parenchyma in T2w images which was most prominent in WT mice whereas reduced in α7−/− mice (Fig. 4B). T2w MR image-based quantitative volumetric SNR analyses of the lungs showed the highest SNR in the lungs of control mice (71.5 ± 15.07), followed by those of α7−/− (67.99 ± 14.88) and WT mice (59.4 ± 10.37).
These data show that WT lung parenchyma is more damaged in WT mice than in α7−/− mice.
Progeny virus was most abundantly detected in areas of infected bronchi.
Next, we analyzed physical sections of the lungs described above by fluorescence light microscopy, ESEM, and correlative light-electron microscopy in order to assess whether areas of virus infection and replication in the lungs correlate with areas of virus production (Fig. 5).
FIG 5.
Correlative imaging of influenza virus-infected WT and importin-α7−/− mouse lungs. On day 3 p.i., lungs of PR8:NS1-GFP virus-infected WT (A, B, E, and F) or α7−/− (C, D, G, and H) mice were fixed with PFA and removed for vibratome sectioning. Without any further treatment, sections of infected lungs were visualized with a wide-field fluorescence microscope (Nikon AZ100) (E and G). Sections of infected lungs were stained with CellMask orange plasma membrane stain (in red) (Life Technologies, Carlsbad, CA) and visualized with a Zeiss LSM 510 Meta confocal microscope (A and C) and a Philips XL30 ESEM after staining with 1% OsO4 (B and D). Biopsy specimens were taken in preselected areas (white circles in panels E and G), processed, and embedded in Epon resin for TEM (F and H) (FEI Tecnai G20 equipped with an Eagle 4k charge-coupled device camera and operated at 80 kV). Virus particles were detected upon consecutive magnifications of the areas indicated.
Fluorescence microscopy imaging of the lungs revealed that virus infection and replication occurred predominantly in the bronchial tubes of α7−/− mouse lungs, in contrast to WT mouse lungs, where, in addition, alveolar cells were abundantly infected. However, the density of GFP+ cells in the bronchi was higher in α7−/− than in WT mice (Fig. 5A and C). ESEM revealed severely damaged parenchymal areas in WT mouse lungs, in contrast to α7−/− mouse lungs (Fig. 5B and D). These findings correlate with the observations made in the histological sections (Fig. 2) and MRI analysis (Fig. 4).
Next, we preselected infected areas with a light microscope and further processed small circular biopsy specimens by TEM (Fig. 5E and G). Upon progressive magnification of these areas, we could detect virus particles especially in areas of infected bronchial epithelia. By inspection of random fields, we found a higher content of virus particles especially around bronchial epithelial cells than in the alveoli of both α7−/− and WT mice (Fig. 5F and H).
These observations indicate that primary virus production most likely occurs in infected bronchial epithelium. Furthermore, importin-α7 is required particularly for alveolar replication, which seems to be essential for severe lung damage and enhanced pathogenicity in mice.
Cytokine and chemokine levels are reduced in influenza virus-infected α7−/− mice.
We next analyzed whether infection of differential respiratory cells in WT and α7−/− mice might affect cytokine and chemokine responses which are associated with lung damage upon influenza virus infection. Therefore, we measured the innate immune response in the lungs of PR8:NS1-GFP (H1N1) influenza virus-infected mice by using an antibody array for 22 different cytokines and chemokines.
Here, we show only the analytes that revealed significant differences between WT and α7−/− mice upon influenza virus infection. In general, chemokine and cytokine levels were considerably more elevated during the course of infection in WT than in α7−/− mice (Fig. 6). The only cytokines that were an exception to this were IL-9 and IL-12, with higher levels in α7−/− than WT mice throughout the course of infection (Fig. 6H and I).
FIG 6.
Chemokine and cytokine responses in lungs of influenza virus-infected WT and importin-α7−/− mice. Lungs of WT (black bars) and α7−/− (red bars) mice infected with 106 PFU of PR8:NS1-GFP virus were harvested on days 1, 3, and 6 p.i. (n = 3 to 5). Homogenates of these lungs were assessed for chemokine and cytokine responses with mouse cytokine antibody array membranes (RayBiotech, Inc.). Shown here are the levels of the following cytokines and chemokines, which revealed significant differences between WT and α7−/− mice upon influenza virus infection: IL-2 (A), IL-13 (B), TNF-α (C), MCP-1 (D), RANTES (E), IL-6 (F), IL-10 (G), IL-9 (H), and IL-12 (I). Relative expression levels were calculated in relation to the standardized array positive control, which was set 100%. Baseline cytokine levels of PBS-inoculated control mice are shown as dashed lines in the cytokine graphs (black lines for WT mice, red lines for α7−/− mice).
These data suggest that the presence of importin-α7 generally correlates with elevated cytokine and chemokine responses in the lung.
Cellular immune responses are not significantly altered in WT and α7−/− mice.
In order to identify infected immune cell subsets that could potentially contribute to reduced cytokine and chemokine responses in α7−/− infected mice, we quantified the cellular immune subpopulations in the lungs upon infection with PR8:NS1-GFP on day 3 p.i. by multicolor flow cytometry (Fig. 7 and 8).
FIG 7.
Immune cell subpopulations in WT and importin-α7−/− mice upon influenza virus infection. WT (black bars) or α7−/− (red bars) mice were infected with 106 PFU of PR8:NS1-GFP (n = 6). Immune cell populations in lung homogenates (A to E) or BAL fluid (F to E) were analyzed at day 3 p.i. with a BD LSR II flow cytometer. Cell frequencies were determined within the total leukocytes (CD45+) (graphs with black and red bars) and within GFP+ live cells (graphs with green bars). Frequencies of granulocytes (CD11b+, CD11c−, F4/80−, Gr-1+) (A and F), macrophages (CD11b+, CD11c−, F4/80+, Gr-1+) (B and G), alveolar macrophages (CD11b−, CD11chigh) (C and H), and DCs (CD11b+, CD11chigh) (D and I) were determined. For a description of the gating strategy used, see Fig. 10. Cell numbers from PBS-treated mice are shown in gray columns. Representative dot plots of GFP+ cells are shown to the right of the corresponding graphs. (E and J) Representative plots of the flow-cytometric analysis. *, P < 0.05; **, P < 0.01.
FIG 8.
Additional immune cell subpopulations in WT and importin-α7−/− mice upon influenza virus infection. Mice were infected with 106 PFU of PR8:NS1-GFP (n = 6). Immune cell populations were analyzed at day 3 p.i. with a BD LSR II flow cytometer. Cell frequencies in the lungs (A, C, E, G, and I) and BAL fluid (B, D, F, H, and J) were determined within the total leukocytes (CD45+) (graphs with black and red bars) and within GFP+ live cells (graphs with green bars). Frequencies of macrophages (CD11b+, CD11c−, F4/80+, Gr-1−) (A and B), macrophages (CD11b+, CD11cmid) (C and D), DCs (CD11b−, CD11cmid) (E and F), NK cells (NKp46+, CD3e−) (G and H), and T cells (NKp46−, CD3e+) (I and J) were determined and gated (see Fig. 10). Cell numbers from PBS-treated mice are shown as gray columns. Representative dot plots of GFP+ cells are shown to the right of the corresponding graphs. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In general, the distribution of immune cell populations in the lungs was similar in uninfected WT and α7−/− control mice, except for dendritic cells (DCs), identified as CD11b+, CD11chigh, and alveolar macrophages (CD11b−, CD11chigh), populations that were considerably larger in uninfected α7−/− controls than in uninfected WT controls (Fig. 7C, D, and I). Upon influenza virus infection, a higher frequency of granulocytes with a CD11b+, CD11c−, F4/80−, Gr-1+ phenotype was present in cell suspensions obtained from α7−/− mouse lungs than in those from WT mouse lungs (Fig. 7A). A similar pattern was seen for lung macrophages, identified as CD11b+, CD11c−, F4/80+, and Gr-1+ (Fig. 7B). Moreover, the presence of GFP in lung granulocytes and macrophages was significantly increased in lung cell suspensions obtained from α7−/− infected mice. The frequency of alveolar macrophages (CD11b−, CD11chigh) and DCs (CD11b+, CD11chigh) was reduced overall upon infection in both WT and α7−/− mice (Fig. 7C and D). Moreover, the frequency of other immune cell subpopulations, including natural killer (NK) and T cells, upon influenza virus infection was not significantly different in the lungs of α7−/− mice than in those of WT mice (Fig. 8A, C, E, G, and I). Flow cytometric analyses of granulocytes, macrophages, alveolar macrophages, and DCs in BAL fluid largely mirrored the findings in lung tissues (Fig. 7F, G, H, and I and 8).
These data indicate that the cellular immune response upon influenza virus infection is similar in the lungs of WT and α7−/− mice with regard to the frequency of alveolar macrophages and DCs. However, the frequencies of granulocytes and macrophages are elevated in the absence of importin-α7.
In vitro primary macrophage function in promoting virus replication or clearance is not significantly altered in WT and α7−/− mice.
To assess the role of an increased macrophage frequency upon infection of α7−/− mice, we performed in vitro macrophage functionality assays (Fig. 9). By this approach, we addressed whether viral RNA-positive macrophages in alveoli, which were reported before to be detected predominantly in infected WT but not α7−/− mouse lungs (6), reflect active virus replication or the remains of virus clearance.
FIG 9.
Role of primary macrophages in virus influenza virus replication and clearance in vitro. Primary macrophages were isolated from WT and α7−/− mice by BAL (AM) or peritoneal lavage (PM). (A) In vitro macrophage infection. Cells were infected 24 h after seeding at an MOI of 1. (B) Virus clearance. MDCK cells were infected at an MOI of 0.01. At 4 h p.i., isolated primary macrophages were added. In both experiments, supernatants were harvested at the time points indicated for subsequent virus titer determination by plaque assay. The virus titer at 0 (A) or 4 (B) h p.i. was set to 100%. The representative results shown are averages of three independent experiments. AMϕ, alveolar macrophages; PMϕ, primary macrophages.
Therefore, we isolated primary macrophages from naive WT and α7−/− mice by two methods, BAL, in order to obtain alveolar macrophages, and peritoneal lavage, in order to obtain interstitial macrophages (Table 2). Enrichment of the macrophage population after seeding was confirmed by flow cytometry as described in Fig. 10. First, we infected these macrophages in vitro at an MOI of 1 and analyzed virus replication at different time points after infection by plaque assay (Fig. 9A). Although virus titers increased by approximately four times at 6 h p.i., virus replication was far less efficient in primary mouse macrophages than in epithelial cells, consistent with previous reports (17, 18).
Additionally, we examined the clearance of virus produced in infected epithelial cells (MDCK) by primary macrophages. MDCK cells infected with PR8:NS1-GFP virus (MOI of 0.01) and cocultured with primary macrophages isolated from BAL fluid or peritoneal lavage fluid at 4 h p.i. Virus replication was then analyzed 7 h later by plaque assay (Fig. 9B). Both WT and α7−/− mouse primary macrophages isolated from BAL fluid and peritoneal lavage fluid were able to reduce virus titers at early but not at later time points (48, 72, and 96 h) p.i., where high virus titers are produced by epithelial cells (data not shown).
Taken together, these data suggest that primary macrophages are not the primary promoters of productive virus replication. Further, primary alveolar and interstitial macrophages did not show any significant difference in the ability to clear a viral infection in vitro.
DISCUSSION
To our knowledge, this is the first report of a cellular factor that contributes to enhanced virus replication in the alveolar epithelium. The pathogenesis of influenza virus-induced ARDS has been recently reported to be centered on the alveolar epithelium. Infection of the alveoli is considered to be a crucial site that decides whether primary viral pneumonia with ARDS might develop or not (8). In general, influenza virus infection and replication start in the respiratory tract in areas close to large conducting airways, followed by bronchioles and deeper areas of the lungs, such as alveoli (19, 20). Influenza viruses target a broad range of epithelial cells in the respiratory tract. Viral replication in the alveolar epithelium, consisting of type I and II pneumocytes, might damage the epithelial-endothelial barrier of the alveoli, leading to fluid leakage into the alveolar lumen and thus ARDS (8).
By combining various ex vivo imaging techniques (MRI, confocal laser scanning microscopy, and correlative TEM), we have shown here that PR8:NS1-GFP influenza virus replication in α7−/− mice is restricted mostly to the bronchial epithelium. In contrast, the bronchial and alveolar epithelium of WT mice is abundantly infected at a time point where overall virus lung titers do not differ (Fig. 1C and 2). Interestingly, GFP signal intensity increased upon multicycle replication in the bronchial epithelium of α7−/− mice without elevated alveolar replication (Fig. 2 and 5). This suggests that influenza virus replication is indeed restricted to the bronchial epithelium, where virus-positive cells increase with the replication cycle and cannot be explained by delayed virus kinetics only. Conversely, virus replication increases in the presence of importin-α7 in WT mice with increased alveolar replication during the course of infection, resulting in alveolar mononuclear infiltration and severe lung damage. Furthermore, the grade of alveolar destruction and pneumonia, as analyzed by MRI, correlates with greater virulence in WT than in α7−/− mice (Fig. 4).
In previous studies where reduced pathogenicity in mice lacking the importin-α7 gene was initially reported, in situ hybridization analysis further revealed that MAC3-positive lung macrophages were less positive for influenza virus RNA in α7−/− than in WT mice (6). To analyze whether this finding is due to improved virus replication in macrophages of WT mice or improved virus clearance properties, we performed in vitro macrophage functionality assays. Since it has been reported that monocyte-derived macrophages are not a good model, as they do not reflect the findings in primary alveolar macrophages (20), we performed all of our studies with primary murine macrophages (Table 2). Both alveolar and interstitial macrophages supported only a low level of virus production at very early time points after infection (Fig. 9A). However, virus titers detected in alveolar or interstitial macrophages did not differ in WT and α7−/− mice, suggesting that importin-α7 is not required for promotion of virus replication in primary macrophages in the lungs. These data are consistent with previous reports that virus replication in primary macrophages is usually not highly productive (17, 20, 21). Furthermore, the viral clearance activity of primary alveolar and interstitial macrophages obtained from WT and α7−/− mice did not significantly differ, suggesting that importin-α7 does not affect the phagocytotic activity of macrophages (Fig. 9B). However, we cannot exclude the possibility that macrophages behave differently in the in vivo setting since the functional assays in this study were performed in vitro.
Electron microscopic analysis further revealed that virus particles could be readily detected budding from the bronchial epithelium but far less from the alveolar epithelium (Fig. 5). This suggests that primary virus production most likely occurs in the bronchi of infected mice. Since our in vitro assays in primary alveolar macrophages did not reveal high-level virus production, it seems that at least these immune cells are not the primary contributors to virus production in the alveoli. These findings suggest that virus production in bronchial epithelia, which is not affected in α7−/− mice, contributes to but is not sufficient for the development of severe lung damage. This further highlights the importance of alveolar virus infection and replication in lung destruction and enhanced virulence.
Importin-α7 has been shown previously to act as a positive cellular factor required for enhanced influenza virus polymerase activity and replication in human cells (5, 6). Human and mammalian influenza viruses seem to have adapted to importin-α7 usage by acquiring host-adaptive mutation 627K or 701N in the polymerase PB2 subunit. Thereby, viral polymerase activity is promoted either by enhanced nuclear import (in the case of PB2 701N) of the polymerase complex or after nuclear entry (in the case of PB2 627K) by an as-yet-unknown mechanism (5, 6). However, importin-α proteins have also been reported to transport key cytokines or transcription factors thereof into the nucleus. IFN-γ was reported to be transported into the nucleus in complex with its receptor, IFN-γ receptor 1, by the importin-α/ß pathway mediated by the nuclear location signal in IFN-γ (22). TNF-α-induced nuclear translocation of NFκB, which is responsible for the expression of many cytokine-encoding genes, was also reported to be mediated by the importin-α/ß pathway (23, 24).
In order to address whether importin-α7 might be involved in displaying differential innate immune responses in influenza virus-infected mice, we have analyzed a panel of cytokines and chemokines in the lungs (Fig. 6). In general, cytokine and chemokine levels were increased to higher levels in infected WT mice than in infected α7−/− mice, with the exception of IL-9 and IL-12. Since neither virus replication in primary murine macrophages nor the cellular host immune response in general was affected, this observation might be predominantly reflecting enhanced virus replication kinetics in epithelial cells. For IL-9 and IL-12, which were considerably elevated in the lungs of α7−/− mice, it was reported that they have protective functions in disease outcome. Thus, for IL-9, it was shown to play an important role in tissue repair after helminth-induced lung inflammation. The IL-9 receptor is involved in upregulation of the antiapoptotic protein BCL-3 in the murine lung. IL-9 receptor-deficient mice fail to restore helminth-induced lung damage (25). IL-12 was reported to contribute to early NK cell-dependent IFN-γ production and to inhibit early influenza virus replication in mice (26). It remains unclear why α7−/− mice display IL-9 and IL-12 levels higher than those of the WT during influenza virus replication in the lungs. Whether importin-α7 is involved in the nuclear import pathway of these cytokines needs future investigation to understand their potentially protective role upon influenza virus infection in α7−/− mice.
Thus, reduced virus replication in the alveoli of importin-α7-deficient mice, as shown in Fig. 2 and 5, seems to lead to a generally reduced innate immune response. This is crucial for a protective disease outcome since hypercytokinemia is known to be detrimental to the clinical outcome, as observed especially upon highly pathogenic avian influenza virus infections in humans (18).
We also analyzed a large variety of immune cell populations of the cellular immune response to study whether these might be affected in importin-α7-deficient mice and additionally contribute to importin-α7-mediated lung pathology. In uninfected α7−/− control lungs, DCs and alveolar macrophages showed slightly higher frequencies than in uninfected WT lungs (Fig. 7). DCs are professional antigen-presenting cells and are the main contributors to antigen transport from the infected lung to the draining lymph nodes, where they activate virus-specific cytotoxic CD8+ T cells (7). DCs also belong to the main cytokine- and chemokine-producing immune cells, especially for IL-12 in response to pulmonary inflammation. This might be a potential explanation for why higher IL-12 levels were detected in α7−/− mice. This would then link the observed differences in the innate immune system to the altered adaptive immune system. Upon infection with highly pathogenic influenza virus strains, macrophages and neutrophils are the prominent cell types associated with severe lung pathology (17, 18, 27). However, infected α7−/− mice exhibited higher levels of granulocytes and macrophages in their lungs and BAL fluid. This might suggest greater recruitment of these immune cells in α7−/− mice or reduced cellular viability in WT mice. Furthermore, a higher frequency of GFP+ immune cells was detected in α7−/− than in WT lungs on day 3 p.i. However, this experimental setting does not allow us to distinguish whether these GFP+ cells reflect active virus replication or are positive because of the uptake of infected cells as a consequence of phagocytosis. In general, flow cytometry studies of infected lungs revealed very similar cellular immune responses in WT and α7−/− mice. The solely elevated GFP+ content in granulocytes and macrophages suggests that importin-α7 does not directly affect the infection or phagocytosis capacity of cells of the innate immune response.
Thus, our findings support the concept that importin-α7 is particularly required for enhanced virus replication in the alveolar epithelium without major involvement of the cellular host response. It has been reported before that importin-α isoforms are expressed at differential levels in different cell types (28). Thus, potential differences in importin-α7 expression levels in alveolar and bronchial epithelial cells might explain its role in promoting virus replication in the respiratory tract. Clearly, further studies are needed to measure importin-α7 expression levels in distinct cell types to understand the mechanism by which virus tropism is affected in the mammalian respiratory tract.
In summary, this study has provided first insights into the role of a cellular factor, importin-α7, in promoting virus replication, particularly in the alveolar epithelium. As a likely consequence of alveolar virus replication, cytokine and chemokine responses are elevated, resulting in alveolar inflammation, pneumonia, and a fatal outcome in mice.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Adolfo García-Sastre (Icahn School of Medicine at Mount Sinai, New York, NY) for kindly providing the PR8:NS1-GFP (H1N1) virus. We are grateful to Enno Hartmann (Institute of Biology, Center for Structural and Cell Biology, University of Lübeck, Lübeck, Germany) and Michael Bader (Max Delbrück Center, Berlin, Germany) for kindly providing the importin-α7−/− mice. We thank Carola Schneider, Gökhan Arman-Kalcek, and Gundula Pilnitz-Stolze for excellent technical assistance. The help from Markus Heine with the preparation of the lungs for correlative imaging is gratefully acknowledged. We also thank the Nikon Application Center and the Flow Cytometry Core Facility.
G.G. is funded by the Emmy-Noether Program of the German Research Foundation. P.R.I. is funded by the Alexander von Humboldt Foundation. This work was further funded by the Foundation for Research and Science Hamburg (to P.A.) and grants from the European Union (FLUPHARM) (to G.G.). The Heinrich Pette Institute, Leibniz Institute for Experimental Virology, is supported by the Free and Hanseatic City of Hamburg and the Federal Ministry of Health. The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 14 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00270-14.
REFERENCES
- 1.Klenk HD, Garten W, Matrosovich M. 2011. Molecular mechanisms of interspecies transmission and pathogenicity of influenza viruses: lessons from the 2009 pandemic. Bioessays 33:180–188. 10.1002/bies.201000118 [DOI] [PubMed] [Google Scholar]
- 2.Resa-Infante P, Gabriel G. 2013. The nuclear import machinery is a determinant of influenza virus host adaptation. Bioessays 35:23–27. 10.1002/bies.201200138 [DOI] [PubMed] [Google Scholar]
- 3.Strambio-De-Castillia C, Niepel M, Rout MP. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11:490–501. 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- 4.Gabriel G, Herwig A, Klenk HD. 2008. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 4:e11. 10.1371/journal.ppat.0040011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudjetz B, Gabriel G. 2012. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-alpha1 and -alpha7. PLoS Pathog. 8:e1002488. 10.1371/journal.ppat.1002488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabriel G, Klingel K, Otte A, Thiele S, Hudjetz B, Arman-Kalcek G, Sauter M, Shmidt T, Rother F, Baumgarte S, Keiner B, Hartmann E, Bader M, Brownlee GG, Fodor E, Klenk HD. 2011. Differential use of importin-alpha isoforms governs cell tropism and host adaptation of influenza virus. Nat. Commun. 2:156. 10.1038/ncomms1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braciale TJ, Sun J, Kim TS. 2012. Regulating the adaptive immune response to respiratory virus infection. Nat. Rev. Immunol. 12:295–305. 10.1038/nri3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Short KR, Kroeze EJ, Fouchier RA, Kuiken T. 2014. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect. Dis. 14:57–69. 10.1016/S1473-3099(13)70286-X [DOI] [PubMed] [Google Scholar]
- 9.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, García-Sastre A. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. U. S. A. 107:11531–11536. 10.1073/pnas.0914994107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez JT, García-Sastre A, Manicassamy B. 2013. Insertion of a GFP reporter gene in influenza virus. Curr. Protoc. Microbiol. Chapter 15:Unit 15G.4. 10.1002/9780471729259.mc15g04s29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rother F, Shmidt T, Popova E, Krivokharchenko A, Hugel S, Vilianovich L, Ridders M, Tenner K, Alenina N, Kohler M, Hartmann E, Bader M. 2011. Importin alpha7 is essential for zygotic genome activation and early mouse development. PLoS One 6:e18310. 10.1371/journal.pone.0018310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabriel G, Klingel K, Planz O, Bier K, Herwig A, Sauter M, Klenk HD. 2009. Spread of infection and lymphocyte depletion in mice depends on polymerase of influenza virus. Am. J. Pathol. 175:1178–1186. 10.2353/ajpath.2009.090339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simionescu N, Simionescu M. 1976. Galloylglucoses of low molecular weight as mordant in electron microscopy. I. Procedure, and evidence for mordanting effect. J. Cell Biol. 70:608–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Goncalves R, Mosser DM. 2008. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. Chapter 14:Unit 14.1. 10.1002/0471142735.im1401s83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujisawa H. 2008. Neutrophils play an essential role in cooperation with antibody in both protection against and recovery from pulmonary infection with influenza virus in mice. J. Virol. 82:2772–2783. 10.1128/JVI.01210-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Juarrero M, Shim TS, Kipnis A, Junqueira-Kipnis AP, Orme IM. 2003. Dynamics of macrophage cell populations during murine pulmonary tuberculosis. J. Immunol. 171:3128–3135. 10.4049/jimmunol.171.6.3128 [DOI] [PubMed] [Google Scholar]
- 17.Cline TD, Karlsson EA, Seufzer BJ, Schultz-Cherry S. 2013. The hemagglutinin protein of highly pathogenic H5N1 influenza viruses overcomes an early block in the replication cycle to promote productive replication in macrophages. J. Virol. 87:1411–1419. 10.1128/JVI.02682-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrone LA, Plowden JK, García-Sastre A, Katz JM, Tumpey TM. 2008. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 4:e1000115. 10.1371/journal.ppat.1000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Riel D, Kuiken T. 2012. The role of cell tropism for the pathogenesis of influenza in humans. Future Virol. 7:295–307. 10.2217/fvl.12.11 [DOI] [Google Scholar]
- 20.van Riel D, Leijten LM, van der Eerden M, Hoogsteden HC, Boven LA, Lambrecht BN, Osterhaus AD, Kuiken T. 2011. Highly pathogenic avian influenza virus H5N1 infects alveolar macrophages without virus production or excessive TNF-alpha induction. PLoS Pathog. 7:e1002099. 10.1371/journal.ppat.1002099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakabe S, Takano R, Nagamura-Inoue T, Yamashita N, Nidom CA, Quynh Le M, Iwatsuki-Horimoto K, Kawaoka Y. 2013. Differences in cytokine production in human macrophages and in virulence in mice are attributable to the acidic polymerase protein of highly pathogenic influenza A virus subtype H5N1. J. Infect. Dis. 207:262–271. 10.1093/infdis/jis523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulcher AJ, Ahmed CM, Noon-Song EN, Kwan RY, Subramaniam PS, Johnson HM, Jans DA. 2008. Interferon gamma is recognised by importin alpha/beta: enhanced nuclear localising and transactivation activities of an interferon gamma mimetic. FEBS Lett. 582:1569–1574. 10.1016/j.febslet.2008.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theiss AL, Jenkins AK, Okoro NI, Klapproth JM, Merlin D, Sitaraman SV. 2009. Prohibitin inhibits tumor necrosis factor alpha-induced nuclear factor-kappa B nuclear translocation via the novel mechanism of decreasing importin alpha3 expression. Mol. Biol. Cell 20:4412–4423. 10.1091/mbc.E09-05-0361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue H, Chen B, Fan Y, Palikhe M, Li Y. 2011. The inhibitory effect of polypeptide cSN50 on alcoholic hepatic injuries through blocking the binding of NF-kappaB to importin alpha. Scand. J. Gastroenterol. 46:931–940. 10.3109/00365521.2011.568516 [DOI] [PubMed] [Google Scholar]
- 25.Turner JE, Morrison PJ, Wilhelm C, Wilson M, Ahlfors H, Renauld JC, Panzer U, Helmby H, Stockinger B. 2013. IL-9-mediated survival of type 2 innate lymphoid cells promotes damage control in helminth-induced lung inflammation. J. Exp. Med. 210:2951–2965. 10.1084/jem.20130071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monteiro JM, Harvey C, Trinchieri G. 1998. Role of interleukin-12 in primary influenza virus infection. J. Virol. 72:4825–4831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damjanovic D, Small CL, Jeyanathan M, McCormick S, Xing Z. 2012. Immunopathology in influenza virus infection: uncoupling the friend from foe. Clin. Immunol. 144:57–69. 10.1016/j.clim.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 28.Yasuhara N, Shibazaki N, Tanaka S, Nagai M, Kamikawa Y, Oe S, Asally M, Kamachi Y, Kondoh H, Yoneda Y. 2007. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat. Cell Biol. 9:72–79. 10.1038/ncb1521 [DOI] [PubMed] [Google Scholar]
- 29.Reed L, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.