ABSTRACT
It remains crucial to develop a laboratory model for studying hepatitis B virus (HBV) chronic infection. We hereby produced a recombinant covalently closed circular DNA (rcccDNA) in view of the key role of cccDNA in HBV persistence. A loxP-chimeric intron was engineered into a monomeric HBV genome in a precursor plasmid (prcccDNA), which was excised using Cre/loxP-mediated DNA recombination into a 3.3-kb rcccDNA in the nuclei of hepatocytes. The chimeric intron was spliced from RNA transcripts without interrupting the HBV life cycle. In cultured hepatoma cells, cotransfection of prcccDNA and pCMV-Cre (encoding Cre recombinase) resulted in accumulation of nuclear rcccDNA that was heat stable and epigenetically organized as a minichromosome. A mouse model of HBV infection was developed by hydrodynamic injection of prcccDNA. In the presence of Cre recombinase, rcccDNA was induced in the mouse liver with effective viral replication and expression, triggering a compromised T-cell response against HBV. Significant T-cell hyporesponsiveness occurred in mice receiving 4 μg prcccDNA, resulting in prolonged HBV antigenemia for up to 9 weeks. Persistent liver injury was observed as elevated alanine transaminase activity in serum and sustained inflammatory infiltration in the liver. Although a T-cell dysfunction was induced similarly, mice injected with a plasmid containing a linear HBV replicon showed rapid viral clearance within 2 weeks. Collectively, our study provides an innovative approach for producing a cccDNA surrogate that established HBV persistence in immunocompetent mice. It also represents a useful model system in vitro and in vivo for evaluating antiviral treatments against HBV cccDNA.
IMPORTANCE (i) Unlike plasmids that contain a linear HBV replicon, rcccDNA established HBV persistence with sustained liver injury in immunocompetent mice. This method could be a prototype for developing a mouse model of chronic HBV infection. (ii) An exogenous intron was engineered into the HBV genome for functionally seamless DNA recombination. This original approach could be also extended to other viral studies. (iii) rcccDNA was substantially induced in the nuclei of hepatocytes and could be easily distinguished by its exogenous intron using PCR. This convenient model system affords the opportunity to test antivirals directly targeting HBV cccDNA.
INTRODUCTION
Viral factors and host immune response have been implicated in the pathogenesis and clinical outcome of hepatitis B virus (HBV) infection. Covalently closed circular DNA (cccDNA) is an essential component of the HBV replication cycle and is regarded as a primary molecular mechanism for HBV persistence. The amount of cccDNA in cells is low, with around 5 to 50 copies in the nucleus. Nevertheless, cccDNA is stable, with a loss rate that correlates with the mitosis or death of infected hepatocytes (1, 2). Current antiviral treatments fail to eliminate the preexisting cccDNA pool that is responsible for viral rebound after therapy cessation. On the other hand, there is still lack of convenient techniques with high sensitivity and specificity to measure the HBV cccDNA pool in the hepatocyte nucleus (3–5).
A laboratory animal model will be crucial for studying chronic HBV infection and disease. Mice are not susceptible to HBV infection because they lack a receptor(s) for viral entry. Even in HBV transgenic (Tg) mice, cccDNA is not formed, for unknown reasons (6, 7). These barriers can be experimentally overcome by hydrodynamic injection of naked plasmid DNA encoding an overlength HBV replicon, which enables intracellular replication of HBV in murine hepatocytes (8–12). However, the HBV replicon-based hydrodynamic injection induces only transient HBV viremia resembling an acute infection in immunocompetent mice. In this regard, a recent model of HBV persistence generated by injection of a pAAV-HBV1.2 plasmid might reflect a role of the adeno-associated virus vector in developing immune tolerance (10).
A double-stranded HBV circular genome can be generated in vitro by PCR amplification with subsequent ligation (13, 14). However, this approach does not generate substantial cccDNA supercoils with high efficiency. In the present study, we produced a recombinant cccDNA (rcccDNA) through site-specific DNA recombination within a precursor plasmid bearing a monomeric HBV genome (i.e., prcccDNA). A loxP-chimeric intron was specially engineered into the viral genome to enable functionally seamless DNA recombination. In the presence of Cre recombinase, rcccDNA was accumulated in the livers of mice by hydrodynamic injection of prcccDNA, resulting in significantly prolonged HBV antigenemia with sustained liver injury. The method used in the study provides an innovative approach to produce a cccDNA surrogate with the unique attribute of establishing HBV persistence in immunocompetent mice. It also represents a useful model system in cell cultures and in vivo for evaluating antiviral treatments against HBV cccDNA.
MATERIALS AND METHODS
DNA constructs.
Plasmid pwtHBV1.3 encoded a 1.3-mer overlength HBV genome (subtype ayw; GenBank accession no. V01460.1). A chimeric intron was introduced between nucleotides (nt) 202 and 203 of the HBV genome by overlapping PCR and subcloning to generate pwtHBV1.3-intron. The insert comprised a 45-bp 5′ donor site (5′-GTAAGTATCAAGGTTACAAGACAGGTTTAAGGAGACCAATAGAAA-3′) and a 45-bp 3′ branch and acceptor site (5′-CACCTATTGGTCTTACTGACATCCACTTTGCCTTTCTCTCCACAG-3′) from the intronic sequence of pCI-neo (Promega). HindIII and PstI sites were designed between the 5′ and 3′ introns for cloning.
pBackbone contained a fragment of nt 3617 to 5391 from pcDNA3.1 (Invitrogen) and a 225-bp bovine growth hormone (BGH) polyadenylation signal and connected head to tail to a multiple-cloning site (HindIII-NcoI-PstI). In the pwtHBV1.3-intron, the NcoI site was in the reading frame of the HBV X (HBx) gene, which was represented twice in the 1.3-mer viral genome. The NcoI (upstream)-HindIII and PstI-NcoI (downstream) fragments from the pwtHBV1.3-intron were ligated to pBackbone to generate a monomeric circular HBV genome containing an intron-like plasmid backbone. To generate prcccDNA, two copies of 34 bp of loxP were inserted at HindIII and PstI sites.
pCMV-Cre carried the Cre recombinase gene from the P1 bacteriophage under the control of the cytomegalovirus (CMV) promoter. pCMV-S2S (15) and pCMV-Core encoded HBV middle and small envelope proteins or HBV core antigen (HBcAg) under the control of CMV promoter, respectively.
Mice.
Alb-Cre transgenic mice (C57BL/6-Tg[Alb-cre]21Mgn/J) were from Jackson Laboratory. DNA immunization was as described previously (16). For hydrodynamic injection, 4 to 16 μg prcccDNA was injected through tail veins in a volume of phosphate-buffered saline (PBS) equivalent to 8% of the mouse body weight (8–10). Injections were finished within 5 to 8 s. All animal studies were approved by the Animal Ethics Committee of Institut Pasteur of Shanghai (no. A2012008).
DNA and RNA hybridization.
Transfected cells or injected mouse livers were lysed in TBS (10 mM Tris-HCl [pH 7.0], 150 mM NaCl) containing 0.5% Nonidet P-40. Nuclei were pelleted by brief centrifugation. To purify HBV DNA from intracellular core particles, cytoplasmic lysates were treated with micrococcal nuclease (Amersham Biosciences) to remove input plasmid DNA. Viral DNA was ethanol precipitated after protease K (Calbiochem) digestion in the presence of 1% SDS for 2 h at 55°C. To purify rcccDNA from nuclei, Hirt extraction was used (17). The nuclear pellet was resuspended in 1 ml SDS lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 150 mM NaCl, and 0.5% SDS), mixed with 0.25 ml of 2.5 M KCl, and incubated at 4°C with gentle rotation overnight. The lysate was centrifuged at 14,000 × g for 20 min. rcccDNA was further extracted with phenol and chloroform, followed by precipitation using ethanol. RNA was extracted with TRIzol reagent (Invitrogen). Hybridization was performed using a 32P-labeled probe of the full-length HBV genome.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (18). Briefly, at 2 days after transfection, Huh-7 cells were fixed in 1% formaldehyde. Isolated cross-linked nuclei were sheared by sonication in 1% SDS lysis buffer. Chromatin was immunoprecipitated for 14 to 16 h at 4°C using antibodies against H4 (Upstate), AcH3 (Upstate), AcH4 (Upstate), HDAc1 (Upstate), HBcAg (Dako), or HBx (Thermo). Immunoprecipitation with nonspecific IgG (Beyotime) was the negative control. Antibody-DNA-protein complexes were precipitated with protein A/G-conjugated agarose beads and digested with RNase A and proteinase K. Reverse cross-linking was performed at 65°C for at least 6 h. DNA was predigested with Plasmid-Safe DNase (Epicentre Biotechnologies) before PCR analysis.
Detection of rcccDNA by PCR.
rcccDNA was extracted using the Hirt extraction procedure (17) from cultured cell lines or mouse livers. DNA was predigested with Plasmid-Safe DNase. HBV-specific primers flanking the chimeric insertion in prcccDNA were used to PCR amplify rcccDNA and nascent wild-type (wt) cccDNA templates (P1, 5′-GTATTTCCCTGCTGGTGGC-3′ [nt 49 to 67]; P2, 5′-GGTGAG TGATTGGAGGTTG-3′ [nt 321 to 339]). Nucleotide positions are according to the sequence of the HBV ayw subtype. Immunoprecipitated rcccDNA was quantified by real-time PCR using a SYBR green real-time PCR master mix kit (Toyobo, Osaka, Japan). Primers P3 (5′-CAAGACAGGTTTAAGGAGAC-3′) and P4 (5′-GAGAGAAAGGCAAAGTGGAT-3′) were from the 5′ donor site and the 3′ acceptor site of the loxP-chimeric intron, allowing PCR amplification from rcccDNA templates only.
Lymphocyte preparation and flow cytometry analysis.
Splenic and hepatic lymphocytes were isolated from mice as described previously (16). Isolated cells were suspended in PBS containing 1% bovine serum albumin (BSA) and labeled with anti-CD3ε (145-2C11), anti-CD8a (53-6.7), anti-gamma interferon (anti-IFN-γ) (XMG1.2), and anti-tumor necrosis factor alpha (anti-TNF-α) (MP6-XT22) (eBioscience). H-2Kb pentamer conjugated with HBV-derived peptide ENV353 (VWLSVIWM) was from ProImmune. Intracellular cytokines were detected as described previously (16) with peptide stimulation (2 μg/ml) in the presence of anti-CD28 (37.51) and anti-CD49d (R1-2) (eBioscience). After fixation and permeabilization, cells were stained with labeled antibodies. Data were acquired using an LSRII fluorescence-activated cell sorter (FACS) (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Statistics.
Data are expressed as means ± standard errors of the means (SEM). Unpaired Student t tests were performed with GraphPad Prism software.
RESULT
Construction of rcccDNA using Cre/loxP-mediated DNA recombination.
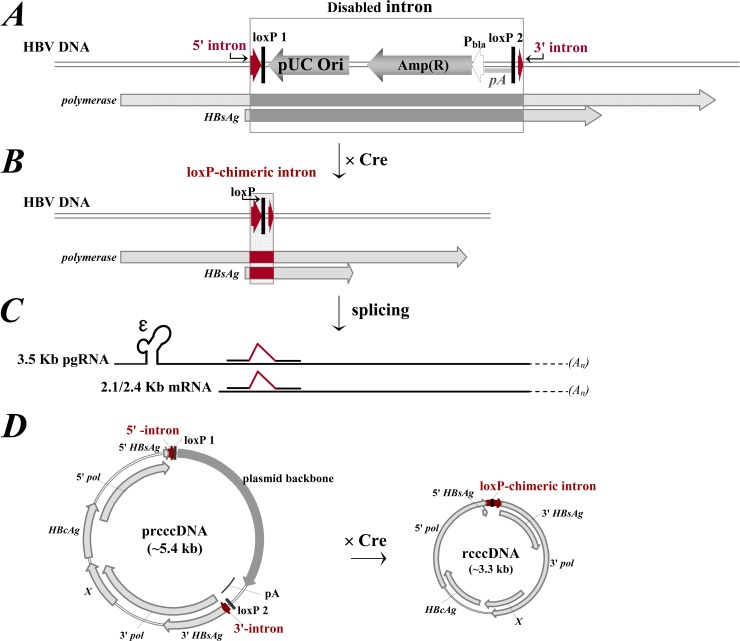
Technically, rcccDNA could be produced from a precursor plasmid containing a floxed HBV monomer genome through Cre/loxP-mediated DNA excision. Upon recombination, however, a residual 34-bp loxP site may disrupt the highly compact HBV genome. We thus designed a loxP-chimeric intron within the viral genome for functionally seamless DNA recombination. The exogenous insert should be removed from viral transcripts through RNA splicing (Fig. 1A to C).
FIG 1.
Schematic illustration of Cre/loxP-mediated rcccDNA production. (A) Overlapping reading frames of the HBsAg and polymerase genes interrupted by the insertion of an exogenous sequence between nucleotides 202 and 203 in the HBV DNA genome. The insert is a plasmid backbone between a 5′ intron (a 45-bp donor site from the first intron of the human β-globin gene) and a 3′ intron (a 45-bp branch/acceptor site from the intron of an immunoglobulin gene). Two directly repeated loxP sites generated a floxed plasmid backbone with a pUC replication origin, an ampicillin resistance gene (bla), and a BGH polyadenylation signal (pA) for eukaryotic transcription termination. (B) Floxed plasmid backbone excised following Cre/loxP-mediated DNA recombination, with a single loxP-chimeric intron in the HBV DNA genome. (C) loxP-chimeric intron spliced from viral transcripts during RNA processing, without disturbing the replication cycle of HBV. (D) Schema of Cre/loxP-mediated recombination of a precursor plasmid (i.e., prcccDNA) excising an rcccDNA episome with a loxP-chimeric intron.
A survey of the HBV genome identified several conserved 5′-(A/C)AG|G-3′ quadruplets (19) as potential exon/exon boundaries. We chose insertion between nt 202 and 203 in the early open reading frame of the HBV surface antigen (HBsAg). The insert comprised a 5′ intronic donor site from the human β-globin gene and a 3′ intronic branch/acceptor site from an immunoglobulin gene. A loxP site was introduced between the two segments to generate a chimeric intron that was spliced from RNA as demonstrated with a 1.3-mer HBV replicon (data not shown).
For construction of the precursor plasmid prcccDNA, the loxP-chimeric intron was inserted into a circular HBV genome, with the single loxP site replaced by two directly repeated loxP segments flanking a prokaryotic plasmid backbone. Thus, a 3.3-kb rcccDNA bearing a chimeric intron can be produced from prcccDNA by Cre/loxP-mediated DNA excision in the nuclei of hepatocytes (Fig. 1D).
rcccDNA is appropriately produced with high efficiency in a human hepatoma cell line.
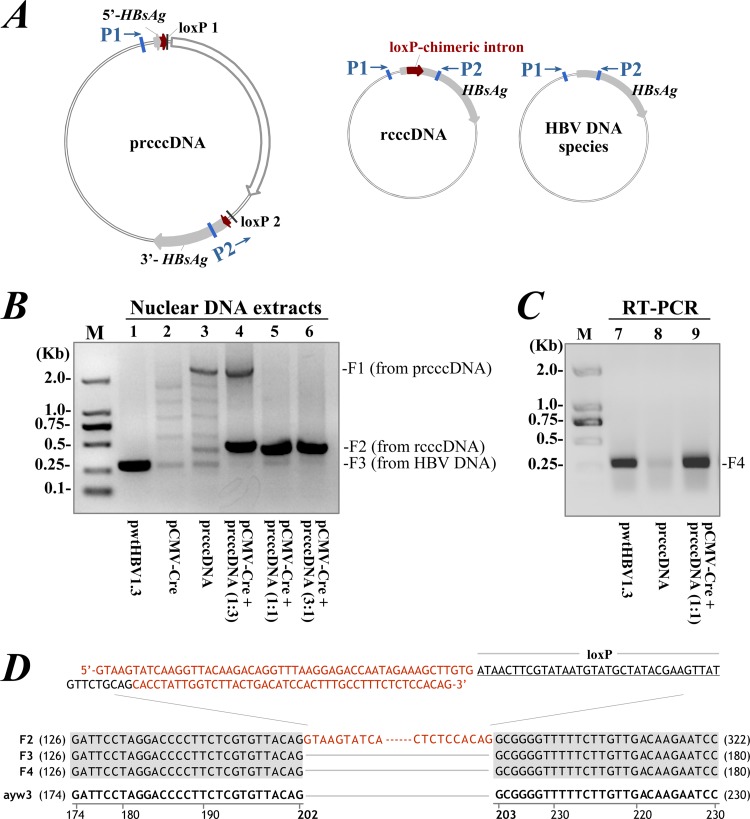
rcccDNA was distinguished from HBV DNA species and input prcccDNA by primer-specific PCR based on exogenous insertion (Fig. 2A). Cotransfection of prcccDNA with pCMV-Cre (encoding Cre) induced rcccDNA in Huh-7 cells, as suggested by the amplification of a 431-bp fragment from nuclear DNA (Fig. 2B). PCR products were verified by sequencing and were consistent with the predicted rcccDNA sequence (Fig. 2D). Cre/loxP-mediated recombination was nearly completed at 3 days after cotransfection of prcccDNA and pCMV-Cre at a 1:1 ratio. Transfection of prcccDNA alone did not induce rcccDNA, with amplification of only a 2,486-bp fragment.
FIG 2.
Predicted rcccDNA is verified by PCR in cultured Huh-7 cells. (A) Primers P1 and P2 flanking the loxP-chimeric intron were used for PCR to identify prcccDNA, rcccDNA, and HBV DNA species, including replicative intermediates and wt cccDNA. (B) PCR amplification of the Hirt DNA extraction from Huh-7 cells transfected with pwtHBV1.3 (lane 1), pCMV-Cre (lane 2), or prcccDNA (lane 3) or cotransfected with pCMV-Cre and prcccDNA at indicated ratios (lanes 4 to 6), measured on day 3 after transfection. F1, F2, and F3 indicate specific PCR products. (C) RT-PCR product of RNA extracted from cells transfected with pwtHBV1.3 (lane 7) or prcccDNA (lane 8) or cotransfected with pCMV-Cre and prcccDNA at a 1:1 ratio (lane 9). PCR was with primers P1 and P2. F4, RT-PCR product. (D) Sequence analysis of the PCR products of F2, F3, and F4. An adjacent HBV sequence (nt 174 to 230, ayw3 genotype) is included for DNA alignment. Top, complete sequence of the loxP-chimeric intron. Red, 5′ intron and 3′ intron.
To assess the efficiency of RNA splicing, cellular RNA was reverse transcribed and analyzed by PCR. The unique 291-bp fragment amplified had the same size as the reverse transcription-PCR (RT-PCR) product from pwtHBV1.3 (with a 1.3-mer linear HBV replicon)-transfected cells (Fig. 2C). Sequencing of the fragment suggested that the loxP-chimeric intron was correctly and efficiently removed from rcccDNA transcripts (Fig. 2D).
rcccDNA is competent for HBV expression and replication in cell culture.
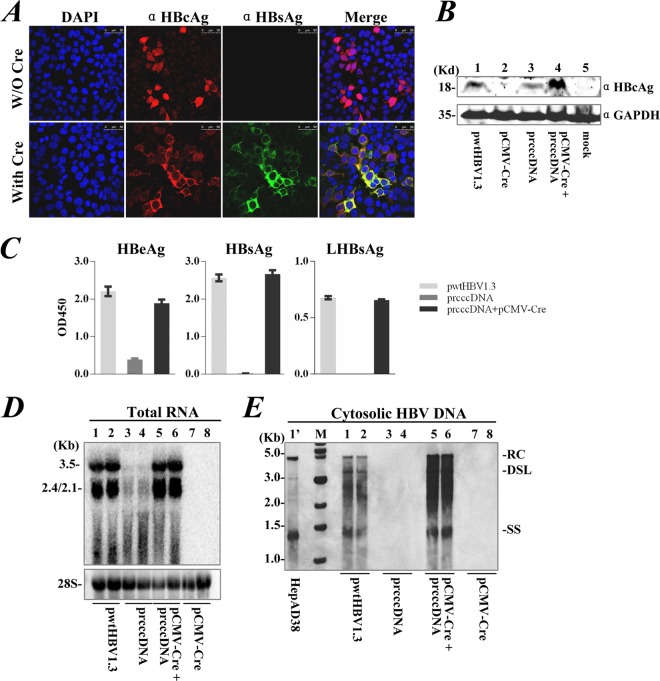
Expression of only HBcAg was detected in Huh-7 cells transfected with prcccDNA (Fig. 3A). Cotransfection of prcccDNA with pCMV-Cre induced HBsAg expression, suggesting that Cre/loxP-mediated recombination of prcccDNA, as well as subsequent RNA splicing, had been functionally achieved. Nucleocytoplasmic translocation of HBcAg was observed and could be directly due to increased HBcAg production (Fig. 3B). This result also likely reflected active HBV replication (20) in these cells. HBV large surface antigen (LHBsAg) was detected in culture medium, with HBV e antigen (HBeAg) substantially increased after DNA recombination (Fig. 3C).
FIG 3.
rcccDNA allows functional expression, transcription, and replication of HBV in cultured cell line. (A) HBsAg (green) and HBcAg (red) expression determined by immunofluorescence staining of Huh-7 cells on day 3 after transfection with prcccDNA or cotransfection with prcccDNA and pCMV-Cre at a 1:1 ratio. (B) HBcAg expression by Western blotting of cells transfected with pwtHBV1.3, pCMV-Cre, prcccDNA, or cotransfected with prcccDNA and pCMV-Cre. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was the loading control. (C) Secretion of HBeAg, HBsAg, and LHBsAg into the medium after cotransfection of prcccDNA and pCMV-Cre, as detected by antibody-specific enzyme-linked immunosorbent assay (ELISA). Control, transfection with pwtHBV1.3 or with prcccDNA alone. (D) Viral transcription detected by Northern blotting of Huh-7 cells cotransfected with prcccDNA and pCMV-Cre (lanes 5 and 6). Cells transfected with pwtHBV1.3 (lanes 1 and 2), prcccDNA (lanes 3 and 4), or pCMV-Cre (lanes 7 and 8) were controls. (E) Intracellular viral replication detected by Southern blotting. Positions of relaxed circular (RC) and double-stranded linear (DSL) forms are indicated. Lane 1′, HBV DNA species extracted from HepAD38 cells (Tet-off).
A BGH polyadenylation signal was designed in prcccDNA, directly downstream of the plasmid backbone, for transcription termination. A transcript under the control of the HBV basal core promoter of prcccDNA was estimated to be at least 3.6 kb. However, this predicted RNA was not readily detectable in prcccDNA-transfected cells by Northern blotting, although specific transcription was identified using RT-PCR (with a reverse primer from the plasmid backbone) (data not shown). Cotransfection of pCMV-Cre and prcccDNA significantly restored HBV transcripts in Huh-7 cells (Fig. 3D). This result probably reflects a role of the HBV 3′-terminal noncanonical polyadenylation signal in enhancing viral transcription (21). Following Cre/loxP-mediated recombination, intracellular viral replication was induced at a higher level than that observed after transfection of pwtHBV1.3 (Fig. 3E). These data clearly showed that rcccDNA with a chimeric intron was competent for expression and replication of HBV in cultured hepatoma cells.
rcccDNA is a surrogate for the natural HBV cccDNA minichromosome.
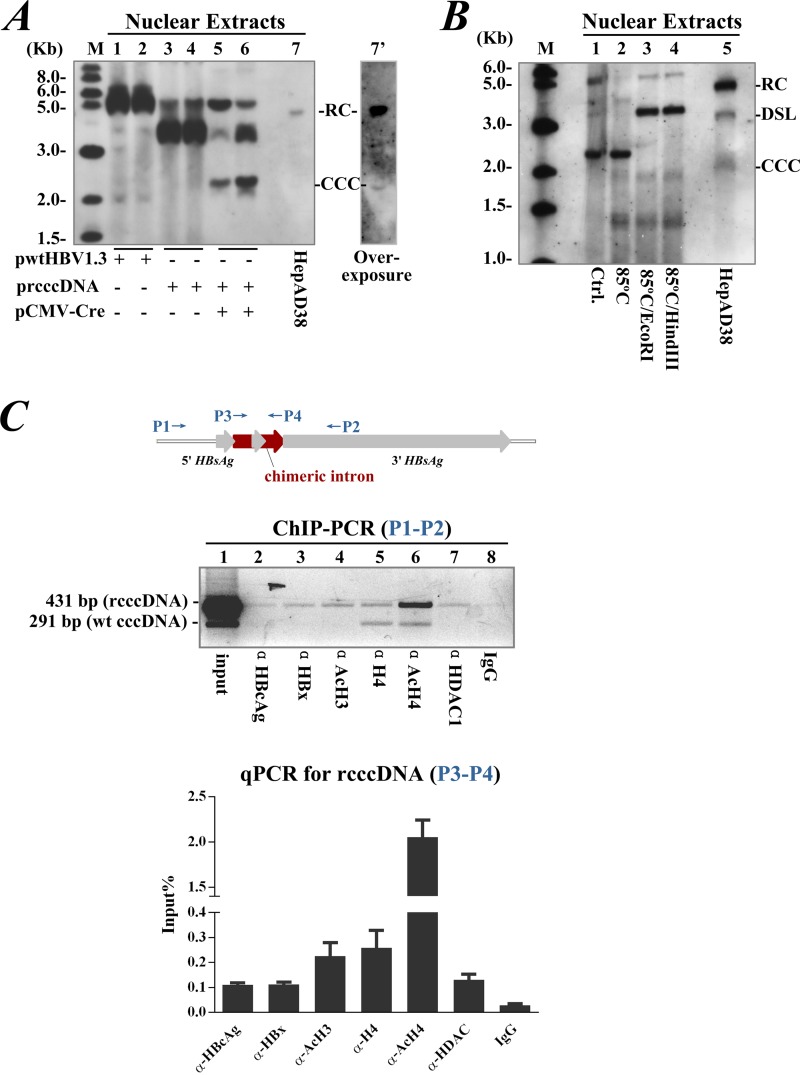
Cotransfection of pCMV-Cre with prcccDNA in Huh-7 cells led to substantial nuclear accumulation of rcccDNA that had an electrophoretic mobility similar to the mobility of supercoiled wt cccDNA (Fig. 4A). The rcccDNA was stable and resistant to linearization after being heated to up to 85°C (22, 23). Digestion of rcccDNA with EcoRI or HindIII led to a comigration with the double-stranded linear (DSL) form of HBV DNA (Fig. 4B).
FIG 4.
Nuclear rcccDNA is epigenetically organized as a minichromosome. (A) rcccDNA determined by Southern blotting from nuclear extracts of Huh-7 cells cotransfected with prcccDNA and pCMV-Cre at a 1:1 ratio (lanes 5 and 6). Cells transfected with pwtHBV1.3 (lanes 1 and 2) or with prcccDNA alone (lanes 3 and 4) were controls. Lane 7, DNA extracted from Hirt supernatant of HepaAD38 cells (Tet-off). Lane 7′, overexposure of lane 7. Experiments were performed as early as 36 h after transfection to detect most DNA species (i.e., processed and unprocessed). (B) rcccDNA from Huh-7 cells, measured on day 3 after cotransfection with prcccDNA and pCMV-Cre (lane 1), was resistant to denaturation at 85°C (lane 2). Lanes 3 and 4, digestion with EcoRI (within the HBV genome) or HindIII (within the chimeric intron) linearized rcccDNA. Lane 5, DNA extracted from Hirt supernatant of HepaAD38 cells (Tet-off). (C) Upper panel, schematic diagram of primer pairs used for ChIP. Middle panel, ChIP of Huh-7 cells cotransfected with prcccDNA and pCMV-Cre using antibodies to HBcAg, HBx, AcH3, H4, AcH4, HDAc1, or control IgG. Primers P1 and P2 were used for PCR amplification of a 431-bp product from rcccDNA. A 291-bp product corresponded to wt cccDNA in transfected Huh-7 cells (18, 24). Similar results were obtained from at least 3 independent experiments. Lower panel, quantitation of immunoprecipitated rcccDNA by real-time PCR using primers P3 and P4. Data (mean ± SEM) are expressed as percentage of input.
A ChIP assay (18, 24) was designed to investigate the epigenetic modification of rcccDNA. Accordingly, acetylated histone H3 (AcH3), histone H4 and its acetylated form (AcH4), the HDAc1 histone deacetylase, and the nonhistone HBcAg and HBx all bound to rcccDNA (Fig. 4C). Using primers P1 and P2, H4- and AcH4-bound wt cccDNA (or other nuclear DNA species of HBV [17, 25, 26]) were identified at low abundance following rcccDNA-based HBV replication (Fig. 4C, middle panel). The increased ratio of wt cccDNA to rcccDNA following anti-H4 immunoprecipitation suggested a better affinity of H4 for wt cccDNA, probably reflecting a difference in epigenetic modification between rcccDNA and the nascent wt cccDNA. Immunoprecipitated rcccDNA was also quantified by real-time PCR using primers P3 and P4 from the 5′ intron and 3′ intron, which amplified a 119-bp fragment from rcccDNA only (Fig. 4C, lower panel). Collectively, these data showed that rcccDNA was supercoiled, heat resistant, and epigenetically organized as a minichromosome similar to the natural cccDNA of HBV.
HBV replicates in mouse liver after hydrodynamic injection of rcccDNA.
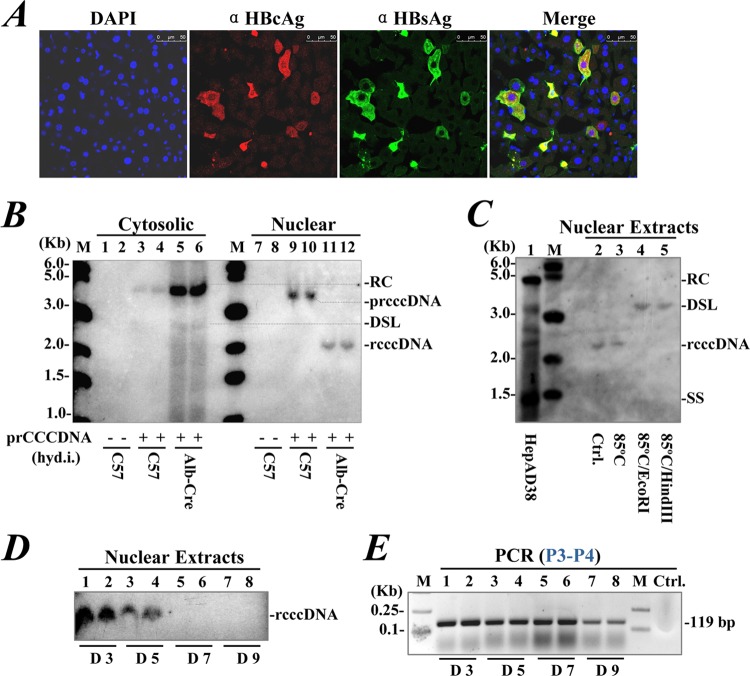
We used hydrodynamic injection to deliver prcccDNA to mice. DNA recombination was induced in Alb-Cre transgenic (Tg) mice that constitutively expressed Cre in the liver. Around 4% of hepatocytes were transfected as determined by immunostaining for HBcAg and HBsAg at 3 days after injection (Fig. 5A). HBV DNA species were detected intracellularly, with rcccDNA identified in nuclear extracts of the liver tissue by Southern blotting (Fig. 5B). The identity of rcccDNA in mouse livers was confirmed by heat denaturation and restriction digestion assays (Fig. 5C).
FIG 5.
Induction of rcccDNA in mouse liver through hydrodynamic injection. (A) Immunofluorescence for HBsAg (green) and HBcAg (red) in frozen liver sections of Alb-Cre Tg mice on day 3 after hydrodynamic injection of prcccDNA (8 μg). (B) Southern blot of cytosolic HBV replicative intermediates (lanes 5 and 6) or nuclear rcccDNA (lanes 11 and 12) extracted from mouse livers on day 3 after prcccDNA injection. Control, non-Tg C57BL/6 mice mock injected with pCDNA3.1 (lanes 1, 2, 7, and 8), or with prcccDNA only (lanes 3, 4, 9, and 10). (C) The rcccDNA (lane 2) was resistant to denaturation at 85°C (lane 3) and could be linearized to a DSL form by digestion with EcoRI (lane 4) or HindIII (lane 5). Lane 1, HBV DNA species extracted from HepAD38 cells (Tet-off). (D) Detection of rcccDNA in liver tissues of non-Tg C57BL/6 mice coinjected with prcccDNA (8 μg) and pCMV-Cre (4 μg) by Southern blotting on days 3 to 9 after injection. (E) PCR with primers P3 and P4 for nuclear rcccDNA from livers of non-Tg mice coinjected with prcccDNA and pCMV-Cre as described in for panel D.
rcccDNA was also induced in the livers of non-Tg mice by coinjection of prcccDNA and pCMV-Cre (Fig. 5D), by which the Cre recombinase was highly expressed in cotransfected cells albeit not constitutively. Primer-specific PCR was designed for a rapid detection of rcccDNA in liver tissue. Using primers P3 and P4 (Fig. 5C), rcccDNA was identified in the mouse liver up to days 7 and 9 after injection (Fig. 5E), which was virtually not observed by the Southern blot assay. Thus, we developed an in vivo model system of rcccDNA-based HBV replication with a convenient measurement.
rcccDNA induces a compromised liver T-cell response.
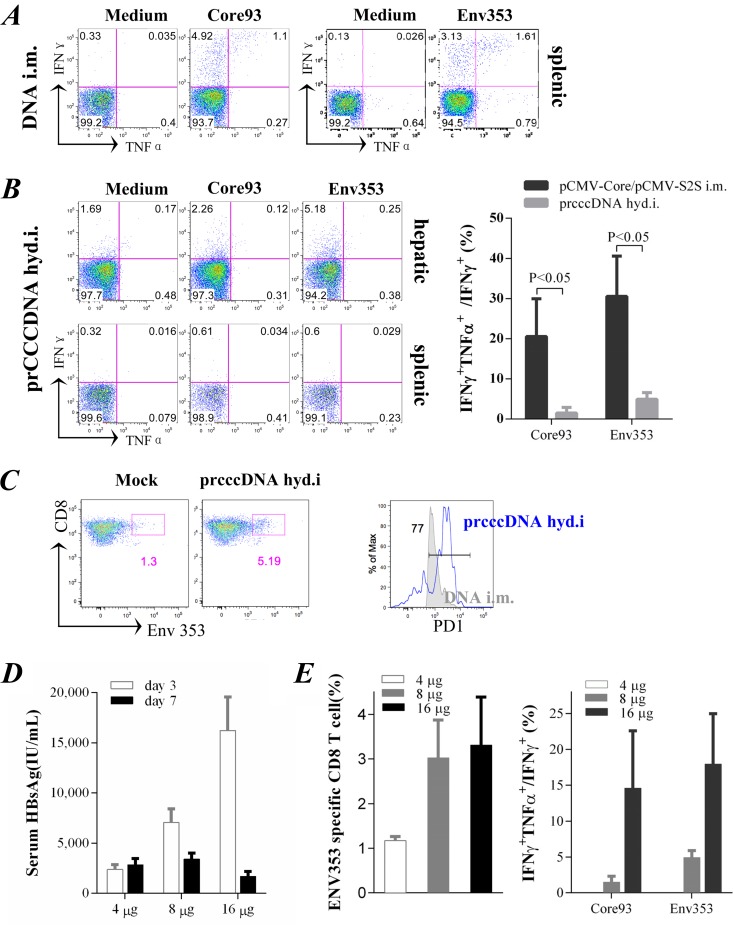
Intramuscular injection with plasmids encoding HBV antigens induced functional T effector cells in the periphery (Fig. 6A). Despite the dominant immunogenicity of viral antigens, hydrodynamic injection of prcccDNA (8 μg) did not trigger a significant T-cell response in the spleens of Alb-Cre Tg mice (Fig. 6B, left lower panels) although substantial antigenemia of HBsAg was induced. Liver-infiltrating lymphocytes were slightly increased (4 million to 6 million) as measured on day 14 after injection. IFN-γ+ CD8+ T cells in the liver were detected upon peptide stimulation. However, they produced little TNF-α (Fig. 6B, left upper panels), suggesting impaired effector function (27). Increased expression of programmed death 1 (PD-1), a marker for T-cell exhaustion, was also revealed on ENV353 pentamer-specific CD8+ T cells in the liver compared with T effector cells in the periphery induced by DNA immunization (Fig. 6C).
FIG 6.
rcccDNA induces a compromised T-cell response in mouse liver (A) CD8+ T cells from mouse spleens gated for intracellular IFN-γ and TNF-α staining upon stimulation with H-2Kb restricted peptide core93 (MGLKFRQL) or ENV353 (VWLSVIWM), as measured on day 14 after intramuscular (i.m.) injection with pCMV-Core or pCMV-S2S. (B) Left, intracellular staining of CD8+ T cells from the livers or spleens of Alb-Cre Tg mice on day 14 after hydrodynamic injection (hyd.i) of prcccDNA (8 μg). Right, ratio of IFN-γ and TNF-α double-positive T cells to IFN-γ single-positive cells in the liver. Mice injected (i.m.) with pCMV-Core or pCMV-S2S were controls for cytokine profiles of fully activated T-cell responses in the periphery. (C) ENV353 pentamer-specific CD8+ T cells in the livers of Alb-Cre Tg mice were gated (left panels) for PD-1 staining (right panel, blue) as measured on day 14 after injection of prcccDNA (8 μg). Control, staining of ENV353-specific T effector cells in the spleens of mice injected (i.m.) with pCMV-S2S. (D) Serum HBsAg detected on days 3 and 7 after injection of prcccDNA at 4, 8, or 16 μg in non-Tg C57BL/6 mice. pCMV-Cre (4 μg) was coinjected in these mice. (E) Left, ENV353 pentamer-specific CD8+ T cells measured in mouse livers on day 14 as depicted in panel D. Right, ratio of IFN-γ and TNF-α double-positive T cells to IFN-γ single-positive cells in the liver after stimulation with core93 or ENV353.
We further investigated the dose dependence of rcccDNA-based immunization. Non-Tg mice were coinjected with 4 μg pCMV-Cre and 4 to 16 μg of prcccDNA. In this respect, the highly expressed Cre appeared to be more efficient than that of Alb-Cre Tg mice in inducing rcccDNA (data not shown). HBsAg antigenemia was induced in a dose-dependent manner as observed on day 3 after injection. At 8 to 16 μg of prcccDNA, however, mice underwent a rapid decline in HBsAg titers from day 3 to 7 (Fig. 6D), with ENV353-specific CD8+ T cells frequently observed in mouse livers as determined on day 14 (Fig. 6E). At 4 μg of prcccDNA, in contrast, the circulating HBsAg remained increased on day 7. Few specific T cells were detected in the liver, and they failed to produce detectable IFN-γ and TNF-α upon peptide stimulation (Fig. 6E). These data indicated that production of rcccDNA in mouse livers induced a compromised T-cell response that was critically influenced by the dosage of the injected plasmid.
rcccDNA induces a prolonged HBV antigenemia in mice.
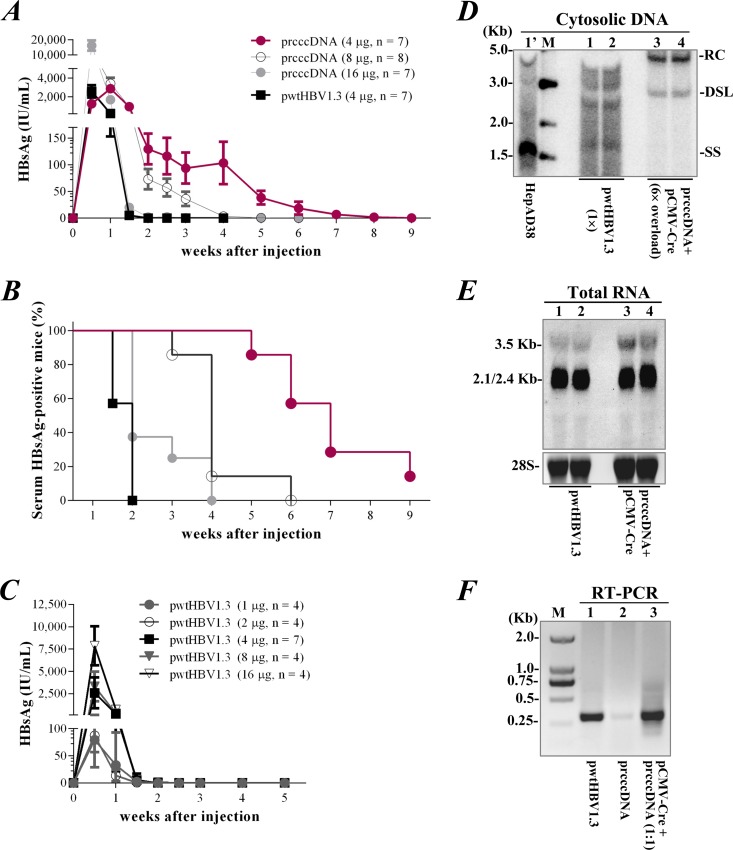
We investigated whether rcccDNA induced HBV persistence in immunocompetent mice. Consistent with the significantly compromised T-cell response, injection of 4 μg prcccDNA (coinjected with pCMV-Cre) resulted in HBsAg antigenemia with delayed clearance over up to 9 weeks (Fig. 7A and B). In contrast, the circulating HBsAg was mainly cleared within 2 to 4 weeks in mice receiving 8 to 16 μg of prcccDNA.
FIG 7.
rcccDNA induces prolonged HBsAg antigenemia in immunocompetent mice. (A) Kinetics of serum HBsAg at the indicated time points in non-Tg C57BL/6 mice (4 weeks old, male) hydrodynamically injected with prcccDNA (4, 8, and 16 μg) or pwtHBV1.3 (4 μg). rcccDNA was induced by coinjection with pCMV-Cre (4 μg). (B) Positive rate of serum HBsAg in mice as depicted in panel A. (C) Kinetics of serum HBsAg at the indicated time points in mice injected with pwtHBV1.3 at 1 to 16 μg, with 4 μg of coinjected pCMV-Cre. (D) Southern blot of HBV replication in the livers of mice on day 3 after injection of 4 μg pwtHBV1.3 (lanes 1 and 2) or 4 μg prcccDNA (lanes 3 and 4). pCMV-Cre (4 μg) was coinjected in these mice. A concentrated DNA extract from 600 mg liver from mice injected with prcccDNA/pCMV-Cre was used for hybridization and compared with extract from 100 mg of liver from mice injected with pwtHBV1.3, which might cause the DNA migration difference. Lane 1′, HBV DNA species extracted from HepAD38 cells (Tet-off) as control. (E) Northern blot of HBV transcripts in the livers of mice injected as described for panel D. (F) RNA was reverse transcribed from livers of mice injected with 4 μg pwtHBV1.3 (lane 1) or 4 μg prcccDNA (lane 2) or coinjected with 4 μg prcccDNA and 4 μg pCMV-Cre (lane 3). PCR used primers P1 and P2. RT-PCR products were verified by DNA sequencing (data not shown).
Rapid HBsAg clearance was also observed in mice within 2 weeks after injection of a plasmid bearing a linear HBV replicon (pwtHBV1.3), although a T-cell hyporesponsiveness was similarly induced with 4 μg of the plasmid (data not shown). In a titration study, injection of pwtHBV1.3 at all doses tested (1 to 16 μg) failed to induce prolonged HBsAg persistence in the mouse model (Fig. 7C). The clearance of serum HBsAg started from day 3 after pwtHBV1.3 injection, suggesting that an early antiviral mechanism was involved. rcccDNA-based HBsAg expression appeared to be less affected by this mechanism, since the antigenemia remained increased at day 7 after injection (for 4 μg only). We therefore investigated rcccDNA-based HBV replication in the liver tissue on day 3 (early phase) after injection, which was substantially lower (6-fold) than in mice injected with pwtHBV1.3 (Fig. 7D), although a similar level of viral transcription was observed (Fig. 7E). rcccDNA induced HBV transcription with efficient, correct RNA splicing in mouse livers as indicated by RT-PCR analysis (Fig. 7F). Note that a higher level of HBV replication was seen with in vitro transfection of cultured cells with prcccDNA/pCMV-Cre than with pwtHBV1.3 (Fig. 3E). Collectively, these data revealed a unique attribute of the cccDNA surrogate in establishing HBV persistence in immunocompetent mice.
rcccDNA induces sustained liver inflammation in immunocompetent mice.
Injection of pwtHBV1.3 resulted in increased serum alanine transaminase (sALT) activity that returned to the baseline level within 3 weeks after injection. In contrast, continuously elevated sALT activity (>60 U/liter) was observed in mice with rcccDNA-based viral persistence (Fig. 8A). Interleukin-6 and TNF-α were modestly increased, around 3- to 5-fold, in liver tissues as measured on days 28 and 35 by quantitative PCR (data not shown). Histological staining revealed increased mononuclear infiltrates in liver sections (Fig. 8B, days 35 and 63), suggesting a sustained inflammatory response. Dividing hepatocytes were frequently identified, indicative of increased hepatocellular regeneration in mouse livers. Collectively, these data showed that rcccDNA induced prolonged HBV antigenemia with sustained liver inflammation in an immunocompetent mouse model.
FIG 8.
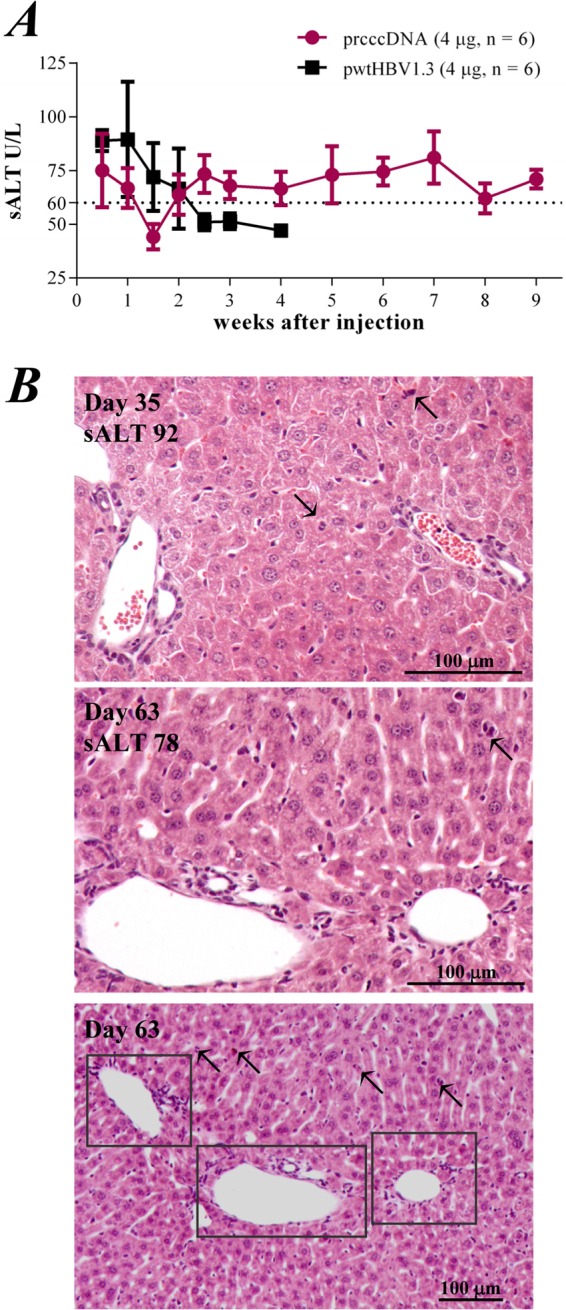
rcccDNA induces sustained liver injury in a mouse model. (A) Serum alanine aminotransferase (sALT) activities (U/liter) at the indicated time points in mice injected with 4 μg pwtHBV1.3 or 4 μg prcccDNA. pCMV-Cre (4 μg) was coinjected in these mice. (B) Hematoxylin-eosin staining of liver sections of mice on day 35 (upper panel) or day 63 (middle panel) after coinjection of prcccDNA and pCMV-Cre. Lower panel, inflammatory foci (framed) in the liver section from day 63 at a lower magnification. Arrows, evidence of hepatocytes undergoing degeneration or dividing. sALT activity and scale are indicated.
DISCUSSION
HBV DNA can be circularized at high efficiency by Cre/loxP-mediated site-specific recombination from plasmids bearing the HBV genome. A key issue is accommodating a residual 34-bp loxP site without disturbing the highly compact HBV genome. Although the biological significance remains unclear, HBV synthesizes multiple spliced transcripts from its pregenomic RNA (28–30). In this study, we introduced a loxP-chimeric intron into the HBV genome. The intron-based loxP insertion was functionally seamless and resulted in effective transcription, expression, and replication of HBV. Future work will further refine the chimeric sequence so that the size of the rcccDNA is closer to that of the natural HBV genome (3,182 bp for the ayw subtype).
Nuclear rcccDNA was heat stable and was epigenetically organized as a minichromosome. rcccDNA could therefore be used as a cccDNA surrogate for HBV studies. The inserted foreign sequence of rcccDNA can be used to discriminate rcccDNA from the precursor plasmid and from DNA replication intermediates of HBV using PCR. Since the chimeric intron was within the HBsAg gene reading frame, HBsAg induction could be a convenient readout for rcccDNA formation in the nuclei of hepatocytes. In this regard, the titer of circulating HBsAg might reflect the copy number or transcriptional regulation of nuclear rcccDNA in the liver.
A major obstacle to in vivo studies of HBV infection is the lack of suitable laboratory models. Hydrodynamic injection of HBV DNA is a convenient way to produce surrogate viral infection in mice. However, immunization with naked plasmid-based DNA might increase the virus-specific T-cell response. Natural infection of HBV is believed to be stealthy, without alerting the innate immune system (31). T-cell activation is significantly delayed even if self-limited acute hepatitis progresses (22). In our study, mice receiving 16 μg of prcccDNA via hydrodynamic injection (coinjected with pCMV-Cre) developed a functional intrahepatic T-cell response against HBV, consistent with a previous report by Huang et al. (32). The immune response was largely compromised at 4 μg prcccDNA. The mice displayed hyporesponsiveness against HBV although with significant viral antigenemia, similar to the HBV tolerance during natural infection.
The HBV-specific T-cell response plays a major role in viral clearance and the associated liver diseases. In the NOD/scid T-cell defective mouse model, hydrodynamic injection of HBV DNA resulted in persistent viral replication and expression (9). However, although T-cell hyporesponsiveness was developed, rapid HBV clearance was observed in mice receiving a low-dose injection of the pwtHBV1.3 plasmid with the linear HBV replicon (Fig. 7C). A possible explanation is that the virus-specific CD8+ T cells induced in the mouse liver might not be fully dysfunctional. In addition, antiviral innate immunity might also be primarily involved in early-stage viral clearance after DNA hydrodynamic injection. Interestingly, rcccDNA-based HBV expression appeared to be less sensitive to these potential mechanisms.
The plasmid pwtHBV1.3 can be regarded as a type of cccDNA. In fact, previous studies on the morphology of HBV cccDNA were based on a surrogate plasmid that displayed a beads-on-string supercoil structural organization (33). pwtHBV1.3 has a bacterial plasmid backbone, which is likely to be a major difference from rcccDNA as well as the HBV cccDNA. Pathogen-associated molecular patterns such as CpG motifs are present in the bacterial plasmid backbone, resulting in high immunogenicity (34). This might explain the rapid viral clearance after pwtHBV1.3-based hydrodynamic injection. However, pwtHBV1.3 prepared from a JM110 Escherichia coli strain, which lacks DNA adenine methylation and DNA cytosine methylation activities, did not cause prolonged viral persistence in a mouse model (unpublished data). It is also noted that both prcccDNA and the coinjected pCMV-Cre also have a plasmid backbone.
Increased HBV replication was observed for rcccDNA compared to the linear HBV replicon in in vitro transfection assays. In the mouse liver, however, rcccDNA-based viral replication was significantly lower than that of pwtHBV1.3 at day 3 after the hydrodynamic injection, although levels of viral transcription were similar. We propose that the pwtHBV1.3 plasmid triggered a stronger innate immunity in vivo than rcccDNA did. In this regard, stimulation by small amounts of type I IFN may temporarily increase HBV viral replication, as reported by Tian et al. (11), which is likely to increase the immune activation and subsequent viral clearance. In fact, an onset of massive viral replication did not always sustain a persistent infection of HBV in the host (35).
In summary, our study provides an innovative approach to produce an HBV cccDNA surrogate that established HBV persistence in immunocompetent mice, which gives the first indication of the difference between an overlength HBV genome and cccDNA. This method is a prototype for developing a mouse model of chronic HBV infection and could be used to develop cccDNA-based antiviral strategies.
ACKNOWLEDGMENTS
This work was supported by grants from the National Science and Technology Major Projects (2008ZX10002007), the Natural Science Foundation (81171566), and the National Key Basic Research Program of China (2012CB519000).
We thank Yongxiang Wang and Weiya Bai for the DNA hybridization assay and Fudi Wang and Guangxun Meng for providing Cre transgenic mice.
Footnotes
Published ahead of print 7 May 2014
REFERENCES
- 1.Lutgehetmann M, Volz T, Kopke A, Broja T, Tigges E, Lohse AW, Fuchs E, Murray JM, Petersen J, Dandri M. 2010. In vivo proliferation of hepadnavirus-infected hepatocytes induces loss of covalently closed circular DNA in mice. Hepatology 52:16–24. 10.1002/hep.23611 [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y, Yamamoto T, Cullen J, Saputelli J, Aldrich CE, Miller DS, Litwin S, Furman PA, Jilbert AR, Mason WS. 2001. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J. Virol. 75:311–322. 10.1128/JVI.75.1.311-322.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong DK, Yuen MF, Yuan H, Sum SS, Hui CK, Hall J, Lai CL. 2004. Quantitation of covalently closed circular hepatitis B virus DNA in chronic hepatitis B patients. Hepatology 40:727–737. 10.1002/hep.20353 [DOI] [PubMed] [Google Scholar]
- 4.Singh M, Dicaire A, Wakil AE, Luscombe C, Sacks SL. 2004. Quantitation of hepatitis B virus (HBV) covalently closed circular DNA (cccDNA) in the liver of HBV-infected patients by LightCycler real-time PCR. J. Virol. Methods 118:159–167. 10.1016/j.jviromet.2004.02.006 [DOI] [PubMed] [Google Scholar]
- 5.Zoulim F. 2006. Assessment of treatment efficacy in HBV infection and disease. J. Hepatol. 44:S95–S99. 10.1016/j.jhep.2005.11.020 [DOI] [PubMed] [Google Scholar]
- 6.Guidotti LG, Matzke B, Schaller H, Chisari FV. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69:6158–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raney AK, Eggers CM, Kline EF, Guidotti LG, Pontoglio M, Yaniv M, McLachlan A. 2001. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. J. Virol. 75:2900–2911. 10.1128/JVI.75.6.2900-2911.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang PL, Althage A, Chung J, Chisari FV. 2002. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc. Natl. Acad. Sci. U. S. A. 99:13825–13830. 10.1073/pnas.202398599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang PL, Althage A, Chung J, Maier H, Wieland S, Isogawa M, Chisari FV. 2010. Immune effectors required for hepatitis B virus clearance. Proc. Natl. Acad. Sci. U. S. A. 107:798–802. 10.1073/pnas.0913498107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang LR, Wu HL, Chen PJ, Chen DS. 2006. An immunocompetent mouse model for the tolerance of human chronic hepatitis B virus infection. Proc. Natl. Acad. Sci. U. S. A. 103:17862–17867. 10.1073/pnas.0608578103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Y, Chen WL, Ou JH. 2011. Effects of interferon-alpha/beta on HBV replication determined by viral load. PLoS Pathog. 7:e1002159. 10.1371/journal.ppat.1002159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takehara T, Suzuki T, Ohkawa K, Hosui A, Jinushi M, Miyagi T, Tatsumi T, Kanazawa Y, Hayashi N. 2006. Viral covalently closed circular DNA in a non-transgenic mouse model for chronic hepatitis B virus replication. J. Hepatol. 44:267–274. 10.1016/j.jhep.2005.07.030 [DOI] [PubMed] [Google Scholar]
- 13.Qin Y, Zhang J, Garcia T, Ito K, Gutelius D, Li J, Wands J, Tong S. 2011. Improved method for rapid and efficient determination of genome replication and protein expression of clinical hepatitis B virus isolates. J. Clin. Microbiol. 49:1226–1233. 10.1128/JCM.02340-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunther S, Li BC, Miska S, Kruger DH, Meisel H, Will H. 1995. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J. Virol. 69:5437–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel ML, Davis HL, Schleef M, Mancini M, Tiollais P, Whalen RG. 1995. DNA-mediated immunization to the hepatitis B surface antigen in mice: aspects of the humoral response mimic hepatitis B viral infection in humans. Proc. Natl. Acad. Sci. U. S. A. 92:5307–5311. 10.1073/pnas.92.12.5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng Q, Mancini-Bourgine M, Zhang X, Cumont MC, Zhu R, Lone YC, Michel ML. 2009. Hepatitis B virus as a gene delivery vector activating foreign antigenic T cell response that abrogates viral expression in mouse models. Hepatology 50:1380–1391. 10.1002/hep.23150 [DOI] [PubMed] [Google Scholar]
- 17.Gao W, Hu J. 2007. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J. Virol. 81:6164–6174. 10.1128/JVI.02721-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. 2006. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 130:823–837. 10.1053/j.gastro.2006.01.001 [DOI] [PubMed] [Google Scholar]
- 19.Breathnach R, Benoist C, O'Hare K, Gannon F, Chambon P. 1978. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc. Natl. Acad. Sci. U. S. A. 75:4853–4857. 10.1073/pnas.75.10.4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melegari M, Wolf SK, Schneider RJ. 2005. Hepatitis B virus DNA replication is coordinated by core protein serine phosphorylation and HBx expression. J. Virol. 79:9810–9820. 10.1128/JVI.79.15.9810-9820.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paran N, Ori A, Haviv I, Shaul Y. 2000. A composite polyadenylation signal with TATA box function. Mol. Cell. Biol. 20:834–841. 10.1128/MCB.20.3.834-841.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825–829. 10.1126/science.284.5415.825 [DOI] [PubMed] [Google Scholar]
- 23.Sprinzl MF, Oberwinkler H, Schaller H, Protzer U. 2001. Transfer of hepatitis B virus genome by adenovirus vectors into cultured cells and mice: crossing the species barrier. J. Virol. 75:5108–5118. 10.1128/JVI.75.11.5108-5118.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M, Levrero M. 2012. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Invest. 122:529–537. 10.1172/JCI58847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT. 2007. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J. Virol. 81:12472–12484. 10.1128/JVI.01123-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kock J, Rosler C, Zhang JJ, Blum HE, Nassal M, Thoma C. 2010. Generation of covalently closed circular DNA of hepatitis B viruses via intracellular recycling is regulated in a virus specific manner. PLoS Pathog. 6:e1001082. 10.1371/journal.ppat.1001082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wherry EJ, Ahmed R. 2004. Memory CD8 T-cell differentiation during viral infection. J. Virol. 78:5535–5545. 10.1128/JVI.78.11.5535-5545.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soussan P, Garreau F, Zylberberg H, Ferray C, Brechot C, Kremsdorf D. 2000. In vivo expression of a new hepatitis B virus protein encoded by a spliced RNA. J. Clin. Invest. 105:55–60. 10.1172/JCI8098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosmorduc O, Petit MA, Pol S, Capel F, Bortolotti F, Berthelot P, Brechot C, Kremsdorf D. 1995. In vivo and in vitro expression of defective hepatitis B virus particles generated by spliced hepatitis B virus RNA. Hepatology 22:10–19. 10.1016/0270-9139(95)90346-1 [DOI] [PubMed] [Google Scholar]
- 30.Gunther S, Sommer G, Iwanska A, Will H. 1997. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology 238:363–371. 10.1006/viro.1997.8863 [DOI] [PubMed] [Google Scholar]
- 31.Wieland SF, Chisari FV. 2005. Stealth and cunning: hepatitis B and hepatitis C viruses. J. Virol. 79:9369–9380. 10.1128/JVI.79.15.9369-9380.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang LR, Gabel YA, Graf S, Arzberger S, Kurts C, Heikenwalder M, Knolle PA, Protzer U. 2012. Transfer of HBV genomes using low doses of adenovirus vectors leads to persistent infection in immune competent mice. Gastroenterology 142:1447–1450. 10.1053/j.gastro.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 33.Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. 2001. Structural organization of the hepatitis B virus minichromosome. J. Mol. Biol. 307:183–196. 10.1006/jmbi.2000.4481 [DOI] [PubMed] [Google Scholar]
- 34.Klinman DM, Yamshchikov G, Ishigatsubo Y. 1997. Contribution of CpG motifs to the immunogenicity of DNA vaccines. J. Immunol. 158:3635–3639 [PubMed] [Google Scholar]
- 35.Asabe S, Wieland SF, Chattopadhyay PK, Roederer M, Engle RE, Purcell RH, Chisari FV. 2009. The size of the viral inoculum contributes to the outcome of hepatitis B virus infection. J. Virol. 83:9652–9662. 10.1128/JVI.00867-09 [DOI] [PMC free article] [PubMed] [Google Scholar]