ABSTRACT
Host cell tRNAs are recruited for use as primers to initiate reverse transcription in retroviruses. Human immunodeficiency virus type 1 (HIV-1) uses tRNALys3 as the replication primer, whereas Rous sarcoma virus (RSV) uses tRNATrp. The nucleic acid (NA) chaperone function of the nucleocapsid (NC) domain of HIV-1 Gag is responsible for annealing tRNALys3 to the genomic RNA (gRNA) primer binding site (PBS). Compared to HIV-1, little is known about the chaperone activity of RSV Gag. In this work, using purified RSV Gag containing an N-terminal His tag and a deletion of the majority of the protease domain (H6.Gag.3h), gel shift assays were used to monitor the annealing of tRNATrp to a PBS-containing RSV RNA. Here, we show that similar to HIV-1 Gag lacking the p6 domain (GagΔp6), RSV H6.Gag.3h is a more efficient chaperone on a molar basis than NC; however, in contrast to the HIV-1 system, both RSV H6.Gag.3h and NC have comparable annealing rates at protein saturation. The NC domain of RSV H6.Gag.3h is required for annealing, whereas deletion of the matrix (MA) domain, which stimulates the rate of HIV-1 GagΔp6 annealing, has little effect on RSV H6.Gag.3h chaperone function. Competition assays confirmed that RSV MA binds inositol phosphates (IPs), but in contrast to HIV-1 GagΔp6, IPs do not stimulate RSV H6.Gag.3h chaperone activity unless the MA domain is replaced with HIV-1 MA. We conclude that differences in the MA domains are primarily responsible for mechanistic differences in RSV and HIV-1 Gag NA chaperone function.
IMPORTANCE Mounting evidence suggests that the Gag polyprotein is responsible for annealing primer tRNAs to the PBS to initiate reverse transcription in retroviruses, but only HIV-1 Gag chaperone activity has been demonstrated in vitro. Understanding RSV Gag's NA chaperone function will allow us to determine whether there is a common mechanism among retroviruses. This report shows for the first time that full-length RSV Gag lacking the protease domain is a highly efficient NA chaperone in vitro, and NC is required for this activity. In contrast to results obtained for HIV-1 Gag, due to the weak nucleic acid binding affinity of the RSV MA domain, inositol phosphates do not regulate RSV Gag-facilitated tRNA annealing despite the fact that they bind to MA. These studies provide insight into the viral regulation of tRNA primer annealing, which is a potential target for antiretroviral therapy.
INTRODUCTION
Retroviruses encode their genetic material in two single-stranded genomic RNAs (gRNA), which are packaged into the virion by a direct interaction with the retroviral Gag protein (1). The RNA genome contains a region known as the primer binding site (PBS), which is complementary to the 3′ 18 nucleotides (nt) of a specific host cell tRNA that acts as a primer for reverse transcription (2). In human immunodeficiency virus type 1 (HIV-1), the primer is tRNALys3 (3, 4), and in Rous sarcoma virus (RSV), it is tRNATrp (5, 6). The retroviral Gag polyprotein is present in immature virions and is made up of three major domains: matrix (MA), capsid (CA), and nucleocapsid (NC). In the case of RSV, there are additional sequences, including p2 and p10 between MA and CA, SP between CA and NC, and protease (PR) at the C terminus of Gag. In HIV-1, Gag contains the C-terminal p6 domain, which is critical for release of viral particles from the plasma membrane (PM). During virus particle assembly, MA serves as the membrane-targeting and binding domain (7–9), CA facilitates Gag multimerization (10–13), and NC binds the Ψ packaging signal in the viral RNA for encapsidation of the viral genome and contributes to the formation of Gag-Gag interactions (1, 14–17). During or just after budding, PR cleaves Gag to form the mature virion.
Gag facilitates tRNA primer annealing to the PBS for HIV-1 (18) and RSV (19). The NC domain of HIV-1 Gag, also referred to as NCp7, is required for this activity (20). HIV-1 Gag and NCp7 are chaperones that remodel nucleic acids (NAs) into energetically more favorable conformations through helix destabilization and NA aggregation activities to promote annealing (21–26). Although well known for its role in recruiting Gag to the PM, HIV-1 MA is also able to bind NA with mid- to low-nM affinity in vitro (20, 27–32). This MA-RNA interaction can be abrogated by phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2]-containing liposomes (30). It has been proposed that RNA inhibits MA-lipid interactions and acts as a regulator of Gag-membrane binding (33). It has also been shown in vitro that the tRNA annealing function of HIV-1 Gag is modulated by MA domain binding to inositol hexakisphosphate (IP6) (20), which is a mimic of PI(4,5)P2 found at the PM (34).
In vitro studies have demonstrated that RSV NC, also referred to as NCp12, has NA chaperone activity comparable to that of HIV-1 NC (35); however, little is known about the NA chaperone activity of the RSV Gag polyprotein. The goal of this work was to characterize the NA chaperone activity of RSV Gag by monitoring protein-facilitated annealing of tRNATrp to the PBS in vitro. To this end, we compared the annealing activity of highly purified recombinant RSV Gag lacking most of the PR domain (H6.Gag.3h), deletion mutants (H6.ΔNCΔSP and H6.ΔMA.3h), and the purified NCp12 and MA domains. We found that RSV H6.Gag.3h is an effective NA chaperone protein. Both the chaperone function and the RNA binding affinity were strongly dependent on NC and are largely independent of MA, which bound NA only weakly. In contrast to HIV-1, RSV Gag-facilitated RNA annealing was not stimulated by IP6; however, when RSV MA was replaced with the HIV-1 MA domain, annealing was stimulated in the presence of IP6. Taken together, these results suggest that mechanistic differences between RSV and HIV-1 Gag NA binding and tRNA annealing activity can be attributed to the MA domain.
MATERIALS AND METHODS
Preparation of proteins and nucleic acids.
All recombinant proteins were expressed and purified from Escherichia coli. Purified RSV NC was a gift from Robert J. Gorelick (AIDS and Cancer Virus Program, Leidos Biomedical Research, Inc., Frederick National Laboratory for Cancer Research). The H6.Gag.3h construct, derived from the Prague C strain of RSV (36, 37), contains an N-terminal His tag, a V314A substitution, and 7 amino acids of the PR sequence and was purified as previously described (38). The plasmid encoding HIV-1 GagΔp6 was a gift from Alan Rein (HIV Drug Resistance Program, Center for Cancer Research, National Cancer Institute). HIV-1 GagΔp6 was prepared using established methods (39). RSV H6.MA was prepared as described previously (38), and the N-terminal His tag was cleaved using tobacco etch virus protease (40) during the dialysis step prior to gel filtration chromatography. The plasmid encoding His-tagged HIV-1 MA (HIV-1 H6.MA) in a pET16B vector was a gift from Louis M. Mansky (University of Minnesota) and was purified using an established protocol (41), with additional steps of polyethylenimine precipitation followed by ammonium sulfate precipitation to remove NAs.
PCR-based mutagenesis was used to obtain RSV H6.Gag.3h truncations H6.ΔNCΔSP and H6.ΔMA.Gag.3h using the plasmid encoding H6.Gag.3h as a template (38). H6.Gag.ΔNCΔSP was prepared using a modification of the RSV H6.MA purification protocol (38). Briefly, protein expression was induced using 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 37°C for 4 h; protein pellets were resuspended in binding buffer (50 mM sodium phosphate, pH 8, 300 mM NaCl, 10 mM imidazole, 3 mM β-mercaptoethanol [βME]), sonicated, and loaded onto a His-Select nickel column (Sigma-Aldrich). Proteins were eluted with binding buffer containing increasing amounts of imidazole (10 mM to 100 mM). Fractions containing H6.Gag.ΔNCΔSP were combined and dialyzed into 2× buffer A (20 mM sodium phosphate, 3 mM potassium phosphate, pH 8, 6 mM KCl, 280 mM NaCl). After dialysis, the protein was further purified on a Superdex 200 gel filtration column (GE Healthcare) in 2× buffer A. Fractions containing pure H6.Gag.ΔNCΔSP were pooled, concentrated, and diluted to 1× buffer A with 80% glycerol prior to storage at −20°C.
RSV H6.ΔMA.Gag.3h expression was induced using 1 mM IPTG at 37°C for 4 h, and the protein was purified as described for H6.Gag.3h (38) with the following two additional steps to solubilize the protein. First, following centrifugation of the lysed cells, the pellet containing protein was resuspended in denaturing buffer (8 M urea, 500 mM NaCl, 25 mM Tris-HCl, pH 8, 10% glycerol, 1 mM βME, and 1 μM ZnCl2) using a homogenizer and then loaded onto a His-Select nickel column. Second, H6.ΔMA.Gag.3h was renatured on the column with five sequential washes of refolding buffer (500 mM NaCl, 25 mM Tris-HCl, pH 8, 10 mM imidazole, 5% glycerol, 1 mM βME, and 1 μM ZnCl2) containing decreasing amounts of urea (8 M, 6 M, 4 M, 2 M, and 1 M). Following elution from the nickel resin using imidazole, the preparation followed the protocol previously established for RSV H6.Gag.3h (38).
The previously described RSV/HIV-1 chimera H32R (42) (Fig. 1B) was cloned into pET-28TEV (43) using standard PCR methods. The gene encoding chimera H132R (Fig. 1B) was synthesized and inserted into pET15B (Novagen) by Genewiz, Inc. (South Plainfield, NJ). This construct encodes an N-terminal His tag followed by HIV-1 MA and then p2 through the C terminus of H6.Gag.3h. The H6.H32R.3h and H6.H132R.3h chimeras were purified essentially as described for H6.Gag.3h, except that proteins were induced using 1 mM IPTG at 37°C for 4 h. Following elution from the nickel resin, fractions containing protein were further purified on a Superdex 200 column (GE Healthcare) run in 28 mM Tris-HCl, pH 7.5, 1 μM ZnCl2, 1 mM βME, and 0.5 mM NaCl. All protein concentrations were determined from the absorbance at 280 nm using the following molar extinction coefficients: 72,880 M−1 cm−1 (H6.H32R.3h), 17,022 M−1 cm−1 (HIV-1 H6.MA), 63,090 M−1 cm−1 (HIV-1 GagΔp6), 65,890 M−1 cm−1 (RSV H6.Gag.3h), 44,920 M−1 cm−1 (RSV H6.ΔMA.Gag.3h), 8430 M−1 cm−1 (RSV NCp12), 57,660 M−1 cm−1 (RSV H6.Gag.ΔNCΔSP), 21,095 M−1 cm−1 (RSV MA), 5,690 M−1 cm−1 (HIV-1 NCp7), and 59,275 M−1 cm−1 (H6.H132R.3h).
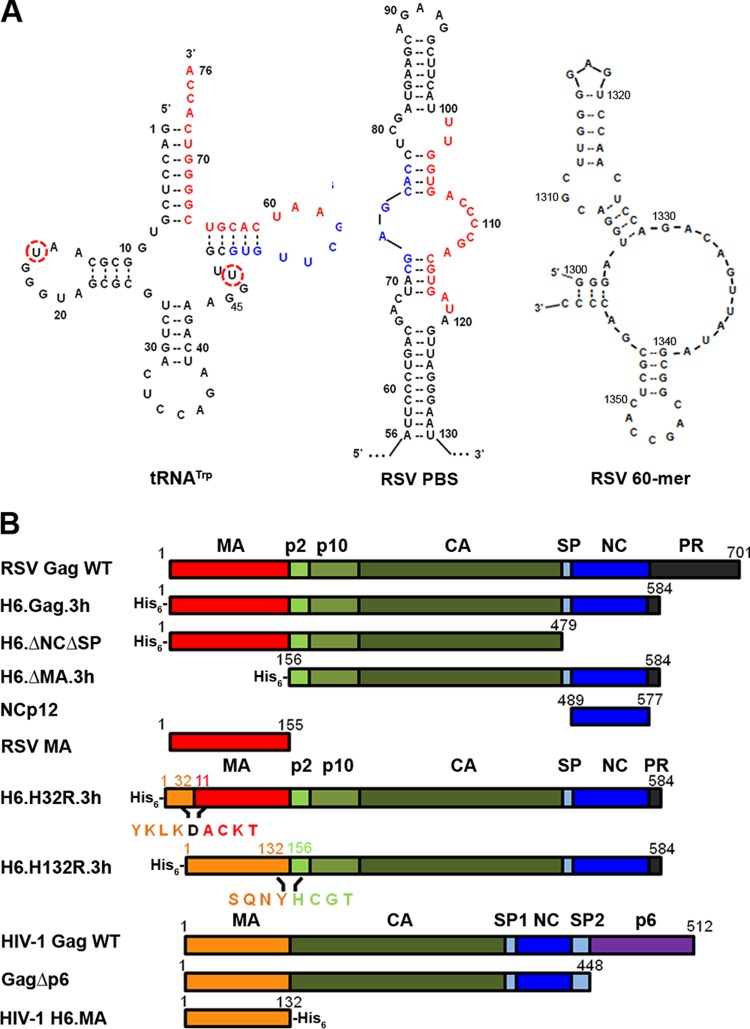
FIG 1.
(A) Predicted secondary structures of unmodified bovine tRNATrp, RSV PBS, and RSV 60-mer. The circled nucleotides differ between bovine (U16, U47) and chicken tRNATrp (C16, C47). The PBS-containing structure shown is derived from positions 56 to 130 of the RSV genome, which is part of the 180-mer PBS construct used in this work (see Materials and Methods). Shown in red and blue are two sets of complementary sequences that base pair during tRNA primer annealing as described previously (6). The RSV 60-mer construct is derived from nt 1300 to 1360 of the RSV genome, with nt 1300 changed from T to G to facilitate in vitro transcription. (B) WT RSV and HIV-1 Gag constructs and variants used in this work. All RSV Gag variants are derived from the previously described genomic sequence of pRC.V8 (45). All HIV-1 Gag variants are derived from HIV-1 isolate BH10. Amino acid residue numbers are shown above constructs. In chimeras, the color indicates whether the domain is from RSV (red, green, blue, and black) or HIV-1 Gag (orange). H6.H32R.3h is a previously described chimera in which the first 10 amino acids of RSV MA are replaced with the first 32 amino acids of HIV-1 MA (42). H6.H132R.3h consists of the entire HIV-1 MA domain fused to RSV ΔMA.3h. The amino acids flanking the junction region of each chimera are shown explicitly below each construct. In the core of H6.H132R.3h, an extra D residue is present at the junction (black).
All RNA constructs were in vitro transcribed from digested plasmids or synthetic DNA oligonucleotides using T7 RNA polymerase and previously established methods (44). The RSV RNA 60-mer construct was derived from nt 1300 to 1360 of the RSV RC.V8 (45) genome, with nt 1300 changed from T to G to promote transcription (Fig. 1A). DNA oligonucleotides encoding the T7 polymerase promoter followed by the RSV 60-mer sequence were purchased from IDT (5′-TGCAGTAATACGACTCACTATAGGGATGGACGCTTGGGGAGTCCAACTCCAGACATTATAGCGGCAGCCACTCGCGACCCC-3′ and 5′-GGGGTCGCGAGTGGCTGCCGCTATAACTGTCTGGAGTTGGACTCCCCAAGCGTCCATCCCTATAGTGAGTCGTATTACTGCA-3′). The DNA was amplified by PCR and ligated into BamHI and EcoRI sites of pUC19 (New England BioLabs). Bovine tRNATrp was in vitro transcribed from pJL122 (a gift from Paul Schimmel, The Scripps Research Institute). Unmodified bovine [32P]tRNATrp used in gel-shift annealing assays was internally labeled using [α-32P]GTP during in vitro transcription (44). The 180-nt RNA construct containing the RSV PBS (Fig. 1A) was in vitro transcribed using Sau3AI-digested plasmid pGEM.RSVLTR/MA (46), encoding sequences derived from the 5′-untranslated region (UTR) of the RSV genome. The RSV PBS 180-mer contains 21 nt of pGEM.RSVLTR/MA followed by nt 1 to 159 of the RC.V8 sequence. RNA concentrations were determined by measuring the absorbance at 260 nm and the following molar extinction coefficients: 134.9 × 104 M−1 cm−1 (RSV PBS), 60.4 × 104 M−1 cm−1 (tRNATrp), and 51.5 × 104 M−1 cm−1 (RSV 60-mer).
Annealing assays.
Gel-shift RNA annealing assays were performed using previously established methods (20), except that annealing assays contained 50 mM HEPES, pH 7.4, 5 mM dithiothreitol, 1 mM MgCl2, 50 mM NaCl, 10 nM bovine tRNATrp spiked with trace amounts of [32P]tRNATrp, and 25 nM RSV PBS. In assays testing the effect of IPs, 5 μM IP6 was also included. Single-time-point titration assays were incubated at room temperature for 30 min using protein concentrations ranging from 0 to 4.5 μM. Time course kinetic assays were carried out at room temperature using a single protein concentration between 0.5 and 3.0 μM. Aliquots were removed at the indicated time points, and annealing data were fit to single-exponential curves using the following equation: A(t) = A∞ − ΔAe−kt, where t is time in min, A(t) is percent tRNATrp annealed as a function of time, A∞ is the equilibrium final percentage of tRNATrp annealed, ΔA is the change in percentage of tRNATrp annealed, and k is the annealing rate. The scaled annealing rate, k′, was obtained by multiplying k by the final fraction of tRNATrp annealed.
FA binding assays.
Fluorescence anisotropy (FA) was used to determine the binding affinity of Gag and Gag variants for NA as described previously (20). RSV 60-mer RNA was labeled with fluorescein-5-isothiocyanate (FITC) at the 3′ end (3′-FITC-RSV 60) using established protocols (47). The concentration and labeling efficiency were determined by measuring the absorbance at 260 nm and 495 nm and using the following molar extinction coefficients: ε495 = 8.5 × 104 M−1 cm−1 (fluorescein) and ε260 = 51.5 × 104 M−1 cm−1 (RSV 60-mer). FA assays were performed in 20 mM Tris-HCl, pH 7.9, 1 mM MgCl2, and 50 mM monovalent ions using 3′-FITC-RSV 60. Final RNA concentrations used were generally 10- to 100-fold below the determined dissociation constant (Kd) value: 20 nM RNA was used for RSV MA, 5.0 nM RNA was used for RSV H6.Gag.3h, RSV H6.Gag.ΔNCΔSP, RSV H6.ΔMA.Gag.3h, and HIV-1 H6.MA, and 1.5 nM RNA was used for HIV-1 GagΔp6, H6.H132R.Gag.3h, and H6.H32R.Gag.3h. Binding affinities were determined by fitting FA data to a 1:1 binding model as described previously (35). Due to aggregation, the dissociation constant of HIV-1 H6.MA was calculated by measuring fluorescence intensity rather than FA, as previously described (20). FA competition assays using IP6 were performed essentially as previously described (20). In these assays, RSV MA (20 μM) or RSV H6.Gag.3h (400 nM) was prebound with 20 nM or 1.5 nM 3′-FITC-RSV 60, respectively. Following a 30-min incubation at room temperature, increasing concentrations of IP6 were added and incubated with the protein/RNA complexes for an additional 30 min prior to FA measurements. All fluorescence measurements were performed on a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA).
RESULTS
The annealing activities of RSV Gag and NC are salt dependent and equivalent under saturating conditions.
All RSV Gag variants used in this work are derived from Prague C strain pRC.V8 (45). H6.Gag.3h contains only the first seven amino acids of the protease domain (38, 48), and previous work demonstrated that the Gag.3h protein forms virus-like particles efficiently in COS-1 cells (48). To begin to characterize the NA chaperone activity of RSV H6.Gag.3h (here referred to as RSV Gag), single-time-point gel-shift tRNA annealing assays were performed using in vitro-transcribed bovine tRNATrp, which differs from chicken tRNATrp in only two nucleotides (Fig. 1A, C16U in the D loop and C47U in the variable loop) and a 180-nt RNA construct derived from the PBS region of the RSV gRNA (RSV PBS) (Fig. 1A).
RSV Gag mediated the annealing of tRNATrp to the RSV PBS (Fig. 2). At 50 mM NaCl, the extent of annealed product observed at a given protein concentration was significantly greater for Gag than for RSV NCp12. Moreover, in the case of Gag, the maximum extent of annealing (∼50%) was observed at 1 μM protein, whereas for NCp12, 10% annealing was achieved at 1 μM and only 35% annealing was observed at 4.5 μM protein. RSV Gag- and NCp12-facilitated tRNA annealing were both inhibited at higher concentrations of NaCl. The annealing activities of RSV Gag and NCp12 were reduced when NaCl was increased from 50 mM to 150 mM by 50% and 80%, respectively (Fig. 2A).
FIG 2.
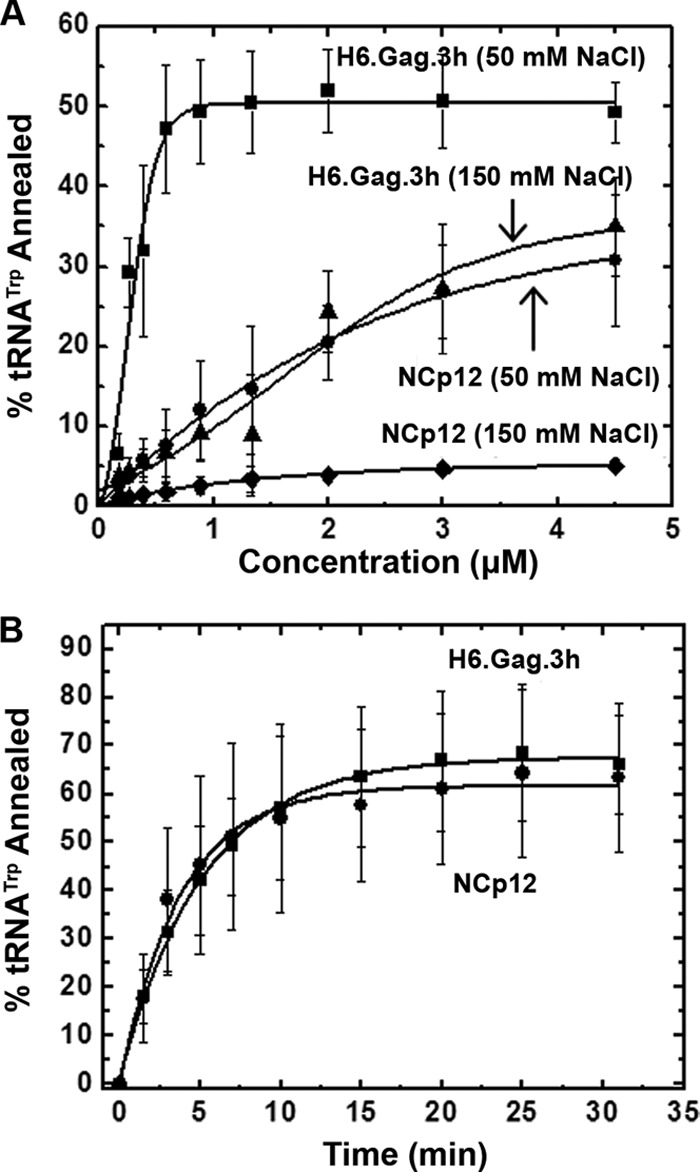
(A) Single-time-point (30 min) gel-shift annealing assays comparing RSV H6.Gag.3h and NCp12 in the presence of the indicated concentration of NaCl. (B) Annealing time course assays for RSV H6.Gag.3h and NCp12 using 3 μM protein in 50 mM NaCl. The curves are single-exponential fits to the averages from three or more trials with standard deviations indicated.
Time course assays next were performed at 0.5, 1.0, 2.0, and 3.0 μM protein concentrations to compare the annealing rates of RSV Gag and NCp12 below and at protein saturation (Table 1). At 0.5 μM protein, RSV Gag and NC demonstrate a similar annealing rate, but in the presence of Gag, an ∼3-fold greater total percent tRNA annealed was observed. At protein concentrations approaching saturation (1 and 2 μM), Gag and NCp12 displayed more similar annealing rates and total percentages of tRNA annealed. At protein saturation (3 μM), both proteins resulted in ∼65% total tRNA annealed and exhibited very similar scaled annealing rates of 0.13 min−1 (Gag) and 0.16 min−1 (NCp12) (Fig. 2B and Table 1).
TABLE 1.
RSV H6.Gag.3h- and NCp12-facilitated annealing of tRNATrp to RSV PBSa
| Protein (concn, μM) | kb (min−1) | tRNATrp annealedc (%) | k′d (min−1) |
|---|---|---|---|
| NCp12 (0.5) | 0.11 ± 0.05 | 15 ± 4 | 0.02 |
| NCp12 (1.0) | 0.09 ± 0.01 | 50 ± 15 | 0.05 |
| NCp12 (2.0) | 0.15 ± 0.04 | 74 ± 19 | 0.11 |
| NCp12 (3.0) | 0.25 ± 0.07 | 63 ± 15 | 0.16 |
| H6.Gag.3h (0.5) | 0.08 ± 0.04 | 49 ± 22 | 0.04 |
| H6.Gag.3h (1.0) | 0.16 ± 0.07 | 70 ± 17 | 0.11 |
| H6.Gag.3h (2.0) | 0.18 ± 0.05 | 73 ± 12 | 0.13 |
| H6.Gag.3h (3.0) | 0.19 ± 0.07 | 66 ± 10 | 0.13 |
All experiments were performed in 50 mM HEPES, pH 7.4, 5 mM dithiothreitol, 1 mM MgCl2, and 50 mM NaCl using 10 nM bovine tRNATrp and 25 nM RSV PBS. All reported values represent the averages from three or more trials with standard deviations reported.
k is the annealing rate constant obtained from single-exponential fits of time course annealing assays.
The overall amount of tRNATrp annealed after 30 min at room temperature.
k′ is the scaled annealing rate obtained by multiplying k by the final fraction of tRNATrp annealed.
NA chaperone activity of RSV Gag is dependent on NC but not on MA.
It was recently reported that the differences in HIV-1 GagΔp6 and NCp7 NA chaperone activities are due to properties of the MA domain, which inhibits Gag-facilitated RNA annealing in vitro (20). To gain further insights into the contributions of NCp12 and MA to RSV Gag's NA chaperone function, purified H6.Gag.3h truncation mutants and the MA domain were tested for annealing activity using gel-shift RNA annealing assays (Fig. 3A). For the H6.Gag.ΔNCΔSP (here called ΔNCΔSP) and MA proteins, no substantial annealing was observed at concentrations of up to 4.5 μM, suggesting that NCp12 is critical for RSV Gag chaperone activity (Fig. 3A). This finding is consistent with previous work demonstrating that NCp7 is required for HIV-1 Gag-facilitated tRNA annealing (20). RSV H6.ΔMA.3h (here referred to as ΔMA) exhibited chaperone activity similar to that of 3 μM Gag (Table 1), with a scaled annealing rate of 0.15 min−1 and 62% ± 8% tRNA annealed (Fig. 3B). Thus, MA does not inhibit annealing of RSV Gag, in contrast to the inhibitory effect of HIV-1 MA on the chaperone activity of HIV-1 GagΔp6 (20).
FIG 3.
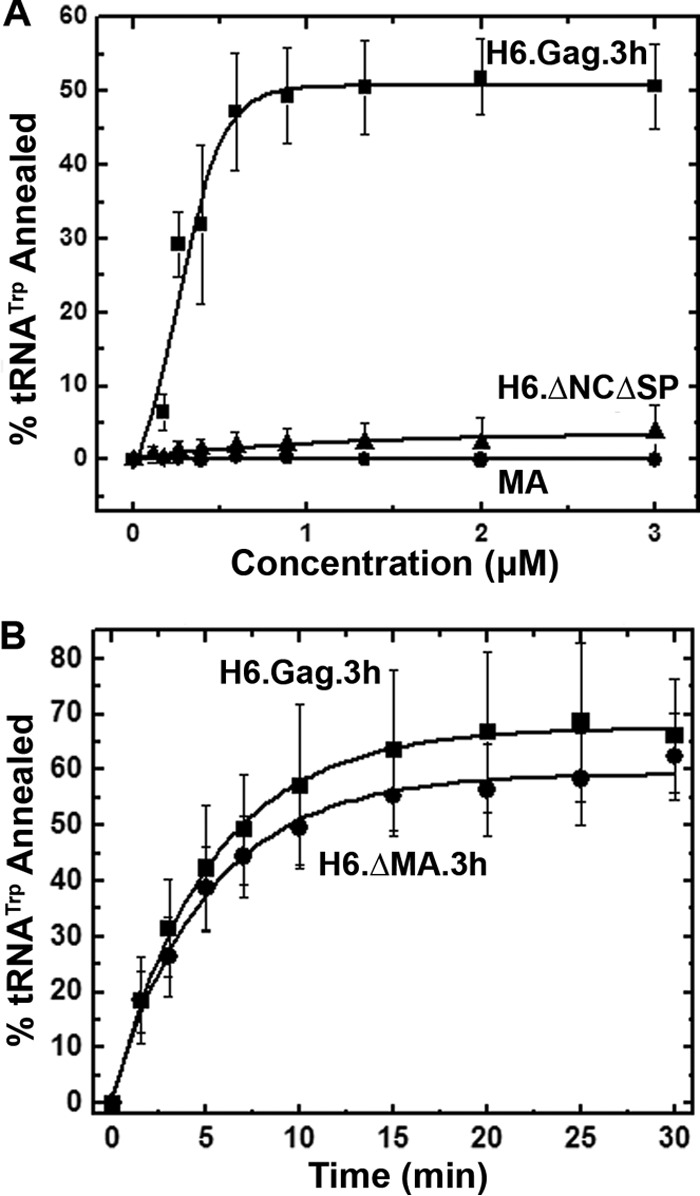
(A) Single-time-point (30 min) gel-shift annealing assays comparing RSV H6.Gag.3h, H6.GagΔNCΔSP, and MA. (B) Annealing time course assays comparing H6.Gag.3h and H6.ΔMA.3h using 3 μM protein. The curves are single-exponential fits to the averages from three or more trials with standard deviations indicated.
RSV Gag NA chaperone activity is not stimulated by IP6.
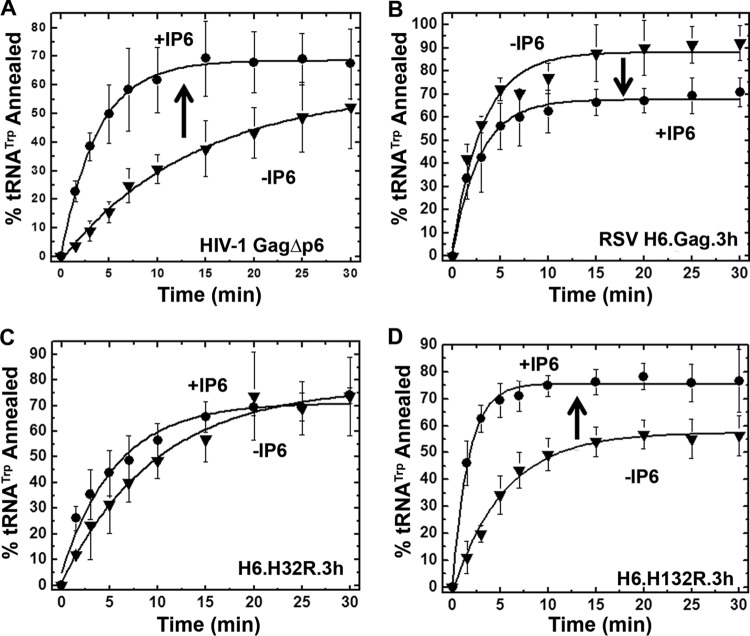
In HIV-1, IP6 stimulates the NA chaperone activity of Gag by binding the inhibitory MA domain and displacing it from NA (20). We hypothesized that because the RSV MA domain does not inhibit annealing of Gag, IP6 binding was unlikely to stimulate RSV Gag NA chaperone activity. To test this hypothesis, Gag-facilitated single-time-point gel-shift RNA annealing assays were performed in the absence and presence of 5 μM IP6 (Fig. 4). As expected, tRNA annealing by HIV-1 GagΔp6 (here referred to as HIV-1 Gag) was stimulated by 15%, and the scaled annealing rate increased 4-fold in the presence of IP6 (Fig. 4A). In contrast, RSV Gag showed a 20% decrease in tRNA annealing and no change in annealing rate in the presence of IP6 (Fig. 4B and Table 2). We attribute this decrease in percent tRNA annealed to inhibition of the interaction of NCp12 and RNA by high concentrations of IP6, as we observed this effect only at IP6 concentrations of >5 μM (data not shown). Importantly, the lack of IP6-dependent stimulation in tRNA annealing by RSV Gag was observed under a variety of conditions, including kinetic annealing assays performed at a range of concentrations of RSV Gag (0.5 to 3.0 μM) and using kinetic and single-time-point assays at 1 to 200 μM IP6 and 250 to 700 μM PI(4,5)P2 (data not shown).
FIG 4.
Annealing time course assays for HIV-1 GagΔp6 (A), RSV H6.Gag.3h (B), H6.H32R.3h (C), and H6.H132R.3h (D) using 0.8 μM protein in the absence or presence of 5 μM IP6. The curves are single-exponential fits to the averages from three or more trials with standard deviations indicated.
TABLE 2.
RSV H6.Gag.3h-, HIV-1 GagΔp6-, and RSV/HIV-1 chimera-facilitated annealing of tRNATrp to RSV PBSa
| Protein (concn, μM) | IP6 | kb (min−1) | tRNATrp annealedc (%) | k′d (min−1) |
|---|---|---|---|---|
| HIV-1 GagΔp6 | − | 0.08 ± 0.03 | 52 ± 15 | 0.04 |
| HIV-1 GagΔp6 | + | 0.26 ± 0.05 | 67 ± 12 | 0.17 |
| RSV H6.Gag.3h | − | 0.35 ± 0.14 | 92 ± 8 | 0.32 |
| RSV H6.Gag.3h | + | 0.37 ± 0.17 | 71 ± 6 | 0.26 |
| H6.H32R.3h | − | 0.11 ± 0.04 | 73 ± 15 | 0.08 |
| H6.H32R.3h | + | 0.14 ± 0.05 | 73 ± 3 | 0.10 |
| H6.H132R.3h | − | 0.18 ± 0.02 | 56 ± 7 | 0.10 |
| H6.H132R.3h | + | 0.59 ± 0.06 | 76 ± 12 | 0.44 |
All experiments were performed in 50 mM HEPES, pH 7.4, 5 mM dithiothreitol, 1 mM MgCl2, and 50 mM NaCl using 10 nM bovine tRNATrp, 25 nM RSV PBS, and 800 nM Gag in the absence or presence of 5 μM IP6. All reported values represent the averages from three or more trials with standard deviations reported.
k is the annealing rate constant obtained from single-exponential fits of time course annealing assays.
The overall amount of tRNATrp annealed after 30 min at room temperature.
k′ is the scaled annealing rate obtained by multiplying k by the final fraction of tRNATrp annealed.
To determine whether the HIV-1 MA domain would confer IP6-dependent stimulation of annealing to RSV Gag, we tested the effect of IP6 on the NA chaperone function of RSV/HIV-1 Gag chimeras H6.H32R.3h and H6.H132R.3h (referred to as H32R and H132R, respectively) which contain either the membrane binding domain (MBD) of HIV-1 MA (H32R) or the entire HIV-1 MA domain (H132R) (Fig. 1B). Chimera H32R annealed ∼70% of the tRNA with a reduced rate of annealing; however, there was no significant difference in the annealing rate in the absence or presence of IP6 (Fig. 4C and Table 2). The overall reduction in the annealing rate might be due to interference of the chimeric MA domain with the chaperone function of NC or a global change in conformation of the chimeric protein. Therefore, the HIV-1 MA MBD alone was not sufficient to trigger IP6-dependent stimulation of tRNA annealing. In contrast, chimera H132R exhibited a 20% stimulation of tRNA annealing (Fig. 4D) and a 4-fold stimulation in annealing rate in the presence of IP6 (Table 2). Taken together, these results show that the complete HIV-1 MA domain is required to confer IP6-dependent stimulation of annealing on RSV Gag, suggesting that residues outside the HIV-1 MBD are needed for this effect.
RSV NCp12 is required for high-affinity binding to NA.
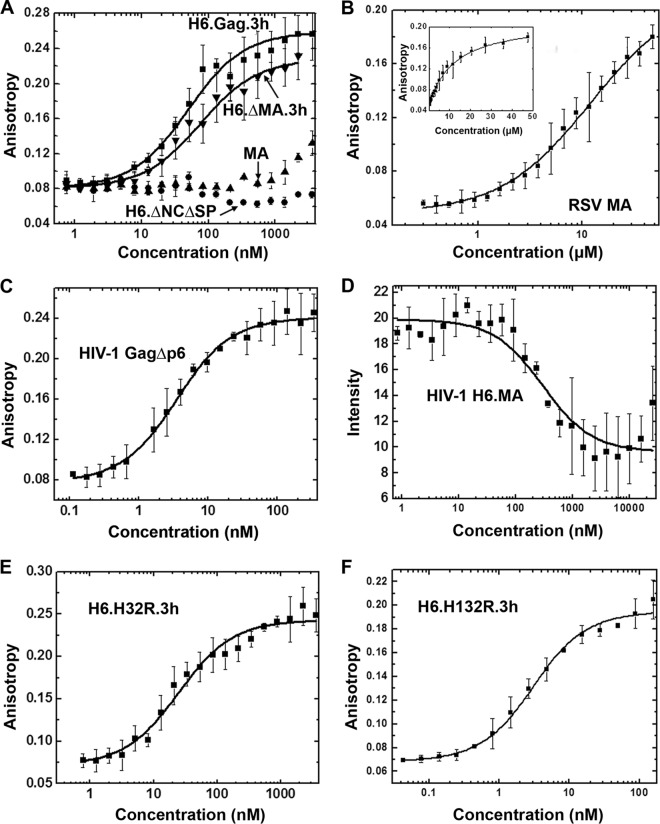
We hypothesize that HIV-1 and RSV Gag NA chaperone activities differ due to differences in MA-NA binding affinities. Thus, FA was used to determine the relative binding affinities of RSV and HIV-1 Gag variants to a nonspecific RNA. For these studies, we chose to use a 60-mer RNA derived from the RSV Gag coding sequence, which is predicted by m-fold to possess both single-stranded and double-stranded regions (Fig. 1A). For HIV-1 MA, NA binding resulted in a significant decrease in fluorescence intensity, making the dissociation constant (Kd) derived from FA less accurate. Therefore, in this case the binding affinity was determined from the fluorescence intensity, as previously described (20).
HIV-1 GagΔp6 (Kd, ≤3 nM) bound to the 60-mer RNA with at least 100-fold higher affinity than HIV-1 MA (Kd, 354 nM) (Table 3). Binding affinities of RSV Gag and ΔMA for NA were similar (Kd, ∼70 nM) to each other, although both bound the 60-mer RNA with at least 20-fold weaker affinity than HIV-1 GagΔp6. Interestingly, RSV Gag derivative ΔNCΔSP and RSV MA bound with low affinity to the RSV 60-mer (Kd of ≥10 μM and 17 μM, respectively) (Fig. 5A and B and Table 3). A previous study of the NA binding properties of RSV MA estimated a Kd in the range of ∼10−3 M, which is even weaker than our data indicated (49). Additionally, the RSV/HIV-1 Gag chimera H32R bound with ∼3-fold higher affinity (Kd, ∼20 nM) (Fig. 5E) than RSV Gag but more weakly than HIV-1 Gag (Table 3), demonstrating that the HIV-1 MBD alone is unable to increase the binding affinity of RSV Gag to the level of HIV-1 Gag. In contrast, H132R, which contains the entire HIV-1 MA domain, exhibited a Kd of ≤3 nM for binding to RSV 60-mer RNA, a value equivalent to the upper limit measured for binding by HIV-1 GagΔp6. In summary, the apparent Kd values decrease in this order: RSV MA ∼ ΔNCΔSP ≫ HIV-1 MA > RSV Gag ∼ RSV ΔMA > H32R > H132R ∼ HIV-1 Gag. These results, together with the similar binding affinities previously determined for HIV-1 NCp7 and RSV NCp12 (35), suggest the ability of HIV-1 Gag to bind NA more tightly than RSV Gag is due to the substantial increase in NA binding affinity of HIV-1 MA relative to RSV MA.
TABLE 3.
Apparent dissociation constants of RSV and HIV-1 Gag for 3′-FITC-RSV 60 determined by FAa

All experiments were performed using 1.5–20 nM 3′-FITC-RSV 60, 20 mM Tris-HCl, pH 7.9, 1 mM MgCl2, and 50 mM monovalent ions.
The detection limit of the FA binding assay allowed only an upper limit for the Kd to be determined.
FIG 5.
Fluorescence measurements to monitor binding of 3′-FITC-RSV 60 to RSV and HIV-1 Gag variants. (A) FA binding assays for RSV Gag variants. (B) Binding of RSV MA performed at higher protein concentrations. The inset shows the same data plotted on a linear rather than log 10 scale. FA binding assays were performed with HIV-1 GagΔp6 (C), HIV-1 H6.MA (D), and RSV/HIV-1 Gag chimeras (E and F). The FA data for RSV Gag variants, HIV-1 GagΔp6, and the chimeras were fit to 1:1 binding curves as described previously (35). The fluorescence intensity change of HIV-1 H6.MA was fit as previously described (20). All graphs represent the averages from three or more trials with standard deviations indicated.
IP6 binds RSV MA and disrupts the MA-NA interaction.
The annealing activity of RSV Gag was not stimulated by IP6 unless the entire MA domain was replaced by HIV-1 MA. To determine whether RSV MA itself could bind IP6, FA competition assays were performed. RSV Gag or RSV MA was prebound to 3′-FITC-RSV 60. We observed that IP6 began to compete with the 3′-FITC-RSV 60-mer for binding to MA at 6 μM, and complete displacement of RNA was observed at 135 μM IP6 (Fig. 6). In contrast, RSV Gag remained bound to 3′-FITC-RSV 60 even at the highest concentrations of IP6, with no detectable competition observed. This result suggests that the MA protein binds IP6, and this binding event is strong enough to displace NA; however, the 3′-FITC-RSV 60 was not displaced from RSV Gag due to the strong affinity of the NC domain for NA. To rule out any effect of the His tag, IP6 competition assays were also performed with RSV GagΔPR, a construct that lacks the N-terminal His tag and the complete protease domain, and similar results were obtained (data not shown).
FIG 6.
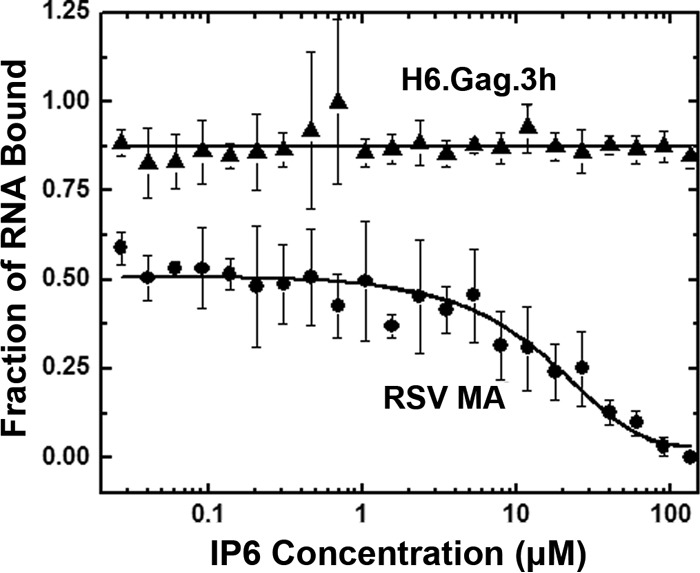
FA measurements to monitor competition of IP6 with 3′-FITC-RNA 60 for binding to RSV H6.Gag.3h and RSV MA. Graphs represent the averages from three or more trials with standard deviations indicated.
DISCUSSION
In this work, we demonstrated that the RSV Gag polyprotein is an effective NA chaperone and that NCp12 is required for this activity, consistent with previous studies performed with HIV-1 GagΔp6 (20). Whereas RSV Gag was a much better NA chaperone on a molar basis than NCp12, at protein saturation, RSV Gag and NCp12 had comparable NA chaperone activities. The observation that at lower protein concentrations the extent of tRNA annealing facilitated by Gag is greater than the extent of annealing facilitated by NCp12 is an indication that other domains of Gag contribute to NA chaperone activity. Although the NC domain is the primary NA interaction domain for both proteins, the Gag polyprotein is able to multimerize due to the CA domain (13, 50), and it is possible that these intermolecular interactions lead to an increase in the total percentage of tRNA annealed at low protein concentrations. This result differs from observations in the HIV-1 system, where at protein saturation GagΔp6-facilitated tRNALys,3 annealing is 11-fold slower than NCp7-facilitated annealing (20). In addition, whereas HIV-1 MA inhibits the NA chaperone function of HIV-1 GagΔp6, the presence of the RSV MA domain had no significant effect on the activity of RSV Gag in vitro annealing. Annealing activities of RSV Gag or chimera H32R were not stimulated by IP6; however, IP-dependent stimulation was observed when using HIV-1 GagΔp6 or the chimera H132R, in which the RSV MA domain was replaced with the entire HIV-1 MA sequence. Furthermore, RSV MA binds NA very weakly; therefore, RSV Gag and ΔMA share equivalent binding affinities for RNA.
We observed that the tRNA annealing activities of RSV Gag and NCp12 both were highly sensitive to monovalent ions in the range of 50 to 150 mM. This finding differs from those for HIV-1, for which reports have shown that NC is salt sensitive (51–53), but Gag-facilitated annealing is much less sensitive to salt in this range (20, 54). Experiments demonstrating that aggregation of NA by RSV NCp12 is abrogated by increasing concentrations of NaCl are consistent with our observation that the NA chaperone activity of NCp12 was salt dependent (55).
The relative binding affinities reported here are generally consistent with predictions of binding based on calculated isoelectric points (pI; Scripps protein calculator). The pI values of RSV NCp12 and HIV-1 NCp7 (BH10) are the same, with a calculated value of 10.02. However, the pI values for Gag differ, with values of 8.51 and 9.25 calculated for RSV H6.Gag.3h and HIV-1 GagΔp6, respectively. The less basic character of RSV H6.Gag.3h relative to HIV-1 GagΔp6 is reflected in our FA studies showing that RSV Gag exhibits at least 22-fold weaker binding to NA than HIV-1 Gag. The different binding capabilities of the Gag proteins likely are due to the different pI values of the MA domains, which are 7.7 (RSV) and 9.2 (HIV-1). The lower overall net positive charge of RSV MA is also consistent with our observation that RSV MA has a very weak binding affinity for RNA relative to HIV-1 H6.MA (Table 3). Interestingly, the pI of the human T-cell leukemia virus type 2 (HTLV-2) MA protein (9.51) is even higher than that of HIV-1 MA, and we recently showed that, similar to bovine leukemia virus MA (56), this deltaretroviral MA protein binds RNA oligonucleotides derived from HTLV-2 gRNA with even higher affinity (∼70 nM at 50 mM NaCl) than HTLV-2 NC (57). Thus, retroviruses appear to possess a wide spectrum of MA-NA binding capabilities, which may have evolved due to distinct mechanisms of action during virus assembly and/or postentry.
In contrast to HIV-1, where it is widely accepted that Gag is localized to the PM, at least in part, via an interaction between MA and PI(4,5)P2 (30, 34, 58, 59), the mechanism by which RSV Gag is targeted to the PM is controversial, as both PI(4,5)P2-dependent and -independent modes of PM localization have been suggested. Early reports showed that RSV MA interacts with negatively charged lipids in the PM due to electrostatic interactions, which is magnified by Gag oligomerization during virus assembly (60, 61). However, more recent studies reported conflicting results regarding whether PI(4,5)P2 is required for PM localization of RSV Gag. One report concluded that PM localization of RSV Gag does not require PI(4,5)P2 in chicken fibroblasts based on the finding that membrane localization of RSV Gag-GFP and release of virus-like particles were unaffected by expression of inositol polyphosphate 5-phosphatase type IV, which reduces intracellular levels of PI(4,5)P2 and phosphatidylinositol-(3,4,5)-triphosphate [PI(3,4,5)P3] (12). However, a subsequent study showed that reduction of intracellular levels of PI(4,5)P2 and PI(3,4,5)P3 prevents PM localization of RSV Gag and results in a 40% reduction in viral budding efficiency in quail fibroblasts (62). Supporting the importance of PI(4,5)P2 in RSV Gag membrane localization, Akira Ono and coworkers found that an HIV-1 Gag construct with RSV MA substituted for HIV-1 MA and a leucine zipper in place of the NC domain binds to PI(4,5)P2-containing liposomes in vitro (Akira Ono, personal communication). In this RSV/HIV chimeric Gag protein, the contribution of NC to liposome binding was negated by the zipper domain, allowing RSV MA sensitivity to PI(4,5)P2 binding to be measured independently. Whether the putative role of PI(4,5)P2 in PM binding by RSV Gag represents a general requirement for charge-mediated interactions between acidic phospholipids in the PM with basic regions of Gag or whether there is indeed a specific binding pocket for PI(4,5)P2 remains uncertain.
IP6 has been proposed to stimulate HIV-1 Gag chaperone activity by displacing the MA domain from bound NA, presumably allowing more effective NA binding and enhanced chaperone function of the NC domain (20). The failure of IP6 to stimulate RSV Gag chaperone function likely is due to the weak binding of the MA domain to NA rather than to lack of MA-IP interaction based on our finding that IP6 competes with RNA for binding to MA. Thus, similar to other retroviral MA domains (20, 56, 57), RSV MA is capable of binding IPs, but unlike the case in HIV-1, this binding does not stimulate chaperone function because MA is so weakly bound to NA in the first place. Furthermore, finding that the RSV Gag-NA interaction was not disrupted by IP6 in vitro suggests that Gag is so tightly bound to NA via the NC domain that IP6 is not an effective competitor.
Exactly when in the retroviral life cycle and where in the cell tRNA primer annealing occurs are open questions. In HIV-1 and RSV, PR-negative virus particles have a significant amount of annealed tRNA, suggesting that tRNA annealing occurs prior to virus maturation (18, 19). It was proposed that for HIV-1, the interaction between MA and RNA inhibits premature tRNA primer annealing by Gag prior to MA-PI(4,5)P2 interaction and assembly (20). In RSV, the Gag protein undergoes nuclear localization due to the nuclear localization signals in the MA and NC domains and a CRM1-dependent nuclear export signal in the p10 domain (63, 64). RSV Gag may have evolved to develop effective IP-independent NA chaperone activity because of the local environment in which primer annealing occurs. Whether tRNA annealing to the PBS occurs in the nucleus is unknown, and future studies will address this question.
ACKNOWLEDGMENTS
We thank Robert J. Gorelick for providing RSV NCp12 and Christopher P. Jones and Ioulia Rouzina for advice and helpful discussions.
This work was supported by NIH grant R01 GM065056 (to K.M.-F.) and by award number R01 CA076534, including supplemental ARRA funds from the NCI (to L.J.P.). T.D.R.-M. was supported by NIH T32-GM008512 and S.N.-H. by NIH T32 CA060395. This project was funded in part by a grant from the Pennsylvania Department of Health using Tobacco CURE Funds.
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCI or the National Institutes of Health. The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Footnotes
Published ahead of print 30 April 2014
REFERENCES
- 1.Sakalian M, Wills JW, Vogt VM. 1994. Efficiency and selectivity of RNA packaging by Rous sarcoma virus Gag deletion mutants. J. Virol. 68:5969–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleiman L, Jones CP, Musier-Forsyth K. 2010. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 584:359–365. 10.1016/j.febslet.2009.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang M, Mak J, Ladha A, Cohen E, Klein M, Rovinski B, Kleiman L. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 67:3246–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleiman L. 2002. tRNA(Lys3): the primer tRNA for reverse transcription in HIV-1. IUBMB Life 53:107–114. 10.1080/15216540211469 [DOI] [PubMed] [Google Scholar]
- 5.Waters LC, Mullin BC, Ho T, Yang WK. 1975. Ability of tryptophan tRNA to hybridize with 35S RNA of avian myeloblastosis virus and to prime reverse transcription in vitro. Proc. Natl. Acad. Sci. U. S. A. 72:2155–2159. 10.1073/pnas.72.6.2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris S, Leis J. 1999. Changes in Rous sarcoma virus RNA secondary structure near the primer binding site upon tRNATrp primer annealing. J. Virol. 73:6307–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou W, Parent LJ, Wills JW, Resh MD. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verderame MF, Nelle TD, Wills JW. 1996. The membrane-binding domain of the Rous sarcoma virus Gag protein. J. Virol. 70:2664–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ono A, Orenstein JM, Freed EO. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855–2866. 10.1128/JVI.74.6.2855-2866.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganser-Pornillos BK, Yeager M, Sundquist WI. 2008. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18:203–217. 10.1016/j.sbi.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2:a006924. 10.1101/cshperspect.a006924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan J, Dick RA, Vogt VM. 2011. Rous sarcoma virus gag has no specific requirement for phosphatidylinositol-(4,5)-bisphosphate for plasma membrane association in vivo or for liposome interaction in vitro. J. Virol. 85:10851–10860. 10.1128/JVI.00760-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spidel JL, Wilson CB, Craven RC, Wills JW. 2007. Genetic studies of the beta-hairpin loop of Rous sarcoma virus capsid protein. J. Virol. 81:1288–1296. 10.1128/JVI.01551-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derdowski A, Ding L, Spearman P. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 78:1230–1242. 10.1128/JVI.78.3.1230-1242.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Summers MF, Karn J. 2011. Special issue: structural and molecular biology of HIV. J. Mol. Biol. 410:489–490. 10.1016/j.jmb.2011.05.001 [DOI] [PubMed] [Google Scholar]
- 16.Zhou J, Bean RL, Vogt VM, Summers M. 2007. Solution structure of the Rous sarcoma virus nucleocapsid protein: muPsi RNA packaging signal complex. J. Mol. Biol. 365:453–467. 10.1016/j.jmb.2006.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weldon RA, Jr, Wills JW. 1993. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J. Virol. 67:5550–5561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Wang J, Shalom A, Li Z, Khorchid A, Wainberg MA, Kleiman L. 1997. Primer tRNA3Lys on the viral genome exists in unextended and two-base extended forms within mature human immunodeficiency virus type 1. J. Virol. 71:726–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart L, Schatz G, Vogt VM. 1990. Properties of avian retrovirus particles defective in viral protease. J. Virol. 64:5076–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones CP, Datta SA, Rein A, Rouzina I, Musier-Forsyth K. 2011. Matrix domain modulates HIV-1 Gag's nucleic acid chaperone activity via inositol phosphate binding. J. Virol. 85:1594–1603. 10.1128/JVI.01809-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. 2007. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 4:118–130. 10.4161/rna.4.3.5445 [DOI] [PubMed] [Google Scholar]
- 22.Rein A, Henderson LE, Levin JG. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297–301. 10.1016/S0968-0004(98)01256-0 [DOI] [PubMed] [Google Scholar]
- 23.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80:217–286. 10.1016/S0079-6603(05)80006-6 [DOI] [PubMed] [Google Scholar]
- 24.Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. 2010. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 7:754–774. 10.4161/rna.7.6.14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. 1995. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 254:523–537. 10.1006/jmbi.1995.0635 [DOI] [PubMed] [Google Scholar]
- 26.Rein A. 2010. Nucleic acid chaperone activity of retroviral Gag proteins. RNA Biol. 7:700–705. 10.4161/rna.7.6.13685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purohit P, Dupont S, Stevenson M, Green MR. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7:576–584. 10.1017/S1355838201002023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lochrie MA, Waugh S, Pratt DG, Jr, Clever J, Parslow TG, Polisky B. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 25:2902–2910. 10.1093/nar/25.14.2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parent LJ, Gudleski N. 2011. Beyond plasma membrane targeting: role of the MA domain of Gag in retroviral genome encapsidation. J. Mol. Biol. 410:553–564. 10.1016/j.jmb.2011.04.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alfadhli A, Still A, Barklis E. 2009. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 83:12196–12203. 10.1128/JVI.01197-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott DE, Coren LV, Gagliardi TD. 2005. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 79:13839–13847. 10.1128/JVI.79.22.13839-13847.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramalingam D, Duclair S, Datta SA, Ellington A, Rein A, Prasad VR. 2011. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J. Virol. 85:305–314. 10.1128/JVI.02626-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chukkapalli V, Oh SJ, Ono A. 2010. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. U. S. A. 107:1600–1605. 10.1073/pnas.0908661107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 101:14889–14894. 10.1073/pnas.0405596101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K. 2008. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J. Virol. 82:10129–10142. 10.1128/JVI.01169-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz DE, Tizard R, Gilbert W. 1983. Nucleotide sequence of Rous sarcoma virus. Cell 32:853–869. 10.1016/0092-8674(83)90071-5 [DOI] [PubMed] [Google Scholar]
- 37.Katz RA, Omer CA, Weis JH, Mitsialis SA, Faras AJ, Guntaka RV. 1982. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned unintegrated avian tumor virus DNA: structure of large terminal repeats in circle junctions. J. Virol. 42:346–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gudleski N, Flanagan JM, Ryan EP, Bewley MC, Parent LJ. 2010. Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA. Proc. Natl. Acad. Sci. U. S. A. 107:9358–9363. 10.1073/pnas.1000304107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datta SA, Rein A. 2009. Preparation of recombinant HIV-1 gag protein and assembly of virus-like particles in vitro. Methods Mol. Biol. 485:197–208. 10.1007/978-1-59745-170-3_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tropea JE, Cherry S, Waugh DS. 2009. Expression and purification of soluble His(6)-tagged TEV protease. Methods Mol. Biol. 498:297–307. 10.1007/978-1-59745-196-3_19 [DOI] [PubMed] [Google Scholar]
- 41.Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244:198–223. 10.1006/jmbi.1994.1719 [DOI] [PubMed] [Google Scholar]
- 42.Parent LJ, Wilson CB, Resh MD, Wills JW. 1996. Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J. Virol. 70:1016–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonanno JB, Edo C, Eswar N, Pieper U, Romanowski MJ, Ilyin V, Gerchman SE, Kycia H, Studier FW, Sali A, Burley SK. 2001. Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 98:12896–12901. 10.1073/pnas.181466998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15:8783–8798. 10.1093/nar/15.21.8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Craven RC, Leure-duPree AE, Erdie CR, Wilson CB, Wills JW. 1993. Necessity of the spacer peptide between CA and NC in the Rous sarcoma virus gag protein. J. Virol. 67:6246–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garbitt RA, Albert JA, Kessler MD, Parent LJ. 2001. Trans-acting inhibition of genomic RNA dimerization by Rous sarcoma virus matrix mutants. J. Virol. 75:260–268. 10.1128/JVI.75.1.260-268.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones CP, Saadatmand J, Kleiman L, Musier-Forsyth K. 2013. Molecular mimicry of human tRNALys anti-codon domain by HIV-1 RNA genome facilitates tRNA primer annealing. RNA 19:219–229. 10.1261/rna.036681.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weldon RA, Jr, Erdie CR, Oliver MG, Wills JW. 1990. Incorporation of chimeric gag protein into retroviral particles. J. Virol. 64:4169–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steeg CM, Vogt VM. 1990. RNA-binding properties of the matrix protein (p19gag) of avian sarcoma and leukemia viruses. J. Virol. 64:847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma YM, Vogt VM. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 76:5452–5462. 10.1128/JVI.76.11.5452-5462.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fisher RJ, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, Shoemaker R, McNitt KL, Johnson DG, Hixson CV, Gorelick RJ, Fabris D, Henderson LE, Rein A. 2006. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Res. 34:472–484. 10.1093/nar/gkj442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Athavale SS, Ouyang W, McPike MP, Hudson BS, Borer PN. 2010. Effects of the nature and concentration of salt on the interaction of the HIV-1 nucleocapsid protein with SL3 RNA. Biochemistry 49:3525–3533. 10.1021/bi901279e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. 2009. Effect of Mg(2+) and Na(+) on the nucleic acid chaperone activity of HIV-1 nucleocapsid protein: implications for reverse transcription. J. Mol. Biol. 386:773–788. 10.1016/j.jmb.2008.12.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webb JA, Jones CP, Parent LJ, Rouzina I, Musier-Forsyth K. 2013. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: implications for viral genomic RNA packaging. RNA 19:1078–1088. 10.1261/rna.038869.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sykora KW, Moelling K. 1981. Properties of the avian viral protein p12. J. Gen. Virol. 55:379–391. 10.1099/0022-1317-55-2-379 [DOI] [PubMed] [Google Scholar]
- 56.Qualley DF, Lackey CM, Paterson JP. 2013. Inositol phosphates compete with nucleic acids for binding to bovine leukemia virus matrix protein: implications for deltaretroviral assembly. Proteins 81:1377–1385. 10.1002/prot.24281 [DOI] [PubMed] [Google Scholar]
- 57.Sun M, Grigsby IF, Gorelick RJ, Mansky LM, Musier-Forsyth K. 2014. Retrovirus-specific differences in matrix and nucleocapsid protein-nucleic acid interactions: implications for genomic RNA packaging. J. Virol. 88:1271–1280. 10.1128/JVI.02151-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shkriabai N, Datta SA, Zhao Z, Hess S, Rein A, Kvaratskhelia M. 2006. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 45:4077–4083. 10.1021/bi052308e [DOI] [PubMed] [Google Scholar]
- 59.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. U. S. A. 103:11364–11369. 10.1073/pnas.0602818103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dalton AK, Murray PS, Murray D, Vogt VM. 2005. Biochemical characterization of Rous sarcoma virus MA protein interaction with membranes. J. Virol. 79:6227–6238. 10.1128/JVI.79.10.6227-6238.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Callahan EM, Wills JW. 2000. Repositioning basic residues in the M domain of the Rous sarcoma virus gag protein. J. Virol. 74:11222–11229. 10.1128/JVI.74.23.11222-11229.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nadaraia-Hoke S, Bann DV, Lochmann TL, Gudleski-O'Regan N, Parent LJ. 2013. Alterations in the MA and NC domains modulate phosphoinositide-dependent plasma membrane localization of the Rous sarcoma virus Gag protein. J. Virol. 87:3609–3615. 10.1128/JVI.03059-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. U. S. A. 99:3944–3949. 10.1073/pnas.062652199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parent LJ. 2011. New insights into the nuclear localization of retroviral Gag proteins. Nucleus 2:92–97. 10.4161/nucl.2.2.15018 [DOI] [PMC free article] [PubMed] [Google Scholar]