ABSTRACT
Syncytin genes are fusogenic envelope protein (env) genes of retroviral origin that have been captured for a function in placentation. Within rodents, two such genes have previously been identified in the mouse-related clade, allowing a demonstration of their essential role via knockout mice. Here, we searched for similar genes in a second major clade of the Rodentia order, the squirrel-related clade, taking advantage of the complete sequencing of the ground squirrel Ictidomys tridecemlineatus genome. In silico search for env genes with full coding capacity identified several candidate genes with one displaying placenta-specific expression, as revealed by quantitative reverse transcription-PCR analysis of a large panel of tissues. This gene belongs to a degenerate endogenous retroviral element, with recognizable hallmarks of an integrated provirus. Cloning of the gene in an expression vector for ex vivo cell-cell fusion and pseudotype assays demonstrated fusogenicity on a large panel of mammalian cells. In situ hybridization on placenta sections showed specific expression in domains where trophoblast cells fuse into a syncytiotrophoblast at the fetomaternal interface, consistent with a role in syncytium formation. Finally, we show that the gene is conserved among the tribe Marmotini, thus dating its capture back to about at least 25 million years ago, with evidence for purifying selection and conservation of fusogenic activity. This gene that we named syncytin-Mar1 is distinct from all seven Syncytin genes identified to date in eutherian mammals and is likely to be a major effector of placentation in its related clade.
IMPORTANCE Syncytin genes are fusogenic envelope genes of retroviral origin, ancestrally captured for a function in placentation. Within rodents, two such genes had been previously identified in the mouse-related clade. Here, in the squirrel-related rodent clade, we identified the envelope gene of an endogenous retrovirus with all the features of a Syncytin: it is specifically expressed in the placenta of the woodchuck Marmota monax, at the level of cells fusing into a syncytium; it can trigger cell-cell and virus-cell fusion ex vivo; and it has been conserved for >25 million years of evolution, suggesting an essential role in its host physiology. Remarkably, syncytin-Mar1 is unrelated to all other Syncytin genes identified thus far in mammals (primates, muroids, carnivores, and ruminants). These results extend the range of retroviral envelope gene “domestication” in mammals and show that these events occurred independently, on multiple occasions during evolution to improve placental development in a process of convergent evolution.
INTRODUCTION
Syncytins are genes of retroviral origin that have been co-opted by their host for a function related to placentation. They correspond to the Envelope gene (env) of ancestral retroviruses that entered the germ line of evolutionarily distant animals and were endogenized (reviewed in references 1, 2, and 3). Two such genes have already been identified in simians, namely, syncytin-1 (4, 5) and syncytin-2 (6, 7), as well as two distinct, unrelated ones in muroids, syncytin-A and syncytin-B (8), one in leporids, syncytin-Ory1 (9), one in carnivores, syncytin-Car1 (10), and more recently one in ruminants, syncytin-Rum1 (11). Their canonical characteristic features, allowing them to be named “Syncytin genes,” comprise (i) placenta-specific expression, (ii) cell-cell fusion activity, and (iii) conservation in evolution of mammalian species for extended periods of time (e.g., >10 million years). Syncytin proteins are expected to participate in the formation of the placental syncytiotrophoblast at the fetomaternal interface, via fusion of the mononucleated cytotrophoblasts (3). Some of them additionally possess an immunosuppressive activity, as classically observed for infectious retroviral envelope glycoproteins, which may be involved in fetomaternal tolerance (12). The direct involvement of Syncytin genes in placentation has been recently demonstrated unambiguously through the generation of knockout mice for syncytin-A and -B (13, 14), whose embryonic placenta displayed defects in cell-cell fusion, resulting in decreased fetomaternal exchange and impaired embryo survival. Interestingly, other captured env genes have been reported to be specifically expressed in the placenta (e.g., syncytin-like env-Cav1 [15], bos-env4/fematrin-1 [11, 16], and enJSRV env [1] genes) and for the latter to be involved in peri-implantation placental morphogenesis (17). Therefore, it appears that on several occasions in the course of mammalian evolution, env genes from endogenous retroviruses have been “co-opted” by their host to participate in the formation of the placenta.
Rodentia is a highly represented order which comprises three major clades: the mouse-related clade Myomorpha, the guinea-pig-related clade Ctenohystrica, and the squirrel-related clade Sciuromorpha (Fig. 1). In fact, Syncytin genes have already been identified within species of the first two clades: namely, the murine syncytin-A and -B which entered the mouse-related clade approximately 20 million years ago (Mya) (8) and can be found in all of the Muroidea, and the guinea pig syncytin-like env-Cav1, which entered the genome of a Ctenohystrica member ∼30 Mya and can be found in all Caviomorpha (15). We searched for Syncytin genes in the third major clade of rodents, and more precisely in the Marmota monax species, for which we could recover placental tissues from pregnant females. We identified a new Syncytin, specifically expressed in the placenta, and conserved over >25 million years of evolution. This gene is distinct from those found in the other rodent clades and corresponds to an independent capture from a distinct ancestral retrovirus that was exapted for a placental function.
FIG 1.
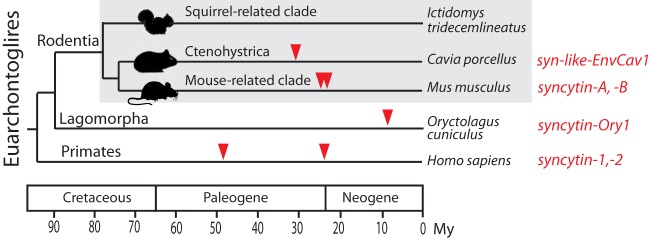
Phylogeny of Euarchontoglires and previously identified Syncytin genes. Euarchontoglires can be grouped into three major orders: Rodentia, Lagomorpha, and Primates (dates are from reference 36). Rodentia can be further subdivided into three major clades: the squirrel-related clade, the mouse-related clade and the guinea-pig-related clade Ctenohystrica (37). Branch length is proportional to time (in million years [My]), and the families where syncytins have been identified to date are indicated with the corresponding gene name.
MATERIALS AND METHODS
Biological samples.
A female woodchuck (Marmota monax) was trapped near Ithaca, NY, during April 2012. The woodchuck was anesthetized (5 mg of xylazine and 100 mg of ketamine/kg administered intramuscularly), midterm pregnancy (normal gestation period, 31 days) was confirmed by abdominal palpation, and the woodchuck was euthanized using a pentobarbital combination (Beuthanasia-D Special; Intervet, Inc., Merck Animal Health, Summit, NY) according to American Veterinary Medical Association Guidelines for the Euthanasia of Animals (2013 edition). A postmortem examination was performed and the uteroplacental unit of two embryonic vesicles, as well as tissues from the adult female (liver, kidney, spleen, heart, lung, pancreas, intestine, ovary, and uterus), were collected. For in situ hybridization, the whole uteroplacental unit of one embryonic vesicle was immediately fixed in 4% paraformaldehyde and then rinsed in 0.1 M phosphate buffer, dehydrated in a graded ethanol series, and embedded in paraffin. For RNA extraction, the placenta, dissected from the uteroplacental unit of the second embryonic vesicle, and the other tissues of the mother were immersed in RNAlater, and total RNA was extracted using an RNeasy RNA isolation kit (Qiagen).
Genomic DNA from Marmota monax was extracted from tissues of the pregnant female. Genomic DNAs from Otospermophilus beecheyi and Urocitellus parryii were extracted from tissues provided by M. A. Buendia (Institut Pasteur, Paris, France). Genomic DNA from Tamias speciosus sequoiensis (MVZ224083) and T. umbrinus inyoensis (MVZ222170) were extracted from tissues provided by the Museum of Vertebrate Zoology (Berkeley, CA). Genomic DNA from Urocitellus richardsonii and Callosciurus prevostii was purified from blood samples collected by B. Mulot and R. Potier (ZooParc de Beauval and Beauval Nature, Saint Aignan, France) using a DNA Blood Kit II (PaxGene). Genomic DNAs from Marmota sibirica, Spermophilus erythrogenys, Spermophilus fulvus, Ictidomys mexicanus, Xerospermophilus spilosoma, Ammospermophilus leucurus, Sciurus aestuans, Glaucomys volans, Aplodontia rufa, Eliomys quercinus, Muscardinus avellanarius, and Glis glis were extracted from alcohol-conserved tissues (Collection de Tissus de Mammifères, Montpellier, France [F. Catzeflis]). Genomic DNA from Sciurus vulgaris was extracted from tissues of an accidentally dead wild individual.
Database screening and sequence analyses.
Retroviral endogenous env gene sequences were searched for by BLAST on the ground squirrel genome (assembly of the Ictidomys tridecemlineatus genome [NCBI/Broad Institute SpeTri2.0, November 2011]). Sequences containing an open reading frame (ORF) longer than 400 amino acids (from start to stop codons) were extracted from the SpeTri2.0 genomic database by using the getorf program of the EMBOSS package (http://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html) and translated into amino acid sequences. These sequences were evaluated using BLAST against the transmembrane (TM) subunit amino acid sequences of 35 retroviral envelope glycoproteins (from representative endogenous retroviruses [ERVs], including known syncytins, and infectious retroviruses) using the tBLASTN program of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST). Putative envelopes were then selected based on the presence of a hydrophobic domain (i.e., a transmembrane domain) located 3′ relative to a highly conserved C-X5,6,7-C motif. The identified Env-encoding sequence coordinates are listed in Materials and Methods.
The ground squirrel genome was secondarily screened with the identified envelope glycoprotein sequences using the BLAST programs from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST). Multiple alignments of amino acid sequences were carried out using the Seaview program under the CLUSTAL W protocol. Maximum-likelihood phylogenetic trees were constructed with RaxML 7.3.2 (18), with bootstrap percentages computed from 1,000 replicates using the GAMMA+GTR model for the rapid bootstrapping algorithm. PAML4 (19) was used to obtain the nonsynonymous/synonymous mutation ratio (dN/dS) and run site-specific selection tests for syncytin-Mar1 sequences. The PAML models analyzed assumed no molecular clock (clock = 0) and a single dN/dS for all tree branches (model = 0), and we used likelihood ratio tests to compare the improvement in likelihood for a model (M8) allowing for positive selection compared to a model (M7) that does not. Each analysis ran until convergence and the control file is available upon request. HyPhy was used on the Datamonkey web server (www.datamonkey.org) to run site-specific random-effect-likelihood (REL) and fixed-effect-likelihood (FEL) tests.
The guinea pig (Cavia porcellus, UCSC/Broad cavPor3, 2008, 6.76X coverage), the naked mole rat ( Heterocephalus glaber, UCSC/Broad hetGla2, 2012), the rabbit (Oryctolagus cuniculus, UCSC/Broad oryCun2.0, 2009, 7.48X coverage), the rat (Rattus norvegicus, UCSC/Baylor HSGC RGSC_v3.4, 2004), the mouse (Mus musculus, UCSC GRCm38, 2011), the human (Homo sapiens, UCSC GRCh37/hg19, 2009), the dog (Canis lupus familiaris, UCSC/Broad CanFam3.1, 2011), the cow (Bos taurus, UCSC/Baylor bosTau7, 2011), and the horse (Equus caballus, UCSC/Broad EquCab2.0, 2007) were also screened for the presence of the identified syncytin-Mar1-containing provirus sequence, using syntenic genomic regions from the UCSC genome browser (http://genome.ucsc.edu/). Analyses of the syntenic conserved sequences in each genome were performed using the MultiPipMaker synteny building tool (http://pipmaker.bx.psu.edu) (20) with the ground squirrel genome sequence as a reference.
Ictidomys tridecemlineatus envelope protein coding sequence coordinates.
Coding sequence locations were as follows (the scaffold number, strand orientation, and coding sequence coordinates are indicated): env1 (JH393329, –, 6382478 to 6380709), env2 (JH393505, +, 347917 to 349131), env3 (JH393317, +, 10462093 to 10463793), env4 (JH393411, –, 2329614 to 2328043), env5 (JH393288, +, 77450 to 79315; JH393282, –, 11072410 to 11070575; JH393300, +, 16577127 to 16578389), env6 (JH393392, +, 309136 to 311655), env7 (JH393506, +, 397561 to 399417), env8 (JH393533, +, 1245254 to 1246675; JH393388, +, 2782355 to 2785516; JH393705, –, 629534 to 627801; JH393770, –, 256210 to 253868; JH393324, +, 11013298 to 11015568; JH393427, –, 989558 to 987774), and env9 (JH393369, –, 1455142 to 1453040).
qRT-PCR.
env1 to env9 mRNA expression was determined by quantitative real-time reverse transcription-PCR (qRT-PCR). Reverse transcription was performed with 1 μg of DNase-treated RNA as described previously (21). Real-time quantitative PCR was with 5 μl of diluted (1:15) cDNA in a final volume of 25 μl by using SYBR green PCR master mix (Applied Biosystems). PCR was carried out using an ABI Prism 7000 sequence detection system. The primers are listed in Table 1. The transcript levels were normalized relative to the amount of the RPL19 gene mRNA encoding ribosomal protein L19. Samples were assayed in duplicate.
TABLE 1.
Primers used in this study
| Method and primer | Sequence (5′–3′) |
|---|---|
| RT-qPCR | |
| Env1-S | CATTGGGAAAGGACTTGAT |
| Env1-R | CCCCTTCCTCAGCCAGTAG |
| Env2-S | CTCGCCACACCAGACAGC |
| Env2-R | GCAGCCATGAGCATATCTAGC |
| Env3-S | CTCTTGTCTCCCCCAGTGTG |
| Env3-R | AGTCTATAGCTCGTTGATTTTGAA |
| Env4-S | ATTACACAGGCACAGGACCAA |
| Env4-R | GGCCAGCAGATAATCCAGAG |
| Env5-S | GGAGCAGTAGGAGTGGGGAC |
| Env5-R | ATCCTTCCTGTACTTTGGCTAT |
| Env6-S | TTTGAAGGCTTAGTAGGGGG |
| Env6-R | ATCTAACCCTCGCCTATTCT |
| Env7-S | CCCAGGCTACTCTATTATTCTACA |
| Env7-R | AAAGCAGTTACAGAGGTTTCTATT |
| Env8-S | TAGCTGCYAACCAAAGAATAGA |
| Env8-R | ATCATTAAATCYRACAGAGGTTAT |
| Env9-S | CCTCTTTTGTTTTATTACCTGCTA |
| Env9-R | GCAGAGCCTTGGAAGTATTT |
| RPL19-F | GAAGGTCTGGTTGGACCCCA |
| RPL19-R | GTATTTTTCCGGCATCGAG |
| 5′ RACE | |
| Race R | TTCCCCCCTTTAATATTGTGCTC |
| 3′ RACE | |
| Race F | TTTATAGGTTTACTCGGGTTTGTTTG |
| In situ hybridization probe synthesis | |
| Syncytin-Mar1-ISH-F1 | ATTTAGTTTGGTTTCCTTTGA |
| Syncytin-Mar1-ISH-R1 | CCACATGAGTCTTATTCCACA |
| Syncytin-Mar1-ISH-F2 | ACTTGGTAAGGAATGTATTGG |
| Syncytin-Mar1-ISH-R2 | CTAGATCAAGTCCTTTCCCAA |
| Syncytin-Mar1-ISH-F3 | GTTGGGGCTATAGAACTCCTA |
| Syncytin-Mar1-ISH-R3 | TTCAATAGCTGCTTCCATAGT |
| Amplification of genomic syncytin-Mar1 complete sequence | |
| Forward primer | TAGGTTTACTCGGGTTTGTTTG |
| Reverse primer downstream of the env (M.sibirica) | TTGTGGGCTGAGGATATAGTTC |
| Reverse primer downstream of the env (Tamias) | CCCCACAATTTAATCTTTTAC |
| Reverse primer downstream of the 3′LTR | GTTCCAGAATAGGCAGACAA |
| Amplification of genomic syncytin-Mar1 internal sequence | |
| Syncytin-Mar1-S | AGTGGAAATCATGTGAACCAT |
| Syncytin-Mar1-R | ACACACCCAACAGTTTTTAAC |
In situ hybridization.
Placenta from Marmota monax was fixed in 4% (wt/vol) paraformaldehyde and embedded in paraffin. Serial sections (7 μm) were either stained with hematoxylin and eosin or used for in situ hybridization. For syncytin-Mar1, three PCR-amplified fragments of 561, 473, and 458 bp, respectively (primers in Table 1) were cloned into pGEM-T Easy (Promega). In vitro synthesis of the antisense and sense riboprobes was performed with SP6 or T7 RNA polymerase and digoxigenin 11-UTP (Roche Applied Science) after DNA template amplification. Sections were processed, hybridized at 42°C overnight with the pooled riboprobes (1 μg of each riboprobe/ml), and incubated further overnight at 4°C with alkaline phosphatase-conjugated anti-digoxigenin antibody Fab fragments (Roche Applied Science). Staining was achieved with the nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) phosphatase alkaline substrates, as indicated by the manufacturer (Roche Applied Science).
Search for syncytin-Mar1 homologous genes in other species.
PCRs were performed on 200 ng of genomic DNA, using Accuprime Taq DNA polymerase (Invitrogen) for 10 cycles (30 s at 94°C, 30 s from 60°C to 50°C, and 2 min at 68°C), followed by 40 cycles (30 s at 94°C, 30 s at 50°C, and 2 min at 68°C). All genomic DNAs from Sciuridae were amplified with primers located within the syncytin-Mar1 ORF (internal sequence primers; Table 1) or with a forward primer located 5′ relative to the start codon of the syncytin-Mar1 ORF and a reverse primer either downstream the env ORF or downstream of the provirus 3′ long terminal repeat (3′LTR; complete sequence primers, Table 1). PCR products were directly sequenced without cloning to avoid background mutations introduced by PCR.
syncytin-Mar1 expression vector and ex vivo cell fusion and pseudotype assays.
syncytin-Mar1 fragments PCR amplified from the genomic DNA of each indicated Marmotini species were cloned into the phCMV-G expression vector (GenBank accession number AJ318514, a gift from F.-L. Cosset, Ecole Normale Supérieure, Lyon, France). Cell-cell fusion assays were performed by cotransfecting cells (1 × 105 to 2 × 105 cells per well) with syncytin-Mar1-expressing vectors, along with a vector expressing an nls-LacZ gene (0.5 to 1 μg at a ratio of 1:1) using a Lipofectamine LTX and Plus reagent transfection kit (Invitrogen). At 24 to 48 h posttransfection, cells were fixed and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) stained for syncytium detection. For the pseudotype assay, murine leukemia virus (MLV) virions pseudotyped with Syncytin-Mar1 were produced by cotransfecting 8 × 105 293T cells with 2.25 μg of pGagPolMLV (expression vector for the retroviral proteins except Env), 2.25 μg of a corresponding lacZ-marked defective retroviral vector (MFG-nlsLacZ), and 0.5 μg of syncytin-Mar1 expression vector, using a Lipofectamine LTX transfection kit (Invitrogen). Supernatants from the transfected cells were harvested 48 h after transfection, filtered through 0.45-μm-pore-size polyvinylidene difluoride membranes, supplemented with Polybrene (8 μg/ml), and transferred to target cells seeded in 24-well plates (5 × 104 to 8 × 104 cells per well) the day before infection, followed by spinoculation at 1,200 × g for 2 h 30 min at room temperature (22). X-Gal staining was performed 3 days postinfection. All cell lines used above were described previously (8, 9) and grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (Invitrogen) and antibiotics (100 U of penicillin and 100 mg of streptomycin/ml; Invitrogen).
RESULTS
In silico search for retroviral env genes within the ground squirrel (Ictidomys tridecemlineatus) genome.
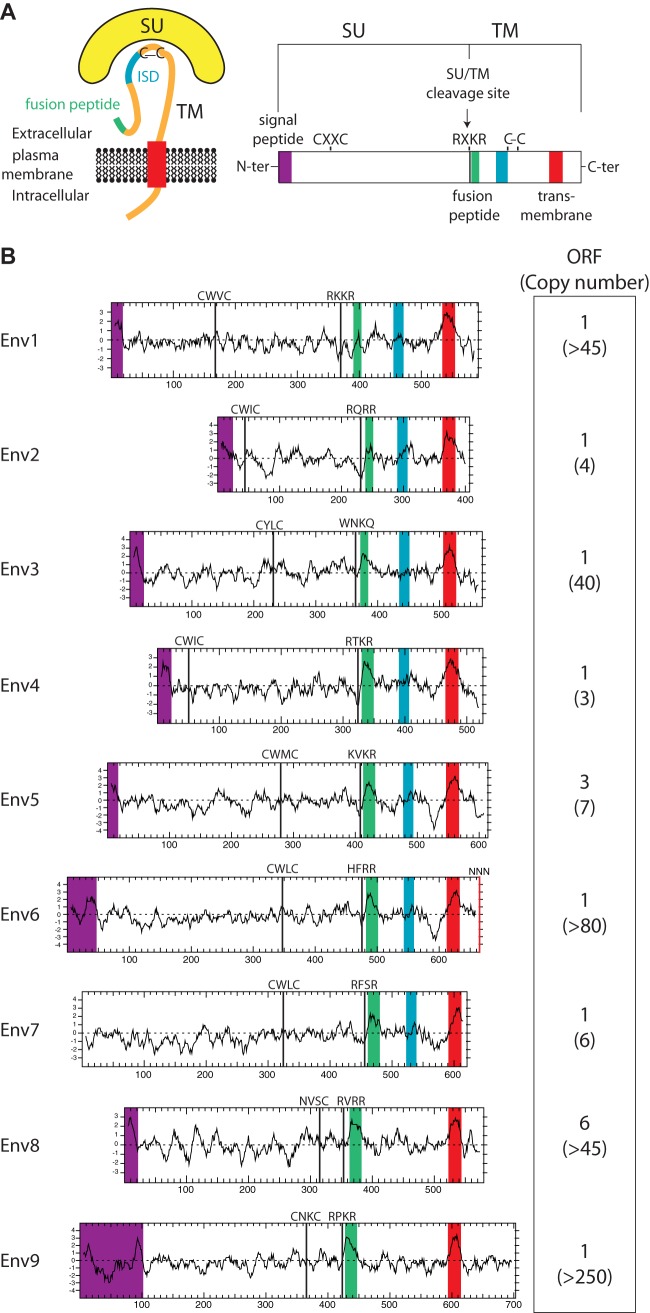
To identify putative env-derived Syncytin genes in the squirrel-related clade, we made use of the available thirteen-lined ground squirrel (Ictidomys/Spermophilus tridecemlineatus) genome sequence (Assembly of the Ictidomys tridecemlineatus genome, NCBI/Broad Institute SpeTri2.0, Nov 2011) and of the method that we previously devised to screen the cow genome for such genes (11). Basically, a BLAST search for ORFs (from the Met start codon to the stop codon) longer than 400 amino acids was performed using a selected series of Env sequences, including all currently identified Syncytin genes (see Materials and Methods). This resulted in a series of sequences that were further selected for the presence of a hydrophobic domain >20 amino acids located 3′ relative to a C-X5,6,7-C motif, corresponding to a highly conserved motif of retroviral envelopes (C-C and transmembrane domain; see the scheme in Fig. 2). This analysis yielded 16 sequences, incorporated into the phylogenetic Env tree shown in Fig. 3. Some of the sequences can be grouped into single families, resulting finally in nine families that we named Env1 to Env9 (Fig. 2).
FIG 2.
Structure of a canonical retroviral envelope protein and characterization of the identified Ictidomys tridecemlineatus candidates. (A) Schematic representation of a retroviral envelope protein, with the surface (SU) and transmembrane (TM) subunits delineated, and the furin cleavage site (consensus, R/K-X-R/K-R) between the two subunits together with the C-X-X-C domain involved in SU-TM interaction indicated; the hydrophobic signal peptide (purple), fusion peptide (green), transmembrane domain (red), and putative immunosuppressive domain (ISD) (blue) are also indicated. (B) Characterization of the thirteen-lined ground squirrel candidate envelope proteins. (Left) The hydrophobicity profile for each candidate is shown with the positioning of the canonical structural features schematized in panel A, when present (same color code). (Right) Number of full-length env gene ORFs within each family of element and total number of genomic copies (in parentheses).
FIG 3.
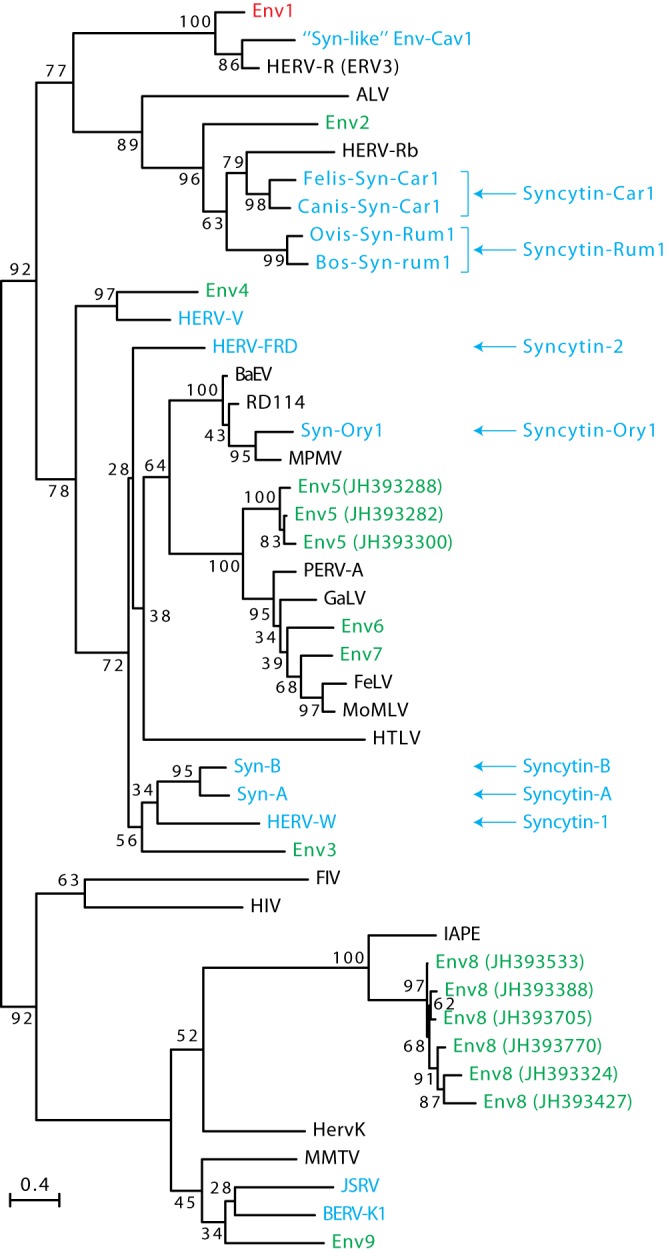
Retroviral envelope protein-based phylogenetic tree with the identified Ictidomys tridecemlineatus Env protein candidates. The maximum-likelihood tree inferred with the RaxML software was constructed using envelope amino acid sequences from mammalian endogenous retroviruses and from a series of infectious retroviruses. The horizontal branch length is proportional to the percentage of amino acid substitutions from the node (scale bar on the left), and the percent bootstrap values obtained from 1,000 replicates are indicated at the nodes. ALV, avian leukosis virus; BaEV, baboon endogenous virus; BERV, bovine endogenous retrovirus; FeLV, feline leukemia virus; FIV, feline immunodeficiency virus; GaLV, gibbon ape leukemia virus; HERV, human endogenous retrovirus; HIV, human immunodeficiency virus; HTLV, human T cell leukemia virus; JSRV, Jaagsiekte sheep retrovirus; MMTV, murine mammary tumor virus; IAPE, intracisternal A-type particle with an envelope gene; MoMLV, Moloney murine leukemia virus; MPMV, Mason-Pfizer monkey virus; PERV, porcine endogenous retrovirus; RD114, feline endogenous type-C retrovirus.
Analysis of the overall structure of the nine identified Env families (Fig. 2) strongly suggests that they indeed correspond to bona fide retroviral Env proteins, with all or most of their characteristic features (23, 24), including the presence of a predicted signal peptide sequence at the N terminus, a putative furin cleavage site delineating a surface (SU) and a transmembrane (TM) subunit (R/K-X-R/K-R), and a CXXC motif in the SU subunit corresponding to a binding domain between the two subunits. Hydrophobicity plots identify the hydrophobic transmembrane domain within the TM subunits required for anchoring the Env protein within the plasma membrane and a putative hydrophobic fusion peptide at the TM subunit N terminus. Some of them contain a canonical immunosuppressive domain (ISD) (12).
To analyze the placenta-specific expression of the candidate env genes, we only had access to placental tissues from a woodchuck species (Marmota monax, which diverged from Ictidomys tridecemlineatus ca. 12 Mya [25]), so we tried to amplify these genes from genomic DNA of Marmota monax to identify homologous relatives in this species. To improve the amplification efficiency, a highly sensitive touchdown PCR was performed (see Materials and Methods) using pairs of primers designed in the regions most highly conserved among all env genes (namely, the furin cleavage site and the C-X5,6,7-C motif). This resulted in the amplification of a PCR product of the expected size for eight of the nine families, and its sequencing confirmed significant sequence identity with the corresponding env genes in Ictidomys tridecemlineatus (data not shown). This indicates the presence of a homologous sequence for these eight env genes in the Marmota monax genome. No PCR amplification product could be obtained for the env6 gene family. Although we cannot formally exclude that the sequences may be too divergent to allow primer annealing and PCR amplification, our findings suggest the absence of this gene in the Marmota monax genome, or at least a low level of sequence conservation. Hence, this gene was not considered further in the present study.
Identification of a placenta-specific env gene.
Quantitative RT-PCR analysis of transcript levels for each candidate env gene was then performed using the primers that could successfully amplify each of them in the Marmota monax genome. Placenta samples were analyzed at midgestation (the gestation period is 31 days). As shown in Fig. 4, only env1 showed the characteristic property expected for a Syncytin gene, with high-level expression in the placenta and limited expression in other tissues. Expression of env1 is indeed highly restricted to the placenta, with <0.5% of the placental level found in all of the other organs tested. Other candidate genes showed only limited expression in the placenta (at least 20-fold lower) or no detectable expression. This low level of expression could not be attributed to primer defects, since all primer pairs showed expected amplification efficiency when tested on a DNA calibration curve (data not shown). Altogether, in silico analyses in Ictidomys tridecemlineatus combined with qRT-PCR assays for Marmota monax retroviral env genes clearly identify env1 as a putative Syncytin gene.
FIG 4.
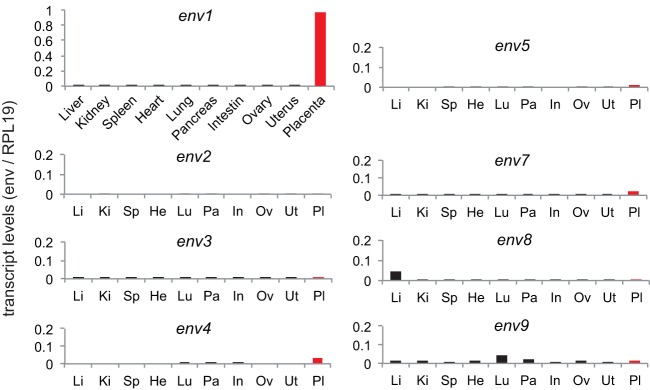
Real-time qRT-PCR analysis of the candidate env gene transcripts from Marmota monax. Transcript levels in the indicated tissues are expressed as the ratio of the expression level of each env gene to that of the RPL19 control gene (see Materials and Methods). Tissue expression of the placenta-specific env1 gene is displayed in the top left panel; expression in the same tissues (abbreviated, displayed in the same order) is shown for the seven other env gene candidates.
Characterization of the env1 candidate gene.
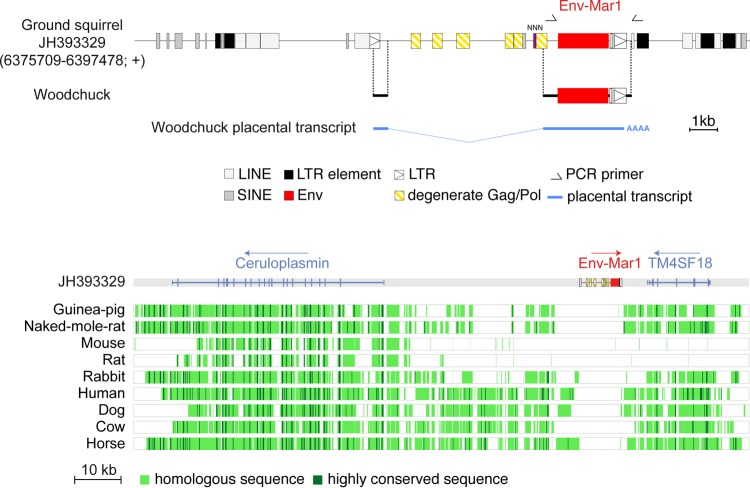
Examination of the Ictidomys tridecemlineatus genomic sequence reveals that env1 is part of a proviral structure, with degenerate, but still identifiable, flanking LTRs and small internal fragments with homology to gag and pol sequences, as detected by RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) and BLAST (Fig. 5). A 5′LTR truncated by a LINE retroelement insertion can be identified, followed by a primer binding site sequence complementary to the arginine tRNA(CCG), according to the Genomic tRNA Database (http://lowelab.ucsc.edu/GtRNAdb/). The 3′LTR displays insertion of a SINE retroelement and shows evidence for a polyadenylation signal. The provirus is integrated between the predicted Ceruloplasmin and TM4SF18 genes, in the antisense orientation as commonly observed for endogenous retroviruses (Fig. 5). Of note, env1 is the only coding env gene within a multicopy family disclosing >45 highly degenerate env1-related sequences and with only four of them being full length (Fig. 2).
FIG 5.
Characterization of the thirteen-lined ground squirrel (Ictidomys tridecemlineatus) and woodchuck (Marmota monax) env-Mar1-containing endogenous retrovirus and of its integration site. (Upper panel) Structure of the env-Mar1-containing ERV and orthology between the ground squirrel and woodchuck env-Mar1 sequences. Repeated mobile elements (gray), proviral LTRs, and the degenerate gag-pol genes, as identified by the RepeatMasker and BLAST web programs, are positioned (the symbols used are given below the panel). Undetermined sequence is indicated by NNN. PCR primers used to identify the env-Mar1 orthologous copy in the woodchuck are indicated (black half arrows), and the regions of the determined sequences homologous to those of the ground squirrel are aligned. The spliced env subgenomic transcript as determined by 5′- and 3′-RACE of woodchuck placental RNA is indicated. (Lower panel) Absence of the env1-containing ERV in the genomes of distant mammalian lineages. The genomic locus of the Ictidomys tridecemlineatus env-Mar1-containing provirus (env in red), along with its surrounding Ceruloplasmin and TM4SF18 genes, was recovered from the UCSC Genome Browser (http://genome.ucsc.edu/), together with the syntenic loci of the indicated species genomes; the positions of exons (vertical lines) of the resident Ceruloplasmin and TM4SF18 genes and the sense of transcription (arrows) are indicated. Homology of the syntenic loci was analyzed using the MultiPipMaker alignment building tool. Homologous regions are shown as pale green boxes, and highly conserved regions (more than 100 bp without a gap displaying at least 70% identity) are shown as dark green boxes.
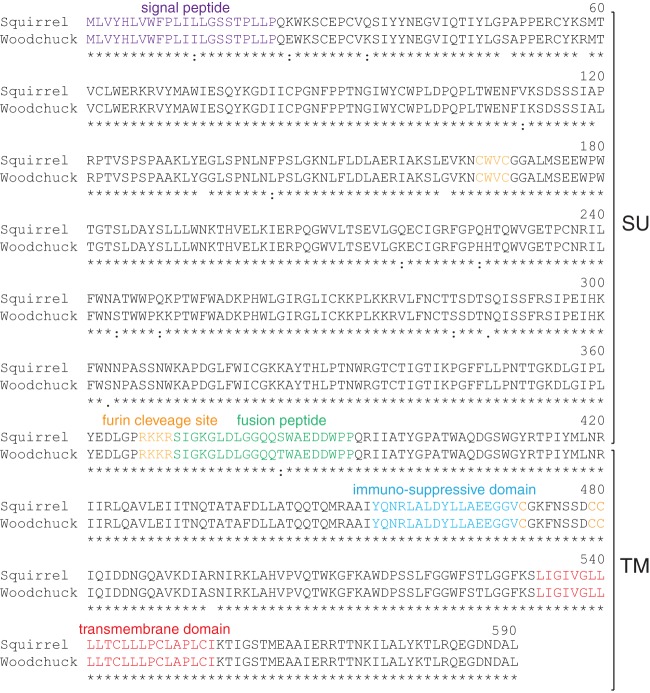
With the aim of identifying the whole gene orthologous to env1 in Marmota monax genomic DNA (only a small amplicon had been previously amplified [see above]), PCR analyses were carried out with primers designed from the Ictidomys tridecemlineatus env sequence, with the forward primer located at the 5′ end of the env gene and the reverse primer positioned 120 bp downstream of the 3′LTR, in the flanking integration site (Fig. 5). This resulted in the amplification of a sequence with a full-length env gene ORF and 3′LTR sequences, followed by a 120-bp flanking sequence, with high homology to that of Ictidomys tridecemlineatus. The SINE retroelement inserted within the 3′LTR is conserved in Marmota monax, indicating that this element integrated prior to the divergence between the two species. Comparison of the ground squirrel and woodchuck env sequences discloses 98.5% nucleotide identity (96.8% amino acid identity), with high conservation of all the canonical sites and domains characteristic for a retroviral protein (Fig. 6). These results thus identify the ortholog of env1 in the Marmota monax genome, which will now be referred to as env-Mar1. 5′- and 3′-RACE (rapid amplification of cDNA ends) RT-PCR experiments conducted on Marmota monax placental RNA with primers allowing full-length amplification of the env ORF led to the amplification of single PCR products, whose direct sequencing revealed the sole env-Mar1 env ORF (no superimposed peaks in the sequence). The env-Mar1 transcript is initiated within the 5′LTR, spliced between a donor and an acceptor splice site located downstream of the 5′LTR and upstream of the env initiation codon, respectively (as classically observed for retroviruses), and stopped at the putative polyadenylation signal in the 3′LTR (Fig. 5). Of note, the sequence of this transcript is 100% identical to the env-Mar1 genomic sequence amplified from the Marmota monax genome, strongly suggesting that it is expressed from this definite locus.
FIG 6.
Primary sequences and alignment of the homologous ground squirrel and woodchuck Env-Mar1 protein. Primary amino acid sequence and characteristic structural features of the homologous Env-Mar1 proteins from the ground squirrel Ictidomys tridecemlineatus and from the woodchuck Marmota monax. The same color code and abbreviations as in Fig. 2 are used. Asterisks indicate amino acid identity, and colons indicate amino acid similarity.
The syntenic genomic loci corresponding to the intergenic region between the Ceruloplasmin and TM4SF18 genes were recovered from the UCSC genome database for other representatives of Rodentia (guinea pig, naked mole rat, mouse, and rat), for the rabbit, and for the human, as well as for representatives of Laurasiatheria (dog, cow, and horse). As shown in Fig. 5, a genomic alignment using the PipMaker synteny building tool (http://pipmaker.bx.psu.edu/cgi-bin/multipipmaker [20]) revealed that env-Mar1 and the associated provirus sequences are missing at the orthologous syntenic locus in all of these species.
Env-Mar1 is a fusogenic retroviral envelope protein.
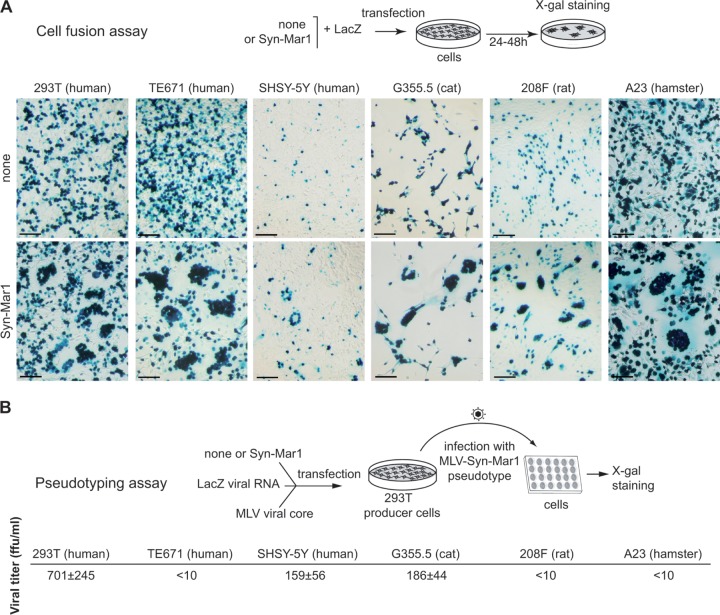
The functionality of Env-Mar1 as an ancestral, retrovirally derived, fusogenic envelope protein was determined by ex vivo assays in cell cultures for the formation of syncytia (cell-cell membrane fusion) and the generation of infectious pseudotypes (virus-cell membrane fusion), as previously described for other syncytins (10, 11). The env-Mar1 coding sequence was PCR amplified from genomic DNA of Marmota monax and inserted into a cytomegalovirus (CMV) promoter-containing expression vector (see Materials and Methods). The plasmids were sequenced, and those containing a full-length env gene ORF 100% identical to the env-Mar1 coding sequence were assayed. As illustrated in Fig. 7A for a series of cell lines, namely, the human 293T embryonic kidney derived cells, the human TE671 rhabdomyosarcoma-derived cells, the human SHSY-5Y neuroblast cells, the cat G355.5 astrocyte cells, the rat 208F fibroblast cells, and the hamster A23 fibroblast cells, transient transfection with the Env-Mar1-expressing vector triggers cell-cell fusion and results in the formation of multiple syncytia (not observed with an empty “none” vector) in all of the cell lines tested. It is noteworthy that no cell line was found to be negative for syncytium formation, a rather uncommon feature among the previously identified syncytins and strongly suggesting that the associated cellular receptor for Env-Mar1 is highly conserved among species and widespread, although its expression in vivo is probably tightly spatially and temporally regulated in order to finely regulate the fusion process, as observed for previously identified Syncytin receptors (26). Env-Mar1 can also form infectious pseudotypes, as expected from its retroviral origin. As illustrated in Fig. 7B, pseudotypes generated with an MLV core are able to infect human (293T and SHSY-5Y) and feline (G355.5) cells with significant virus titers from 150 to 700 focus-forming units/ml (and yet no titer was obtained using an HIV or SIV core [data not shown]). Such low values have already been reported for some other identified syncytins (e.g., Syncytin-2 [7]) and might be due to the fact that env-Mar1 is no more a “retroviral” gene but has adapted to a cellular “lifestyle,” with possibly a reduced ability to be pseudotyped in the viral cores currently assayed. Altogether, these experiments establish that env-Mar1 is a fusogenic gene, and it accordingly has now been named syncytin-Mar1.
FIG 7.
Syncytin-Mar1 is a fusogenic retroviral envelope protein. (A) Assay for cell-cell fusion mediated by Syncytin-Mar1. The indicated cell lines were transfected with an expression vector for Syncytin-Mar1 or an empty vector (none), together with a LacZ expression vector. Cells were cultured for 1 to 2 days after transfection, fixed, and stained with X-Gal. Syncytia were detected in the syncytin-Mar1-transfected cells, with only mononucleated cells visible using the empty vector. Scale bar, 100 μm. (B) Assay for cell infection mediated by Syncytin-Mar1-pseudotyped virus particles. Pseudotypes were produced by cotransfection of human 293T cells with expression vectors for the MLV core, the Syncytin-Mar1 protein (or an empty vector), and a lacZ-containing retroviral transcript. Supernatants were used to infect the indicated target cells, which were X-Gal stained 3 days after infection. Virus titers assayed on a panel of target cells from human (293T, TE671, and SHSY-5Y), cat (G355.5), or rodent (208F and A23) expressed as focus-forming units (FFU) per ml ± the standard errors of the mean are corrected for the background values of control particles without an Env protein and are means from three independent experiments.
In situ hybridization on placenta sections.
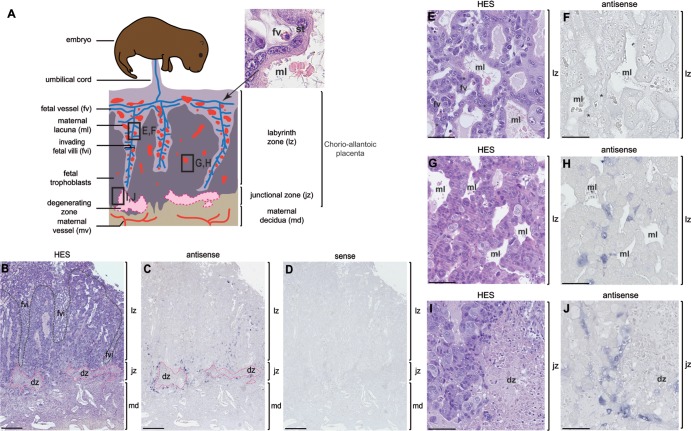
Among the Sciuridae, within the squirrel-related clade, the global architecture and morphogenesis of the Ictidomys tridecemlineatus placenta has been documented (27). In Marmota monax, the only available description of the placenta has been mainly focused on the fetomaternal interface composed, as observed by electron microscopy analyses, of a single layer of syncytial trophoblast (28). Here, the Marmota monax placenta was analyzed in detail and yet taking into consideration these former descriptions. As in the Ictidomys tridecemlineatus species, we observed that the placenta of Marmota monax is a chorioallantoic, deciduous, discoidal, labyrinthine placenta. As schematized in Fig. 8A, it is composed, from fetus to mother, of (i) the labyrinth zone, with fetal villi having penetrated and extending into the maternal tissue to constitute an exchange area between maternal and fetal blood through the formation of a fetal syncytiotrophoblast (28); (ii) the junctional zone, where the front-line fetal villi are invading the maternal tissue; and (iii) the maternal decidua. During the formation of the chorioallantoic placenta, the invasive front of fetal trophoblasts causes a degeneration of the maternal decidua (delineated in pink in Fig. 8A). This destruction provides a pathway to the fetal trophoblasts to intrude into the maternal tissue, leaving only a few isolated decidua cells or some islets of decidua cells. At the tip and lateral sides of the fetal villi, fetal trophoblasts differentiate into a multinucleated syncytial layer, the syncytiotrophoblast, via cell-cell fusion. Accordingly, a labyrinthine zona progressively substitutes for the maternal decidua allowing formation, between the fetal vessels and the maternal lacunae, of a fetal syncytium that mediates nutrient, gas, and waste exchanges between the maternal and fetal blood. On the paraffin sections of Marmota monax placentae obtained from midgestation pregnant females, we could observe—after hematoxylin-eosin-saffron (HES) staining (Fig. 8B)—the large labyrinth zone (indicated in purple in panels B and E), the junctional zone with the degenerative region (indicated in pink in panels B and I), and the decidual zone. In the well-developed labyrinth zone, we observed that maternal lacunae are in close contact with the fetal vessels, with the presence of a syncytiotrophoblast located at the fetomaternal interface (cf. the HES staining in panels A and E).
FIG 8.
Structure of the placenta and in situ hybridization for syncytin-Mar1 expression on woodchuck placental sections. (A) Schematic representation (based on panel B) of the woodchuck placenta with, from fetus to mother, the labyrinthine zona (lz), the junctional zone (jz), and the maternal decidua (md). An enlarged view of a fetal villus tissue section stained with hematoxylin-eosin-saffron (HES) (upper right) shows the multinucleated syncytiotrophoblast (st) generated by fusion from the underlying mononucleated cytotrophoblast cells forming the interface between the fetal vessel (fv) and the maternal lacuna (ml). (B to J) Sections of placenta from woodchuck at midgestation, with the positions of the E to J panels indicated in panel A. (B, E, G, and I) HES staining of placental sections with the three layers (lz, jz, and md) indicated; in panel B the fetal villi (fvi) are delineated with a black dotted line, and the degenerating zone (dz) with a pink dotted line. (C, D, F, H, and J) In situ hybridization on serial sections observed at different magnifications, using digoxigenin-labeled antisense (C, F, H, and J) or sense (negative control [D]) riboprobes revealed with an alkaline phosphatase-conjugated anti-digoxigenin antibody. (E and F) Intermediate magnification of the labyrinth zone in the fetal villi delineated in panel A, with no detectable labeling. (G and H) Intermediate magnification of the labyrinth zone in formation delineated in panel A, with labeling close to the maternal lacunae. (I and J) Intermediate magnification of the junctional zone delineated in panel A, with labeling abutting the degenerating zone on the right. Scale bars: 0.5 mm for the serial sections (B to D), 50 μm for panels E and F, and 100 μm for panels G to J.
To assess the physiological relevance of syncytin-Mar1 expression, we performed in situ hybridization experiments. Specific digoxigenin-labeled antisense probes were synthesized for the detection of woodchuck syncytin-Mar1, and the corresponding sense probes were used as negative controls. As shown in Fig. 8C and D, labeling was only observed with the antisense probe. syncytin-Mar1 expression was mainly localized in the junctional zone, surrounding the degenerative zone (Fig. 8C and J) and, to a lesser extent, between the fetal villi near the maternal lacunae present in the labyrinth zone in formation (Fig. 8C and H). Thus, labeling was predominantly observed in regions where cellular fusion processes take place. However, in the well-established central part of the fetal villi, where the syncytiotrophoblast is already formed, no syncytin-Mar1 expression signal was detectable (Fig. 8C and F). Of note, since the RACE PCR experiments performed on placenta RNA only disclosed the syncytin-Mar1 sequence (see above), the findings strongly suggest that the in situ hybridization signal is specific to this gene. Conclusively, the syncytin-Mar1 expression profile is consistent with a role in triggering the fusion of trophoblasts to form the fetal syncytiotrophoblast.
Search for syncytin-Mar1 in other members of the squirrel-related clade.
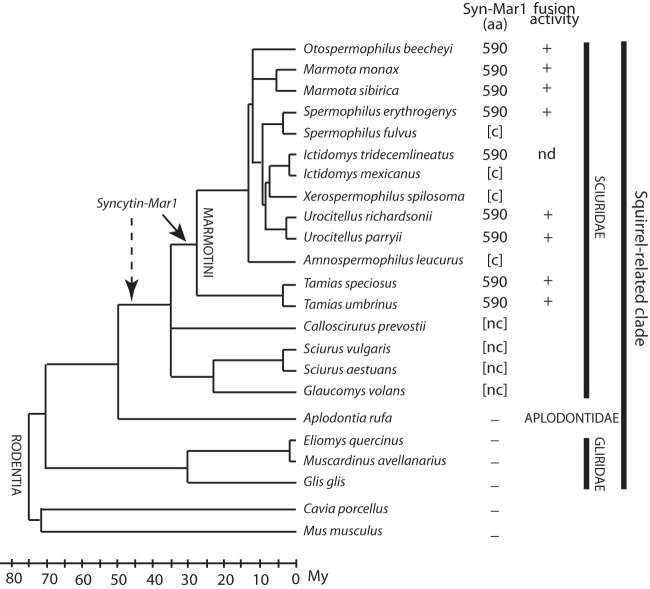
To further characterize syncytin-Mar1 and determine its status and evolution, we searched for the orthologous gene in other representatives of the tribe Marmotini (which contains the woodchuck and ground squirrel species), as well as within the larger families Sciuridae, Aplodontidae, and Gliridae within the squirrel-related clade (Fig. 9). Locus-specific pairs of PCR primers (a forward primer upstream of syncytin-Mar1 and a reverse primer downstream of the provirus in the 3′-flanking sequence [see Table 1]) were used to tentatively amplify genomic DNA from a series of representative species. In most of the Marmotini members tested, the expected PCR amplification product was obtained with a conserved size, strongly suggesting the presence of the orthologous syncytin-Mar1 in these species. This was confirmed by sequencing the PCR products (GenBank sequence accession numbers KJ145238 to KJ145244, KJ670373, and KJ670374), which revealed the presence of a syncytin-Mar1 gene encoding a full-length ORF (590 amino acids long). For Marmota sibirica and Tamias species, using a primer immediately downstream of the ORF, we could also successfully amplify the entire ORF sequence, although without the 3′-flanking sequence. For the other Marmotini species tested, we could only amplify an internal fragment (∼400 bp) but showing significant homology to the syncytin-Mar1 ORF and with coding capacity (primers in Table 1). Finally, in the remaining Sciuridae species, PCR using the same primers yielded a fragment with significant homology to syncytin-Mar1 but with loss of coding capacity due to stop codons or frameshifts. These data indicate the presence of copies belonging to the syncytin-Mar1 env gene family in all Sciuridae species, but the existence of an orthologous env gene with a full-length ORF could only be formally assessed for Marmotini species. Finally, DNAs from Aplodontia rufa, Eliomys quercinus, Muscardinus avellanarius, Glis glis (belonging to the families Aplotontidae and Gliridae, the closest outgroups of Sciuridae), and Mus musculus and Cavia porcellus (belonging to the mouse-related and the Ctenohystrica clades, respectively) were found negative using the same primers (Fig. 9), although an actin fragment could be successfully amplified from these DNAs as a control. Although we cannot formally exclude that sequences may be too divergent to allow primer annealing and PCR amplification, for representatives of the two latter families an in silico search of the corresponding Ensembl genomic databases using the BLAST program confirmed that no env gene with significant homology (>50%) could be found. These data suggest (i) that the ancestral infectious retrovirus that provided the env gene founder of the syncytin-Mar1 env family entered the genome of a Sciuridae ancestor 35 to 50 Mya and (ii) that the unique syncytin-Mar1 locus was then recruited—and thereafter conserved during the course of evolution—by an ancestor of the tribe Marmotini at least 25 Mya (29–31) (Fig. 9).
FIG 9.
Status of syncytin-Mar1 during the radiation of the Sciuridae. Phylogenetic tree of the squirrel-related clade, with the families Sciuridae, Aplodontidae, and Gliridae, as well as the outgroup species Cavia porcellus and Mus musculus (dates and branching positions are from references 29, 30, 31, and 38), with the time scale at bottom left. Branches of the family Sciuridae whose interrelationships are still unresolved are represented as polytomies. The names of the species tested for the presence of the syncytin-Mar1 gene are indicated, together with the length (in amino acids) of the Syncytin-Mar1 proteins that were identified for each species (all of the sequences were deposited in GenBank under accession numbers KJ145238 to KJ145244, KJ670373, and KJ670374). Brackets indicate that only partial sequences could be retrieved. aa, amino acids; [c], coding sequence; [nc], noncoding sequence; –, no syncytin-Mar1 homologous sequence identified, by either PCR amplification or database search; nd, not done. The fusogenic activity of each cloned gene, as determined by the fusion assay in Fig. 7, is indicated.
Purifying selection and functional conservation of syncytin-Mar1 in the tribe Marmotini.
Sequence analysis of the nine syncytin-Mar1 genes with a complete ORF demonstrates very high sequence similarities, with 87 to 95% identity in amino acids, as expected for a bona fide cellular gene (Fig. 10). Interestingly, the syncytin-Mar1-based phylogenetic tree is highly congruent with the Marmotini phylogenetic tree.
FIG 10.
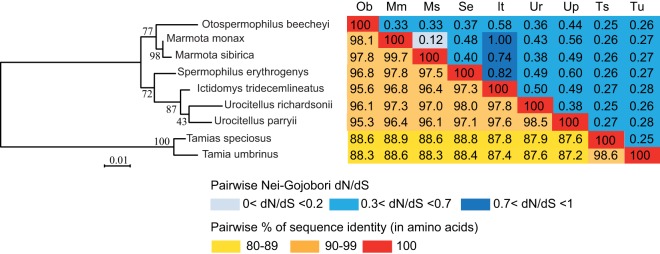
Sequence conservation and evidence for purifying selection of syncytin-Mar1 in the tribe Marmotini. (Left) Maximum-likelihood phylogenetic tree determined using amino acid alignment of the Syncytin-Mar1 proteins identified in Fig. 9 and inferred with the RaxML software. The horizontal branch length and scale indicate the percentage of amino acid substitutions. The percent bootstrap values obtained from 1,000 replicates are indicated at the nodes. (Right) Double-entry table for the pairwise percentage of amino acid sequence identity between the Syncytin-Mar1 proteins among the indicated species (lower triangle) and the pairwise Nei-Gojobori dN/dS ratio (upper triangle). A color code is provided below the table for both series of values.
To further characterize the conservation/evolution of the syncytin-Mar1 gene among Marmotini, analysis of the nonsynonymous/synonymous mutation ratio (dN/dS) between all pairs of species sequences was performed, using the Nei-Gojobori method (Fig. 10) (32). Accordingly, the entire env gene shows purifying selection between all pairs of species, with dN/dS ratios being lower than unity (between 0.12 and 0.6)—with the exception of three pairs of species with a ratio between 0.7 and 1 due to low rates of overall substitutions and hence biased dN/dS ratios. This pattern of dN/dS ratio is classically observed for cellular genes with a physiological function, in which nonsynonymous mutations are strongly selected against.
Finally, we performed a more refined analysis of the sequences, using methods allowing differences in selection pressure between different domains of the proteins to be revealed (site-specific selection). Such an analysis, using the PAML package (19), provided support for a model (model M7) in which most of the codons are under purifying selection (dN/dS < 0.31, 63% of the codons) and some under neutral selection (dN/dS = 1.0, 37% of the codons). There is no significant support for a positive selection model (model M8 versus M7: χ2 = 4.6, df = 2, P = 0.097), suggesting that no sites are under positive selection. Analyses using the HyPhy package (33), with slightly different site-specific models (REL and FEL) lead to similar conclusions. Conclusively, the syncytin-Mar1 genes are under strong purifying selection, as expected for a cellular gene with a physiological function.
To determine whether the selective pressure exerted on the syncytin-Mar1 gene correlates with the conservation of its functional properties, an ex vivo cell-cell fusion assay, as illustrated in Fig. 7, was performed. The syncytin-Mar1 genes from the species in Fig. 9 with a complete ORF were PCR amplified (except for Ictidomys tridecemlineatus [no DNA was available]), cloned into the same eukaryotic expression vector, and assayed using 293T and/or TE671 human cells. As indicated in Fig. 9, all of them (8/8) were determined to be positive in this assay. Altogether, the data suggest that syncytin-Mar1 is a bona fide cellular gene, co-opted for a physiological role in placentation.
DISCUSSION
We identified syncytin-Mar1, the env gene from an endogenous retrovirus which has integrated into the genome from a common ancestor of Marmotini before the radiation of this tribe more than 25 Mya (29, 31). This gene has been maintained as a functional retroviral env gene since then, being conserved in all of the Marmotini species from which it could be PCR amplified. It displays all of the canonical characteristics of a Syncytin gene: (i) it exhibits fusogenic activity, since it can mediate cell-cell fusion in an ex vivo assay and can further functionally replace a present-day retroviral env gene within a recombinant infectious retrovirus; (ii) it has been subject to purifying selection in the course of evolution, displaying low rates of nonsynonymous to synonymous substitutions and conservation of its fusogenic property; and (iii) it is specifically expressed in the placenta, as evidenced by both qRT-PCR analyses and in situ hybridization of woodchuck placental tissue sections. The in situ hybridization experiments using syncytin-Mar1 sequences as a probe clearly show that expression takes place both at the level of the invading fetal villi in close apposition to the degenerating maternal zone and between the fetal villi in the labyrinthine zona in formation, near the maternal lacunae onto which the fetal vessels will abut and where the fetomaternal syncytialized interface will be formed (28). Clearly, labeling is observed in regions where cellular fusion processes are under way and is not observed in the already formed fetal villi. The sites of syncytin-Mar1 expression are therefore consistent with a direct role of this fusogenic gene in syncytiotrophoblast formation.
syncytin-Mar1 adds to the two primate syncytin-1 and syncytin-2 genes first identified (4, 5, 6), to the two syncytin-A and syncytin-B genes later found in Muroidea (8), and to the syncytin-Ory1 (9), syncytin-Car1 (10), and syncytin-Rum1 (11) genes recently identified. Importantly, all eight Syncytin genes are unrelated, although some of them are more closely related to one another, e.g., syncytin-Car1 and syncytin-Rum1, possibly due to infection/endogenization of closely related retroviruses (Fig. 3). However, they all correspond to independent captures at different periods of evolution and in different mammalian branches. In the case of the syncytin-A and -B genes, knockout mice demonstrated that they are absolutely required for trophoblast cell fusion and syncytiotrophoblast formation in vivo, with evidence for placenta-dependent embryonic lethality for at least one of them (13, 14). It can therefore be proposed that the other identified syncytins, including the newly discovered Marmotini syncytin-Mar1, are likely to play a similar role in placentation by being involved in syncytiotrophoblast formation. This is consistent with the fact that species in which the above-mentioned Syncytin genes have been discovered and whose placentas are of the hemochorial, endotheliochorial, or synepitheliochorial type all possess multinucleated syncytia formed at the fetomaternal interface (28, 34, 35).
Although a functional syncytin-Mar1 gene has been dated back to 25 Mya, it remains possible that the corresponding ERV family entered the rodents earlier, since we provide evidence here that noncoding or partially coding sequences with a high level of similarity to syncytin-Mar1 were already present in Sciuridae, corresponding to a date of initial insertion of 35 to 50 Mya (29, 31). Accordingly, it is not excluded that whole-genome sequencing of new species—together with refined in situ analyses of placental tissue of the corresponding species—will lead in the future to the extension of the prevalence of syncytin-Mar1 in all Sciuridae. However, it is also clear that this family of elements could not be found in the Aplondontidae or Gliridae family (or in other Euarchontoglires and Laurasiatherians), indicating that syncytin-Mar1 and the other identified Syncytin genes—and especially the Muroidea syncytin-A and syncytin-B genes—never coexisted in a common rodent ancestor. This would be consistent with the model that we previously proposed, in which the currently identified Syncytin genes are “recent” gene acquisitions that have replaced a more “primitive” retroviral envelope gene capture that has been pivotal for the emergence of placental mammals from egg-laying species (3).
A final important outcome of the present investigation within rodents is that the discovery of syncytin-Mar1 gene extends the presence of Syncytin genes outside the mouse-related clade, where the previously identified murine syncytins had been found (8) to include members from the squirrel-related clade. This also adds to the Syncytin-like gene that we previously identified within the Ctenohystrica clade, a gene conserved in the evolution of Caviomorpha and also specifically expressed in the placenta, at the level of the invading syncytial streamers (15). Altogether, these results show that Syncytin gene capture has been a widespread process that has taken place independently and on several occasions even within a single order (i.e., rodents) over the course of evolution.
ACKNOWLEDGMENTS
We thank C. Conroy (Museum of Vertebrate Zoology, Berkeley, CA) for the gift of Tamias tissues, M. A. Buendia (Pasteur Institute, Paris, France) for the gift of tissues from two Marmotini species, P. Opolon and O. Bawa for their contribution to the histological analyses, S. Souquère and G. Pierron for their contribution in placenta structure analyses, and D. Schlafer (Cornell University, Ithaca, NY), S. J. Steppan (Florida State University, Tallahassee, FL), and A. C. Enders (University of California, Davis, CA) for helpful discussions. We thank C. Lavialle for comments and critical reading of the manuscript.
This study was supported by the CNRS and by grants to T.H. from the Ligue Nationale contre Le Cancer (Equipe Labelisée) and the ANR (Retro-Placenta).
Footnotes
Published ahead of print 30 April 2014
REFERENCES
- 1.Black S, Arnaud F, Palmarini M, Spencer T. 2010. Endogenous retroviruses in trophoblast differentiation and placental development. Am. J. Reprod. Immunol. 64:255–264. 10.1111/j.1600-0897.2010.00860.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupressoir A, Lavialle C, Heidmann T. 2012. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta 33:663–671. 10.1016/j.placenta.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 3.Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T. 2013. Paleovirology of ‘syncytins,' retroviral env genes exapted for a role in placentation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120507. 10.1098/rstb.2012.0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321–3329. 10.1128/JVI.74.7.3321-3329.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mi S, Lee X, Li X, Veldman G, Finnerty H, Racie L, LaVallie E, Tang X, Edouard P, Howes S, Keith JJ, McCoy J. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 17:785–789 [DOI] [PubMed] [Google Scholar]
- 6.Blaise S, de Parseval N, Bénit L, Heidmann T. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin-2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. U. S. A. 100:13013–13018. 10.1073/pnas.2132646100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaise S, Ruggieri A, Dewannieux M, Cosset F-L, Heidmann T. 2004. Identification of an envelope protein from the FRD family of human endogenous retroviruses (HERV-FRD) conferring infectivity on retroviral particles and functional conservation among simians. J. Virol. 78:1050–1054. 10.1128/JVI.78.2.1050-1054.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupressoir A, Marceau G, Vernochet C, Bénit L, Kanellopoulos C, Sapin V, Heidmann T. 2005. syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. U. S. A. 102:725–730. 10.1073/pnas.0406509102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidmann O, Vernochet C, Dupressoir A, Heidmann T. 2009. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new “syncytin” in a third order of mammals. Retrovirology 27:107. 10.1186/1742-4690-6-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis G, Heidmann O, Bernard-Stoecklin S, Reynaud K, Véron G, Mulot B, Dupressoir A, Heidmann T. 2012. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc. Natl. Acad. Sci. U. S. A. 109:E432–E441. 10.1073/pnas.1115346109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis G, Heidmann O, Degrelle S, Vernochet C, Lavialle C, Letzelter C, Bernard-Stoecklin S, Hassanin A, Mulot B, Guillomot M, Hue I, Heidmann T, Dupressoir A. 2013. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc. Natl. Acad. Sci. U. S. A. 110:E828–E837. 10.1073/pnas.1215787110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangeney M, Renard M, Schlecht-Louf G, Bouallaga I, Heidmann O, Letzelter C, Richaud A, Ducos B, Heidmann T. 2007. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl. Acad. Sci. U. S. A. 104:20534–20539. 10.1073/pnas.0707873105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T. 2009. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. U. S. A. 106:12127–12132. 10.1073/pnas.0902925106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupressoir A, Vernochet C, Harper F, Guégan J, Dessen P, Pierron G, Heidmann T. 2011. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl. Acad. Sci. U. S. A. 108:1164–1173. 10.1073/pnas.1012185108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vernochet C, Heidmann O, Dupressoir A, Cornelis G, Dessen P, Catzeflis F, Heidmann T. 2011. Syncytin-like endogenous retrovirus envelope gene of the guinea pig specifically expressed in the placenta junctional zone and conserved in Caviomorpha. Placenta 32:885–892. 10.1016/j.placenta.2011.08.006 [DOI] [PubMed] [Google Scholar]
- 16.Nakaya Y, Koshi K, Nakagawa S, Hashizume K, Miyazawa T. 2013. Fematrin-1 is involved in fetomaternal cell-to-cell fusion in Bovinae placenta and contributed to diversity of ruminant placentation. J. Virol. 87:10563–10572. 10.1128/JVI.01398-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, Spencer TE. 2006. Endogenous retroviruses regulate peri-implantation placental growth and differentiation. Proc. Natl. Acad. Sci. U. S. A. 103:14390–14395. 10.1073/pnas.0603836103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 19.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- 20.Schwartz S, Zhang Z, Frazer K, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. 2000. PipMaker-A web server for aligning two genomic DNA sequences. Genome Res. 10:577–586. 10.1101/gr.10.4.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Parseval N, Lazar V, Bénit L, Casella J, Heidmann T. 2003. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 77:10414–10422. 10.1128/JVI.77.19.10414-10422.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavillette D, Marin M, Ruggieri A, Mallet F, Cosset FL, Kabat D. 2002. The envelope glycoprotein of human endogenous retrovirus type W uses a divergent family of amino acid transporters/cell surface receptors. J. Virol. 76:6442–6452. 10.1128/JVI.76.13.6442-6452.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanstrom R, Wills JW. 1997. Synthesis, assembly, and processing of viral proteins, p 263–334 In Coffin JM, Hughes SH, Varmus HE. (ed), Retroviruses. Cold Spring Harbor Laboratory Press, New York, NY: [PubMed] [Google Scholar]
- 24.Vogt VM. 1997. Retroviral virions and genome, p 27–69 In Coffin JM, Hughes CL, Varmus HE. (ed), Retroviruses. Cold Spring Harbor Laboratory Press, New York, NY: [PubMed] [Google Scholar]
- 25.Steppan S, Kenagy G, Zawadzki C, Robles R, Lyapunova E, Hoffmann R. 2011. Molecular data resolve placement of the Olympic marmot and estimate dates of trans-Beringian interchange. J. Mammal. 92:1028–1037. 10.1644/10-MAMM-A-272.1 [DOI] [Google Scholar]
- 26.Esnault C, Priet S, Ribet D, Vernochet C, Bruls T, Lavialle C, Weissenbach J, Heidmann T. 2008. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc. Natl. Acad. Sci. U. S. A. 105:17532–17537. 10.1073/pnas.0807413105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mossman H, Weisfeldt L. 1939. The fetal membranes of a primitive rodent, the thirteen-striped ground squirrel. Am. J. Anat. 64:59–109. 10.1002/aja.1000640104 [DOI] [Google Scholar]
- 28.Enders AC, Carter AM. 2004. What can comparative studies of placental structure tell us? A review. Placenta 25(Suppl A):S3–S9. 10.1016/j.placenta.2004.01.011 [DOI] [PubMed] [Google Scholar]
- 29.Mercer J, Roth L. 2003. The effects of cenozoic global change on squirrel phylogeny. Science 299:1568–1572. 10.1126/science.1079705 [DOI] [PubMed] [Google Scholar]
- 30.Steppan S, Storz B, Hoffmann R. 2004. Nuclear DNA phylogeny of the squirrels (Mammalia: Rodentia) and the evolution of arboreality from c-myc and RAG1. Mol. Phylogenet. Evol. 30:703–719. 10.1016/S1055-7903(03)00204-5 [DOI] [PubMed] [Google Scholar]
- 31.Fabre P, Hautier L, Dimitrov D, Douzery E. 2012. A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC Evol. Biol. 12:88. 10.1186/1471-2148-12-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 33.Kosakovsky Pond S, Frost S. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208–1222. 10.1093/molbev/msi105 [DOI] [PubMed] [Google Scholar]
- 34.Leiser R, Kaufmann P. 1994. Placental structure: in a comparative aspect. Exp. Clin. Endocrinol. 102:122–134. 10.1055/s-0029-1211275 [DOI] [PubMed] [Google Scholar]
- 35.Wooding P, Burton GJ. 2008. Comparative placentation: structures, functions, and evolution. Springer-Verlag, Berlin, Germany [Google Scholar]
- 36.Meredith R, Janečka J, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simão TL, Stadler T, Rabosky DL, Honeycutt RL, Flynn JJ, Ingram CM, Steiner C, Williams TL, Robinson TJ, Burk-Herrick A, Westerman M, Ayoub NA, Springer MS, WJM. 2011. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334:521–524. 10.1126/science.1211028 [DOI] [PubMed] [Google Scholar]
- 37.Blanga-Kanfi S, Miranda H, Penn O, Pupko T, DeBry R, Huchon D. 2009. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol. Biol. 9:71. 10.1186/1471-2148-9-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montgelard C, Forty E, Arnal V, Matthee C. 2008. Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evol. Biol. 8:321. 10.1186/1471-2148-8-321 [DOI] [PMC free article] [PubMed] [Google Scholar]