ABSTRACT
The RV144 ALVAC/AIDSVax HIV-1 vaccine clinical trial showed an estimated vaccine efficacy of 31.2%. Viral genetic analysis identified a vaccine-induced site of immune pressure in the HIV-1 envelope (Env) variable region 2 (V2) focused on residue 169, which is included in the epitope recognized by vaccinee-derived V2 monoclonal antibodies. The ALVAC/AIDSVax vaccine induced antibody-dependent cellular cytotoxicity (ADCC) against the Env V2 and constant 1 (C1) regions. In the presence of low IgA Env antibody levels, plasma levels of ADCC activity correlated with lower risk of infection. In this study, we demonstrate that C1 and V2 monoclonal antibodies isolated from RV144 vaccinees synergized for neutralization, infectious virus capture, and ADCC. Importantly, synergy increased the HIV-1 ADCC activity of V2 monoclonal antibody CH58 at concentrations similar to that observed in plasma of RV144 vaccinees. These findings raise the hypothesis that synergy among vaccine-induced antibodies with different epitope specificities contributes to HIV-1 antiviral antibody responses and is important to induce for reduction in the risk of HIV-1 transmission.
IMPORTANCE The Thai RV144 ALVAC/AIDSVax prime-boost vaccine efficacy trial represents the only example of HIV-1 vaccine efficacy in humans to date. Studies aimed at identifying immune correlates involved in the modest vaccine-mediated protection identified HIV-1 envelope (Env) variable region 2-binding antibodies as inversely correlated with infection risk, and genetic analysis identified a site of immune pressure within the region recognized by these antibodies. Despite this evidence, the antiviral mechanisms by which variable region 2-specific antibodies may have contributed to lower rates of infection remain unclear. In this study, we demonstrate that vaccine-induced HIV-1 envelope variable region 2 and constant region 1 antibodies synergize for recognition of virus-infected cells, infectious virion capture, virus neutralization, and antibody-dependent cellular cytotoxicity. This is a major step in understanding how these types of antibodies may have cooperatively contributed to reducing infection risk and should be considered in the context of prospective vaccine design.
INTRODUCTION
Development of a preventive HIV-1 vaccine is a global priority. The Thai RV144 vaccine efficacy trial used an ALVAC-HIV (vCP1521) prime and AIDSVax B/E boost and demonstrated an estimated 31.2% protection from infection (1). An analysis of immune correlates of infection risk revealed an inverse correlation between the levels of IgG antibodies (Abs) against the first and second variable domains (V1 and V2) of HIV gp120 envelope (Env) protein and the risk of infection (2). A viral genetic analysis of RV144 breakthrough infections found a vaccine-induced site of immune pressure associated with vaccine efficacy at V2 amino acid position 169 (3). V2 monoclonal antibodies (MAbs) CH58 and CH59 were isolated from an RV144 vaccinee, and cocrystal structures of the MAbs and V2 peptides determined that Ab contacts centered on K169 (4). Moreover, CH58 MAb bound with the clade B gp70V1/V2 CaseA2 fusion protein used to identify V2-binding as a correlate of infection risk (2). MAbs CH58 and CH59 do not capture or neutralize difficult-to-neutralize (tier 2) viruses that were tested, but they do bind to the surface of tier 2 HIV-1-infected CD4+ T cells and mediate antibody-dependent cellular cytotoxicity (ADCC) (4).
Analysis of the secondary immune correlates of the RV144 clinical trial revealed reduced risk of infection in vaccine recipients with low levels of plasma anti-HIV-1 Env IgA Abs and high levels of ADCC activity (2). We have previously reported that HIV-1 Env constant 1 (C1) region Ab responses constitute the dominant ADCC Ab response in RV144 vaccine recipients and have isolated several MAbs from RV144 vaccine recipients that represent this group of Ab specificities (5).
A crucial limitation of studies conducted with individual MAbs is that they fail to represent the complex interactions present in polyclonal Ab responses in vivo. Abs of similar specificities may compete for binding to their target epitopes, resulting in antagonism of Ab antiviral effector functions as observed for ADCC activities of C1 IgG Abs in the presence of C1-specific IgA (6). Alternatively, Abs of different specificities may have additive effects or may synergize as described for some HIV-1 neutralizing antibodies (7–16).
Based on the observations that C1 and V2 Abs contributed to RV144 ADCC responses (4, 5), we have asked if vaccine-induced V2 and C1 Ab specificities interact in mediation of anti-HIV-1 effector functions. We found that vaccine-induced C1 and V2 MAbs synergistically mediate neutralization and virus capture against tier 1 viruses and, importantly, ADCC against tier 2 virus-infected CD4+ T cells.
MATERIALS AND METHODS
Plasma and cellular samples from vaccine recipients.
Plasma samples were obtained from volunteers receiving the prime-boost combination of vaccines containing ALVAC-HIV (vCP1521) (Sanofi Pasteur) and AIDSVax B/E (Global Solutions for Infectious Diseases). Vaccine recipients were enrolled in the phase I/II clinical trial (17) and in the community-based, randomized, multicenter, double-blind, placebo-controlled phase III efficacy trial (1).
Peripheral blood mononuclear cells (PBMCs) from five HIV-1-uninfected vaccine recipients enrolled in the phase II (recipient T141449) and phase III (recipients 347759, 210884, 200134, and 302689) trials whose plasma showed ADCC activity were used for isolation of memory B cells and production of MAbs (5).
All trial participants gave written informed consent as described for both studies. Samples were collected and tested according to protocols approved by Institutional Review Boards at each site involved in these studies.
Isolation of ADCC-mediating monoclonal antibodies.
Monoclonal antibodies were isolated from subjects 210884 (CH54), 347759 (CH57, CH58, and CH59) and 200134 (HG107) by culturing IgG+ memory B cells at near clonal dilution for 14 days (4, 5, 18) followed by sequential screenings of culture supernatants for HIV-1 gp120 Env binding and ADCC activity as previously reported (5). The MAbs CH90 and HG120 were isolated from subjects T141449 and 302689, respectively, by flow cytometry sorting of memory B cells that bound to HIV-1 group M consensus gp140Con.S Env as previously described and with subsequent modification (5).
Epitope mapping of V2 and C1 MAbs from ALVAC/AIDSVax vaccines.
The characteristics of the V2 and C1 MAbs generated from ALVAC/AIDSVax vaccinees have been previously reported (4–6) and are summarized in Table 1. Among the C1 MAbs, CH57 was isolated from the same vaccine recipient (347759) used to generate MAbs CH58 and CH59. The gp120 C1 A32 Fab fragment (5, 19) blocked the ADCC activity of CH57, and CH57 was itself able to block binding of another ALVAC/AIDSVax ADCC MAb, CH20, that was not blocked by A32 (5, 20). The differential abilities of CH57 and A32 to inhibit binding of CH20 suggest that they recognize overlapping but not identical epitopes. This difference is also supported by the inability of CH57 to reciprocally block A32 in Env-binding assays. The second MAb of interest, CH54, was isolated from vaccinee 210884. CH54 displayed a similar cross-clade ADCC profile to A32, and the A32 Fab was able to block its activity. CH54 could reciprocally block 30% of A32 binding to HIV Env, but was unable to inhibit binding of CH20. Finally, CH90 is an ADCC-mediating A32-blockable MAb generated from vaccinee T141449. This MAb blocked 20% of A32 binding, and it displayed a different cross-clade ADCC profile from A32.
TABLE 1.
Kd and ADCC EC of MAbs in this study
| IgG MAb | Specificity | ka, (M−1 s−1) × 103a | kd, (s−1) × 10−3b | Kd (nM)c | ADCC EC (μg/ml)d |
|---|---|---|---|---|---|
| A32 | Anti-C1 | 223.0 | 0.15 | 0.7 | 0.003 |
| CH54 | A32-blockable | 26.4 | 4.92 | 188.0 | 0.330 |
| CH57 | A32-blockable | 13.6 | 15.6 | 115.0 | 0.090 |
| CH90 | A32-blockable | 62.8 | 34.2 | 547.0 | 4.650 |
| CH58 | Anti-V2 | 226 | 0.23 | 1.0 | 18.260 |
| CH59 | Anti-V2 | 9.9 | 0.13 | 1.3 | 1.050 |
| HG107 | Anti-V2 | 12.0 | 0.16 | 1.3 | 5.260 |
| HG120 | Anti-V2 | 67.1 | 0.06 | 1.2 | 5.410 |
ka, association rate constant.
kd, dissociation rate constant.
Kd was calculated for binding to AE.A244Δ11 gp120. The data shown are means of three independent experiments, except for CH57 data, which are representative of two experiments.
The ADCC EC was calculated for AE.CM235-infected target cells by 3-h luciferase ADCC.
Generation of MAb F(ab) and F(ab′)2 fragments.
F(ab) and F(ab′)2 fragments were produced by papain or pepsin digestion, respectively, of recombinant IgG1 MAbs using specific fragment preparation kits (Pierce Protein Biology Products, Rockford, IL) according to the manufacturer's instructions. The resulting fragments were extensively characterized and purified by Coomassie brilliant blue staining and size exclusion by standard procedures.
SPR kinetics and Kd measurements.
The Env gp120 binding dissociation constant (Kd) and rate constant for IgG MAbs were calculated on BIAcore 3000 instruments using an anti-human Ig Fc capture assay as described earlier (21, 22). The humanized monoclonal antibody (IgG1k) directed to an epitope in the A antigenic site of the F protein of respiratory syncytial virus, Palivizumab (MedImmune, LLC, Gaithersburg, MD), was purchased from the manufacturer and used as a negative control. For surface plasmon resonance (SPR), Palivizumab was captured on the same sensor chip as a control surface. Nonspecific binding of Env gp120 to the control surface and/or blank buffer flow was subtracted for each MAb-gp120 binding interaction. All curve-fitting analyses were performed using global fit of multiple titrations to the 1:1 Langmuir model. The mean and standard deviation (SD) of rate constants and Kd were calculated from at least three measurements on individual sensor surfaces with equivalent amounts of captured antibody. All data analysis was performed using the BIAevaluation 4.1 analysis software (GE Healthcare).
SPR antibody synergy assay.
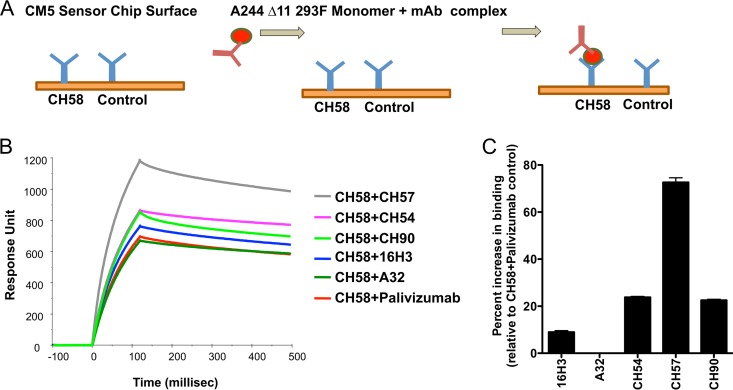
SPR antibody synergy of monoclonal antibody binding was measured on BIAcore 4000 instruments by immobilizing the test V2 MAb (IgG) on a CM5 sensor chip to about 5,000 to 6,000 response units (RU) using standard amine coupling chemistry. C1 MAbs (A32, CH57, CH90, and 16H3) at 40 μg/ml were preincubated with Env gp120 (20 μg/ml) in solution and then injected over the CH58 immobilized surface. Env gp120-MAb complexes were injected at 10 μl/min for 2 min, and the dissociation was monitored for 5 min. Following each binding cycle, surfaces were regenerated with a short injection (10 to 15 s) of glycine-HCl (pH 2.0). Enhancement of binding was calculated from binding responses measured in the early dissociation phase and the percentage of enhancement was calculated from the ratio of binding response as follows: % enhancement = [1 – (response with gp120 + upregulating Ab – response with gp120 + control MAb/response with gp120 + control MAb)] × 100. A schematic of this method is provided in Fig. 1.
FIG 1.
Synergy of MAb binding to monomeric gp120 by SPR. (A) Schematic of the SPR assay utilized to test the presence of synergy between the V2 and C1 MAbs for binding to the recombinant AE.244Δ11 gp120 according to the procedure reported in Materials and Methods. (B) SPR of binding of the CH58 MAb in combination with negative-control MAb Palivizumab and the C1 MAbs A32, 16H3, CH57, CH54, and CH90. The y axis represents the response unit values, and the x axis represents the time in milliseconds. (C) Fold increase of the V2 MAb CH58 binding to the recombinant AE.A244Δ11 gp120. The data are reported as percentage of increase compared to the binding of the CH58 MAb to gp120 incubated with Palivizumab used as negative control. The results represent the average ± standard error of the mean (SEM) of the results obtained in three separate experiments.
IMC.
The HIV-1 reporter virus used was a replication-competent infectious molecular clone (IMC) designed to encode the CM235 (subtype A/E) env genes in cis within an isogenic backbone that also expresses the Renilla luciferase reporter gene and preserves all viral open reading frames (23, 24). The Env-IMC-LucR virus used was the NL-LucR.T2A-AE.CM235-ecto (IMCCM235) (GenBank accession no. AF259954.1; plasmid provided by Jerome Kim, U.S. Military HIV Research Program). Reporter virus stocks were generated by transfection of 293T cells with proviral IMC plasmid DNA, and virus titer was determined on TZM-bl cells for quality control (25).
Infection of the CEM.NKRCCR5 cell line and primary CD4+ T cells with HIV-1 IMC.
Primary CD4+ T cells used in surface staining assays were activated, isolated, and infected with uncloned HIV-1 92TH023 virus or IMCCM235 by spinoculation as previously described (19). For ADCC assays, IMCCM235 was titrated in order to achieve maximum expression within 36 h postinfection as determined by detection of luciferase activity and intracellular p24 expression. We infected 1 × 106 CEM.NKRCCR5 cells with 1 50% tissue culture infective dose (TCID50)/cell IMCCM235 by incubation for 0.5 h at 37°C and 5% CO2 in the presence of DEAE-dextran (7.5 μg/ml). The cells were subsequently resuspended at 0.5 × 106/ml and cultured for 36 h in complete medium containing 7.5 μg/ml DEAE-dextran. For each ADCC assay, we monitored the frequency of infected target cells by intracellular p24 staining. Assays performed using the infected target cells were considered reliable if the percentage of viable p24+ target cells on the assay day was ≥20%.
Binding of MAbs to the surface of HIV-1-infected primary CD4+ T cells.
The staining of infected CD4+ T cells was performed as a modification of the previously published procedure (19). Briefly, the A32 MAb and vaccine-induced C1 A32-blockable MAbs were preincubated with the infected cells for 15 min at 37°C in 5% CO2 prior to addition of the vaccine-induced V2 MAb CH58. The V2 MAb CH58 was conjugated to Alexa Fluor 488 (Invitrogen, Carlsbad, CA) using a monoclonal antibody conjugation kit per the manufacturer's instructions (Invitrogen). Both the C1-specific and V2-specific MAbs were used at a final concentration of 10 μg/ml. The combined MAbs were incubated with the infected cells for 2 to 3 h at 37°C in 5% CO2, after which the cells were stained with a viability dye and for intracellular expression of p24 by standard methods.
Virion capture assay.
V2 MAbs CH58 and CH59 were mixed with 2 × 107 RNA copies/ml AE.92TH023 HIV-1 viral stock at final concentration of 10 μg/ml in 300 μl with or without the presence 10 μg/ml A32 antibody. The MAbs and virus-immune complex mixtures were prepared in vitro and absorbed on a protein G MultiTrap 96-well plate as described previously (26–28). The viral particles in the flowthrough or captured fraction were measured by detection of viral RNA with HIV-1 gag real-time reverse transcription (RT)-PCR. The infectious virus in the flowthrough was measured by infecting the TZM-bl reporter cell line. Briefly, 25 μl flowthrough was used to infect TZM-bl cells. Each sample was run in triplicate. Infection was measured by a firefly luciferase assay at 48 h postinfection as described previously. One hundred microliters of supernatant was removed, and 100 μl Britelite (PerkinElmer, Waltham, MA) was added to each well. After 2 min of incubation, 150 μl of lysate was used to measure HIV-1 replication as expressed as relative luciferase units (RLU). The infectious virion capture index of antibodies (IVCI) (28), representing the fold change of the ratio of infectious virions relative to the total viral particles in the flowthrough fraction after passing through protein G plates, was calculated according to the following formula:
In this analysis, IVCI values of <1 indicate selection bias for binding to noninfectious virions and IVCI values of >1 indicate selection bias for binding to infectious virions. The MAbs that do not capture HIV-1 virions or have no selection bias should exhibit an ICVI value of 1, as indicated for the virus-only negative control.
Neutralization assays.
Neutralizing antibody assays in TZM-bl cells were performed as described previously (29). The neutralizing activities of V2 MAbs CH58 and CH59 were tested in serial 3-fold dilutions starting at a 50-μg/ml final concentration against 5 pseudotyped HIV-1 viruses, including tier 1 viruses B.MN and AE.92TH023 and tier 2 virus AE.CM244, from which RV144 vaccine immunogens (4) were derived, as well the tier 2 CRF01-AE-derived circulating HIV-1 strains AE.427299 and AE.703357, isolated from Thailand. HIV-1 AE.703357 was isolated from a breakthrough HIV-1-infected RV144 vaccine recipient. Each MAb was tested alone or in combination with A32 MAb at concentrations of 50, 25, or 5 μg/ml. The data were calculated as a reduction in luminescence compared with the fluorescence in the control wells and are reported as MAb 50% inhibitory concentration (IC50) in μg/ml.
Luciferase ADCC assay.
We utilized a modified version of our previously published ADCC luciferase procedure (4). Briefly, CEM.NKRCCR5 cells (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH from Alexandra Trkola) (30) were used as targets for ADCC luciferase assays after infection with subtype AE HIV-1 IMCCM235. The target cells were incubated in the presence of 50, 5, or 1 μg/ml of vaccine-induced V2 MAb CH58 and C1 MAb CH90 alone and in all possible combinations. Purified CD3− CD16+ NK cells were obtained from an HIV-seronegative donor with the low-affinity 158F/F Fcγ receptor IIIa phenotype (31). The NK cells were isolated from cryopreserved PBMCs by negative selection with magnetic beads (Miltenyi Biotec GmbH, Germany) after resting overnight. The NK cells were used as effector cells at an effector-to-target ratio of 5:1. The effector cells, target cells, and Ab dilutions were plated in opaque 96-well half-area plates and were incubated for 3 h at 37°C in 5% CO2. The final readout was the luminescence intensity generated by the presence of residual intact target cells that have not been lysed by the effector population in the presence of ADCC-mediating MAb. The percentage of killing was calculated using the formula
In this analysis, the RLU of the target plus effector wells represents spontaneous lysis in the absence of any source of Ab. The RSV-specific MAb Palivizumab was used as a negative control.
We also evaluated synergy between V2 MAbs (CH58, CH59, HG107, and HG120) and C1 MAbs (CH54, CH57, and CH90) or F(ab′)2 fragment of CH90 at equivalent (1:1) concentrations across a range of 5-fold serial dilutions beginning at 50 μg/ml. From the ADCC activity curves, we interpolated the endpoint concentration (EC) in μg/ml and calculated the combination index (CI) as described previously (32). The CIEC was calculated according to the equation
where ECC1 MAb alone and ECV2 MAb alone are the EC in μg/ml of each MAb when tested alone, and ECcombination is the EC in μg/ml of the MAbs when used in combination. From the ADCC activity curves, we also determined the concentration at which 75% of the maximum killing activity (MK75) was reached for each V2 and C1 MAb and the concentration at which the same killing activity would be reached when the V2 and C1 MAbs were tested in combination. The CI values for MK75 (CIMK75) were then calculated according to the equation
where MK75C1 MAb alone and MK75V2 MAb alone are the MK75 in μg/ml of each MAb when tested alone. MK75C1 MAb in combination is the concentration at which 75% of the maximum activity of the C1 MAb alone is reached when tested in combination with the V2 MAb, and MK75V2 MAb in combination is the concentration at which 75% of the peak activity of the V2 MAb alone is reached when tested in combination with the C1 MAb. In all cases, both mutually exclusive (β = 0) and mutually nonexclusive (β = 1) CI values were determined. Synergy is indicated by CI values of <1, additivity by CI values = 1, and antagonism by CI values of >1.
RESULTS
V2 MAbs from ALVAC/AIDSVax vaccine recipients.
Vaccine-induced V2 MAbs CH58, CH59, HG107, and HG120 recognize Env V2 residues within positions 168 to 183 and mediate ADCC (Table 1). The CH58 MAb bound the gp70 V1/V2 antigen associated with reduced risk of infection in RV144 (2) and thus represents the specificity of V2 Abs that may be related to decreased transmission risk. CH58, CH59, HG107, and HG120 ALVAC/AIDSVax vaccine-induced MAbs all recognize V2 epitopes that center on amino acid residue K169 and thus are candidates to mediate immune pressure (3, 4).
C1 MAbs from ALVAC/AIDSVax vaccine recipients.
Of the 19 A32-blockable anti-C1 ADCC MAbs originally generated from the ALVAC/AIDSVax vaccine recipients (5), three (CH54, CH57 and CH90) were of particular interest because they likely recognize distinct overlapping epitopes of the Env C1 A32-blockable region (see reference 5 and Materials and Methods). Therefore, they were selected as representative of vaccine–induced C1 Ab responses and were tested for their ability to synergize with the V2 MAb CH58 for enhanced recognition of HIV envelope and antiviral effector functions. A32 was included to represent the overall C1 Ab responses.
Synergy of V2 and C1 MAb for binding to monomeric recombinant AE.A244 Δ11 gp120.
To test the hypothesis that the V2 MAb CH58 could synergize with the A32-blockable C1 MAbs, we performed surface plasmon resonance (SPR) analysis of binding of CH58 MAb to the recombinant AE.A244Δ11 gp120 as representative of the vaccines used in the ALVAC/AIDSVax clinical trial. The CH58 MAb was captured on the SPR sensor chip by an anti-Fc antibody along with anti-RSV MAb Palivizumab as a negative control. The A32, CH54, CH57, and CH90 MAbs or the murine HIV-1 C1-specific MAb 16H30 were incubated with AE.A244Δ11 gp120 and then flowed over the CH58 MAb bound onto the chip (Fig. 1A). The amount of C1 MAb/gp120/V2 complex was measured by SPR (Fig. 1B). In Fig. 1C, the data are expressed as the percentage of increase in specific binding relative to the binding of gp120 in complex with negative-control MAb Palivizumab. No increase in binding of MAb CH58 was observed when tested in combination with Palivizumab or with the C1 MAb A32. In contrast, ALVAC/AIDSVax vaccine-induced MAbs CH54, CH57, and CH90 cooperatively increased the binding of MAb CH58 to recombinant HIV-1 gp120 by 24, 73, and 25%, respectively. The murine-derived C1 MAb, 16H3, increased CH58 binding by a modest 9%. Thus, the V2 MAb binding to the monomeric gp120 can be enhanced by preincubation with C1 MAbs, indicating that the C1 MAbs likely induce a conformational change in the gp120 allowing for synergy among Abs with these specificities.
Synergy of V2 and C1 MAbs for binding to Env expressed on the surface of HIV-1-infected CD4+ T cells.
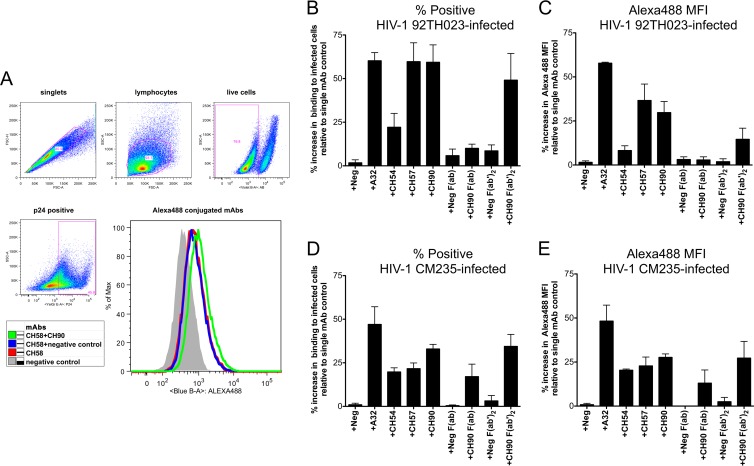
We next evaluated whether V2 and C1 MAbs can act in synergy for the recognition of HIV-1-infected cells. Activated primary blood CD4+ T cells isolated from an HIV-seronegative donor were infected with HIV-1 subtypes AE.92TH023 and CM235, representing tier 1 and tier 2 neutralization-sensitive isolates, respectively. The V2 MAb CH58 was conjugated with Alexa Fluor 488, allowing for direct flow cytometric analysis of its ability to recognize Env on the surface of HIV-1-infected CD4+ T cells. Coincubation with unconjugated anti-C1 MAbs (10 μg/ml each) was used to identify binding synergy. The gating strategy used to identify live HIV-1-infected cells (intracellular p24+) and representative histograms of CH58 surface staining and CH90-induced synergy within the p24+ cell populations are shown in Fig. 2A. Mock-infected cells were used to establish the cutoff for the p24+ gate. The incubation of directly conjugated CH58 MAb with AE.92TH023-infected CD4+ T cells in combination with the unconjugated nonfluorescent A32, CH57, and CH90 MAbs resulted in 60, 60, and 59% increases, respectively, in the frequency of HIV-1-infected cells recognized by the CH58 MAb compared to the frequency of HIV-1-infected cells recognized by CH58 MAb alone (Fig. 2B). The mean fluorescent intensity (MFI) of the CH58-stained cells was concomitantly increased (Fig. 2C). In contrast, we observed a modest increase (22%) in binding of CH58 to 92TH023-infected cells in the presence of MAb CH54. The incubation of AE.CM235-infected cells (Fig. 2D and E) with CH58 in the presence of A32 revealed a similar (>45%) increase in both the frequency of infected cells recognized by CH58 and the MFI of the cells. Enhancement of CH58 binding to CM235-infected cells was also detected with CH54 (20% MFI increase), CH57 (23% MFI increase), and CH90 (28% MFI increase). These data demonstrate that C1 Abs can enhance the binding of V2 Abs to HIV-infected CD4 cells. However, we observed differences in the magnitude of synergy between C1 MAbs and CH58 for AE.92TH023-infected cells compared to CM235-infected cells. This difference suggested that the envelopes of these two HIV-1 isolates differed in their expressed conformations and thus differed in the abilities of the Abs to synergize in the recognition of cells infected with the two virus strains. The impact of differences in Env structures on the potential for C1 and V2 antibody synergy was further supported by the discordance between the synergy observed with A32 and CH58 in binding HIV-infected cells and the lack of synergy between these two MAbs in binding to A244Δ11 gp120 monomer as detected by SPR.
FIG 2.
Synergy of MAb for binding to the infected CD4 T cells. Primary CD4+ T cells were activated and infected with HIV-1 AE.92TH023 (A to C) and AE.CM235 (D and E) for 72 h. Cells were stained with viability dye and anti-p24 Ab to identify viable (live) infected (p24+) cells using the gating strategy shown by the dot plot in panel A. The CH58 MAb was conjugated with Alexa Fluor 488 fluoropohore. The other MAbs and MAb Fab fragments (Palivizumab [Neg], A32, CH54, CH57, and CH90) were used as nonconjugated reagents. The histogram in panel A is representative of the CH58 Alexa Fluor-488 staining of infected cells (p24+) observed with individual and combined MAbs. (B to E) The infected CD4+ T cells were stained with CH58 Alexa Fluor 488 in combination with the MAbs or Fab fragments indicated on the x axes at 10 μg/ml each. The y axes represent the percentage of increase of stained cells (B and D) and mean fluorescent intensity (MFI) (C and E) for each combination of MAb or Fab fragment relative to the staining of cells observed when the CH58 MAb was used alone. The results represent the average ± SEM of the results obtained in four experiments.
We next utilized F(ab) and F(ab′)2 fragments of MAb CH90 to determine if synergy for binding HIV-1-infected cells was mediated by events associated with a monovalent or bivalent interaction between the Env epitope and the Ab antigen-binding region as represented by the F(ab) and F(ab′)2, respectively, or whether the whole Ab with Fc region was required. Interestingly, almost no enhancement of binding (<10% with AE.92TH023 and 15% with AE.CM235) was observed for CH58 in the presence of CH90 F(ab). However, binding of CH58 was increased in the presence of CH90 F(ab′)2 to levels comparable to those observed with whole CH90 IgG (Fig. 2B to E). These data indicated that the Fc portion of MAb CH90 was not required for synergy in the recognition of infected cells with MAb CH58, but bivalent binding of the hinged antigen-binding region of C1 MAb CH90 was necessary to induce improved recognition of cell-surface Env by the V2 MAb, CH58.
Synergy of V2 and C1 MAbs for HIV-1 neutralization.
Next, the ability of C1 and V2 MAbs to synergize in the neutralization of HIV-1 was investigated against a panel of viruses that represented HIV-1 tier 1 (B.MN and AE.92TH023), tier 2 (C.TV-1 and AE.CM244), and subtype CRF_01-AE transmitted/founder isolates (AE.427299 and AE.703357) using the TZM-bl pseudovirus neutralization assay. None of the C1 MAbs displayed neutralizing activity when tested alone against any of the HIV-1 isolates as previously reported for the prototype C1 MAb, A32. The V2 MAbs also lacked neutralizing activity against HIV-1 strains B.MN, C.TV-1, AE.427299, AE.703357, and AE.CM244 (IC50, >50 μg/ml). All four V2 MAbs were able to neutralize the tier 1 HIV-1 AE.92TH023. Thus, we focused on determining if the C1 MAbs and V2 MAbs synergize for neutralization of this isolate. The 50% inhibitory concentrations (IC50) of each V2 MAb alone or in combination with 50 μg/ml of each C1 MAb are shown in Table 2. We observed no neutralization synergy between the V2 MAbs and C1 MAbs CH54, CH57, and CH90. However, the IC50s of the V2 MAbs decreased 20- to 65-fold when tested in combination with the C1 MAb A32.
TABLE 2.
Neutralizing activities of MAbs
| MAb | HIV-1 92TH023.6 IC50 (μg/ml)a |
||||
|---|---|---|---|---|---|
| V2 MAb alone | V2 MAb + A32 | V2 MAb + CH54 | V2 MAb + CH57 | V2 MAb + CH90 | |
| CH58 | 28.14 | 0.86 | 22.37 | NTb | 35.52 |
| CH59 | 8.95 | 0.15 | 8.24 | 5.54 | 26.23 |
| HG107 | 6.23 | 0.28 | 7.66 | 8.42 | 9.99 |
| HG120 | 5.92 | 0.09 | 4.06 | 7.33 | 8.46 |
Data for CH58 and CH59 are reported as the average of four experiments; all other conditions were tested in duplicate.
NT, not tested.
Synergy of V2 and C1 MAbs for ADCC.
The ability of C1 and V2 MAbs to synergize in the recognition of HIV-1-infected cells suggested that that these Ab specificities may also synergize in their ability to mediate ADCC. We focused on ADCC directed against target cells infected with the HIV-1 AE.CM235 virus, since this isolate is representative of tier 2 neutralization-sensitive phenotype CRF_01-AE HIV-1 isolates that are responsible for the vast majority of transmission events in the RV144 trial (33).
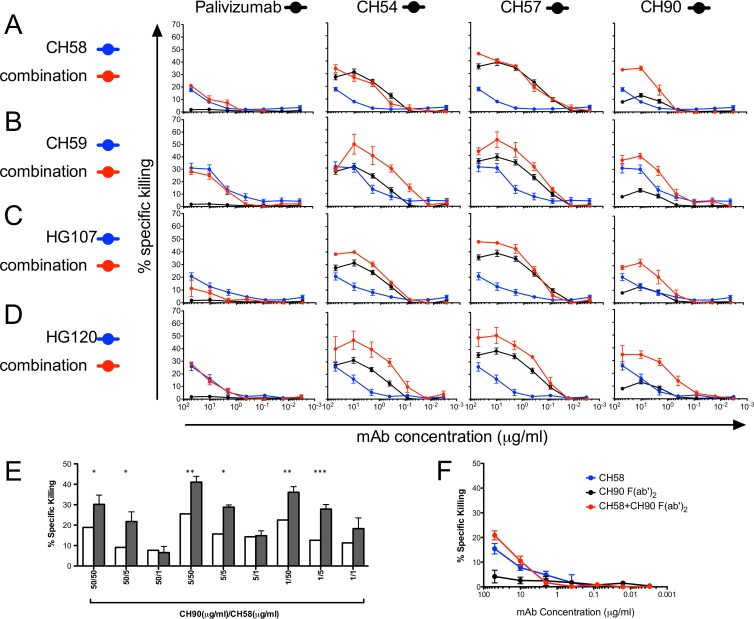
To evaluate and quantitate synergy of V2 and C1 MAbs for ADCC, we measured the activities of 5-fold serial dilutions of each antibody alone or in equimolar combinations against AE.CM235-infected target cells (Fig. 3). All four ALVAC/AIDSVax vaccine recipient V2-specific MAbs, CH58, CH59, HG107, and HG120, were included in this study to characterize the potential for synergistic ADCC interactions between a number of V2 and C1 Ab combinations. The ADCC activity curves were used to interpolate the endpoint concentrations (EC) and the concentrations (μg/ml) at which 75% of the maximum killing activity (MK75) of the V2 or C1 specific MAb was reached (Table 3). The EC and MK75 concentrations were used to calculate the combination index (CI) for the MAb pair (Table 3) (32).
FIG 3.
ADCC synergy with combinations of V2 and C1 MAbs. (A to E) ADCC synergy with equimolar combinations of V2 and C1 MAbs. Shown is the percentage of specific killing observed by V2 MAbs CH58 (A), CH59 (B), HG107 (C), and HG120 (D) alone and in combination with negative-control Palivizumab or C1 MAbs CH54, CH57, and CH90 at a 1:1 ratio over 5-fold serial dilutions in the luciferase ADCC assay with CM235-infected targets. Each MAb and the combinations were tested in duplicate wells. The results represent the average ± SEM of the results obtained in a minimum of two independent experiments. (E) ADCC synergy with nonequimolar combinations of V2 and C1 MAbs. The percentage of specific killing was detected by incubating individual MAbs and the combinations indicated with HIV-1 AE.CM235-infected CEM.NKRCCR5 target cells for 3 h in the luciferase ADCC assay. The expected ADCC activities for an additive effect are represented by white bars. The actual observed activities are represented by filled bars. Results represent the mean and SEM of two experiments, each tested in duplicate. Significantly different results between the expected and observed mean activities by t test are indicated (*). (F) Synergy of CH58 anti-V2 IgG and CH90 anti-C1 F(ab′)2 for ADCC. The graph represents the percentage of specific killing observed by V2 MAb CH58 and CH90 C1 F(ab′)2 alone and in combination at a 1:1 ratio over 5-fold serial dilutions in the luciferase ADCC assay with CM235-infected targets. The graphs represent the average ± SEM of the results obtained in three experiments with each run in duplicate.
TABLE 3.
CI values for ADCC activities of vaccine-induced C1 and V2 MAbs
| MAb | MAb condition(s)a | % maximum killing | ECb (μg/ml) | 75% maximum killing | MK75 concnc (μg/ml) | CI value |
|||
|---|---|---|---|---|---|---|---|---|---|
| Mutually exclusive (β = 0) |
Mutually nonexclusive (β = 1) |
||||||||
| EC | MK75 | EC | MK75 | ||||||
| CH58 | CH54 (C1) | 31.3 | 0.33 | 23.5 | 1.95 | 2.376 | 2.213 | 2.474 | 2.289 |
| CH58 (V2) | 17.8 | 18.26 | 13.3 | 31.75 | |||||
| CH54 when comb. with CH58 | 34.1 | 0.77 | 23.5 | 4.25 | |||||
| CH58 when comb. with CH54 | 13.3 | 1.11 | |||||||
| CH57 (C1) | 38.8 | 0.09 | 29.1 | 1.25 | 1.005 | 1.092 | 1.010 | 1.099 | |
| CH58 (V2) | 17.8 | 18.26 | 13.3 | 31.75 | |||||
| CH57 when comb. with CH58 | 45.7 | 0.09 | 29.1 | 1.36 | |||||
| CH58 when comb. with CH57 | 13.3 | 0.20 | |||||||
| CH90 (C1) | 13.1 | 4.65 | 10.0 | 4.65 | 0.356 | 0.336 | 0.377 | 0.350 | |
| CH58 (V2) | 17.8 | 18.26 | 13.3 | 31.75 | |||||
| CH90 when comb. with CH58 | 34.5 | 1.32 | 10.0 | 1.32 | |||||
| CH58 when comb. with CH90 | 13.3 | 1.63 | |||||||
| CH59 | CH54 (C1) | 31.3 | 0.33 | 23.5 | 1.95 | 0.231 | 0.179 | 0.241 | 0.184 |
| CH59 (V2) | 31.1 | 1.05 | 23.3 | 6.73 | |||||
| CH54 when comb. with CH59 | 48.8 | 0.06 | 23.5 | 0.27 | |||||
| CH59 when comb. with CH54 | 23.3 | 0.27 | |||||||
| CH57 (C1) | 38.8 | 0.09 | 29.1 | 1.25 | 0.724 | 0.328 | 0.762 | 0.339 | |
| CH59 (V2) | 31.1 | 1.05 | 23.3 | 6.73 | |||||
| CH57 when comb. with CH59 | 52.4 | 0.06 | 29.1 | 0.36 | |||||
| CH59 when comb. with CH57 | 23.3 | 0.26 | |||||||
| CH90 (C1) | 13.1 | 4.65 | 10.0 | 4.65 | 0.455 | 0.310 | 0.486 | 0.329 | |
| CH59 (V2) | 31.1 | 1.05 | 23.3 | 6.73 | |||||
| CH90 when comb. with CH59 | 40.8 | 0.39 | 10.0 | 0.39 | |||||
| CH59 when comb. with CH90 | 23.3 | 1.53 | |||||||
| HG107 | CH54 (C1) | 31.3 | 0.33 | 23.5 | 1.95 | 0.824 | 0.655 | 0.862 | 0.665 |
| HG107 (V2) | 20.7 | 5.26 | 15.5 | 24.09 | |||||
| CH54 when comb. with HG107 | 39.9 | 0.26 | 23.5 | 1.25 | |||||
| HG107 when comb. with CH54 | 15.5 | 0.39 | |||||||
| CH57 (C1) | 38.8 | 0.09 | 29.1 | 1.25 | 1.695 | 0.585 | 1.743 | 0.590 | |
| HG107 (V2) | 20.7 | 5.26 | 15.5 | 24.09 | |||||
| CH57 when comb. with HG107 | 47.8 | 0.15 | 29.1 | 0.72 | |||||
| HG107 when comb. with CH57 | 15.5 | 0.24 | |||||||
| CH90 (C1) | 13.1 | 4.65 | 10.0 | 4.65 | 0.316 | 0.226 | 0.341 | 0.236 | |
| HG107 (V2) | 20.7 | 5.26 | 15.5 | 24.09 | |||||
| CH90 when comb. with HG107 | 32.3 | 0.78 | 10.0 | 0.78 | |||||
| HG107 when comb. with CH90 | 15.5 | 1.40 | |||||||
| HG120 | CH54 (C1) | 31.3 | 0.33 | 23.5 | 1.95 | 0.257 | 0.164 | 0.261 | 0.165 |
| HG120 (V2) | 26.0 | 5.41 | 19.5 | 23.64 | |||||
| CH54 when comb. with HG120 | 47.5 | 0.08 | 23.5 | 0.30 | |||||
| HG120 when comb. with CH54 | 19.5 | 0.24 | |||||||
| CH57 (C1) | 38.8 | 0.09 | 29.1 | 1.25 | 0.734 | 0.268 | 0.743 | 0.270 | |
| HG120 (V2) | 26.0 | 5.41 | 19.5 | 23.64 | |||||
| CH57 when comb. with HG120 | 51.3 | 0.07 | 29.1 | 0.33 | |||||
| HG120 when comb. with CH57 | 19.5 | 0.18 | |||||||
| CH90 (C1) | 13.1 | 4.65 | 10.0 | 4.65 | 0.108 | 0.099 | 0.111 | 0.102 | |
| HG120 (V2) | 26.0 | 5.41 | 19.5 | 23.64 | |||||
| CH90 when comb. with HG120 | 34.8 | 0.27 | 10.0 | 0.27 | |||||
| HG120 when comb. with CH90 | 19.5 | 0.99 | |||||||
“when comb. with” refers to the first MAb when in combination with the second MAb.
EC, endpoint concentration.
MK75, concentration at which 75% of the maximum observed killing activity is reached for each individual V2 and C1 MAb, or the concentration at which the 75% of the maximum killing activity of each individual MAb is reached when tested in combination.
The C1 and V2 MAbs recognize different regions of the HIV-1 Env, thus fulfilling the criteria of mutual exclusivity. However, C1 and V2 MAbs also fulfill the criteria of nonmutual exclusivity because they act together to mediate a single antiviral effector function (ADCC). Table 3 presents both the mutually and nonmutually exclusive CI values (32). By these methods, CI values between 0 and 1 indicate a synergistic interaction, and the closeness to 0 provides an indication of the magnitude of synergy. Most combinations of vaccine-induced V2 and C1 MAbs resulted in synergy for ADCC. For MAb CH58, synergy was observed only when tested in combination with C1 MAb CH90 (Fig. 3A; Table 3). Synergy for ADCC was observed between MAb CH59 and MAbs CH54, CH57, and CH90 (Fig. 3B; Table 3). For HG107, EC and MK75 synergy was observed when tested in combination with CH54 and CH90 (Fig. 3C; Table 3), while for the V2 MAb HG120, strong synergy was observed with CH54, CH57, and CH90 (Fig. 3D; Table 3). Thus, only one C1 MAb, CH90, was found to work in synergy with all four V2 MAbs. As indicated in Table 3, the CI values predominately indicated a greater degree of synergy for MK75 compared to EC, suggesting that the synergistic ability of combinations of MAb to mediate ADCC is limited by a minimum threshold concentration of Ab required to activate Fcγ receptor signaling on NK effector cells. We observed no examples of contradiction between the mutually exclusive and nonmutually exclusive methods when applied to our data set. In addition, we observed no enhancement of ADCC activity when any of the V2 MAbs were tested against HIV-1 AE.CM235-infected target cells in combination with the negative-control MAb Palivizumab (Fig. 3A to D) or the C1 MAb A32 (not shown).
We also tested vaccine-induced CH58 and CH90 MAbs individually at three different concentrations of 50, 5, and 1 μg/ml as well as in combination. Based on the individual testing of the MAbs, we calculated the percentage of specific killing we expected to be observed for an additive effect of each combination of MAbs and defined this percentage of specific killing as the “expected activity” (Fig. 3E; white bars). The “expected activity” was compared to the “observed” activity after the actual testing of each combination of MAbs (Fig. 3E; filled bars). The anti-RSV MAb Palivizumab was used as a negative control, and its combination with CH58 represents the negative control for MAb combinations. There was no observable synergistic increase in ADCC activity directed against HIV-1 AE.CM235-infected target cells when CH58 was combined with Palivizumab. In contrast, we observed a significant synergistic effect when CH58 was tested in combination with the C1 MAb CH90 (Fig. 3E). The average increase over the observed ADCC activity over the expected ADCC activity of CH58 and CH90 combinations was 65% (range, 0 to 140%).
C1 Ab regions involved in V2/C1 ADCC synergy.
ADCC is an Ab effector function that requires two concurrent interactions: recognition of antigen by the Ab Fab region and binding of the Ab Fc region with Fcγ receptor on the surface of cytotoxic effector cells. We used the F(ab′)2 fragment of MAb CH90 to evaluate the contributions of Fab and Fc regions to the ADCC synergy observed with MAbs CH90 and CH58. ADCC activity was measured using equimolar serial dilutions of both the CH90 F(ab′)2 and CH58 MAb. Modest synergy between the CH90 F(ab′)2 and CH58 MAbs was only evident at combinations of 50- and 10-μg/ml concentrations of F(ab′)2 and MAb resulting in a 35% increase in specific killing (Fig. 3F). This was in contrast to the ADCC synergy observed between whole MAb CH90 and MAb CH58 that was observed at all concentrations above the positive response threshold (Fig. 3A). It should be noted that CI values for this combination were not calculated because the absence of ADCC mediated by the CH90 F(ab′)2 limits the utilization of the formula described to calculate the CI. Thus, these data suggest that the synergy observed for ADCC is a consequence of both enhanced recognition of Env on the surface of HIV-1-infected cells and increased cross-linking of Fcγ receptors on ADCC effector cells.
We have previously investigated the relative concentration of CH58-like Ab in the plasma of ALVAC/AIDSVax vaccine recipients by using an SPR-based blocking assay. We determined that the average concentration of the vaccine-induced CH58-blockable Ab in vaccinee plasma was 3.6 ± 3.2 μg/ml (4). At this concentration, the CH58 MAb alone had no detectable neutralization or ADCC activities (4). The data collected in the present study indicated that the CH58-CH90 combination reduced the required functional concentration of CH58 for mediation of ADCC to 1.32 μg/ml, a concentration within the levels of CH58-like Abs detected in the plasma of vaccine recipients.
Synergy for infectious virion capture.
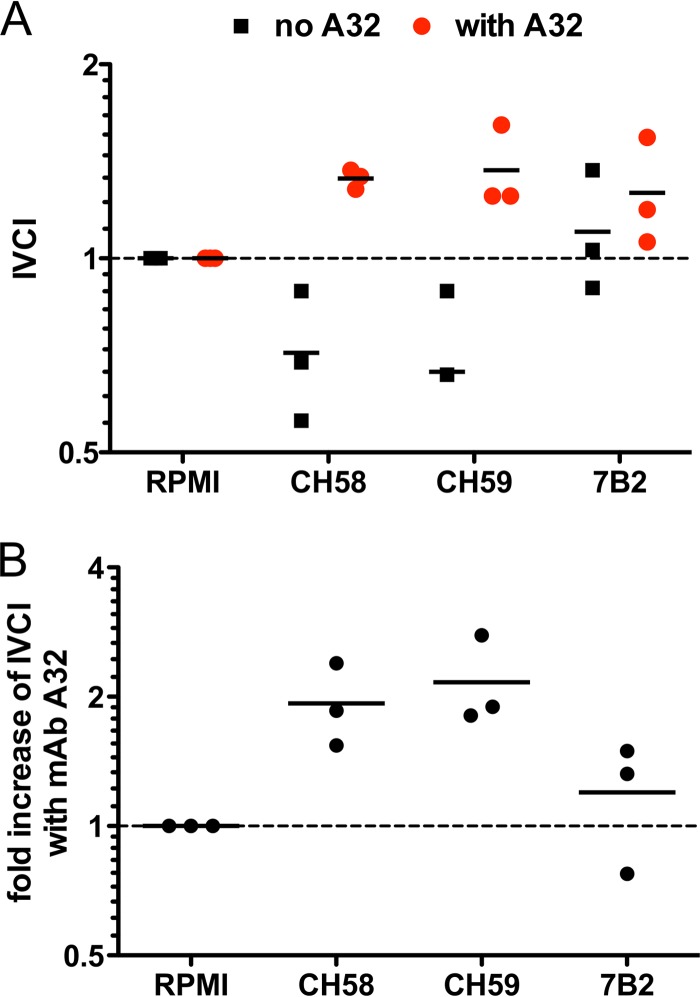
Finally, we investigated whether the C1 and V2 Abs can synergize for the capture of infectious virions. V2 MAbs CH58 and CH59, or the gp41 MAb 7B2 included as a positive control, were mixed with AE.92TH023 HIV-1 viral stock with or without C1 MAb A32 at equivalent concentrations. The capture of infectious and noninfectious virions was measured by the protein G/TZM-bl/real-time PCR combined virion capture assay as described previously (26–28). To quantify the selective ability to capture infectious virions, we calculated the infectious virion capture index (IVCI) of each MAb and MAb combination as described previously (28). The V2 MAbs CH58 and CH59 MAb captured a low level of infectious virions (4), and the IVCI values suggest a bias toward capture of noninfectious virions (IVCI, <1) (Fig. 4A). In contrast, in the presence of MAb A32, both CH58 and CH59 demonstrated increased capture of infectious AE.92TH023 HIV-1 virions (IVCI, >1), observed as a 2-fold increase in IVCI values (Fig. 4A and B). These data suggest that the synergistic effect of V2 and C1 MAbs may act in binding to both Env on the surface of HIV-1-infected CD4+ T cells and Env expressed on the surface of HIV-1 tier 1 isolates of HIV-1 virions, confirming the results observed with the neutralization assay.
FIG 4.
V2 MAbs synergize with C1 MAb for enhanced selective capture of infectious HIV-1 AE.92TH023 virions. IVCI (i.e., the fold change of the ratios of infectious HIV-1 AE.92TH023 virions to total virion particles) was used to quantify the selective capture of infectious versus noninfectious virions as described in Materials and Methods. (A) IVCI values of V2 MAbs CH58 and C59 and the positive-control gp41 MAb 7B2 when tested alone (black squares) or in combination with the C1 MAb A32 (red circles). (B) Fold increase of IVCI values observed in the presence of C1 MAb A32. The line represents the mean of the results obtained in three experiments.
DISCUSSION
In this article, we have found that V2 and C1 MAbs isolated from ALVAC/AIDSVax B/E vaccinees showed synergy for monomeric Env gp120 recognition and for recognition of Env trimer expressed on the surface of HIV-1-infected cells. Moreover, both neutralizing activity and infectious virion capture against the tier 1 isolate AE.92TH023 and ADCC directed against the tier 2 HIV-1 CM235 isolate were also increased when V2 antibodies were tested in the presence of C1 MAbs.
The analysis of V2 responses has revealed differences between responses induced by the vaccine regimen used in the RV144 ALVAC/AIDSVax B/E clinical trial and natural HIV-1 infection. V2 responses were elicited in 97% of the Thai vaccine recipients, whereas they have only been detected in 50% of the HIV-1 CRF01-AE-infected Thai individuals (34). Moreover, the CH58, CH59, HG107, and HG120 MAbs that represent the vaccine-induced V2 responses recognized a linear V2 peptide comprised of amino acid residues 168 to 183 and are able to mediate ADCC (4), whereas the V2 697-D MAb isolated from an HIV-infected individual recognizes a glycosylation-dependent conformational V2 region epitope and does not mediate ADCC (35, 36).
Rolland and collaborators recently reported that a genetic sieve analysis of viral sequences of RV144 clinical trial infections demonstrated that the presence of a lysine at amino acid residue 169 in the V2 region was associated with vaccine efficacy (3). Liao and collaborators demonstrated that binding, neutralizing, and ADCC activities of RV144 vaccinee-derived MAbs CH58 and CH59 were decreased by mutations at position K169. In contrast, the activities of V2 MAbs isolated from infected individuals were moderately or not at all affected by the presence of these mutations (4). Thus, ALVAC/AIDSVax B/E vaccine-induced V2 Ab responses were indeed different than those elicited by HIV-1 infection and also had different immune effector functions.
In this study, we have identified synergy between V2 and C1 MAbs in binding to monomeric gp120, in binding to HIV-infected cells, for virus neutralization and infectious virion capture, and for the ability to mediate ADCC. Interestingly, we also observed unique profiles of synergy provided by different V2/C1 MAb combinations. We hypothesize that these profiles likely reflect differences in the overlapping C1 epitopes recognized by these MAbs as demonstrated by different levels of cross-inhibition among the C1 MAbs (5). The recognition of the overlapping C1 epitopes may lead to different outcomes of the observed synergistic effects uniquely related to each antiviral function: i.e., synergy in neutralization or virus capture versus ADCC. Reaction kinetics of the interactions between MAbs and their epitopes may also impact the cooperative potential of Abs. A32 has the fastest monomeric gp120 binding on rate and did not enhance binding of CH58 to gp120. In contrast, CH57 indicated the slowest on rate and highest magnitude of synergy for monomeric gp120 binding. However, it is important to note that the unique profiles of synergy observed for binding to monomeric gp120 or to infected cells did not consistently predict synergy for ADCC. This finding is in partial agreement with a previous study that described no correlation between the ability of Env-specific MAbs to bind to infected cells and to mediate ADCC, although a direct correlation was observed between binding of Env and ADCC activity for polyclonal IgG preparations (37). Our data suggest that polyclonal IgG preparations reflect a repertoire of antigen specificities that cannot be entirely recapitulated by the study of single MAbs or limited combinations of MAbs.
In the absence of a well-defined binding site for MAb A32 and each of the three anti-C1 RV144 MAbs, we cannot fully define the Env conformations that may be induced by these MAbs that result in increasing both the binding to Env and enhancing the antiviral functions of the V2 MAb CH58. Furthermore, differences observed between synergistic binding to the surface of CD4 T cells infected with the tier 1 HIV-1 isolate AE.92TH023 and the tier 2 isolate AE.CM235, as demonstrated for the combination of CH58 and CH54, suggest that synergy is influenced by both the conformation of epitopes recognized by the combination of MAbs and structural differences between envelopes of HIV-1 isolates. Crystallization studies will be needed to resolve the fine details of these molecular interactions.
We utilized MAb F(ab) and F(ab′)2 fragments to identify Ab regions involved in binding synergy and ADCC synergy. These experiments demonstrated that F(ab′)2 but not F(ab) fragments were sufficient to induce the conformational changes in Env expressed on the surface of HIV-1-infected cells that allow for enhanced recognition by MAb CH58. Using F(ab′)2 fragments, we also determined that the ability of these non-Fc-bearing fragments to enhance binding can result in ADCC synergy. However, the most potent synergy was observed when whole C1 and V2 MAbs were used in combination. This suggests that ADCC synergy is to a large part dependent on augmented Fcγ-receptor and Ab-Fc interactions that are facilitated by multivalent recognition of Env and the extent to which the amount of Ab bound to the cell surface forms an immunocomplex capable of cross-linking the Fcγ receptor on the membrane of the effector cells. The influence of the size of the immunocomplex and the glycosylation profile of Ab have been recently reported as contributing factors for binding to the membrane-bound Fcγ receptor (38).
Importantly, synergy for ADCC was observed for most combinations of C1 and V2 MAbs against the tier 2 neutralization-sensitive isolate AE.CM235, whereas this synergy was not observed for either neutralizing or virion capture activities. Transmitted/founder viruses isolated from infected vaccine recipients regarding neutralization are also tier 2 viruses. Our findings support the hypothesis that these types of synergistic interactions could be related to the ability of the immune system to reduce the risk of infection as observed in the RV144 vaccine trial. Moreover, we observed that the V2/C1 synergistic activity was capable of increasing CH58 MAb-mediated ADCC at concentrations of CH58 MAb that are lower than the average concentrations of CH58-like antibodies detected in the plasma of RV144 vaccine recipients (4).
Several studies have described the ability of HIV-1 Abs to synergize for improved virus neutralization (7–16). Overall, our observations indicate for the first time that synergistic mechanisms of action exist for functional nonneutralizing Ab responses correlated to the reduced of risk of HIV-1 infection. These synergistic interactions may more closely represent the composition of plasma Ab specificities and account for the difficulty to recapitulate the polyclonality of protective ADCC responses, such as those observed in the ALVAC/AIDSVax trial, when using combination of MAbs (2, 37). Passive protection studies in nonhuman primate models with simian-human immunodeficiency virus (SHIV) challenges can inform whether Ab-mediated ADCC synergy can be a mechanistic correlate of protection. Moreover, these data raise the hypothesis that C1 and V2 Env-specific Ab-mediated ADCC synergy could correlate with decreased HIV-1 risk, a hypothesis that can be tested in upcoming human HIV-1 efficacy trials.
ACKNOWLEDGMENTS
This work was supported by the Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery (CHAVI-ID) and CHAVI (UM-AI100645 and U01 AI067854), National Institutes of Health (NIH/NIAID/DAIDS), Bill and Melinda Gates Foundation's Collaboration for AIDS Vaccine Discovery Grants (Haynes B Cell Lineage [1033098] and Haynes IgA [1040758], Montefiori CA-VIMC [1032144], Koup CT-VIMC [1032325]), and the Duke University Center for AIDS Research (CFAR) Grant (P30 AI 64518). In addition, funding was provided by Interagency Agreement Y1-AI-2642-12 between U.S. Army Medical Research and Material Command (USAMRMC) and the National Institutes of Allergy and Infectious Diseases through a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense (DOD). J.P. was supported by NIH, NIAID grant AI07392.
The views expressed in this article are those of the authors and should not be construed as official or as representing the views of the Department of Health and Human Services, the National Institute of Allergy and Infectious Diseases (NIAID), the Department of Defense, or the Department of the Army. Trade names are used for identification purposes only and do not imply endorsement.
We are indebted to the volunteers and clinical staff who participated in the RV144 vaccine trial. We thank Kelly A. Soderberg and Charla Andrews for clinical trial and or project management. We also thank Kaylan Whittaker for technical assistance, Marcella Sarzotti-Kelsoe for quality assurance oversight, and Nathan Vandergrift and Sheetal Sawant for statistical support.
M.B., M.A.M., J.H.K., N.L.M., H.-X.L., B.F.H., and G.F. have filed patent applications on some of the RV144 MAbs and related antigens used in this study.
Footnotes
Published ahead of print 7 May 2014
REFERENCES
- 1.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, MOPH-TAVEG Investigators. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220. 10.1056/NEJMoa0908492 [DOI] [PubMed] [Google Scholar]
- 2.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao H-X, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286. 10.1056/NEJMoa1113425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, Decamp AC, Carrico C, Menis S, Magaret CA, Ahmed H, Juraska M, Chen L, Konopa P, Nariya S, Stoddard JN, Wong K, Zhao H, Deng W, Maust BS, Bose M, Howell S, Bates A, Lazzaro M, O'Sullivan A, Lei E, Bradfield A, Ibitamuno G, Assawadarachai V, O'Connell RJ, deSouza MS, Nitayaphan S, Rerks-Ngarm S, Robb ML, McLellan JS, Georgiev I, Kwong PD, Carlson JM, Michael NL, Schief WR, Gilbert PB, Mullins JI, Kim JH. 2012. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature 490:417–420. 10.1038/nature11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao H-X, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang K-K, Chen X, Tsao C-Y, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang Z-Y, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. 2013. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 38:176–186. 10.1016/j.immuni.2012.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonsignori M, Pollara J, Moody MA, Alpert MD, Chen X, Hwang K-K, Gilbert PB, Huang Y, Gurley TC, Kozink DM, Marshall DJ, Whitesides JF, Tsao C-Y, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Kim JH, Michael NL, Tomaras GD, Montefiori DC, Lewis GK, DeVico A, Evans DT, Ferrari G, Liao H-X, Haynes BF. 2012. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J. Virol. 86:11521–11532. 10.1128/JVI.01023-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomaras GD, Ferrari G, Shen X, Alama SM, Liao H-X, Pollara J, Bonsignori M, Moody MA, Fong Y, Chen X, Poling B, Nicholson CO, Zhang R, Lu X, Parks R, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Gilbert PB, Kim JH, Michael NL, Montefiori DC, Haynes BF. 2013. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proc. Natl. Acad. Sci. U. S. A. 110:9019–9024. 10.1073/pnas.1301456110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ketas TJ, Holuigue S, Matthews K, Moore JP, Klasse PJ. 2012. Env-glycoprotein heterogeneity as a source of apparent synergy and enhanced cooperativity in inhibition of HIV-1 infection by neutralizing antibodies and entry inhibitors. Virology 422:22–36. 10.1016/j.virol.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hrin R, Montgomery DL, Wang F, Condra JH, An Z, Strohl WR, Bianchi E, Pessi A, Joyce JG, Wang Y-J. 2008. In vitro synergy between peptides or neutralizing antibodies targeting the N- and C-terminal heptad repeats of HIV type 1 gp41. AIDS Res. Hum. Retroviruses 24:1537–1544. 10.1089/aid.2008.0129 [DOI] [PubMed] [Google Scholar]
- 9.Zwick MB, Wang M, Poignard P, Stiegler G, Katinger H, Burton DR, Parren PWHI. 2001. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 75:12198–12208. 10.1128/JVI.75.24.12198-12208.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verrier F, Nadas A, Gorny MK, Zolla-Pazner S. 2001. Additive effects characterize the interaction of antibodies involved in neutralization of the primary dualtropic human immunodeficiency virus type 1 isolate 89.6. J. Virol. 75:9177–9186. 10.1128/JVI.75.19.9177-9186.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W, Smith-Franklin BA, Li PL, Wood C, He J, Du Q, Bhat GJ, Kankasa C, Katinger H, Cavacini LA, Posner MR, Burton DR, Chou TC, Ruprecht RM. 2001. Potent neutralization of primary human immunodeficiency virus clade C isolates with a synergistic combination of human monoclonal antibodies raised against clade B. J. Hum. Virol. 4:55–61 [PubMed] [Google Scholar]
- 12.Vijh-Warrier S, Pinter A, Honnen WJ, Tilley SA. 1996. Synergistic neutralization of human immunodeficiency virus type 1 by a chimpanzee monoclonal antibody against the V2 domain of gp120 in combination with monoclonal antibodies against the V3 loop and the CD4-binding site. J. Virol. 70:4466–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mascola JR, Louder MK, VanCott TC, Sapan CV, Lambert JS, Muenz LR, Bunow B, Birx DL, Robb ML. 1997. Potent and synergistic neutralization of human immunodeficiency virus (HIV) type 1 primary isolates by hyperimmune anti-HIV immunoglobulin combined with monoclonal antibodies 2F5 and 2G12. J. Virol. 71:7198–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laal S, Burda S, Gorny MK, Karwowska S, Buchbinder A, Zolla-Pazner S. 1994. Synergistic neutralization of human immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J. Virol. 68:4001–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchbinder A, Karwowska S, Gorny MK, Burda ST, Zolla-Pazner S. 1992. Synergy between human monoclonal antibodies to HIV extends their effective biologic activity against homologous and divergent strains. AIDS Res. Hum. Retroviruses 8:425–427. 10.1089/aid.1992.8.425 [DOI] [PubMed] [Google Scholar]
- 16.Tilley SA, Honnen WJ, Racho ME, Chou T-C, Pinter A. 1992. Synergistic neutralization of HIV-1 by human monoclonal antibodies against the V3 loop and the CD4-binding site of gp120. AIDS Res. Hum. Retroviruses 8:461–467. 10.1089/aid.1992.8.461 [DOI] [PubMed] [Google Scholar]
- 17.Nitayaphan S, Pitisuttithum P, Karnasuta C, Eamsila C, de Souza M, Morgan P, Polonis V, Benenson M, VanCott T, Ratto-Kim S, Kim J, Thapinta D, Garner R, Bussaratid V, Singharaj P, El-Habib R, Gurunathan S, Heyward W, Birx D, McNeil J, Brown AE, Thai AIDS Vaccine Evaluation Group. 2004. Safety and immunogenicity of an HIV subtype B and E prime-boost vaccine combination in HIV-negative Thai adults. J. Infect. Dis. 190:702–706. 10.1086/422258 [DOI] [PubMed] [Google Scholar]
- 18.Bonsignori M, Hwang K-K, Chen X, Tsao C-Y, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, Sinangil F, Pancera M, Yongping Y, Zhang B, Zhu J, Kwong PD, O'Dell S, Mascola JR, Wu L, Nabel GJ, Phogat S, Seaman MS, Whitesides JF, Moody MA, Kelsoe G, Yang X, Sodroski J, Shaw GM, Montefiori DC, Kepler TB, Tomaras GD, Alam SM, Liao H-X, Haynes BF. 2011. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 85:9998–10009. 10.1128/JVI.05045-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, Kappes JC, Shaw GM, Hoxie JA, Robinson JE, Haynes BF. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 85:7029–7036. 10.1128/JVI.00171-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollara J, Bonsignori M, Moody MA, Pazgier M, Haynes BF, Ferrari G. 2013. Epitope specificity of human immunodeficiency virus-1 antibody dependent cellular cytotoxicity [ADCC] responses. Curr. HIV Res. 11:378–387. 10.2174/1570162X113116660059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, Camacho ZT, Gewirth D, Kelsoe G, Chen P, Haynes BF. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178:4424–4435. 10.4049/jimmunol.178.7.4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alam SM, Scearce RM, Parks RJ, Plonk K, Plonk SG, Sutherland LL, Gorny MK, Zolla-Pazner S, Vanleeuwen S, Moody MA, Xia S-M, Montefiori DC, Tomaras GD, Weinhold KJ, Karim SA, Hicks CB, Liao H-X, Robinson J, Shaw GM, Haynes BF. 2008. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J. Virol. 82:115–125. 10.1128/JVI.00927-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edmonds TG, Ding H, Yuan X, Wei Q, Smith KS, Conway JA, Wieczorek L, Brown B, Polonis V, West JT, Montefiori DC, Kappes JC, Ochsenbauer C. 2010. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology 408:1–13. 10.1016/j.virol.2010.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125. 10.1128/JVI.79.16.10108-10125.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P, Overman RG, Yates NL, Alam SM, Vandergrift N, Chen Y, Graw F, Freel SA, Kappes JC, Ochsenbauer C, Montefiori DC, Gao F, Perelson AS, Cohen MS, Haynes BF, Tomaras GD. 2011. Dynamic antibody specificities and virion concentrations in circulating immune complexes in acute to chronic HIV-1 infection. J. Virol. 85:11196–11207. 10.1128/JVI.05601-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu P, Yates NL, Shen X, Bonsignori M, Moody MA, Liao HX, Fong Y, Alam SM, Overman RG, Denny T, Ferrari G, Ochsenbauer C, Kappes JC, Polonis VR, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Montefiori DC, Gilbert P, Michael NL, Kim JH, Haynes BF, Tomaras GD. 2013. Infectious virion capture by HIV-1 gp120-specific IgG from RV144 vaccinees. J. Virol. 87:7828–7836. 10.1128/JVI.02737-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu P, Williams LD, Shen X, Bonsignori M, Vandergrift NA, Overman RG, Moody MA, Liao H-X, Stieh DJ, McCotter KL, French AL, Hope TJ, Shattock R, Haynes BF, Tomaras GD. 2014. Capacity for infectious HIV-1 virion capture differs by envelope antibody specificity. J. Virol. 88:5165–5170. 10.1128/JVI.03765-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montefiori DC. 2009. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 485:395–405. 10.1007/978-1-59745-170-3_26 [DOI] [PubMed] [Google Scholar]
- 30.Trkola A, Matthews J, Gordon C, Ketas T, Moore JP. 1999. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. J. Virol. 73:8966–8974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehrnbecher T, Foster CB, Zhu S, Leitman SF, Goldin LR, Huppi K, Chanock SJ. 1999. Variant genotypes of the low-affinity Fcgamma receptors in two control populations and a review of low-affinity Fcgamma receptor polymorphisms in control and disease populations. Blood 94:4220–4232 [PubMed] [Google Scholar]
- 32.Chou T-C, Talalay P. 1984. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22:27–55. 10.1016/0065-2571(84)90007-4 [DOI] [PubMed] [Google Scholar]
- 33.Shaw GM, Hunter E. 2012. HIV transmission. Cold Spring Harb. Perspect. Med. 2:a006965. 10.1101/cshperspect.a006965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karasavvas N, Billings E, Rao M, Williams C, Zolla-Pazner S, Bailer RT, Koup RA, Madnote S, Arworn D, Shen X, Tomaras GD, Currier JR, Jiang M, Magaret C, Andrews C, Gottardo R, Gilbert P, Cardozo TJ, Rerks-Ngarm S, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Paris R, Greene K, Gao H, Gurunathan S, Tartaglia J, Sinangil F, Korber BT, Montefiori DC, Mascola JR, Robb ML, Haynes BF, Ngauy V, Michael NL, Kim JH, de Souza MS, MOPH TAVEG Collaboration. 2012. The Thai phase III HIV type 1 vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res. Hum. Retroviruses 28:1444–1457. 10.1089/aid.2012.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorny MK, Moore JP, Conley AJ, Karwowska S, Sodroski J, Williams C, Burda S, Boots LJ, Zolla-Pazner S. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J. Virol. 68:8312–8320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forthal DN, Landucci G, Gorny MK, Zolla-Pazner S, Robinson WE. 1995. Functional activities of 20 human immunodeficiency virus type 1 (HIV-1)-specific human monoclonal antibodies. AIDS Res. Hum. Retroviruses 11:1095–1099. 10.1089/aid.1995.11.1095 [DOI] [PubMed] [Google Scholar]
- 37.Smalls-Mantey A, Doria-Rose N, Klein R, Patamawenu A, Migueles SA, Ko S-Y, Hallahan CW, Wong H, Liu B, You L, Scheid J, Kappes JC, Ochsenbauer C, Nabel GJ, Mascola JR, Connors M. 2012. Antibody-dependent cellular cytotoxicity against primary HIV-infected CD4+ T cells is directly associated with the magnitude of surface IgG binding. J. Virol. 86:8672–8680. 10.1128/JVI.00287-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lux A, Yu X, Scanlan CN, Nimmerjahn F. 2013. Impact of immune complex size and glycosylation on IgG binding to human Fc Rs. J. Immunol. 190:4315–4323. 10.4049/jimmunol.1200501 [DOI] [PubMed] [Google Scholar]