Abstract
Given the unprecedented tools now available for rapidly comparing genomes, the identification and study of genetic and genomic changes unique to our species has accelerated, and we are entering a golden age of human evolutionary genomics. Here we provide an overview of these efforts, highlighting important recent discoveries, examples of the different types of human-specific genomic and genetic changes identified, and salient trends such as the localization of evolutionary adaptive changes to complex loci that are highly enriched for disease associations. Lastly, we discuss the remaining challenges, such as the incomplete nature of current genome sequence assemblies, and difficulties in linking human-specific genomic changes to human-specific phenotypic traits.
Introduction
Many phenotypic traits unique to the human lineage likely resulted from selective pressures on our genome and the unique demographic history since our divergence from the Pan lineage approximately 6 million years ago (mya) (Box 1). A fundamental question related to the origin of our species is which genomic sequences contributed to the unique evolutionary trajectory taken by the human lineage. With rapid advances in genomic technologies facilitating comparison of numerous genomes within and between species, we are in an unprecedented era of advancement in comparative genomics. With the availability of draft genomes for nine primate species including all of the “great apes” (chimpanzee, bonobo, gorilla, and orangutan), and the ability to rapidly sequence the genomes of multiple individuals within each species, we have never been in a better position to evaluate what genetic changes contributed towards making us human.
Box 1. Examples of human lineage specific traits and potential forces shaping them.
Human lineage specific (HLS) traits are phenotypes of the human lineage that arose since the split from the Pan lineage. A substantial number of forces likely contributed to the development and maintenance of these traits, and several examples are listed here. Plausible forces commonly discussed are macro- and micro-level climate changes that occurred frequently over the course of human evolution1, which may have selected for rapid HLS changes to survive novel climatic challenges. While an enhanced cognitive capacity would clearly be beneficial in dealing with such extreme and abrupt environmental changes, other important HLS phenotypic changes were also occurring. For example, anatomical and physiological changes associated with endurance running, such as HLS changes in the musculo-skeletal system and in energy utilization and metabolism, may have enabled novel hunting practices, e.g. persistence hunting, to emerge. These in turn allowed the energetic benefits of meat to be increasingly incorporated into the human dietary regimen2–4. An incomplete but representative list of traits identified as unique to the human lineage is shown, along with possible selective advantages2–7. A more complete list can be found in the Matrix of Comparative Anthropogeny (see Box 5).
| Phenotypic Feature (Examples) | Human Lineage Specific Trait | Possible Evolutionary Advantages |
|---|---|---|
| Brain growth trajectory | Prolonged postnatal brain growth and delayed myelinization period; Enhanced cognition | Allowed creation of novel solutions to survival threats, increased the critical period of learning new skills, and facilitated emergence of uniquely human cognitive skills |
| Brain size | Increased brain-body size ratio; Enhanced cognition | Allowed creation of novel solutions to survival threats and improved social cognition. |
| Descended Larynx | Portion of tongue resides in throat at level of pharynx. Larynx descended into throat | Helped enable spoken language |
| Eccrine sweat gland density | Higher density of eccrine glands; enhanced sweating capacity | Enhanced cooling ability; protection of heat-sensitive tissues (e.g. brain) against thermal stress. Facilitated endurance running. |
| Endurance running | Improved energy utilization during periods of high energy demand; Increased capacity to transfer energy (glycerol) from fat stores to muscle; Anatomical changes relating to running ability | Allowed persistence hunting to emerge as a viable strategy for accessing the benefits of increased meat consumption Increased range of food sources Improved diet may have facilitated brain evolution |
| Labor | Earlier onset and longer duration of labor | Partially protected child/mother from damage due to increased head circumference |
| Lacrimation | Emotional lacrimation (Crying) | Enhance emotional communication within social groups Increased affective communication |
| T cell function | Relative T cell hyperreactivity | Enhanced immune function |
| Thumb | Increased length, more distally placed, larger associated muscles | Created more detailed tools Manipulated objects on finer scale |
A major goal of studies that identify human lineage specific (HLS) genomic changes is to correlate genotype with phenotype; however, this task also remains most formidable. As direct human experimentation is not possible, researchers have historically relied upon naturally occurring variation and disease to understand HLS implications. A further limitation is the comparatively limited information about non-human primate phenotypes. This type of observational reliance frequently makes particular changes difficult to interpret. While variation and disease are still heavily relied upon, emergent technologies, like heterologous expression of human regulatory regions in mice8,9, are allowing for evolutionary hypotheses to be tested in ways previously not possible. Work in this field has significantly advanced in recent years, with the number of gene-to-phenotype candidates having more than doubled since the topic was previously covered10,11.
This article discusses current knowledge of the genetic and genomic changes that make Homo sapiens different from other primates, with particular emphasis on recent advancements. We explore genetic changes that may have contributed to human specific traits and where applicable the hypothesized evolutionary pressures, such as accelerated evolution (Box 2) and positive selection underlying these changes in human characteristics. We examine HLS genomic changes with a brief look at the technologies that made these discoveries possible and explore a representative group of HLS gene changes along with their associated phenotypes. We then address the growing number of HLS genetic and genomic changes connected to disease, including the correlation between complex loci and multiple disease associations. We conclude with a look at the future challenges in compiling a comprehensive list of HLS changes and their associated traits.
Box 2. Accelerated evolution.
There are a number of genomic regions that have undergone substantial alteration of sequence or re-arrangement in the human lineage. Accelerated evolution refers to situations in which sequence changes occur at a rate greater than the neutral mutation rate.
Accelerated evolution implies that the changes have been selected due to their advantageous nature and thus underwent rapid fixation. Identification of these regions relies on multiple methods and differs depending on whether the change is at the coding sequence, non-coding sequence, copy number or other structural level.
At the protein coding sequence level, a comparison of nonsynonymous (Ka) to synonymous (Ks) substitutions in sequence is often used. Most gene coding regions will have Ka/Ks ratios well below 1.0 due to the effects of purifying selection. In contrast, coding regions under positive selection will exhibit a higher frequency of nonsynonymous changes and, as a result, a higher Ka/Ks ratio. Via this method, studies have identified accelerated evolution in the human lineage of a number of genes, one example being genes involved in nervous system function12. However, these estimates can be confounded by gene conversion events that erase evidence of selection due to the creation of stretches of identical nucleotide sequences between homologous genes13.
Evaluation of non-coding sequences is not as straight forward due to difficulty in interpreting the significance of a change. Thus studies identifying regions of accelerated evolution in non-coding regions have relied upon looking for human-specific mutations in sequences highly conserved across mammals. An example of this is the identification of human accelerated region 1 forward (HAR1f)14. HAR1f is a non-coding RNA expressed in fetal brain that colocalizes with reelin, a protein important to cortical development.
Accelerated evolution may also occur at the level of whole genes or genomic regions in the form of copy number variations and structural rearrangements. Identification of such regions typically involves looking for HLS sequence copy number expansions and contractions which can range from as small as a few nucleotides to as large as segmental duplications identifiable by FISH. Unlike identification of accelerated evolution at the single nucleotide level, there are no rigorous statistical tests for these types of changes and are mainly observational.
Uniquely human genome changes
The ability to identify genomic changes unique to humans is dependent on the definition of human lineage-specific (HLS) events. In addition to the genome sequence of Homo sapiens, there are two ancient Hominin lineages, Neandertals15 and Denisovans16, for which draft genome sequences are currently available. As these two genome sequences are far less accurate and complete than the human assembly, this review will not rely on them unless the ancient hominin sequencing data for a particular sequence is of high quality. Our working definition of HLS therefore requires that changes found in the Homo sapiens genome be uniquely different from those found in other extant primates. As this review requires that the changes be well established as HLS and based on multiple out-group comparisons, it excludes studies reporting on human and chimpanzee differences alone.
Identification of HLS genomic changes can be complicated by several factors that can potentially occlude accurate comparisons of gene and genomic sequences. Gene annotation is often imprecise and can change between different genome builds, making it difficult to determine if a change is real or represents computational and/or assembly error. Sample size of individuals sequenced can also be an impediment as an apparent HLS change may only be polymorphic in the human population. In addition, the lack of sufficient individuals from the other primates sequenced can make the ancestral state difficult to determine. This is exacerbated by the fact that, with the possible exception of bonobos (P. paniscus), all great ape species harbor far more sequence diversity than humans17. Finally, it is important to have an effective number of primate outgroups to determine HLS status. For example, comparison of gene copy number changes between human and the great apes found that 57% of genes increased in copy number in human versus chimpanzee are not HLS18. With these criteria defined, we discuss examples of HLS genomic changes below.
Large scale changes
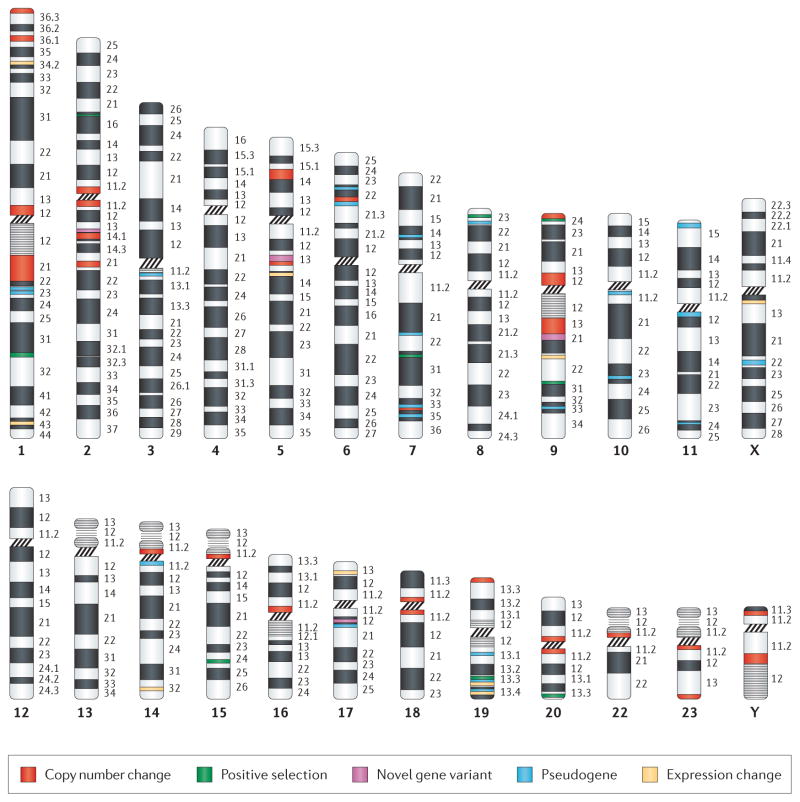
Large-scale genomic differences between humans and our closest primate relatives have been noted since the 1970s, when chromosomes were examined using chromatin-stained banding techniques19. Given technological limitations, observable differences were restricted to detection of: change in haploid number from 24 to 23 chromosomes due to the fusion of two ancestral ape chromosomes, resulting in humans having one large chromosome 2; the addition of human-specific constitutive heterochromatic C-bands on chromosomes 1, 9, 16, and Y; and, human-specific pericentric inversions on chromosomes 1 and 1819. While no conclusive evidence has as yet directly linked these cytogenetically visible events to HLS traits, the genomic regions where these events took place tend to be hotbeds of recent gene duplication, harboring many unique human-specific genes and copy number variations (CNV)3,18 (Box 3). This suggests that selection for these novel genes drove the changes to fixation, although drift cannot be formally excluded as an additional factor. Timing for these events continues to be of interest with a recent paper based on segmental duplications (SD) in chimpanzee and gorilla estimating that the chromosome 2 fusion event occurred ~4–5 mya26.
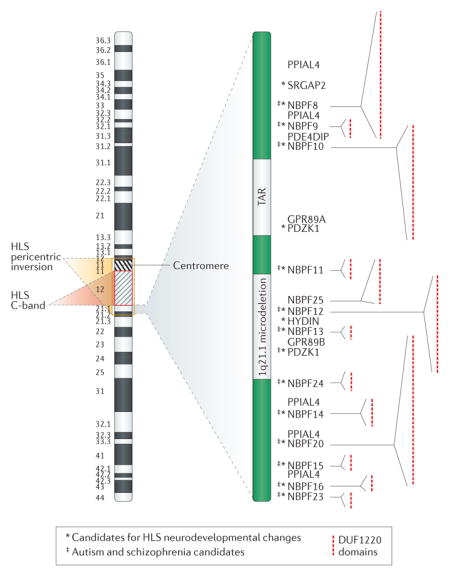
Box 3. 1q21.1 as an example of large scale to small scale genomic changes.
Large cytogenetically visible changes in genome structure were among the first human-specific genomic changes noted between human and great apes. More recently it has been determined that these regions harbor more significance than just being human-specific heterochromatin. Indeed such regions are frequently adjacent to regions greatly enriched for evolutionarily recent gene duplications, and which often function as gene nurseries3,18. For example, the 1q21.1 region of the genome, which lies adjacent to the human-specific 1q12 C-band and within the HLS chromosome 1 pericentric inversion, has undergone significant genomic enlargement due to numerous HLS copy number expansions within the region, as shown by the green bands in the following figure. Striking HLS copy number increases in DUF1220 domains (shown by the red dots in the figure) and the gene family that encodes them (NBPF)18, as well as duplicative transpositions of non-1q21.1 genes to the region, such as SRGAP220 and HYDIN21, have identified the region as being highly enriched for HLS copy number expansion. In addition, all of the above genes mentioned are candidates to underlie both HLS neurodevelopmental changes and cognitive disorders22–25.
In the late 1980’s application of fluorescent in situ hybridization (FISH) to banded chromosomes and its further use in FISH chromosome painting in the 1990s permitted the evaluation of large-scale structural changes between human and great apes not visible with conventional banding techniques27,28. More recently these studies were aided by interspecies BAC-based array comparative genomic hybridization (CGH) experiments which identified over 60 HLS SDs greater than 65 kb in size29,30. In addition, similar efforts identified large genome rearrangements such as those on nearly half of all human telomeres31.
Small scale changes
Small scale changes encompass all differences smaller than those identified by the large scale tests previously discussed, which have a resolution limit of ~20 kb. These include single base pair changes, insertions/deletions (indels) of varying size, and gene copy number differences. The differences can affect coding regions, non-coding regulatory regions, and repetitive sequence content. Strategies for identifying small scale changes often involve scanning the genome for signatures of positive selection when comparing human to non-human primates and rodents. While the results of these studies do not always coincide, there is substantial overlap in the phenotypes implicated, including taste and olfaction, immunity, signal transduction, lipid metabolism, chaperone activity, motor activity and structural support18,32–35,. However, it is likely that many important traits have yet to be identified given the large proportion of genes with no known function.
Initial estimates of sequence divergence between human and chimpanzee were ~1.2%32 based on the number of single nucleotide substitution differences between the two genomes. However these estimates did not account for regions unalignable between species due to structural divergences such as indels, highly duplicated sequences, and CNVs. While more recent divergence estimates reach as high as 5% when taking all types of variability into account36, high confidence divergence estimates remain elusive. For example, the incompleteness of other primate genome assemblies impedes accurate assessment of the uniquely human indel content, for which estimates range from 0.21% to 3%32,37. In addition, the importance of using multiple primate outgroups in making HLS assignments is borne out by recent studies of interspecific copy number variation. For example, these studies show a significant number of genes with elevated copy number across the three African great apes18,38, while the human copy number resembles that of orangutan and other primate lineages.
The first genome-wide, and first gene-based array CGH study comparing humans to all four great ape lineages identified 140 genes with HLS changes (134 and 6 genes that showed HLS copy number gain and loss, respectively)18. The majority of these HLS changes were confirmed after expanding the study to include 10 primate lineages3. Interestingly, these HLS changes showed strong positional biases, frequently clustering within the genome at pericentromeric, subtelomeric, and particularly complex, duplication-rich regions. Indeed, the greatest number of HLS gene copy number increases were found adjacent to the previously mentioned human-specific C-bands on chromosomes 1 and 918.
More recently, sequence read depth has been employed to estimate copy number from next-generation sequencing (NGS) platforms, providing a complement to array-based strategies39. Although NGS has confirmed the findings of many previous interspecific array CGH studies18, as well as identified new HLS candidates40, broad use has been slowed by the short read length capabilities of current NGS platforms (typical NGS read length is 50–150 bp). Short reads of highly duplicated sequences will often lack adjacent single copy sequences that serve to anchor the sequence read within the genome, making it difficult to accurately localize duplicated copies41,42. Utilization of NGS for sequencing of regions containing structural variations (e.g highly similar, duplicated sequences) has thus required implementation of novel bioinformatics methods, delaying their broad use within the field42. For a full review of the subjects see Treangen and Salzberg 201141.
Other major contributors to HLS genomic content are repetitive elements such as transposable element (TE) insertions, which constitute roughly half of the human genome. TEs comprise both DNA transposons and retrotransposons, with retrotransposons subdivided into long terminal repeat (LTR) containing and non-LTR elements. The former include endogenous retroviruses (ERVs) and recent hominid evolution has led to the accumulation of lineage specific subsets of ERV in great apes and humans43. While non-LTRs, such as L1s, Alus, and SVAs (hominid specific SINE-VNTR-Alu composite repetitive elements) encompass seventy-five percent of human repetitive content, it is difficult to determine what percent of these are HLS due to the incomplete nature of other primate genome sequence assemblies and their inability to accurately represent repetitive elements such as TEs44. Among those retrotransposons successfully identified as HLS, L1Hs-Ta1, the youngest of five HLS L1 subfamilies45, is of particular interest. At ~80–100 copies, L1Hs-Ta1 insertions are the only TEs still active within the human genome46. L1Hs-Ta1s are hypothesized to play a pivotal role in human neural plasticity as they are highly active during neurogenesis and contribute to neuron-specific genomic diversity47,48.
Taken together these studies establish that there are far more genomic differences between human and other primate genomes than was originally thought11. Nevertheless it is difficult to precisely estimate what fraction of the genome contains HLS sequences. While the human genome is the most complete and accurate mammalian genome sequence currently available, it still contains many sequence gaps in complex genomic regions, which may harbor important HLS genes49. Confounded by the far more incomplete nature of all other primate genomes, there are likely many HLS changes that have yet to be discovered.
Unique gene differences and associated traits
The number of identified HLS gene differences has rapidly increased in recent years, and this rate of discovery will likely continue in the future. While there is a lag in the number of HLS phenotypes that have been associated with these HLS genomic changes, there is a significant body of data attempting to link the two (See Table 1 for examples and Figure 1 for their genomic location). While roughly half of these associations relate to brain morphology and/or cognition, it is not known whether this tendency is a true representation of genetic change or reflects a bias in research focus. Other areas with a significant number of HLS changes are disease resistance and immunity, metabolism, physiological and anatomical differences, and changes in human reproduction and parturition. Some representative case studies of successful efforts for connecting HLS genotype to phenotype are presented in Box 4.
Table 1.
Partial list of genes and genetic elements showing HLS changes
| Gene/Element Name | Mechanism of Change | Proposed Phenotype | Relative Phenotypic Certainty | Possible Gene-Associated Disease | Refs |
|---|---|---|---|---|---|
| Androgen receptor (AR) | Deletion of regulatory DNA/Expression Change | Loss of sensory vibrissae and penile spines | Likely | Androgen insensitivity, hypospadias 1, spinal and bulbar muscular atrophy of Kennedy, prostate cancer susceptibility | 8 |
| Apolipoprotein C-I (APOC1) | Pseudogene | Unknown | N/A | Alzheimer’s severity, atherosclerosis and coronary heart disease | 50–53 |
| Aquaporin 7 (AQP7) | Copy number increase | Energy (glycerol) storage/transport/utilizat ion; increased sweating | Plausible | Nonfunctional glycerol response to exercise | 2, 3, 18, 40, 54–56 |
| Asp (abnormal spindle) homolog, microcephaly associated (ASPM) | Positive selection | Increased Brain Size | Plausible | Microcephaly | 57, 58 |
| CDK5 regulatory subunit associated protein 2 (CDK5RAP2) | Positive selection | Increased Brain Size | Plausible | Microcephaly | 58, 59 |
| Chemokine (C-C motif) ligand 3-like 1 (CCL3L1) | Novel gene variant | Immune function (chemokine for lymphocytes and macrophages) | Likely | HIV/AIDs susceptibility, Kawasaki Disease, Rheumatoid Arthritis, susceptibility to Chronic hepatitis C infection | 56 |
| Cholinergic receptor, muscarinic 3 (CHRM3) | Alternative splicing/Expression change/Novel exon | Change in human reproduction | Plausible | Eagle-Barrett Syndrome | 60 |
| Cholinergic receptor, nicotinic alpha 7 and FAM7A fusion (CHRFAM7A) | Copy number increase | Higher brain function | Plausible | P50 sensory gating deficit | 18, 40, 56, 61 |
| Creatine kinase brain (CKB) | Expression change | Metabolic changes in brain | Plausible | Multiple sclerosis | 4, 62 |
| Cytidine Monophosphate Acetylneuraminic acid hydroxylase (CMAH) | Pseudogene | Changed sialic acid composition on all cells in the body/Secondary effects in multiple systems | Definite | Duchenne muscular dystrophy, Red-meat related carcinoma risk. | 63, 64 |
| Cytochrome c oxidase subunit Va (COX5A) | Amino acid change/Positive Selection | Mitochondrial metabolism | Plausible | 65 | |
| Dopamine receptor D5 (DRD5) | Copy number increase | Regulation of mood, memory, learning, attention, movement | Likely | DRD5 deficiency, ADHD, primary cervical dystonia | 18, 56 |
| DUF1220/Neuroblastoma breakpoint factor (NBPF) family | Protein domain copy number increase (hyperamplification) | Brain size | Likely | Microcephaly, macrocephaly | 18, 23, 24, 40, 66. 67 |
| Fc fragment of IgG, high affinity 1a, receptor (FCGR1A) | Copy number increase | Immune function | Plausible | IgG receptor I phagocyte deficiency | 40, 56 |
| Follicle stimulating hormone receptor (FSHR) | Positive selection | Decreased gestation/Birth timing | Plausible | Amenorrhea, infertility, ovarian dysgenesis type 1 (premature ovarian failure), ovarian hyperstimulation syndrome | 68, 69 |
| Forkhead box P2 (FOXP2) | Amino acid change/Positive Selection | Language/speech development and increased length of dendrite spines | Definite | Speech-language disorder-1 | 70 |
| Forkhead box D4 (FOXD4) | Novel Gene Variant | Nervous system development | Plausible | Dilated cardiomyopathy, suicidality, OCD | 56 |
| General transcription factor 2-I repeat domain-containing protein 2 (GTF2IRD2) | Copy number increase | Unknown | N/A | Williams-Beuren syndrome | 18, 56 |
| Growth arrest and DNA-damage-inducible, gamma (GADD45G) | Deletion of regulatory DNA/Expression change | Expansion of human forebrain | Plausible | Thyroid carcinoma | 8 |
| Glutamate receptor, ionotropic, N-methyl-D-aspartate 3A (GRIN3A) | Amino acid change/Positive Selection | Learning and memory | Plausible | Unknown | 71 |
| Glutamate receptor, ionotropic, N-methyl-D-aspartate 3B (GRIN3B) | Amino acid change/Positive Selection | Higher brain function | Plausible | Unknown | 71 |
| Human accelerated conserved noncoding region 1 (HACNS1) | Positive Selection | Changes in development of anterior wrist and thumb | Likely | Unknown | 9 |
| Human accelerated region 1 forward (HAR1f) | Positive Selection | Development of neocortex | Plausible | Unknown | 14 |
| Mannose receptor C, type 1 (MRC1) | Novel gene variant | Recovery from inflammation, regulator of glycoprotein homeostasis | Plausible | Leprosy manifestation | 56 |
| Microcephalin 1 (MCPH1) | Positive Selection | Brain Size | Plausible | Microcephaly | 58, 72 |
| Myosin heavy chain 16 (MYH16) | Pseudogene | Cranio-facial musculature and morphology | Plausible | Unknown | 73 |
| Neutrophil cytosolic factor I (NCFI) | Copy number increase | Phagocyte generation of superoxides | Likely | Chronic granulomatous disease, Williams-Beuren syndrome | 56 |
| NLR family, apoptosis inhibitory protein (NAIP) | Copy number increase | Inhibition of apoptosis | Likely | Spinal muscular atrophy | 18, 40, 56 |
| Occludin (OCLN) | Copy number increase | Regulation of TGF-beta, cell migration | Likely | Susceptibility to hepatitis C, Band-like calcification with simplified gyration and polymicrogyria | 18, 56, 74 |
| p21 protein (Cdc42/Rac)-activated kinase 2 (PAK2) | Copy number increase | Neuronal differentiation | Plausible | 3q29 microdeletion syndrome | 18, 75 |
| Peripheral myelin protein 2 (PMP2) | Copy number increase | Myelin stabilization/Protection from demyelination | Plausible | Charcot-Marie-Tooth peroneal muscular atrophy | 18, 76 |
| Phosphodiesterase 4D interacting protein (PDE4DIP) | Copy number increase | Higher brain function | Plausible | Myeloproliferative disorder associated with eosinophilia | 18, 40, 77 |
| Protocadherin 11 X Y linked (PCDH11XY) | Copy number increase/Expression change | Cerebral asymmetry/Language development, neuroendocrine transdifferentiation | Likely | Klinefelter’s syndrome, Alzheimer’s disease, prostate cancer | 18, 78–82 |
| Sialic acid-binding Ig superfamily lectin 5 (SIGLEC5) | Expression change | T-cell hyperactivation due to lowered expression | Likely | Susceptibility to T-cell mediated disease | 64, 83 |
| Sialic acid-binding Ig superfamily lectin 6 (SIGLEC6) | Expression change | Prolonged labor | Plausible | Preeclampsia | 84 |
| Sialic acid-binding Ig superfamily lectin 11 (SIGLEC11) | Gene conversion/Expression change | Alleviation of neurotoxicity from activated microglia. Potential neurotrophic effects. | Likely | Unknown | 85, 86 |
| Sialic acid-binding Ig superfamily lectin 13 (SIGLEC13) | Gene loss | Disease resistance to Sialylated Bacteria | Likely | Unknown | 87 |
| Solute carrier family 2 (facilitated glucose transporter) member 1 (SLC2A1) | Expression change | Metabolic changes in brain and skeletal muscle/Brain size | Plausible | GLUT1 deficiency syndrome 1 and 2, susceptibility to HTLV infection | 88 |
| Solute carrier family 2 (facilitated glucose transporter) member 4 (SLC2A4) | Expression change | Metabolic changes in brain and skeletal muscle/Brain size | Plausible | Noninsulin-dependent diabetes mellitus | 88 |
| Solute carrier family 6 (facilitated glucose transporter) member 13 (SLC6A13) | Copy number increase | Higher brain function | Plausible | Unknown | 18, 89 |
| Survival of motor neurone2, centromeric (SMN2) | Novel gene variant | Motor neuron maintenance, neuronal growth | Likely | Spinal muscular atrophy severity | 18, 56 |
| SLIT-ROBO Rho GTPase activating protein 2 (SRGAP2) | Copy number increase | Increased neuronal branching | Likely | Early infantile epileptic encephalopathy | 18, 20, 25, 40 |
| Sperm protein associated with the nucleus, X-linked family members A/D (SPANXA/D) | Copy number increase | Post-meiotic spermatogenesis | Likely | Unknown | 90, 91 |
| Thrombospondin 4 (THBS4) | Expression change | Synaptic organization and plasticity | Plausible | Familial premature coronary heart disease | 92 |
Figure 1.
Genome positions of HLS gene changes
Box 4. Interpreting HLS change: FOXP2 and HACNS1 as case studies.
In spite of the substantial difficulties involved in determining the function of HLS genetic and genomic changes, a number of encouraging studies have been reported that may serve as models in this challenging arena. One example involved the use of humanized transgenic mice to show that a gain of function HLS phenotype was produced via amino acid change in the forkhead box P2 (FOXP2) gene70. FOXP2 has been implicated in human speech production based primarily on mutations identified in a family with severe speech disabilities94. Further analyses of the gene identified two HLS amino acid substitutions with evidence of positive selection95,96. To investigate the phenotypic change resulting from the two amino acid differences, mice with humanized FOXP2 were generated70, which showed increased neuronal dendritic length and synaptic plasticity and changes in ultrasonic vocalization. These results provide support that the two substitutions in this transcription factor could have affected human speech production capabilities.
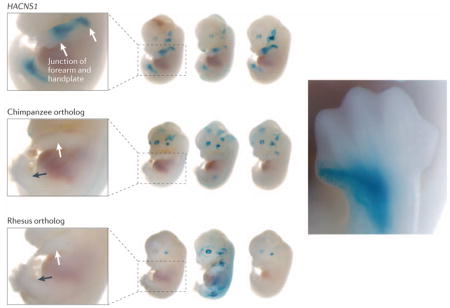
Research examining the function of HACNS1, an enhancer that has undergone accelerated evolution in the human lineage, followed a successful strategy using a series of expression assays driven by the non-coding sequences in mouse embryos9. Mouse embryos injected with a reporter gene construct, driven by the human version of the 546 bp homologous HACNS1 enhancer regions from human, chimpanzee, and macaque showed expression in the anterior developing forelimb and hindlimb, particularly in the forearm, handplate, anterior-most digit, and the corresponding structures in the hindlimb (see figure below). Neither the chimpanzee nor the macaque constructs showed this pattern, suggesting that the region may have played an important role in HLS morphological changes to the hands and feet and making HACNS1 an important candidate for contributions to human bipedalism and tool making. Additional work on chimeric constructs then narrowed the expression pattern change to 13 divergent bases within an 80 bp region, establishing a narrow window for future investigations.
These represent only a sampling of discovered HLS genes and their associated traits; a more comprehensive list can be found in the Genetics and Genomics Domains at the Center for Academic Research and Training in Anthropogeny (CARTA) Matrix of Comparative Anthropogeny (MOCA) resource on human origins. MOCA also includes the ability to link these genetic and genomic changes to many other features of human uniqueness, in domains ranging from molecules to culture (see Box 5).
Box 5. The Matrix of Comparative Anthropogeny (http://carta.anthropogeny.org/moca/domains).
The Matrix of Comparative Anthropogeny (MOCA) of the Center for Academic Training and Research in Anthropogeny (http://http://carta.anthropogeny.org/) is a web-based collection of information comparing humans and our closest evolutionary relatives (chimpanzees, bonobos, gorillas and orangutans i.e, the “great apes”), with a specific emphasis on uniquely human features. Comparisons of these non-human hominids with humans are difficult, as so little is currently known about their phenotypic features (“phenomes”), in contrast to humans30. Ethical, fiscal and practical issues also limit the collection of further information about great apes. MOCA attempts to collect existing information about HLS differences from great apes that is currently scattered in the literature. Having such information in one location could lead to new insights and multi-disciplinary interactions, and to ethically-sound studies for explaining these differences, as well as for understanding of uniquely human specializations. It is for this reason that MOCA is called a “matrix” -- i.e. an arrangement of information from which something else originates, develops, or takes form. MOCA is organized by Domains, each grouping Topics by areas of interest and scientific discipline. MOCA is a work in progress and each topic entry will eventually cover existing information about a particular difference (alleged or documented) between humans and non-human hominids. Topics are also linked across domains, e.g. each genetic HLS topic is linked to phenotypic traits including anatomy, physiology, behavior and even culture to the extent possible.
MOCA is not targeted at experts in specific disciplines, but rather aims to communicate basic information to a broad audience of scientists from many backgrounds, and to the interested lay public. MOCA includes not only aspects wherein there are known or apparent differences between humans and “great apes”, but additionally, topics for which popular wisdom about claimed or assumed differences is not entirely correct.
The MOCA site is being launched at an early stage, so that interested readers with expertise on specific topics can provide feedback. New information and topics will continue to be added as they are identified or discovered.
HLS gene changes discussed in this paper are displayed in their corresponding genomic position across the human karyotype. The changes are divided into five categories that correspond to those listed in Table 1, and each type is color coded. It should be noted that many genes have undergone multiple types of HLS changes, and in this case only one type is shown. For visualization purposes the size of the colored bands are not drawn to scale.
Genes that have associated HLS traits are listed with an assigned level of certainty with regard to their impact on human uniqueness. Certainty ranges from plausible (the association is still hypothetical based upon what is known about the gene), to likely (may have a disease association or animal model evidence to substantiate the claim), or definite (requires multiple lines of supporting evidence). In cases where the gene has been implicated in a disease but a HLS phenotype has not been proposed the certainty column is not applicable and thus N/A is listed. In addition associated disease links are given if known. Disease phenotypes are listed partly based on information in the Online Mendelian Inheritance of Man database93. Details regarding these genes, and additional examples of genes associated with HLS traits can be found in the Genetics Domain of the Matrix of Comparative Anthropogeny (MOCA) (Box 5).
Alteration of gene structure resulting from splicing
Alteration of gene structure provides a major mechanism through which evolutionary adaptive changes can be introduced. A common means of modifying gene structure is by alterations in transcriptional splicing. Several studies have identified genes differentially spliced in the human lineage, including differential expression in the brain97 and significant numbers of genes involved in metabolism and morphological development98. A recent survey of human-specific transcript variants found 112 genes showing differential HLS transcripts as the result of novel promoters, exons, and splicing sites, most of which are the result of TE insertions99. One of the best characterized is the cholinergic receptor, muscarinic 3 (CHRM3), a G protein coupled receptor mediating multiple physiological functions through its control of smooth muscle contraction60. In humans, an L1HS transposon insertion occurred at the 5′ end of the gene, resulting in a novel first exon, promoter, and transcript60,99. The L1HS derived transcript is the only CHRM3 transcript expressed in placenta and could be important for human gestation60.
Alteration of gene structure by protein domain amplification
Another means of gene structure alteration is changes in the number of protein domains within a gene. While the importance of gene duplication to evolutionary change has been emphasized since 1970100, an appreciation for the contributions of protein domain amplification, the process by which a protein domain undergoes a copy number increase, has only recently emerged101. The most striking example of this process has been reported for the DUF1220 protein domain, which shows the largest HLS copy number increase of any protein coding region in the human genome66,67. DUF1220 domains are encoded within the neuroblastoma breakpoint family (NBPF) of genes, and while there are several HLS NBPF genes found in the human genome, the great majority of HLS copies of DUF1220 have arisen by intragenic domain hyper-amplification3,18,67. Humans (272 copies) have more than twice the copy number of chimpanzee (126), the next highest number, while mouse and rat have only 1 copy. It is estimated that, on average, 28 additional copies of DUF1220 have been added specifically to the human genome every million years since the Human and Pan lineages diverged67. Recent correlative data from evolutionary studies and studies of brain size in normal and pathological populations (microcephaly/macrocephaly) support the view that DUF1220 copy number is a general effector of brain size, and may be largely responsible for the dramatic evolutionary expansion in brain size that occurred in the human lineage23,24.
Alteration of gene structure by amino acid change
Gene structure can also be modified by smaller local alterations that result in changes to the amino acid sequence. There are numerous examples of accelerated genome evolution that have been linked to amino acid change34,35 (see Box 2). One gene that may have ramifications for HLS metabolic changes is cytochrome c oxidase subunit Va (COX5A), the fifth of ten nuclear encoded subunits that make up the terminal proteins in the mitochondrial electron transport chain102. The genes encoding these subunits are generally highly conserved but several show an increased rate of non-synonymous substitutions within the anthropoid primate lineages, with COX5A containing two HLS amino acid changes103. While a complete understanding of COX5A function is lacking, its interaction with thyroid hormone T2 suggests that the changes are important in regulating fat metabolism65,104. A second example of functionally significant HLS amino acid change is provided by the Forkhead box P2 (FOXP2) gene that is proposed to have impacted human speech development70 (see Box 4 for more details).
Alteration of gene function by pseudogenisation
Not all gene alterations generate functional variants, and indeed such structural changes often produce non-functional genes. This is the case with pseudogenization, where a sequence alteration renders the gene inactive although the majority of the gene remains intact within the genome. A recent analysis using updated sequencing data, found 38 fixed HLS pseudogenes in the human genome105, with only nine of these being fixed single copy genes. No excess of pseudogene fixation over expected rates was detected (i.e., no evidence in support of the “less is more” hypothesis of Olson106). One noteable pseudogene is apolipoprotein C-I (APOC1), involved in lipoprotein metabolism107. While great apes have two APOC1 genes, encoding negatively and positively charged forms of the protein, a premature stop codon in the gene encoding the negatively charged protein resulted in humans only having the positive APOC1 protein50. While implications of this loss are not currently understood they may be related to human health. Human disease and mouse model studies indicate that polymorphisms in APOC1 are risk factors for more severe forms of Alzheimer’s disease51,108 and for developing atherosclerosis and coronary heart disease52,53. As these diseases appear to be unusually common and severe in humans, it is plausible that this gene loss could be a contributing factor109,110. Other examples of confirmed pseudogenes are cytidine monophospho-N-acetylneuraminic acid hydroxyase (CMAH) and myosin heavy chain 16 (MYH16). The former pseudogenization is responsible for inactivation of biosynthesis of the sialic acid N-glycolylneuraminic and led to a radical reconfiguration of human cell surfaces by changing millions of molecules on the surface of human cells, with major consequences for human specific innate immunity and other systemic roles of sialic acid biology63,64. A further example is the pseudogenization of a myosin filament, one of the basic units of muscle, specifically expressed in primate jaws and thus may have altered HLS cranio-facial musculature and morphology73.
Pseudogenization is also a common trend seen in HLS multigene families. This mechanism is often found in genes coding for proteins of neural sensation such as those for olfaction. Within the olfactory receptor family, over 60% of the genes within the family have been rendered nonfunctional by pseudogenization in the human lineage although some increases in copy number have also been noted, specifically in the OR-A gene105.
Alteration of gene function by gene conversion
Many gene structural changes occur as the result of misalignments during replication. One such example of this is gene conversion, where a portion of one gene is “pasted” onto another gene, and often occurs between members of genes within the same family. While gene conversion events usually lead to pseudogenization, there are cases where the conversion is functionally significant. For example the gene Sialic acid-binding Ig superfamily lectin-11 (SIGLEC11) encoding, a member of a family of cell surface membrane proteins involved in modulating immunity underwent two tandem HLS gene conversion events with an adjacent pseudogene, resulting in it acquiring a novel promoter and N-terminal protein sequences85. This led to a change in the binding specificity of Siglec-11, with recognition of novel ligands and initiation of its expression in microglia, the cells responsible for immune defense and neuroprotection in the central nervous system. Siglec-11 expression alleviates neurotoxicity of microglial cells, which can damage neurons and contribute to neurodegenerative disorders86. More recently, a unique expression pattern of Siglec-11 and its ligands in human ovaries has demonstrated a possible role in HLS ovarian changes111.
Alteration of gene function by changes in gene family size
Gene family size can be altered through the addition of new copies of a gene already present in the lineage. Such duplicates, when functional, can simply confer an increase in dosage or can diverge to take on potentially new functions. The NBPF gene family encodes DUF1220 protein domains, as discussed above, which are thought to play a role in brain size and cognition23,24. This family has undergone both HLS gene copy number expansion, adding an estimated four new human gene copies, and HLS domain copy number hyper-amplification, adding over 160 copies specifically to the human genome67. Another HLS multigene family is the double homeobox (DUX) family, which includes a number of genes involved in transcriptional regulation and embryogenesis. In humans, three of the DUX genes on chromosome Y (DUXY2-4) have undergone neofunctionalization through removal of an ancestral stop codon, although the functional significance of this mutation has yet to be determined112.
Gene families also undergo inactivation events with relative frequency although whole gene deletion is generally rare. A recent example of this kind of loss is Siglec-13113. The SIGLEC13 locus underwent a HLS whole gene deletion mediated by an Alu recombination event87. Expression of the chimpanzee form of Siglec-13 on monocytes affects inflammatory cytokine secretion and sialic acid binding, potentially enhancing susceptibility to infection by sialylated bacterial pathogens. It is hypothesized that SIGLEC13 loss may have been selected in relation to the bottleneck at the origin of modern humans, as it improved fitness in infants who would otherwise be susceptible to these bacteria87.
Copy number change
While the previously mentioned DUF1220 domain sequences show an extreme HLS copy number increase, many additional HLS copy number changes exist and make up a significant proportion of the differences between human and great ape genomes3,18,40. One example is SLIT-ROBO Rho GTPase activating protein 2 (SRGAP2), which has at least one fixed HLS partial duplication20,40. SRGAP2 is a negative regulator of neuronal migration and promotes neurite outgrowth25. It is hypothesized that the partially duplicated protein dimerizes with the full length SRGAP2 protein acting as a dominant partial inhibitor, which presumably leads to neotenous changes including increased density of longer neurite spines25. Another identified HLS copy number increase involves the aquaporin 7 gene (AQP7)3, of which there are several additional copies in the human genome. AQP7 is involved in the transport of water and glycerol, and is responsible for utilization of glycerol (energy) from fat cells especially during fasting and prolonged human exercise54,55. AQP7 amplification is hypothesized to be adaptive for metabolic needs in human endurance running and possibly thermoregulation via increased sweating2,3. Such activities have been proposed to be critical to human’s exceptional persistence hunting capabilities and establishment of humans as diurnal endurance predators3.
The creation of de novo human genes
While copy number change is established as a mechanism resulting in new gene function, it acts on pre-existing genes. In contrast recent evidence suggests that several novel genes appeared in the human genome de novo from previously non-coding DNA. Although once thought to be an extremely rare event in genome evolution, de novo gene generation has become a subject of considerable interest and debate. Three published papers have claimed the identification of HLS de novo genes114–116. None have functional assignments, but this is not surprising given the novelty of the genes identified. However, one gene, CLLU1 (chronic lymphocytic leukemia up-regulated 1), is found to be up-regulated in patients with a particularly aggressive form of chronic lymphocytic leukemia117, lending credence to the claim that the regions identified are actually functional.
The HLS de novo gene publications are the subject of much controversy centered around two key issues. The first relates to the fundamental definition of what is de novo. The Wu et al. dataset allowed human genes with up to 20% of their coding region homologous to a predicted open reading frame in other primates to still be considered de novo, whereas the Knowles and McLysaght and Li et al. papers used stricter criteria. This has generated deliberation as to whether a gene that shares this much of its coding region can truly by called de novo. The other key issue relates to changes in gene annotation. The original set of genes identified in Wu et al. did not include the three genes identified in the Knowles and McLysaght paper, as the human genome build no longer lists them as annotated genes, casting doubt on whether the genes identified in these studies are real. At present it appears that de novo genes may have contributed HLS genes, but to what extent remains to be determined.
Expression changes
Alteration to gene expression is a common mode of evolutionary change and can result from multiple changes at the genetic level, such as changes in regulatory DNA affecting promoters, enhancers, and suppressors, and dosage changes resulting from CNVs. These types of alterations may change gene amounts, timing, or even what tissues gene expression occurs. Advancements in identifying gene expression changes using RNA sequencing technologies are the subject of a recent review by Romero et al. however we will highlight a few examples117.
Identified as a HLS deletion in a bioinformatics survey of regions highly conserved between chimpanzee and macaque, a 60.7 kb deletion upstream of the androgen receptor’s (AR) removed a regulatory region leading to the loss of expression of AR in sensory vibrissae and penile spines8. Expression constructs in embryonic mice and in human foreskin fibroblasts showed the corresponding non-deleted region from chimpanzee controls AR expression. Humans lack sensory vibrissae and penile spines, both of which are found in our closest ape relatives. Such anatomical differences between humans and the great apes lend additional support to the validity of these studies. Another example of a regulatory change is nucleotide changes in the HACNS1 enhancer that may have led to HLS changes in digit and limb development9 (see Box 4).
Copy number changes may alter expression through gene dosage changes like the duplication of Protocadherin 11 from the X chromosome to the Y chromosome (PCDH11XY)78,118. This duplication doubled the gene dosage in humans as genes present on both the X and Y are protected from X-gene inactivation78,79. This change is hypothesized to contribute to cerebral asymmetry and language development79, a claim corroborated by disease findings of severe language impairment associated with PCDH11XY loss80.
Other significant examples of expression change are loss of expression of SIGLEC5 from human T-cells plausibly resulting in T-cell hyperactivation83, and uniquely human SIGLEC6 expression in the placenta84, hypothesized to be implicated in the human specific disease, preeclampsia119. The specific genomic alterations responsible for these expression changes remain unknown.
HLS changes linked with human disease
Human disease has been, and continues to be, one of the few ways to highlight the phenotypic implications of many HLS genetic changes68,70. While the link between human disease and HLS genomic changes has been the subject of recent reviews560,11, an appreciation of this connection has only begun to emerge within the last decade120,121.
Demonstration of causality versus association is difficult, but it is increasingly clear that many regions undergoing HLS change play an important role in human disease (Box 1). Selection on numerous genes involved in innate immunity improved human resistance to particular diseases but apparently did so at the cost of other human-specific impairments, such as an increased propensity for autoimmune disorders including atopic diseases and allergies120. Increased immune response may have arisen during the unique changes to HLS pathogen regimes associated with use of home bases, scavenging or hunting of different prey, and more extensive inter-group contacts. An example of association between improved immunity and autoimmune disorders is seen in the case of the SIGLEC gene family. The levels of many Siglecs are decreased in humans; as most of these have an inhibitory effect on lymphocyte activation, lower Siglec expression leads to increased lymphocyte reactivity64. This hyper-reactivity, while potentially protective against infection, may predispose to autoimmunity. In addition, a number of HLS changes linked to human specific cognitive abilities are also associated with severity of Alzheimer’s disease and dementias51,108. For example, FLJ33706115, a novel human gene highly expressed in the cortex, cerebellum and midbrain, has been reported to show increased expression in Alzheimers brain specimens. The Asp (abnormal spindle) homolog, microcephaly (ASPM) gene, a microcephaly disease gene57,58, reportedly underwent accelerated evolution in the human lineage, although the certainty of this is contested in recent literature123. Thus there are multiple cases in which a region found to have undergone HLS selection is also associated with one or more major disease phenotypes.
Beyond natural selection, unstable genomic architecture is another driving force in human genetic novelty associated with human disease. Certain regions of the genome that are complex and repeat-rich often act as gene nurseries3,17 and might therefore be evolutionarily advantageous to maintain in the population. However, the instability associated with the architectures of the same genomic regions also makes them prone to deleterious copy number gains and losses. Such regions have been linked to numerous genomic disorders, including several neuropsychiatric and neurodegenerative diseases42,124 (Box 3). For example, the 1q21.1 region of the human genome has undergone multiple HLS expansions, including those involving the aforementioned DUF1220 protein domain17,67 and SRGAP2 gene20, two candidates to underlie human brain morphological changes. CNVs in the region have also been implicated in a striking number of recurrent diseases22,24 Therefore, as the study of disease continues to be a resource for understanding the function of HLS genomic change, the identification of HLS changes provides a source of candidate genetic changes contributing to human disease.
Conclusions and prospects
With continued improvements in DNA sequencing technologies the number of new genome sequences available will likely continue to grow exponentially, and this can be expected to further clarify which changes are truly human-lineage specific. However, unless there is a focus on developing technologies to accurately sequence and assemble complex, duplication-rich, genomic regions, such regions will continue to be woefully under-examined. For example recent publication of the last two great apes to be sequenced, bonobo and gorilla, were both sequenced using whole genome shotgun methods that relied heavily on NGS technologies125,126. These methods generate poor coverage and assembly of duplication-rich regions, and therefore are deficient in sequences that are particularly relevant to the identification of genes showing HLS changes in copy number127.
Obtaining accurate sequence from complex genomic regions is important for several reasons: they have been linked to numerous diseases and disorders42,120; they are often sites of rapid evolutionary change18,127; and, they have been proposed as candidates to harbor the “missing genetic heritability” that has eluded many genome-wide disease gene investigations49. Finally, the importance of examining complex genomic regions has been recently borne out by a study of the human-specific SRGAP2 gene duplications. These extra copies were only discovered through the development of a new genome assembly utilizing a haploid human genome resource (hydatidiform mole) and long read sequencing to properly sequence repetitive content20.
Another area of improvement needed is exemplified in de novo sequencing efforts114–116. Inability to verify the results of prior studies due to differences between genome builds only epitomizes the need for solid manual annotation of genes in all genomes. Inconsistencies in the annotation of the human genome, which has the most extensive annotation work of all the mammalian genomes, can lead to reduced confidence in gene calls within other primate genomes. Conclusive identification of a gene’s HLS status requires the availability of accurate genome assemblies from multiple individuals within a lineage, combined with comparison to multiple individuals in multiple primate outgroups.
Despite the difficulties involved, it is reasonable to expect that the process of linking human-specific gene and genomic features to important human-specific traits will accelerate in the next several years. However, challenges in the field continue, such as the very limited data existing on great ape phenotypes as compared to humans128, and the major impact of gene-culture interactions in generating the human phenotype10. While keeping ethical considerations foremost in mind, it is important to gather as much novel information on the phenotypes of these species as possible, an objective that would be much advanced by providing proper medical care to captive great apes in the US and in ape sanctuaries throughout Africa129. Improving transgenic technology in mice potentially affords more functional insights than disease associations alone and, as such, may prove to be a valuable strategy for verifying many HLS traits.
One may anticipate that the identification of key genes and genomic sequences that are responsible for the unusual capacities of human cognition and physiology may lay the groundwork for new fields of study. For example, a new field of neuroscience, evolutionary neurogenomics, may emerge from the application of genomic technologies to the study of human brain evolution11. By providing a novel, biologically unbiased, entry point into the study of the brain, such a discipline may uncover new insights regarding human cognitive function and dysfunction (Box 3). Many of the genetic changes underlying the dramatic change in cognitive capacities in the human lineage are likely to be part of evolutionary trade-offs, where increased mental capacities have come at a cost to other organismal functions. Thus, genotype-to-phenotype associations are likely to have medical relevance as well, showing how evolutionary selective pressures may have produced side effects such as unstable genomic regions enriched for disease-associated variants (e.g. 1q21, 5q13)18,42,130. Finally we can anticipate that the use of genome-based data to help decipher what has made the human species unique will become an increasingly powerful tool for understanding our evolutionary origins (anthropogeny) and the human condition.
Acknowledgments
We would like to thank Sean O’Bleness for editorial comments, Michael Dickens for graphics assistance, and James Noonan for access to published images. We also thank the many student and faculty contributors to the MOCA web site. Work in our laboratories has been supported by the National Institutes of Health and by the Mathers Foundation of New York, which also supports the MOCA web site.
Glossary Terms
- Accelerated Evolution
More nucleotide or copy number changes within a particular region or gene than would be expected from background rates of mutation over time (e.g., COX5A)
- Alternative Splicing
Genetic change that leads to HLS modification of RNA (e.g., CHRM3)
- Amino Acid Change
A DNA change that leads to a change at the protein sequence level (e.g., GRIN3A)
- Array-based Comparative Genomic Hybridization (array CGH)
a microarray-based method for detecting copy number variation in the genome
- Copy Number Change
Increases or decreases in the number of copies of a gene or segment (e.g., SRGAP2)
- De Novo Human Gene
Novel gene arising from formerly non-coding DNA (e.g. CLLU1)
- Domain Amplification
Intragenic copy number increase of a protein domain (e.g., DUF1220)
- Exon Deletion
Loss of one or more exons from a gene (e.g., ELN)
- Expression Pattern Change
Change in timing, level, and/or location of gene expression (e.g., PCDH11XY)
- Fluorescent in Situ Hybridization (FISH)
A technique used to visualize the location of specific DNA sequences on chromosomes
- Gene Conversion
“Pasting” of identity from one homologous gene to another (e.g., SIGLEC11)
- Gene Nursery
A dynamic region of the genome that is capable of undergoing rapid evolutionary change due to a duplication-prone genome architecture, and therefore is a frequent site of production of novel genes via gene duplication
- Human-Specific Disease
Disease that is present only in the human lineage. A number of diseases are thought to be human specific (Alzheimers, myocardial infarction), but proving that such diseases are not present in other species remains a challenging task
- Hydatidiform mole
An abnormal form of pregnancy where a non-viable egg, likely the result of an egg missing a nucleus, is fertilized and becomes a mass on the uterine wall. The resultant growing tissue is haploid in nature due to it only having a paternal genetic contribution
- “Less-is-More” Hypothesis
The hypothesis that gene loss plays a major role in evolution
- Neofunctionalization
A process in which a genetic change in an allele produces a novel protein function (e.g., DUX)
- Polymorphism
Allelic genetic variation within a species (e.g., AMY1)
- Protein Domain
A discrete portion of a protein sequence that is may evolve and function independently of the rest of the protein (e.g., DUF1220)
- Pseudogenization
Loss of gene function while the majority of the gene is retained (e.g., APOC1)
Biographies
Majesta O’Bleness Majesta O’Bleness is a Postdoctoral fellow at the University of Colorado School of Medicine. She conducted her PhD work at the Advanced Center for Genome Technology at the University of Oklahoma. Her current research efforts focus on genomic investigations of genes involved in human cognition, with particular attention on those genes undergoing copy number variation.
Veronica Searles Veronica Searles is pursuing an MD/PhD joint degree at the University of Colorado’s Medical Scientist Training Program. Her previous research includes studying psychiatric genetics at Brown University and population and evolutionary genetics at Stanford University. Recently she has conducted research investigating the cellular role of DUF1220 and examining potential environmental predictors of suicide.
Pascal Gagneux Pascal Gagneux is Associate Professor of Cellular and Molecular Medicine at the University of California San Diego, and Associate Director of the Center for Academic Research and Training in Anthropogeny (CARTA). His group has recently shown that glycan polymorphisms due to loss-of function mutations initially selected by pathogens, can come under sexual selection by female anti-glycan immunity and lead to reproductive incompatibility.
Ajit Varki Ajit Varki is Distinguished Professor at UCSD, co-director of the Glycobiology Research and Training Center, founder and co-director of the UCSD/Salk Center for Academic Research and Training in Anthropogeny (CARTA), and member of the American Academy of Arts and Sciences and the Institute of Medicine. His research focuses on uniquely human aspects of sialic acid biology.
James Sikela Dr. Sikela is a Professor at the University of Colorado School of Medicine. He made several major contributions to the Human Genome project and the area of human evolutionary genomics. His recent work focuses on the extreme human-specific copy number increase of DUF1220 protein domains and their relevance to human brain evolution.
Footnotes
Competing Interest Statement
JMS is founder and shareholder in GATC Science, LLC. AV is Co-Founder and shareholder in Sialix, Inc.
References
- 1.Gamble C, Davies W, Pettitt P, Richards M. Climate change and evolving human diversity in Europe during the last glacial. Philos Trans R Soc Lond B Biol Sci. 2004;359:243–253. doi: 10.1098/rstb.2003.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bramble DM, Lieberman DE. Endurance running and the evolution of Homo. Nature. 2004;432:345–352. doi: 10.1038/nature03052. Provides a well-reasoned argument, based on anatomical and physiological evidence, that endurance running played a major role in human evolution, at the origin of the genus Homo. [DOI] [PubMed] [Google Scholar]
- 3.Dumas L, et al. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17:1266–1277. doi: 10.1101/gr.6557307. The most extensive arrayCGH investigation to date of gene-based copy number change across primate species. Many of the >4,000 genes identified that showed lineage-specific changes in copy number are excellent candidates to underlie lineage-specific traits among these species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfefferle AD, et al. Comparative expression analysis of the phosphocreatine circuit in extant primates: Implications for human brain evolution. J Hum Evol. 2011;60:205–12. doi: 10.1016/j.jhevol.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin RD. The evolution of human reproduction: a primatological perspective. Am J Phys Anthropol Suppl. 2007;45:59–84. doi: 10.1002/ajpa.20734. [DOI] [PubMed] [Google Scholar]
- 6.Fooladi MM. The healing effects of crying. Holist Nurs Pract. 2005;19:248. doi: 10.1097/00004650-200511000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, et al. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 2012;22:611–622. doi: 10.1101/gr.127324.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLean CY, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prabhakar S, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. Demonstrates how a conserved noncoding sequence (HACNS1), which evolved extremely rapidly in humans, may underlie human-specific aspects of limb (hand/foot) development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varki A, Geschwind D, Eichler E. Explaining human uniqueness: genome interactions with environment, behavior, and culture. Nat Rev Genet. 2008;9:749–763. doi: 10.1038/nrg2428. An earlier review on the same subject cautions against a “gene-centric” view of human evolution, and suggests that some aspects of human genome evolution may be due to relaxed selection, resulting from masking by behavior and culture. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sikela JM. The jewels of our genome: the search for the genomic changes underlying the evolutionarily unique capacities of the human brain. PLoS Genet. 2006;2:e80. doi: 10.1371/journal.pgen.0020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorus S, et al. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 13.Chen JM, Cooper DN, Chuzhanova N, Férec C, Patrinos GP. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 14.Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–72. doi: 10.1038/nature05113. Using a genome wide comparison of human and nonhuman genome sequences, this study identified a dramatically changing microRNA-encoding gene, HAR1F, which showed a highly accelerated HLS change in sequence and is highly expressed in the human fetal brain. [DOI] [PubMed] [Google Scholar]
- 15.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enard W, Paabo S. Comparative primate genomics. Annu Rev Genomics Hum Genet. 2004;5:351–78. doi: 10.1146/annurev.genom.5.061903.180040. [DOI] [PubMed] [Google Scholar]
- 18.Fortna A, et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. The first genome-wide and first gene-based array CGH study of lineage specific gene copy number gain and loss among human and great ape lineages. One hundred forty genes were identified that showed HLS changes in copy number including the MGC8902 gene that encodes DUF1220 protein domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yunis JJ, Prakash O. The origin of man: a chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- 20.Dennis MY, et al. Evolution of Human-Specific Neural SRGAP2 Genes by Incomplete Segmental Duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doggett NA, et al. A 360-kb interchromosomal duplication of the human HYDIN locus. Genomics. 2006;88:762–71. doi: 10.1016/j.ygeno.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 22.Brunetti-Pierri N, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumas L, Sikela JM. DUF1220 Domains, Cognitive Disease, and Human Brain Evolution. Cold Spring Harb Symp Quant Biol. 2009;74:375–382. doi: 10.1101/sqb.2009.74.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumas L, et al. DUF1220 domain copy number implicated in human brain size pathology and evolution. Am J Hum Genet. doi: 10.1016/j.ajhg.2012.07.016. Accepted/in print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charrier C, et al. Inhibition of SRGAP2 Function by Its Human-Specific Paralogs Induces Neoteny during Spine Maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ventura M, et al. The evolution of African great ape subtelomeric heterochromatin and the fusion of human chromosome 2. Genome Res. 2012;22:1036–1049. doi: 10.1101/gr.136556.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhatt B, Burns J, Flannery D, McGee J. Direct visualization of single copy genes on banded metaphase chromosomes by nonisotopic in situ hybridization. Nucl Acids Res. 1988;16:3951–3961. doi: 10.1093/nar/16.9.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jauch A, et al. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc Natl Acad Sci USA. 1992;89:8611–8615. doi: 10.1073/pnas.89.18.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson GM, et al. Identification of full-coverage array CGH of human DNA copy number increases relative to chimpanzee and gorilla. Genome Res. 2006;16:173–181. doi: 10.1101/gr.4456006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goidts V, et al. Identification of large-scale human-specific copy number differences by inter-species array comparative genomic hybridization. Hum Genet. 2006;119:185–198. doi: 10.1007/s00439-005-0130-9. [DOI] [PubMed] [Google Scholar]
- 31.Linardopoulou EV, et al. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikkelsen TS, et al. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 33.Clark AG, et al. Inferring Nonneutral Evolution from Human-Chimp-Mouse Orthologous Gene Trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- 34.Berglund J, Pollard KS, Webster MT. Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009;7:e26. doi: 10.1371/journal.pbio.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grossman SR, et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327:883–886. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- 36.Britten RJ. Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels. Proc Natl Acad Sci USA. 2002;99:13633–13635. doi: 10.1073/pnas.172510699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen FC, Chen CJ, Li WH, Chuang TJ. Human-specific insertions and deletions inferred from mammalian genome sequences. Genome Res. 2007;17:16–22. doi: 10.1101/gr.5429606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marques-Bonet T, et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457:877–81. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nature Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudmant PH, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2011;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–76. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khodosevich K, Lebedev Y, Sverdlov E. Comp Funct Genomics. 2002;3:494–8. doi: 10.1002/cfg.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J, et al. Different evolutionary fates of recently integrated human and chimpanzee LINE-1 retrotransposons. Gene. 2007;390:18–27. doi: 10.1016/j.gene.2006.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–31. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singer T, et al. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes. Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eichler EE, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puppione DL, et al. Detection of two distinct forms of apoC-I in great apes. Comp Biochem Physiol. 2010;5:73–79. doi: 10.1016/j.cbd.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lucatelli JF, et al. Genetic influences on Alzheimer’s disease: evidence of interactions between the genes APOE, APOC1 and ACE in a sample population from the South of Brazil. Neurochem Res. 2011;36:1533–1539. doi: 10.1007/s11064-011-0481-7. [DOI] [PubMed] [Google Scholar]
- 52.Hansen JB, et al. The apolipoprotein C-I content of very-low-density lipoproteins is associated with fasting triglycerides, postprandial lipemia, and carotid atherosclerosis. J Lipids. 2011;2011:271062. doi: 10.1155/2011/271062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grallert H, et al. Eight genetic loci associated with variation in lipoprotein-associated phospholipase A2 mass and activity and coronary heart disease: meta-analysis of genome-wide association studies from five community-based studies. Eur Heart J. 2012;33:238–251. doi: 10.1093/eurheartj/ehr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kondo H, et al. Human aquaporin adipose (AQPap) gene. Genomic structure, promoter analysis and functional mutation. Eur J Biochem. 2002;269:1814–26. doi: 10.1046/j.1432-1033.2002.02821.x. [DOI] [PubMed] [Google Scholar]
- 55.Walker CG, Holness MJ, Gibbons GF, Sugden MC. Fasting-induced increases in aquaporin 7 and adipose triglyceride lipase mRNA expression in adipose tissue are attenuated by peroxisome proliferator-activated receptor alpha deficiency. Int J Obes (Lond) 2007;31:1165–71. doi: 10.1038/sj.ijo.0803555. [DOI] [PubMed] [Google Scholar]
- 56.Cooper DN, Kehrer-Sawatzki H. Exploring the potential relevance of human-specific genes to complex disease. Hum Gen. 2011;5:99–107. doi: 10.1186/1479-7364-5-2-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evans PD, et al. Adaptive evolution of ASPM, a major determinant of cerebral cortical size in humans. Hum Molec Genet. 2004;13:489–494. doi: 10.1093/hmg/ddh055. [DOI] [PubMed] [Google Scholar]
- 58.Rimol LM, et al. Sex-dependent association of common variants of microcephaly genes with brain structure. Proc Natl Acad Sci USA. 2010;107:384–388. doi: 10.1073/pnas.0908454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Evans PD, Vallender EJ, Lahn BT. Molecular evolution of the brain size regulator genes CDK5RAP2 and CENPJ. Gene. 2006;375:75–79. doi: 10.1016/j.gene.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 60.Huh JW, et al. Gain of new exons and promoters by lineage-specific transposable elements-integration and conservation event on CHRM3 gene. Mol Cells. 2009;28:111–117. doi: 10.1007/s10059-009-0106-z. [DOI] [PubMed] [Google Scholar]
- 61.Araud T, et al. The chimeric gene CHRFAM7A, a partial duplication of the CHRNA7 gene, is a dominant negative regulator of α7*nAChR function. Biochem Pharmacol. 2011;82:904–914. doi: 10.1016/j.bcp.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steen C, Wilczak N, Hoogduin JM, Koch M, De Keyser J. Reduced creatine kinase B activity in multiple sclerosis normal appearing white matter. PLoS One. 2010;5:e10811. doi: 10.1371/journal.pone.0010811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayakawa T, Aki I, Varki A, Satta Y, Takahata N. Fixation of the human-specific CMP-N-acetylneuraminic acid hydroxylase pseudogene and implications of haplotype diversity for human evolution. Genetics. 2006;172:1139–46. doi: 10.1534/genetics.105.046995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Varki A. Uniquely human evolution of sialic acid genetics and biology. Proc Natl Acad Sci USA. 2010;107:8939–8946. doi: 10.1073/pnas.0914634107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cioffi F, Lanni A, Goglia F. Thyroid hormones, mitochondrial bioenergetics and lipid handling. Curr Opin Endocrinol Diabetes Obes. 2010;17:402–407. doi: 10.1097/MED.0b013e32833cf354. [DOI] [PubMed] [Google Scholar]
- 66.Popesco MC, et al. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. The first report that describes the striking human lineage-specific increase in copy number of DUF1220 protein domains. [DOI] [PubMed] [Google Scholar]
- 67.O’Bleness, et al. Evolutionary History and Genome Organization of DUF1220 Protein Domains. G3 (Bethesda) doi: 10.1534/g3.112.003061. Accepted for publication/in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lussiana C, et al. Mutations and polymorphisms of the FSH receptor (FSHR) gene: clinical implications in female fecundity and molecular biology of FSHR protein and gene. Obstet Gynecol Surv. 2008;63:785–95. doi: 10.1097/OGX.0b013e31818957eb. [DOI] [PubMed] [Google Scholar]
- 69.Plunkett J, et al. An evolutionary genomic approach to identify genes involved in human birth timing. PLoS Genet. 2011;7:e1001365. doi: 10.1371/journal.pgen.1001365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Enard W, et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–71. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 71.Goto H, et al. The identification and functional implications of human-specific “fixed” amino acid substitutions in the glutamate receptor family. BMC Evol Biol. 2009;9:224. doi: 10.1186/1471-2148-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Evans PD, Anderson JR, Vallender EJ, Choi SS, Lahn BT. Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum Molec Genet. 2004;13:1139–1145. doi: 10.1093/hmg/ddh126. [DOI] [PubMed] [Google Scholar]
- 73.Stedmann HH, et al. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature. 2004;428:415–418. doi: 10.1038/nature02358. [DOI] [PubMed] [Google Scholar]
- 74.Ciesek S, et al. Impact of intra- and interspecies variation of occludin on its function as coreceptor for authentic hepatitis C virus particles. J Virol. 2011;85:7613–7621. doi: 10.1128/JVI.00212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shin EY, et al. Phosphorylation of p85 beta PIX, a Rac/Cdc42-specific guanine nucleotide exchange factor, via the Ras/ERK/PAK2 pathway is required for basic fibroblast growth factor-induced neurite outgrowth. J Biol Chem. 2002;277:44417–44430. doi: 10.1074/jbc.M203754200. [DOI] [PubMed] [Google Scholar]
- 76.Majava V, et al. Structural and functional characterization of human peripheral nervous system myelin protein P2. PLoS One. 2010;5:e10300. doi: 10.1371/journal.pone.0010300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hennah W, Porteous D. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS One. 2009;4:e4906. doi: 10.1371/journal.pone.0004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalmady SV, Venkatasubramanian G. Evidence for positive selection on Protocadherin Y gene in Homo sapiens: implications for schizophrenia. Schizophr Res. 2009;108:299–300. doi: 10.1016/j.schres.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 79.Crow TJ. Handedness, language lateralisation and anatomical asymmetry: relevance of protocadherin XY to hominid speciation and the aetiology of psychosis. Point of view. Br J Psychiatry. 2002;181:295–7. doi: 10.1192/bjp.181.4.295. [DOI] [PubMed] [Google Scholar]
- 80.Speevak MD, Farrell SA. Non-syndromic language delay in a child with disruption in the Protocadherin11X/Y gene pair. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:484–9. doi: 10.1002/ajmg.b.31186. [DOI] [PubMed] [Google Scholar]
- 81.Ross NLJ, et al. Methylation of Two Homo sapiens-Specific X-Y Homologous Genes in Klinefelter’s Syndrome (XXY) Am J Med Genet Part B. 2006;141B:544–548. doi: 10.1002/ajmg.b.30339. [DOI] [PubMed] [Google Scholar]
- 82.Carrasquillo MM, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nat Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nguyen DH, Hurtado-Ziola N, Gagneux P, Varki A. Loss of Siglec expression on T lymphocytes during human evolution. Proc Natl Acad Sci U S A. 2006;103:7765–70. doi: 10.1073/pnas.0510484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brinkman-Van der Linden EC, et al. Human-specific expression of Siglec-6 in the placenta. Glycobiology. 2007;17:922–31. doi: 10.1093/glycob/cwm065. [DOI] [PubMed] [Google Scholar]
- 85.Wang X, et al. Evolution of Siglec-11 and Siglec-16 Genes in Hominins. Mol Biol Evol. 2012;29:2073–2086. doi: 10.1093/molbev/mss077. First examples of genes inactivated in relation to the timing of origin of modern humans. The data suggest that infectious agents may have played a role in selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y, Neumann H. Alleviation of neurotoxicity by microglial human Siglec-11. J Neurosci. 2010;30:3482–3488. doi: 10.1523/JNEUROSCI.3940-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang X, et al. Specific Inactivation of Two Immunomodulatory SIGLEC Genes During Human Evolution. Proc Natl Acad Sci USA. 2012;109:9935–40. doi: 10.1073/pnas.1119459109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fedrigo O, et al. A potential role for glucose transporters in the evolution of human brain size. Brain Behav Evol. 2011;78:315–26. doi: 10.1159/000329852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saus E, et al. Comprehensive copy number variant (CNV) analysis of neuronal pathways genes in psychiatric disorders identifies rare variants within patients. J Psychiatr Res. 2010;44:971–978. doi: 10.1016/j.jpsychires.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 90.Kouprina N, et al. The SPANX gene family of cancer-testis specific antigens: rapid evolution, an unusual case of positive selection and amplification in African Great Apes and hominids. Proc Natl Acad Sci USA. 2004;101:3077–3082. doi: 10.1073/pnas.0308532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Westbrook VA, et al. Hominoid-specific SPANXA/D genes demonstrate differential expression in individuals and protein localization to a distinct nuclear envelope domain during spermatid morphogenesis. Mol Hum Reprod. 2006;12:703–716. doi: 10.1093/molehr/gal079. [DOI] [PubMed] [Google Scholar]
- 92.Cáceres M, Suwyn C, Maddox M, Thomas JW, Preuss TM. Increased cortical expression of two synaptogenic thrombospondins in human brain evolution. Cereb Cortex. 2007;17:2312–2321. doi: 10.1093/cercor/bhl140. [DOI] [PubMed] [Google Scholar]
- 93.Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; Baltimore, MD: World Wide Web URL: http://omim.org/ [Google Scholar]
- 94.Lai CS, Gerrelli D, Monaco AP, Fisher SE, Copp AJ. FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain. 2003;126:2455–2462. doi: 10.1093/brain/awg247. [DOI] [PubMed] [Google Scholar]
- 95.Enard W, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. First evidence that the coding sequence of the FOXP2 gene, mutated in a family with a speech disorder, underwent human-specific alterations consistent with positive selection. [DOI] [PubMed] [Google Scholar]
- 96.Zhang J, Webb DM, Podlaha O. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics. 2002;162:1825–1835. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin L, et al. Evolution of alternative splicing in primate brain transcriptomes. Hum Mol Genet. 2010;19:2958–2973. doi: 10.1093/hmg/ddq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim DS, Hahn Y. Identification of human-specific transcript variants induced by DNA insertions in the human genome. Bioinformatics. 2011;27:14–21. doi: 10.1093/bioinformatics/btq612. [DOI] [PubMed] [Google Scholar]
- 100.Ohno S. Evolution by gene duplication. Springer-Verlag; 1970. A classic text that provides some of the first arguments making the case that gene duplication is a major mechanism underlying genome evolution. [Google Scholar]
- 101.Li WH. Molecular Evolution. Sinauer Associates, Inc; 1997. [Google Scholar]
- 102.Rizzuto R, Nakase H, Zeviani M, DiMauro S, Schon EA. Subunit Va of human and bovine cytochrome c oxidase is highly conserved. Gene. 1988;69:245–256. doi: 10.1016/0378-1119(88)90435-0. [DOI] [PubMed] [Google Scholar]
- 103.Uddin M, et al. Molecular evolution of the cytochrome c oxidase subunit 5A gene in primates. BMC Evol Biol. 2008;8:8. doi: 10.1186/1471-2148-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arnold S, Goglia F, Kadenbach B. 3,5-Diiodothyronine binds to subunit Va of cytochrome-c oxidase and abolishes the allosteric inhibition of respiration by ATP. Eur J Biochem. 1998;252:325–330. doi: 10.1046/j.1432-1327.1998.2520325.x. [DOI] [PubMed] [Google Scholar]
- 105.Kim HL, Igawa T, Kawashima A, Satta Y, Takahata N. Divergence, demography and gene loss along the human lineage. Phil Trans R Soc B. 2010;365:2451–2457. doi: 10.1098/rstb.2010.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Olson MV. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pastorcic M, Birnbaum S, Hixson JE. Baboon apolipoprotein C-I: cDNA and gene structure and evolution. Genomics. 1992;13:368–374. doi: 10.1016/0888-7543(92)90255-q. [DOI] [PubMed] [Google Scholar]
- 108.Berbée JF, et al. Apolipoprotein CI knock-out mice display impaired memory functions. J Alzheimers Dis. 2011;23:737–747. doi: 10.3233/JAD-2010-100576. [DOI] [PubMed] [Google Scholar]
- 109.Varki NM, et al. Heart disease is common in humans and chimpanzees, but is caused by different pathological processes. Evol Appl. 2009;2:101–112. doi: 10.1111/j.1752-4571.2008.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Varki NM, Strobert E, Dick EJ, Jr, Benirschke K, Varki A. Biomedical Differences Between Humans and Nonhuman Hominids; potential roles for uniquely human aspects of sialic acid biology. Annu Rev Pathol Mech. 2011;6:365–393. doi: 10.1146/annurev-pathol-011110-130315. Review emphasizes that many human diseases are uniquely human, and relates some of the differences to evidence that genes involved in the biology of sialic acids constitute a “hot-spot” in human evolution. [DOI] [PubMed] [Google Scholar]
- 111.Wang X, et al. Expression of Siglec-11 by human and chimpanzee ovarian stromal cells, with uniquely human ligands: implications for human ovarian physiology and pathology. Glycobiology. 2011;21:1038–1048. doi: 10.1093/glycob/cwr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schmidt J, Kirsch S, Rappoid GA, Schempp W. Complex evolution of a Y-chromosomal double homeobox 4 (DUX4)-related gene family in hominoids. PLoS ONE. 2009;4:e5288. doi: 10.1371/journal.pone.0005288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Crocker PR. Siglecs in innate immunity. Curr Opin Pharmacol. 2005;5:431–437. doi: 10.1016/j.coph.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 114.Knowles DG, McLysaght A. Recent de novo origin of human protein-coding genes. Genome Res. 2009;19:1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li CY, et al. A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput Biol. 2010;6:e1000734. doi: 10.1371/journal.pcbi.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ohno S. Evolution by gene duplication. Springer-Verlag; 1970. [Google Scholar]
- 116.Wu DD, Irwin DM, Zhang YP. De novo origin of human protein-coding genes. PLoS Genet. 2011;7:e1002379. doi: 10.1371/journal.pgen.1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Romero IG, Ruvinsky I, Gilad Y. Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet. 2012;13:505–516. doi: 10.1038/nrg3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Buhl AM, et al. Identification of a gene on chromosome 12q22 uniquely overexpressed in chronic lymphocytic leukemia. Blood. 2006;107:2904–2911. doi: 10.1182/blood-2005-07-2615. [DOI] [PubMed] [Google Scholar]
- 119.Yoshida K, Sugano S. Identification of a novel protocadherin gene (PCDH11) on the human XY homology region in Xq21.3. Genomics. 1999;62:540–3. doi: 10.1006/geno.1999.6042. [DOI] [PubMed] [Google Scholar]
- 120.Winn VD, et al. Severe preeclampsia-related changes in gene expression at the maternal-fetal interface include sialic acid-binding immunoglobulin-like lectin-6 and pappalysin-2. Endocrinology. 2009;150:452–62. doi: 10.1210/en.2008-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. Important paper discussing the connection between complex genomic architecture and recurrent disease and put forth the idea that these regions may play an important role in primate speciation. [DOI] [PubMed] [Google Scholar]
- 122.Olson MV, Varki A. Sequencing the chimpanzee genome: insights into human evolution and disease. Nat Rev Genet. 2003;4:20–28. doi: 10.1038/nrg981. [DOI] [PubMed] [Google Scholar]
- 123.Soto PC, Stein LL, Hurtado-Ziola N, Hedrick SM, Varki A. Relative over-reactivity of human versus chimpanzee lymphocytes: implications for the human diseases associated with immune activation. J Immunol. 2010;184:4185–4195. doi: 10.4049/jimmunol.0903420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mekel-Bobrov N, et al. The ongoing adaptive evolution of ASPM and Microcephalin is not explained by increased intelligence. Hum Mol Genet. 2007;16:600–608. doi: 10.1093/hmg/ddl487. [DOI] [PubMed] [Google Scholar]
- 125.Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Prüfer K, et al. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McConkey EH, Varki A. Genomics. Thoughts on the future of great ape research. Science. 2005;309:1499–501. doi: 10.1126/science.1113863. [DOI] [PubMed] [Google Scholar]
- 129.Altevogt BM, Pankevich DE, Pope AM, Kahn JP. Research agenda. Guiding limited use of chimpanzees in research. Science. 2012;335:41–42. doi: 10.1126/science.1217521. [DOI] [PubMed] [Google Scholar]
- 130.Khaitovich P, et al. Metabolic changes in schizophrenia and human brain evolution. Genome Biol. 2008;9:R124. doi: 10.1186/gb-2008-9-8-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]