Abstract
HIV-1 reverse transcriptase (RT) contributes to the development of resistance to all anti-AIDS drugs by introducing mutations into the viral genome. At the molecular level, mutations in RT result in resistance to RT inhibitors. Eight nucleoside/nucleotide analogs (NRTIs) and five non-nucleoside inhibitors (NNRTIs) are approved HIV-1 drugs. Structures of RT have been determined in complexes with substrates and/or inhibitors, and the structures have revealed different conformational and functional states of the enzyme. Understanding the molecular mechanisms of resistance to NRTIs and NNRTIs, and their complex relationships, may help in designing new drugs that are periodically required to overcome existing as well as emerging trends of drug resistance.
Background
AIDS started spreading silently in the 1970s, and came into the limelight in the early 1980s as a mysterious pandemic that was one of the most serious public health threats in history. In the 1980s, clinical and scientific research communities were confronted with unexpected overwhelming challenges to understand the cause and mystery behind AIDS, and to find prevention and treatment options. Detection of reverse transcription activity in cultures of lymph node cells taken from AIDS patients in the early 1980s [1,2] revealed that AIDS is caused by a retrovirus that was subsequently named the human immunodeficiency virus (HIV).
Reverse transcription is a key step in the life cycle of retroviruses, and the process is responsible for synthesis of double-stranded (ds) DNA from a viral single-stranded (ss) RNA genome. A viral DNA polymerase or reverse transcriptase (RT) enzyme is responsible for synthesis of DNA complementary to an RNA or DNA template. Not surprisingly, the reverse transcription/DNA polymerization step in HIV replication was immediately considered as a prime drug target, and the nucleoside analog AZT (zidovudine, ZDV) [3,4] was approved as the first anti-AIDS drug. Clinical use of AZT revealed that treatment of HIV infection with a single drug was not effective in keeping the viral load down for a prolonged period. Also, it was realized that HIV could not be cleared from an infected individual, and drug-resistant viruses emerged with loss of sensitivity to AZT.
Even today, challenges like discovery of an effective AIDS vaccine, complete cure from HIV infection, and effective ways to overcome drug resistance remain as open questions. However, sincere scientific research commitments initiated and supported by public and private initiatives have led to the development of drugs and treatment strategies that help HIV infected individuals lead a near normal lifespan if the patient complies with treatments to maintain viral load at or near undetected level. HIV-1 is the predominant virus that spreads AIDS. So far 26 individual anti-AIDS drugs have been approved, of which 13 target RT. The remaining drugs target several key steps in the viral lifecycle, namely: (i) the enzyme protease that is responsible for cleaving the viral polyprotein precursors into functional entities and for maturation of virus particles, (ii) the enzyme integrase that integrates the viral dsDNA to the host cell chromosome, and (iii) viral entry/fusion.
HIV-1 exhibits high genetic variability, and thereby, HIV-1 develops resistance to existing drugs and escapes host immune responses elicited by AIDS vaccine candidates. HIV-1 enters a host cell by binding the CD4 receptor on the surface of immune cells and a co-receptor, either CCR5 or CXCR4. Entry and fusion of an HIV-1 particle releases its two copies of the viral ssRNA genome, about fifty copies of RT, and other viral entities into the cytoplasm. RT copies the viral genome into a dsDNA that is subsequently transported into the nucleus of the infected cell and integrated into the host cell chromosome by another viral enzyme, integrase. Recently approved integrase inhibitors (raltegravir and elvitegravir) bind the active site of HIV-1 integrase in a pre-integration complex [5,6] and block the viral DNA integration into host cell chromosomes.
Usually, three or four drugs are combined in a treatment regimen, commonly referred to as highly active antiretroviral therapy (HAART). Some of the widely used treatment regimens include (i) two nucleoside RT inhibitors (NRTIs) + one non-nucleoside RT inhibitor (NNRTI) or protease inhibitor (PI), and (ii) combinations of an integrase or entry inhibitor with RT inhibitors and PIs. Selecting the right treatment strategies and combinations remain challenging due to factors such as: (i) limitations in availability of drugs at different geographic locations and socioeconomic conditions, (ii) compatibilities among individual drugs and adherence, (iii) side-effects from long-term use of drugs, (iv) existence and emergence of drug-resistance profiles, and (v) different responses by treatment-naive or treatment-experienced individuals, etc. However, three decades of extensive research on various aspects of HIV-1 and related viruses have brought success in effectively managing HIV-1 infection, and the scientific knowledge garnered has in many ways blazed the trail for finding treatment solutions for several challenging chronic diseases and emerging drug-resistance problems.
The RT enzyme is a critical molecular machine of HIV-1 that is targeted by the largest number of anti-AIDS drugs. At the same time, RT is responsible for developing resistance to all anti-AIDS drugs (i) directly to RT inhibitors at the molecular level and/or (ii) indirectly as a primary source for introducing genetic variations. In this two-part review, we discuss the critical roles of RT in developing drug resistance, a structural perspective on molecular mechanisms of RT inhibition and drug resistance, and possible molecular bases for synergistic benefits of several RT drug combinations used to combat drug resistance.
RT in generating HIV-1 quasispecies – evolution of drug resistance
Drug resistance evolution is a primary phenomenon that helps a pathogen survive and replicate even under harsh drug pressure. An HIV-1 infected individual harbors a pool of HIV-1 variants generated initially mostly from one (a “founder”) that had transmitted the infection [7]. In general, a virus yields a pool of “quasispecies” [8] by undergoing genetic variations that arise from errors made by HIV-1 while copying its genetic information combined with a very high rate of HIV-1 turnover [9]. RT copies the ~10,000 nucleotide RNA HIV-1 genome [10,11] into dsDNA via RNA- and DNA-dependent DNA polymerization, a process requiring ~20,000 nucleotide incorporation steps. RT is an error-prone enzyme that introduces approximately one misincorporation per 104 nucleotide incorporations at the enzyme level. Apparently, the rate of error is lower in viral replication than that expected based on RT misincorporation data. The in vivo mutation rate in viral replication is ~10-5, a order of magnitude lower than that observed with RT [12]. Apart from RT, the host RNA polymerase may also introduce mutations while transcribing the viral DNA to mRNA copies that are (i) translated into viral polyproteins and (ii) processed into viral ssRNA copies. The errors are random, resulting in random sequence variations in the progeny viruses and translated viral proteins. Many, if not most, of the mutations may be deleterious for the structural assembly or the functions of viral proteins, and therefore the virus can't afford those mutations. Interestingly, HIV-1 can retain its activity despite carrying several mutations, and each of these mutant strains can replicate and generate new variants. Each replication cycle expands heterogeneity in the pool of viruses in an HIV-1 infected individual.
The wild-type variants are well adapted to the natural host environment and replicate as the predominant strains whereas, various quasispecies replicate less efficiently. However, the replication dynamics changes under drug selection pressure. A typical drug is primarily designed to hit the wild-type virus hard. The mutants that are least inhibited by the drugs emerge as the predominant variants. At times, such a drug-resistant variant may be less fit on its replication or transmission capability; however, some of these variants may gain fitness either by accruing compensatory mutations or by adding resistance mutations to an existing variant containing the compensatory mutation background.
Structures and conformations of RT
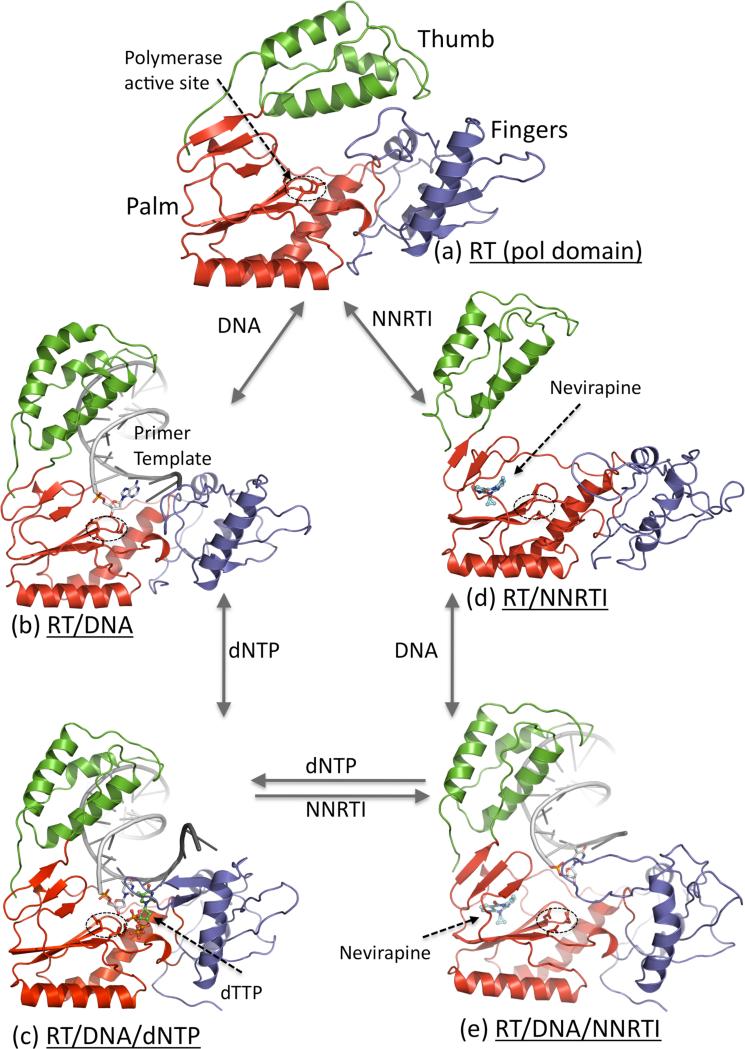
RT is a heterodimer consisting of two polypeptide chains p66 (66kDa; 560 amino acid residues) and p51 (51 kDa; 440 amino acid residues). Large precursor polyproteins translated from the HIV-1 pol gene are cleaved by HIV-1 protease to produce the p66 subunit. The p66 chain contains an N-terminal polymerase domain and a C-terminal RNase H domain. A likely scenario appears to be that two p66 chains dimerize, and HIV-1 protease cleaves the RNase H moiety of one chain to produce a stable p66/p51 heterodimeric RT. The structural architecture of RT (Figure 1A) has been known for two decades [13,14]. The p66 chain contains both the polymerase and RNase H active sites. The polymerase domain of RT (Figure 1B) resembles the shape of a “right hand” with fingers, palm, thumb and connection subdomains; the connection subdomain links the polymerase domain to the RNase H domain. The p51 chain also contains the fingers, palm, thumb, and connection subdomains; however, the subdomains have different spatial arrangements in p51 than in p66. The subdomains in p51 are assembled into a relatively rigid structure that provides structural support to p66. The subdomains in p66 are flexible, and can be spatially rearranged to different conformational states necessary to carry out the functions of RT. Structures representing five conformational/functional states of RT have been determined (Figure 2). Some of the major conformational rearrangements revealed by RT structures are: (i) the thumb lifts up to bind nucleic acid, (ii) the fingers fold down to hold a dNTP substrate in the presence of a nucleic acid, (iii) NNRTI-binding leads to thumb hyperextension even if RT is not bound to a nucleic acid, and (iv) nucleic acid at the polymerase active site is repositioned upon binding of an NNRTI to RT-DNA complex. These structural rearrangements of RT result from inter-subdomain hinge movements and local structural rearrangements while the overall structural folds of individual subdomains remain almost invariant.
Figure 1.
HIV-1 RT structure and sites for common drug resistance mutations. (a) A ribbon representation of the structure of HIV-1 RT-DNA-dNTP complex. (b) Sites of commonly observed NRTI resistance mutations (magenta) and NNRTI resistance mutations (cyan) in the fingers and palm subdomains of HIV-1 RT.
Figure 2.
Five structurally characterized conformational states of HIV-1 RT; only the polymerase domain (fingers (blue), palm (red), and thumb (green)) of RT is shown. (a) Thumb is positioned near the fingers in closed conformation that occupies the nucleic acid binding cleft in apo HIV-1 RT structures [29]. (b) Upon binding of a nucleic acid substrate, the thumb is lifted up and acts as a clamp to hold the nucleic acid [14]; the primer 3′-end is positioned at the polymerase active site (D110, D185, and D186) that is highlighted by a dotted ellipse. (c) Binding of a dNTP to RT-DNA complex results in closing of the fingers to bind a dNTP in a catalytically competent state [30]. (d) Binding of an NNRTI to RT causes several conformational changes; thumb subdomain is lifted to a hyper-extended position and the nucleic acid-binding cleft is open even in absence of a nucleic acid [13]. (e) Binding of an NNRTI to RT-DNA complex resulted in reduction of DNA interactions with the polymerase domain of RT, and repositioning of the primer 3′-end away from the polymerase active site [31••]; the structural study revealed no ordered dNTP binding to RT-DNA-NNRTI complex.
The sites of mutations that confer resistance to either NRTI or NNRTI drugs are primarily located in the polymerase domain (Figure 1B). Routine RT sequencing from clinical isolates were limited to the polymerase domain only where primary NRTI- and NNRTI-resistance mutations occur. Relatively recent sequencing of the complete RT from clinical isolates have shown that mutations in the remote connection subdomain and RNase H domain enhance resistance to both classes of RT drugs [15,16] by indirect mechanisms that are not well understood. This review focuses on the NRTI and NNRTI drugs, and the primary resistance mutations that are confined to the polymerase domain.
Nucleoside analog (NRTI) drugs
Nucleotide misincorporations by RT contribute to generating mutant HIV-1 proteins including mutant RTs, and mutant RTs can develop resistance to RT inhibitors. Drugs targeting HIV-1 RT are either nucleoside RT inhibitors (NRTIs) or non-nucleoside RT inhibitors (NNRTIs). An NRTI (Figure 3A) is converted to a dNTP analog by a phosphorylation cascade performed by cellular kinases, and then RT catalytically incorporates the drug as an NRTI monophosphate (or a nucleotide analog) at the 3’-end of the growing viral DNA primer; pyrophosphate is released as the reaction byproduct. The nucleoside phosphonate analogs like tenofovir (a “nucleotide analog”) require addition of β– and γ–phosphates whereas the other NRTIs are elaborated with α–, β–,γ–phosphates. Efficient intracellular phosphorylation of NRTI to NRTI-triphosphate (TP) is an essential requirement for the efficacy of an NRTI drug. Upon incorporation, an NRTI inhibits the elongation of DNA primer because NRTIs lack a 3′-OH group and/or contain a modified sugar moiety that prevents addition of the next nucleotide. An NRTI-TP does not block the activity of an RT molecule; however, certain RT mutations cause NRTI resistance by discriminating an NRTI-TP from the analogous dNTP substrate. NRTI-resistance mutations primarily appear along the dNTP-binding track extending from the β3–β4 fingers loop region to YMDD residue M184 (Figure 1B). Also, RT has the ability to remove certain NRTIs from the DNA primer (“unblocking”) by reversing the direction of its catalytic reaction of polymerization [17,18]. The molecular mechanisms of individual resistance mutations are unique and relationships among the mutations are complex. Biochemical and structural studies have unraveled some of the unique resistance mechanisms and their relationships that are discussed in Part 2 of this review article.
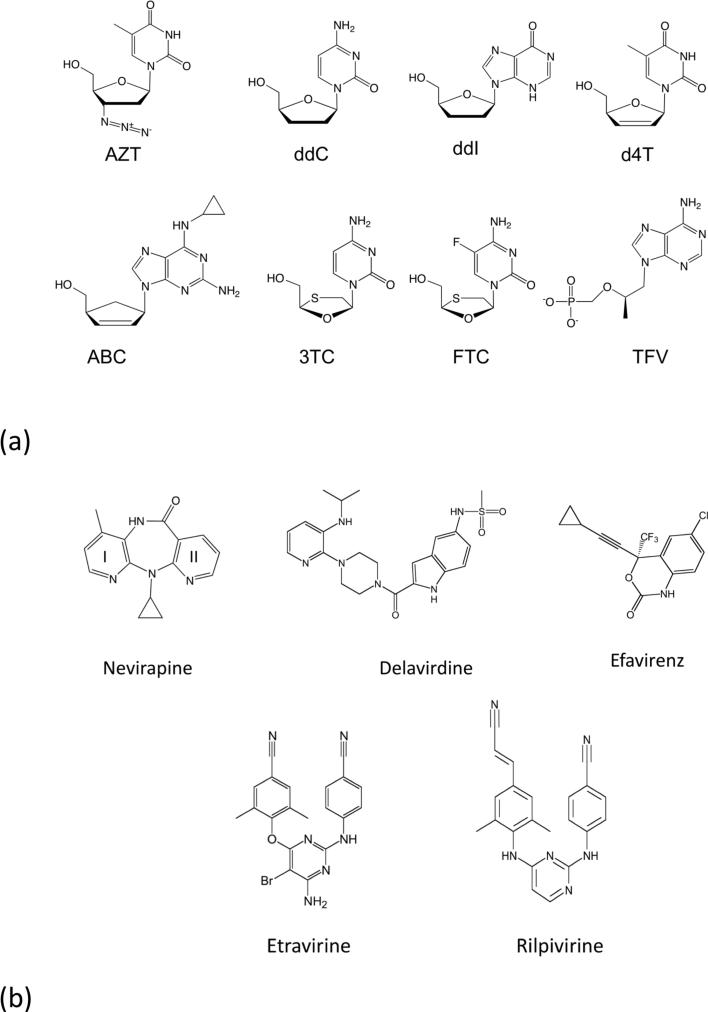
Figure 3.
The chemical structures of anti-AIDS drugs that target HIV-1 RT: (a) The clinically approved NRTIs are: (i) AZT (zidovudine, ZDV, azidothymidine, Retrovir®) has a 3′-azido group substituted for the 3′-OH of dTTP; (ii) ddC (dideoxycytidine, zalcitabine, Hivid®) (iii) ddI (didanosine, Videx®), and (iv) d4T (stavudine, Zerit®) lack 3′-OH groups; (v) ABC (abacavir, Ziagen®) has a cyclopentene ring, (vi) 3TC (lamivudine, Epivir®) has an altered β-L-pseudo-ribose ring, (vii) FTC (emtricitabine, Emtriva®), and (viii) TFV (tenofovir) is a nucleotide analog that has an acyclic methoxypropyl moiety substituted for the deoxyribose ring of dATP; tenofovir is formulated as tenofovir disoproxil fumarate (TDF, PMPA, Viread®). (b) Chemical structures of the five NNRTI drugs approved for treating HIV-1 infections. (i) Nevirapine (NEV, Viramune®), (ii) delavirdine (DLV,Rescriptor®), (iii) etravirine (ETR, Intelence®), and (iv) rilpivirine (RPV, Edurant®).
Non-nucleoside RT inhibitors (NNRTIs)
Unlike NRTIs that do not directly inhibit RT, an NNRTI drug binds to a hydrophobic pocket in the palm subdomain adjacent to the base of the thumb subdomain (Figure 2d) and allosterically inhibits DNA polymerization. The NNRTI pocket permits the design of highly specific inhibitors having low toxicities and minimal side effects. In fact, the NNRTIs are HIV-1 specific and even do not effectively inhibit HIV-2 RT. The NNRTI pocket is not required to be highly conserved for carrying out the enzyme activity unlike the conserved active site or dNTP-binding site of RT. Therefore, HIV-1 has a relatively lower genetic barrier for developing NNRTI-resistance mutations than for NRTI-resistance mutations. Primary NNRTI-resistance mutations appear in and around the NNRTI pocket, i.e., most of the pocket residues can mutate to confer NNRTI resistance. There are five NNRTI drugs approved for treating HIV-1 infections – nevirapine (NEV, Viramune®), efavirenz (EFV, Sustiva®), delavirdine (DLV, Rescriptor®), etravirine (ETR, Intelence®), and rilpivirine (RPV, Edurant®) (Figure 3B). An NNRTI is generally used in combination with NRTIs because the two classes of RT drugs have non-overlapping inhibition mechanisms and resistance mutation sites, and NNRTI-resistance mutations can emerge relatively quickly.
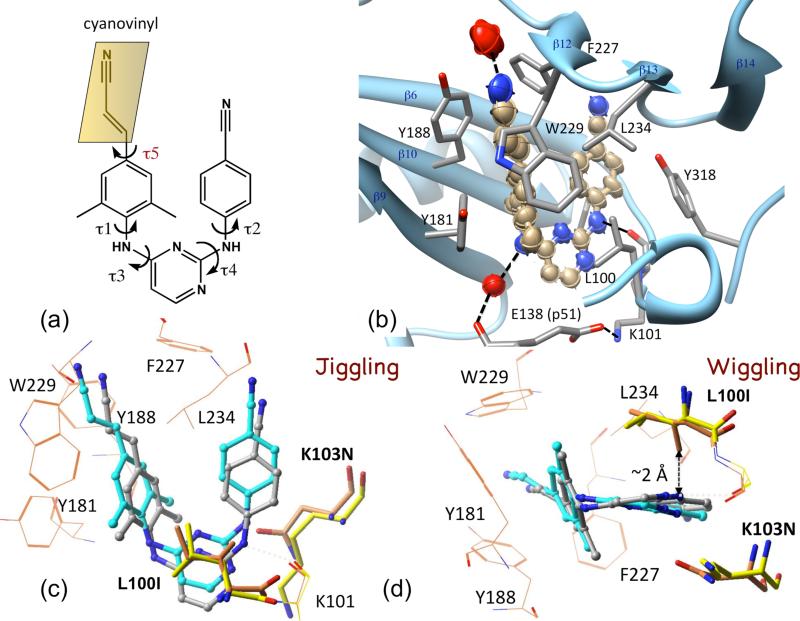
The first-generation NNRTI drug nevirapine invokes resistance mutations even after a single dose that is usually given to pregnant mothers to prevent mother-to-child transmission of HIV-1. An effective NNRTI should overcome the impacts of common drug-resistance mutations – a challenge emerged with the discovery of first-generation NNRTIs in early 1990s. The initial structure of the RT-nevirapine complex [13] revealed the hydrophobic NNRTI-binding pocket which is ~10 Å away from the polymerase active site. The first-generation NNRTIs nevirapine, TIBO, and α-APA were found to assume a common “butterfly-like” binding mode despite their broad chemical diversity [19]. These NNRTIs could be optimized to nanomolar inhibitors of wild-type virus/enzyme; however, they drastically lose their potency against a single common NNRTI-resistance mutation such as K103N or Y181C. The diarylpyrimidine (DAPY) NNRTI drugs ETR and RPV were developed out of a structure-based multidisciplinary approach [20-22], and these drugs inhibited HIV-1 carrying common NNRTI-resistance mutations. The DAPY NNRTIs exhibit conformational flexibility that helps the drugs reorient “wiggle” and reposition “jiggle” to retain binding efficacy to RT even when pocket mutations emerge [20] (Figures 4A & B). Further, the structures of rilpivirine in complexes with wild-type and two double mutant (K103N + Y181C and K103N + L100I) RTs demonstrated how the NNRTI RPV wiggles and jiggles in the pocket to retain its binding affinity [23•] (Figures 4C & D). The predominant inhibitor-protein interactions associated with NNRTI binding are: (i) hydrophobic sandwiches, (ii) a characteristic hydrogen bond with the K101 main-chain carbonyl, and (iii) water-mediated hydrogen bonds. It is important that an NNRTI retains these interactions with RT while it wiggles and jiggles in NNRTI-pocket. The most recent NNRTI drug rilpivirine exhibits the flexibility to wiggle and jiggle by which it retains all of the above interactions including the key hydrogen-bonding interaction with the K101 main-chain carbonyl when it binds wild-type RT or the two double mutants [23•]. A recent study shows that an NNRTI designed to have additional hydrogen bonds with RT while maintaining its flexibility has improved resilience against resistance mutations [24]. The cyanovinyl group of rilpivirine can swivel to maintain interactions with RT (Figure 4A); addition of the cyanovinyl group, i.e., chemical modification from TMC120 → rilpivirine, enhanced the inhibition potency by ~3-fold. A recent collaborative study combining 2D infrared (IR) spectroscopy, high-resolution (1.51 Å) crystal structure, and molecular dynamics simulation of rilpivirine bound to RT demonstrated the involvement of the cyanovinyl group with an invariant water-mediated interaction that may be a critical feature for future NNRTI design considerations (Figure 4B) [25•]. Rilpivirine possesses highly favorable pharmacokinetics, that in combination with its high resilience to drug-resistance mutations demonstrate clinical efficacy even at relatively low doses of 25 mg/day (efavirenz is usually administered at 600 mg/day). This ideal pharmacokinetic characteristic of RPV may correlate with its property to form ordered nanoparticles with diameter ~100 nM, whereas several related compounds including ETV which did not exhibit as favorable pharmacokinetics, were found to form disordered aggregates rather than uniform nanoparticles [26]. However, new drug-resistance patterns emerge from long-term use and combination therapies, discussed later. Understanding the molecular mechanism of NNRTI inhibition and resistance caused by different mutations may help with designing both future drugs and optimal drug combinations.
Figure 4.
Wiggling and jiggling of an NNRTI to retain potency against drug-resistance mutations. (a) Chemical structure of rilpivirine; the five torsionally flexible bonds define the conformational freedom of the NNRTI. (b) Thermal ellipsoid representation of rilpivirine drawn using the anisotropic B-factors of individual atoms from 1.5 Å resolution structure of RT-rilpivirine complex (PDB ID 4G1Q) [25•]. The interacting side chains and water molecules (red) are displayed. (c & d) Comparison of the binding modes of rilpivirine to wild-type RT (gray) vs. L100I + K103N mutant RT (cyan) revealed how the drug jiggle and wiggles, respectively, to evade the effects of drug-resistance mutations [23•]; the mutations modifies the side chains from yellow to orange.
Drug combinations for suppressing antiviral resistance
Fusion of a virus into a host cell starts the infection with the release of two copies of viral RNA and about fifty copies of RT into cytoplasm. Ideally only a single RT molecule could be adequate to carry out the entire reverse transcription process. A complex set of factors determine the clinical efficacy of drugs targeting HIV-1 RT [27••]. It is unlikely that an NNRTI at its IC50 cellular concentration would inhibit all RT molecules for complete blockage of HIV-1 replication; the effective concentration of an NNRTI for 100% inhibition is significantly higher than its IC50 concentration [28], and the effective concentration would be unattainably high if drug-resistant RTs are present. The effective inhibitory concentration for an NRTI in a cell also depends upon the efficiency of the cellular phosphorylation of the NRTI. NRTI-blocked primers are generally repaired by removing the NRTI. These are among several factors that help virus replicate even when attempts are made to block viral replication by multiple drugs.
The next part (Part 2) discusses structural and biochemical understanding of mechanisms of resistance to HIV-1 drugs that RT develops by acquiring various mutations.
Highlights.
HIV-1 RT is targeted by 13 anti-AIDS drugs
Mutations in RT confer resistance to RT inhibitors
Structures have revealed various functional/conformational states of RT
New NNRTIs are adaptive, which enhances their resilience to resistance mutations
Acknowledgments
EA is grateful to the National Institutes of Health for support from R37 AI 27690 (MERIT Award) and P50 GM103368.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Gallo RC, Salahuddin SZ, Popovic M, Shearer GM, Kaplan M, Haynes BF, Palker TJ, Redfield R, Oleske J, Safai B, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 3.Mitsuya H, Weinhold KJ, Furman PA, St Clair MH, Lehrman SN, Gallo RC, Bolognesi D, Barry DW, Broder S. 3′-Azido-3′-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc Natl Acad Sci U S A. 1985;82:7096–7100. doi: 10.1073/pnas.82.20.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischl MA, Richman DD, Grieco MH, Gottlieb MS, Volberding PA, Laskin OL, Leedom JM, Groopman JE, Mildvan D, Schooley RT, et al. The efficacy of azidothymidine (AZT) in the treatment of patients with AIDS and AIDS-related complex. A double-blind, placebo-controlled trial. N Engl J Med. 1987;317:185–191. doi: 10.1056/NEJM198707233170401. [DOI] [PubMed] [Google Scholar]
- 5.Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, Espeseth AS, Wolfe A, Egbertson M, Bourgeois M, Melamed J, et al. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc Natl Acad Sci U S A. 2002;99:6661–6666. doi: 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci U S A. 2010;107:20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489.. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 9.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 10.Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich B, Josephs SF, Doran ER, Rafalski JA, Whitehorn EA, Baumeister K, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 11.Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985;40:9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- 12.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 14.Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr., Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikolenko GN, Palmer S, Maldarelli F, Mellors JW, Coffin JM, Pathak VK. Mechanism for nucleoside analog-mediated abrogation of HIV-1 replication: balance between RNase H activity and nucleotide excision. Proc Natl Acad Sci U S A. 2005;102:2093–2098. doi: 10.1073/pnas.0409823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap SH, Sheen CW, Fahey J, Zanin M, Tyssen D, Lima VD, Wynhoven B, Kuiper M, Sluis-Cremer N, Harrigan PR, et al. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 2007;4:e335. doi: 10.1371/journal.pmed.0040335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer PR, Matsuura SE, So AG, Scott WA. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc Natl Acad Sci U S A. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 19.Ding J, Das K, Moereels H, Koymans L, Andries K, Janssen PA, Hughes SH, Arnold E. Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors. Nat Struct Biol. 1995;2:407–415. doi: 10.1038/nsb0595-407. [DOI] [PubMed] [Google Scholar]
- 20.Das K, Clark J, A.D., Lewi PJ, Heeres J, de Jonge MR, Koymans LMH, Vinkers HM, Daeyaert F, Ludovici DW, Kukla MJ, et al. Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants. J. Med. Chem. 2004;47:2550–2560. doi: 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 21.Janssen PA, Lewi PJ, Arnold E, Daeyaert F, de Jonge M, Heeres J, Koymans L, Vinkers M, Guillemont J, Pasquier E, et al. In search of a novel anti-HIV drug: multidisciplinary coordination in the discovery of 4-[[4-[[4-[(1E)-2-cyanoethenyl]-2,6-dimethylphenyl]amino]-2-pyrimidinyl]amino]benzonitrile (R278474, rilpivirine). J Med Chem. 2005;48:1901–1909. doi: 10.1021/jm040840e. [DOI] [PubMed] [Google Scholar]
- 22.de Bethune MP. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: a review of the last 20 years (1989-2009). Antiviral Res. 2010;85:75–90. doi: 10.1016/j.antiviral.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 23•.Das K, Bauman JD, Clark AD, Jr., Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci U S A. 2008;105:1466–1471. doi: 10.1073/pnas.0711209105. [Structures of wild-type and two clincally relevant RT double mutants in complexes with rilpivirine revealed how the NNRTI wiggles and jiggles to overcome the impact of drug-resistance mutations in the pocket.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chong P, Sebahar P, Youngman M, Garrido D, Zhang H, Stewart EL, Nolte RT, Wang L, Ferris RG, Edelstein M, et al. Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase. J Med Chem. 2012;55:10601–10609. doi: 10.1021/jm301294g. [DOI] [PubMed] [Google Scholar]
- 25•.Kuroda D, Bauman JD, Challa JR, Patel D, Troxler T, Das K, Arnold E, Hochstrasser RM. Snapshot of the equilibrium dynamics of a drug bound to human immunodeficiency virus 1 reverse transcriptase. Nature Chem. 2013 doi: 10.1038/nchem.1559. In press (DOI: 10.1038/NCHEM.1559) [Using two-dimensional infrared spectroscopy (2D IR), molecular dynamics simulations, and crystallography, the study showed that the cinnamonitrile group of rilpivirine maintains a water-mediated interaction with RT. This interaction appears to be important for improved binding of rilpivirine compared to its predecessor analogs. This paper also reports a 1.51 Å structure of RT, the highest resolution reported to date. 2D-IR spectral signatures of the two nitrile groups of rilpivirine bound to RT are similar in crystal and solution, indicating that ligand-protein interactions in the crystal are representative of solution conditions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frenkel YV, Clark AD, Jr., Das K, Wang YH, Lewi PJ, Janssen PA, Arnold E. Concentration and pH dependent aggregation of hydrophobic drug molecules and relevance to oral bioavailability. J Med Chem. 2005;48:1974–1983. doi: 10.1021/jm049439i. [DOI] [PubMed] [Google Scholar]
- 27••.Jilek BL, Zarr M, Sampah ME, Rabi SA, Bullen CK, Lai J, Shen L, Siliciano RF. A quantitative basis for antiretroviral therapy for HIV-1 infection. Nat Med. 2012;18:446–451. doi: 10.1038/nm.2649. [The study developed new methodology for quantitating dose-response curves for HIV-1 drugs from single-round infectivity assays in CD4+ T-cells. The analysis help rationalize several complex relationships among therapeutic combinations of drugs, and the approach developed may be critical in designing optimal drug combinations and in preclinical development of new drugs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen L, Peterson S, Sedaghat AR, McMahon MA, Callender M, Zhang H, Zhou Y, Pitt E, Anderson KS, Acosta EP, et al. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat Med. 2008;14:762–766. doi: 10.1038/nm1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsiou Y, Ding J, Das K, Clark AD, Jr., Hughes SH, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 30.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 31••.Das K, Martinez SE, Bauman JD, Arnold E. HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat Struct Mol Biol. 2012;19:253–259. doi: 10.1038/nsmb.2223. [The study showed that the RT–DNA complex in a crystal could bind an NNRTI or dNTP (NRTI-TP), but not both. The structure of RT-DNA-nevirapine complex illustrated that an NNRTI inhibits DNA polymerization by imposing conformational restrictions on the RT-DNA complex. The 3′-primer terminus is shifted in conjunction with the primer grip by more than 5 Å away from its position in catalytically competent conformations of RT-DNA.] [DOI] [PMC free article] [PubMed] [Google Scholar]