Abstract
To design and build living systems, synthetic biologists have at their disposal an increasingly large library of naturally derived and synthetic parts. These parts must be combined together in particular orders, orientations, and spacing’s to achieve desired functionalities. These structural constraints can be viewed as grammatical rules describing how to assemble parts together into larger functional units. Here, we develop a grammar for the design of synthetic transcription factors (sTFs) in eukaryotic cells and implement it within GenoCAD™, a Computer-Aided Design (CAD) software for synthetic biology. Knowledge derived from experimental evidence was captured in this grammar to guide the user to create designer transcription factors that should operate as intended. The grammar can be easily updated and refined as our experience with using sTFs in different contexts increases. In combination with grammars that define other synthetic systems, we anticipate that this work will enable the more reliable, efficient, and automated design of synthetic cells with rich functionalities.
Keywords: GenoCAD, grammar, synthetic transcription factor, eukaryotic transcription factor, Saccharomyces cerevisiae
Introduction
Synthetic biology aims to rationally create living systems for basic science, biomedical, and biotechnology applications. To do this, one must first understand how to design synthetic networks with which to program these living systems. In order to implement complex synthetic networks, synthetic biologists require a library of well-characterized parts, such as promoters, terminators, transcription factors (TFs) and reporters, as well as rules for assembling these parts into higher-order circuits.
Transcription factors are an important class of parts for synthetic biology. They often form the regulatory links within the networks that synthetic biologists build. Synthetic networks constructed to date have largely relied upon the use of TFs from nature, such as TetR, LacI and AraC1–3. However, the number of well-characterized and orthogonal natural TFs is limited; solely relying on natural TFs therefore imposes limitations on the size of synthetic networks that can be constructed. To overcome this problem, synthetic transcription factors (sTFs) have been created4–18, and a variety of platforms for implementing large libraries of sTFs have been described7,8,12 which remove the constraints imposed by natural TFs.
Substructure of sTFs
Synthetic transcription factors rely on the fact that proteins can be modularized and assembled into interchangeable, and generally quasi-independent, natural protein domains. The term ‘transcription factor’ traditionally refers to any protein that regulates transcription by any means. However, within the context of this discussion, all TFs, whether synthetic or natural, influence gene expression by DNA binding at or near promoters and therefore require DNA-binding domains (DBDs). For instance, the zinc-finger-based class of sTFs uses a series of zinc fingers as DBDs, with each zinc finger (ZF) containing a defined amino-acid sequence, which recognizes a specific DNA triplet code (e.g. CTG). By fusing together multiple zinc fingers, a larger DNA-binding domain that recognizes a longer DNA sequence can be constructed.17,19–22
TFs can generally be divided into two classes; activators, which activate or increase transcription and repressors, which decrease or repress transcription. In yeast, activation and repression is typically mediated by ‘effector domains’, which are fused to DBDs, allowing them to be targeted to specific promoters. A commonly used activation domain in synthetic activators in yeast is the VP16 domain, or its derivative, the VP64 domain (formed from 4 tandem repeats of the VP16 domain)7,8,12, while a commonly used repression domain is the SSN6 domain23. VP16 recruits various transcription factors necessary for transcription and a Histone Acetylase Complex (HAC)24. HACs lead to acetylation of nearby histones, causing chromatin to unwind and allowing access to the promoter by the transcriptional machinery24. Conversely, SSN6 is thought to repress transcription by preventing transcriptional initiation by RNA polymerase and recruiting Histone De-acetylase Complexes (HDACs), which de-acetylate histones, leading to compaction of the chromatin and prevention of further access to the promoter by the transcriptional machinery25. Repression can also be achieved without an effector domain by using DBDs to sterically block initiation of RNA polymerase18.
Rules and Grammars
A grammar is simply a set of design rules, which can be used to guide the design process or enforce standards. This formalism is suited to capturing a domain expertise in a format that constrains non-expert users to produce designs that conform to what is known by expert users (i.e., the experienced synthetic biologist) to typically work. For instance, many synthetic biologists working on various applications use eukaryotic sTFs in their projects. Yet, only a small fraction of the potential users of sTFs are familiar enough with sTF design to take advantage of the rapid progress in this field. The grammar presented here could help transfer the expertise of sTF specialists to those with expertise in other fields.
To someone specializing in the development of the next generation of sTFs, the benefits of constraining the design process may not be immediately apparent since optimal designs are unknown. In this case, grammars are a formal representation of a hypothesis that will be tested experimentally. This formalization effort encourages a thorough analysis of the different aspects of the design process, which can help uncover potential issues before starting the experimental validation. It also supports the articulation of various context-dependencies that may affect the success of a design strategy in different conditions. Furthermore, grammars implemented within computer-aided design tools may help to organize experimental libraries and plans.
Rules for sTF Design
On some level, all biological parts (whether natural or synthetic) conform to certain design rules to varying degrees. For example, E. coli promoters usually require −10 and −35 boxes for RNA polymerase binding to initiate transcription, while proteins require a start codon from where translation is started. The structure of sTFs can also be designed to conform to certain rules. For instance, to design an sTF that behaves as an activator, it should have a DBD fused somehow to an AD. However, just as the structure of an sTF can be more complex than a two-domain fusion, the grammar can also be more complex.
Here, we propose a grammar for the design of sTFs in Saccharomyces cerevisiae. We implement this in GenoCAD, a web-based synthetic-biology CAD software26. GenoCAD was derived from the observation that constructs used in synthetic biology could be generated by context-free grammars27. It is therefore a logical choice for implementing an sTF grammar. GenoCAD includes a system to create and manage libraries of user-defined parts. The GenoCAD design module provides a wizard-like interface which guides users to generate structurally valid constructs, and allows the online design workspace to be customized26. We propose grammars for the design of sTFs based on zinc fingers, Transcription Activator-like Effectors (TALEs), and the recently developed Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas-based system. Our grammar covers the design of sTFs that: 1) use any one of these systems, 2) use effector domains to activate or repress transcription, 3) use fluorescent reporter domains to enable quantification of sTF abundance, and 4) permit the design of sTFs that form dimeric complexes with other sTFs, which can be used to engineer cooperativity between sTF monomers.
We believe our grammar serves as a first attempt to standardize sTF design and create a foundation that can be built upon and refined as experience with designing and using sTFs grows.
The sTF grammar
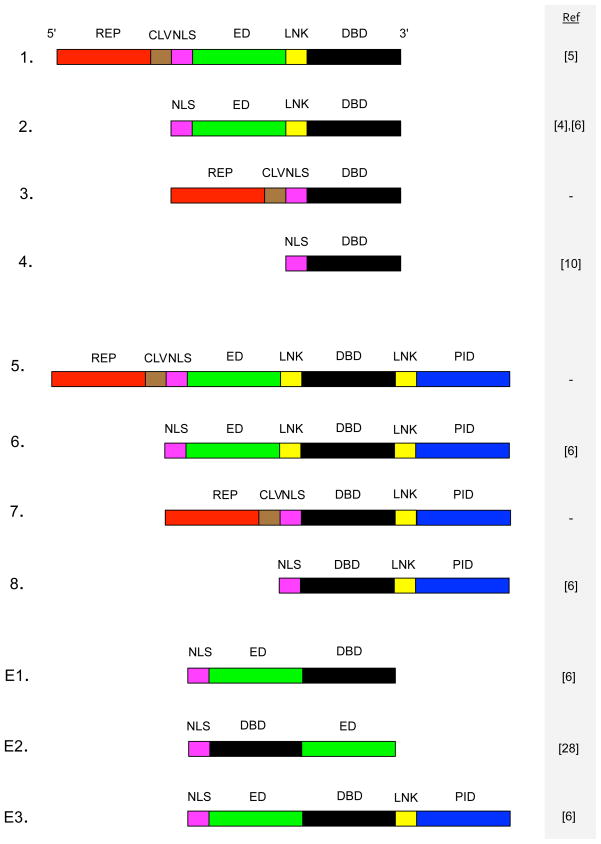
While it would be possible to construct an arbitrarily broad grammar that would allow an expert user to define any combination of protein domains in any order, this defeats the purpose of the grammar in productively constraining non-expert users. Therefore we have opted for a highly constrained grammar based around 11 possible sTF structures (Figure 1). The 11 possible structures are, 5′ to 3′:
Figure 1.
Structures of sTFs allowed within the grammar. Eight possible general structures are allowed within the grammar. In addition, E1–E3 are experimentally verified structures. DBD = DNA Binding Domain, LNK = Linker sequence, ED = Effector Domain, NLS = Nuclear Localization Signal, CLV = Cleavage sequence, REP = REPorter, PID = Protein Interaction Domain. All constructs are oriented from 5′ to 3′. References for structures 1–8 describe studies in which similar structures have been experimentally verified. References for structures E1–E3 denote the study in which the structure was experimentally verified.
-
1
5′-REP-CLV-NLS-ED-LNK-DBD-3′
-
2
5′-NLS-ED-LNK-DBD-3′
-
3
5′-REP-CLV-NLS-DBD-3′
-
4
5′-NLS-DBD-3′
-
5
5′-REP-CLV-NLS-ED-LNK-DBD-LNK-PID−3′
-
6
5′-NLS-ED-LNK-DBD-LNK-PID−3′
-
7
5′-REP-CLV-NLS-DBD-LNK-PID−3′
-
8
5′-NLS-DBD-LNK-PID−3′
-
E1
5′-NLS-ED-DBD-3′
-
E2
5′-NLS-DBD-ED-3′
-
E3
5′-NLS-ED-DBD-LNK-PID−3′
where PID = Protein Interaction Domain, LNK = Linker sequence, ED = Effector Domain, CLV = CLeaVage sequence, REP = REPorter domain, NLS = Nuclear Localization Sequence.
Structures 1–8 shown in Figure 1 allow for the construction of sTFs that can provide either activation through effector domains, or repression by effector domains or steric hindrance of RNA polymerase initiation. The sTF expression levels can be quantified using reporter proteins and sTFs can be made to behave cooperatively when paired with a suitable partner. These structures therefore cover the range of functions that are required by sTFs in the construction of synthetic gene networks. The design of our structures is based on a synthesis of the available experimental evidence. However, many of these structures are themselves novel, and to our knowledge have not yet been experimentally verified. References in Figure 1 denote studies that offer experimental evidence for structures that are similar to the structures presented here. We have also included three structures that have been experimentally verified in S. cerevisiae (E1–E3). These are two variants of an NLS-DBD-ED structure (E1 and E2) and a modification of E1 that allows for protein-protein interactions.
For structures 1–8, the general structural constraints captured in the grammar are as follows:
General structure:
The physical structure of the sTF (i.e., the ordering of the domains) is organized around the position of the DBD.
All domains apart from Protein-Interaction-Domains (PID) (with a LNK domain present between it and the DBD) are built 5′ to the DBD.
PID domains are built 3′ to the DBD.
If a reporter is used, it is at the 5′ terminal domain.
Between a DBD and either a PID or an ED, there must be a linker domain (LD).
Between any domain and the reporter domain, there must be a cleavage domain (CD). The most common cleavage domain, and the one used in the library with our GenoCAD grammar is the 2A sequence. However, this is not a true cleavage domain as no proteolytic cleavage of the protein occurs. Rather, the 2A sequence causes ‘ribosome skipping’ to occur28, whereby the peptide bond formation does not occur, and two separate proteins are therefore produced29. In order to follow the literature on 2A domains, we refer to CDs as ‘cleavage domains’29 to denote that they include any domain which separates proteins, whether it be by true cleavage or not.
A Nuclear Localization Signal (NLS) is added at the 5′ of the protein. If a cleavage domain is present, then the NLS is immediately 3′ to the cleavage domain.
There are many possible variations of the structures 1–8 shown in Figure 1, and the subdomains in these different fusion configurations may have different structures and therefore different activities. For simple fusion proteins, such as the fusion of one domain with a fluorescent protein, it has been suggested that both configurations of the fusion protein be tested30. However, when the number of possible configurations is large, testing all possible configurations is usually impractical. The justifications for these general constraints are as follows:
Both the PDZ and leucine zipper (LZ) domains have been used successfully as a PID domain to enable cooperativity in sTFs8,12. LZ domains have been shown to function when placed internally in the sTF8 and should also function at either terminus. However, the ligand to which the PDZ domain binds must be at the C-terminal of the protein31. To minimize the number of available structures, we therefore constrain both components of a PID−based interaction (the protein and its ligand) to be at the 3′ (C-terminal) end.
Because of the constraint on the PID domain to be at the 3′ end, we therefore constrain all other domains to be 5′ to the DBD. ZF-based sTFs have also been successfully constructed with the effector domain to the 5′ of the DBD12.
Linker domains are routinely used when creating fusion proteins and have been shown to improve folding and stability of fusion proteins, enhance the expression of fusion proteins, and increase the activity of the fusion protein32.
Nuclear localization signals have been placed internal to sTFs7 and as well as at the termini7,12. To our knowledge, there has not yet been a comprehensive study as to if and how the placement and number of NLS sequence(s) affects the characteristics of the sTF. Therefore, here we place the NLS at the N-terminal region with respect to the DBD.
The addition of a reporter domain at the 5′ end of the sTF allows for the concentration of sTFs present within the cell at any given time to be quantified. However, the presence of this additional reporter domain may adversely affect the folding of the rest of the sTF and consequently impair its function. Placing a ‘cleavage’ domain before the reporter may mitigate any such issue. Upon translation, the ‘cleavage’ domain results in the protein sequence being cleaved at a specific position. The efficiency of ‘cleavage’ with 2A domains has been shown to be affected by the sequence of the upstream protein29. However, by simply adding a Gly-Ser-Gly (GSG linker) before the 2A sequence, the efficiency can be increased to ~100% for all upstream proteins tested29. We include this GSG linker as a standard component of the P2A sequence.
PIDs
PID domains can be defined as either homodimerization (e.g. the LZ domain) or heterodimerization domains (e.g. the PDZ domain and its ligand, or heterospecific interactions based on coiled-coils33,34). Heterodimeric PID domains can be further defined as ‘positive (+)’ or ‘negative (−)’. A ‘positive’ PID domain is intended to interact with a complementary ‘negative’ PID domain. As both PID domains are on the 3′ end of the sTF, in the case of PDZ domains one of the sTFs must reverse its direction to allow for interaction between the two sTFs. It is trivial to synthesize the DNA-binding site of the DBD in reverse, so this is where the orientation issue is dealt with, rather than allowing PID domains to be at the 5′ end of the sTF.
DBDs
ZF domains cannot be further sub-defined in the grammar. This is because of apparent interdependencies between the individual ZFs that form a ZF-array DBD, which means that ZFs do not always behave in a truly modular fashion35–37. It is therefore more reliable to use entire ZF-array DBDs that have been verified for specificity, rather than construct them de novo and risk interdependency issues. No such interdependencies are known for the Repeat Variable Domain (RVDs) that form the TALE-based DBDs, and therefore TALEs can be further sub-defined in the design process. A TALE domain must include a 5′ and 3′ TALE region, and >0 repeat variable domains (RVDs) in-between the 5′ and 3′ TALE DBD region. The dCas9 domain cannot be further sub-defined.
gRNA
The CRISPR-TF system comprises a dCas9 domain (optionally fused to an Effector Domain) and a guide RNA (gRNA). dCas9 is a catalytically inactive form of the Cas9 nuclease. The gRNA itself is comprised of a sequence that binds through complementary base pairing to one strand of the DNA target sequence, and a ‘handle’ sequence: a hairpin forming sequence that dCas9 recognizes and binds to. The gRNA therefore ‘guides’ the dCas9 based TF to its target site and determines the DNA-binding specificity of the dCas9:gRNA complex, and therefore its effects on the expression of the target gene10,11,16. Every dCas9 domain should have a gRNA defined for it. We use a single gRNA, where the handle and the targeting sequence are fused, rather than the original ‘dual’ RNA system, where these components were separate and had to interact in vivo for the system to function38,39.
The user is able to define gRNAs within the sTF grammar. As gRNAs are not translated, they do not require either a start or stop codon.
EDs
Effector domains can be designated as either Activator Domains (ADs) or Repressor Domains (RDs).
REP
Reporter domains can either be designated as a fluorescent protein reporter (e.g., GFP or mCherry) or a non-fluorescent protein reporter (e.g., beta-galactosidase)
The GenoCAD implementation
The preceding section described the biological details of the grammar. This section describes the specifics of the implementation of this grammar within GenoCAD.
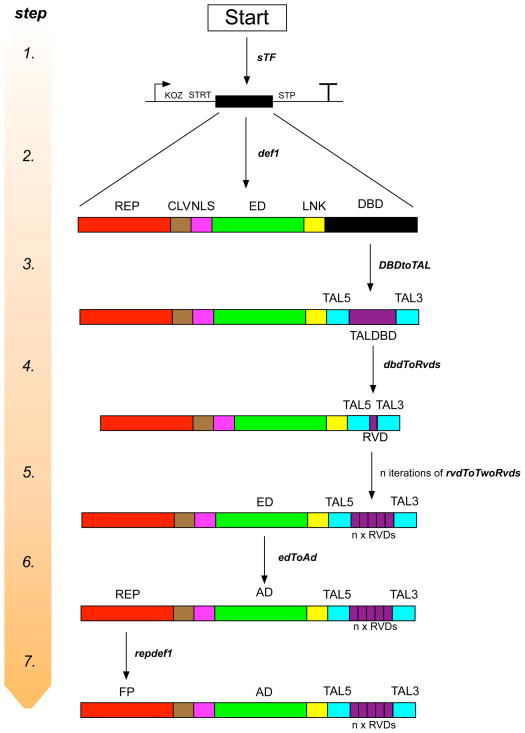
A GenoCAD grammar is defined by categories of genetic parts and transformation rules between these categories. For instance, an ED would be a category, as would an AD and an RD. The transformation rule that links these would be that an ED can be defined as (or ‘transformed’ into) either an AD or an RD. When a user wants to define a genetic construct within GenoCAD, they always begin from the ‘start’ category. From the ‘start’ category they can iteratively transform particular categories into different sub-categories, therefore defining the specifics of the genetic construct. An illustrative example is shown in Figure 2.
Figure 2.
An example design process for an sTF within GenoCAD. The seven steps of the process are oriented from top to bottom. The transformation rules that transform the construct from the ‘Start’ state to the final construct are depicted in bold italics (‘sFT’, ‘def1’, ‘DBDtoTAL’, ‘dbdToRvds’, ‘rvdToTwoRvds’, ‘edToAd’, ‘repdef1’). DBD = DNA Binding Domain, LNK = Linker sequence, ED = Effector Domain, NLS = Nuclear Localization Signal, CLV = Cleavage sequence, REP = reporter, TALBDB = TALE DNA Binding Domain, TAL5 = 5′ domain of the TAL, TAL3 = 3′ domain of the TAL, RVD = Repeat Variable Domain, AD = Activation Domain, FP = Fluorescent Protein. The right-angled arrow and ‘T’ denote the promoter and the terminator, respectively. ‘KOZ’, ‘STRT’ and ‘STP’ denote a Kozak sequence, start codon, and stop codon, respectively. All constructs are oriented from 5′ to 3′.
The categories and transformation rules for the sTF grammar are given in Tables 1 and 2, respectively.
Table 1.
Categories in the sTF grammar. Re-writable categories can be transformed into other categories, while terminal categories cannot.
| Category ID | Description | Type |
|---|---|---|
| S (start) | Start category; the default ‘root’ category of the grammar | Re-writable |
| sTF | The entire sTF (not including the promoter or terminator) | Re-writable |
| DBD | The DBD of the sTF | Re-writable |
| TALDBD | The part of the TAL formed by RVDs. Does not include the 5′ and 3′ TAL ends | Re-writable |
| RVD | An individual RVD that forms part of the TAL DBD | Re-writable |
| ED | The effector domain. Can be either an activation or repression domain | Re-writable |
| REP | The reporter domain | Re-writable |
| PID | A protein interaction domain. Can interact with other protein interaction domains to allow the sTF to form dimers | Re-writable |
| gRNA | The guide RNA | Re-writable |
| PROM | The promoter that drives expression of the sTF | Terminal |
| KOZ | A Kozak sequence | Terminal |
| TERM | The terminator for the sTF | Terminal |
| ZFDBD | The DBD for a ZF based sTF | Terminal |
| DCAS9 | A catalytically inactive Cas9 domain | Terminal |
| TAL5 | The 5′ end of the TAL | Terminal |
| TAL3 | The 3′ end of the TAL | Terminal |
| LNK | A (usually) short linker sequence that joins two domains | Terminal |
| CLV | An amino acid sequence that joins two domains but is ‘cleaved’ during/after translation, separating the domains | Terminal |
| FP | Fluorescent protein that acts as a reporter | Terminal |
| xREP | Any domain that acts as a reporter but is not an FP | Terminal |
| AD | An effector domain that is an activation domain; it causes an increase in the expression of the target promoter | Terminal |
| RD | An effector domain that is a repression domain; it causes a decrease in the expression of the target promoter | Terminal |
| PIDhm | A homodimerizing PID domain | Terminal |
| PIDht+ | A heterodimerizing PID+ domain; will interact (bind to) its corresponding PID− domain | Terminal |
| PIDht− | A heterodimerizing PID− domain; will interact (bind to) its corresponding PID+ domain | Terminal |
| STRT | A start codon | Terminal |
| TRGT | The sequence of the gRNA complementary to the target sequence | Terminal |
| HNDLE | The Cas9 binding domain of the gRNA | Terminal |
| STP | A stop codon | Terminal |
Table 2.
Transformation rules in the sTF grammar.
| Rule Code | Rule | Description |
|---|---|---|
| sTF | S to PROM-STRT-sTF- STP-TERM | Converts the start state to a gene structure containing an sTF |
| def1 | sTF to RP-CLV-NLS- ED-LNK-DBD | Converts the sTF to the 1st structure variant in list Figure 1. |
| def2 | sTF to NLS-ED-LNK- DBD | Converts the sTF to the 2nd structure variant in list Figure 1. |
| def3 | sTF to RP-CLV-NLS- DBD | Converts the sTF to the 3rd structure variant in list Figure 1. |
| def4 | sTF to NLS-DBD | Converts the sTF to the 4th structure variant in list Figure 1. |
| def5 | sTF to RP-CLV-NLS- ED-LNK-DBD-LNK-PID | Converts the sTF to the 5th structure variant in list Figure 1. |
| def6 | sTF to NLS-ED-LNK- DBD-LNK-PID | Converts the sTF to the 6th structure variant in list Figure 1. |
| def7 | sTF to RP-CLV-NLS- DBD-LNK-PID | Converts the sTF to the 7th structure variant in list Figure 1. |
| def8 | sTF to NLS-DBD-LNK- PID | Converts the sTF to the 8th structure variant in list Figure 1. |
| defE1 | sTF to NLS-ED-DBD | Converts the sTF to the structure E1 in Figure 1. |
| defE2 | sTF to NLS-DBD-ED | Converts the sTF to the structure in E2 in Figure 1. |
| defE3 | sTF to NLS-ED-DBD- LNK-PID | Converts the sTF to the structure E3 in Figure 1. |
| DBDtoTAL | DBD to TAL5-TALDBD- TAL3 | Converts the DBD to a TAL DBD including the 5′ and 3′ TAL end regions |
| DBDtoZF | DBD to ZF | Converts the DBD to a Zinc Finger |
| DBDtodcas9 | DBD to dcas9 | Converts the DBD to a dCas9 domain |
| edToAd | ED to AD | Converts the Effector domain to an activation domain |
| edToRd | ED to RD | Converts the effector domain to a repression domain |
| dbdToRvds | TALDBD to RVD | Converts the TAL DBD to an RVD |
| rvdToTwoRvd | RVD to RVD-RVD | Converts one RVD domain to two RVD domains |
| repdef1 | REP to FP | Converts a Reporter to a fluorescent protein |
| repdef2 | REP to xREP | Converts a Reporter to a reporter domain other than a fluorescent protein |
| PIDhm def | PID to PIDhm | Converts a PID to a PIDhm domain |
| PIDht+def | PID to PIDht+ | Converts a PID to a PIDht+ domain |
| PIDht−def | PID to PIDht− | Converts a PID to a PIDht− domain |
| gRNA | S to PROM-gRNA- TERM | Converts the start state to a gene structure containing a gRNA |
| gRNAdef | gRNA to TRGT-HNDLE | Converts the gRNA to a target sequence and a handle sequence |
Future Developments to the Grammar
In this paper, we have presented a grammar for the design of synthetic transcription factors. We have implemented these in GenoCAD, a CAD software that uses grammars to define synthetic constructs. The grammars allow for the construction of 11 different sTF structures based on commonly used components. The DNA-binding domain of the sTF can be defined as zinc fingers, TALEs, or the dCas9 protein, which acts in concert with a gRNA to target specific DNA sequences. Our grammar also allows for the design of cooperative transcription factors through the incorporation of protein interaction domains.
The grammars presented here represent one interpretation of our current experience with sTFs. However, we make two implicit assumptions in defining a grammar: firstly, our grammar is focused on the domain structure of the transcription factors, while ultimately it is the amino-acid sequence of the protein that is important, as it is this sequence which defines how the protein folds, and therefore how it functions. Secondly, although we base the selection of our 11 structures on experimental evidence, we are extrapolating from this evidence to form the structures we described here. Thus, we assume that what has been observed in one context (e.g. the placement of NLS in a particular sTF) will also be applicable in other sTFs.
These rules are intended to allow a user to design sTFs with structures that will be functional. It should be noted that the 8 general structures we present in Figure 1 have not yet been experimentally verified for functionality – although there are similarities to known functional structures. However, there likely exist structures that will have more desirable characteristics that the ones allowed within this grammar. For instance, perhaps using multiple nuclear-localization sequences in various specific positions may increase the rate of nuclear import for a certain sTF40,41 or putting a longer linker in between a ZF DBD and a particular ED may increase the magnitude of the expression change caused by the ED32. These grammars should therefore be revised as our knowledge of sTF design increases.
This grammar could be improved in a number of ways. For example, although our grammar allows for a TALE DBD to be constructed with only a single RVD, in reality, to ensure both sufficient specificity and binding affinity, the number of RVDs would typically be on the order of twenty7. With a single rewriting rule (RVD -> RVD RVD) the grammar can introduce as many RVDs as necessary. However, the process is cumbersome and having many RVD icons in the design is not particularly elegant. A more refined version of the grammar could introduce categories representing blocks of 1, 5, 10 RVDs and the corresponding rules. Future iterations of the grammars will make it possible to quickly generate a broad range of RVDs using a smaller number of icons and rewriting steps. Furthermore, the PID domains are labeled ‘positive’ and ‘negative’, which guide the user somewhat towards permissible pairings of sTFs. However, this is not a constraint, and the user is still able to pair sTFs incorrectly. An improvement would therefore be for the user to ‘pair up’ designed sTFs within GenoCAD, which could be automatically examined for compatibility. Another useful constraint on pairing would be between dCas9 domains and gRNAs.
The current version of the grammar focuses on the design of individual transcription factors. A natural extension of this grammar would be to include rules allowing the design of gene networks derived from these sTF. For instance, one could constrain sTFs to ‘pair’ with promoters that contain sequences that the DBD of the sTF is able to bind to. Adding a network layer to the grammar would make it possible to benefit from GenoCAD simulation environment. As sTF libraries become better characterized with kinetic data, it would be advantageous to be able to incorporate this information into GenoCAD for the purpose of simulating the dynamics of gene networks built from these sTFs. Further integration of synthetic circuit modeling within whole-cell models in GenoCAD could enhance the utility of this approach42.
As the number and complexity of synthetic components engineered by synthetic biologists increases, encapsulating current knowledge by defining standards will become increasingly important. These standards will allow for more reliable construction of synthetic living systems by scientists and engineers with a more wide-ranging level of expertise. We propose that sTF grammars, such as those presented here, begin to be considered as a first step towards the standardization of a broad range of synthetic genetic parts that could be combined in synthetic gene circuit designs.
Acknowledgments
This work was supported by the Defense Advanced Research Projects Agency, the NIH New Innovator Award (1DP2OD008435), the National Science Foundation (1124247 to TKL and 0850100 to JP), and the Office of Naval Research (N00014-13-1-0424).
Footnotes
The sTF grammar is available upon request.
Author Contributions.
O.P. and J.P. implemented the grammars. O.P., J.P., and T.K.L. designed and analysed the grammars and wrote the paper.
[Conflict of Interest]: JP has a financial interest in GenoFAB, LLC
References
- 1.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 2.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 3.Lou C, Liu X, Ni M, Huang Y, Huang Q, Huang L, Jiang L, Lu D, Wang M, Liu C, Chen D, Chen C, Chen X, Yang L, Ma H, Chen J, Ouyang Q. Synthesizing a novel genetic sequential logic circuit: a push-on push-off switch. Mol Syst Biol. 2010;6:350. doi: 10.1038/msb.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F, Crawford GE, Hartemink AJ, Gersbach CA. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Methods. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 7.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lohmueller JJ, Armel TZ, Silver PA. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 2012;40:5180–5187. doi: 10.1093/nar/gks142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, Joung JK, Collins JJ. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morbitzer R, Römer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci U S A. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folcher M, Xie M, Spinnler A, Fussenegger M. Synthetic mammalian trigger-controlled bipartite transcription factors. Nucleic Acids Res. 2013;41:e134. doi: 10.1093/nar/gkt405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geissler R, Scholze H, Hahn S, Streubel J, Bonas U, Behrens SE, Boch J. Transcriptional activators of human genes with programmable DNA-specificity. PLoS One. 2011;6:e19509. doi: 10.1371/journal.pone.0019509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farzadfard F, Perli SD, Lu TK. Tunable and Multifunctional Eukaryotic Transcription Factors Based on CRISPR/Cas. ACS Synth Biol. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beerli RR, Barbas CF. Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 18.Blount BA, Weenink T, Vasylechko S, Ellis T. Rational diversification of a promoter providing fine-tuned expression and orthogonal regulation for synthetic biology. PLoS One. 2012;7:e33279. doi: 10.1371/journal.pone.0033279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q, Segal DJ, Ghiara JB, Barbas CF. Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci U S A. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim JS, Pabo CO. Getting a handhold on DNA: design of poly-zinc finger proteins with femtomolar dissociation constants. Proc Natl Acad Sci U S A. 1998;95:2812–2817. doi: 10.1073/pnas.95.6.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sera T, Uranga C. Rational design of artificial zinc-finger proteins using a nondegenerate recognition code table. Biochemistry. 2002;41:7074–7081. doi: 10.1021/bi020095c. [DOI] [PubMed] [Google Scholar]
- 23.Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 24.Hall DB, Struhl K. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J Biol Chem. 2002;277:46043–46050. doi: 10.1074/jbc.M208911200. [DOI] [PubMed] [Google Scholar]
- 25.Malavé TM, Dent SYR. Transcriptional repression by Tup1--Ssn6. Biochem Cell Biol. 2006;84:437–443. doi: 10.1139/o06-073. [DOI] [PubMed] [Google Scholar]
- 26.Czar MJ, Cai Y, Peccoud J. Writing DNA with GenoCAD™. Nucleic Acids Res. 2009;37:W40–W47. doi: 10.1093/nar/gkp361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Y, Hartnett B, Gustafsson C, Peccoud J. A syntactic model to design and verify synthetic genetic constructs derived from standard biological parts. Bioinformatics. 2007;23:2760–2767. doi: 10.1093/bioinformatics/btm446. [DOI] [PubMed] [Google Scholar]
- 28.Donnelly MLL, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 29.Szymczak-Workman AL, Vignali KM, Vignali DAA. Design and construction of 2A peptide-linked multicistronic vectors. Cold Spring Harb Protoc. 2012;2012:199–204. doi: 10.1101/pdb.ip067876. [DOI] [PubMed] [Google Scholar]
- 30.Snapp E. Curr Protoc Cell Biol. John Wiley & Sons, Inc; Hoboken, NJ, USA: 2005. Design and Use of Fluorescent Fusion Proteins in Cell Biology; pp. 21.4.1–21.4.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Zaro JL, Shen W-C. Fusion protein linkers: Property, design and functionality. Adv Drug Deliv Rev. 2012;65:1357–1369. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson KE, Bashor CJ, Lim WA, Keating AE. SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains. ACS Synth Biol. 2012;1:118–129. doi: 10.1021/sb200015u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinke AW, Grant RA, Keating AE. A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J Am Chem Soc. 2010;132:6025–6031. doi: 10.1021/ja907617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramirez CL, Foley JE, Wright DA, Müller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, Cathomen T, Voytas DF, Joung JK. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam KN, van Bakel H, Cote AG, van der Ven A, Hughes TR. Sequence specificity is obtained from the majority of modular C2H2 zinc-finger arrays. Nucleic Acids Res. 2011;39:4680–4690. doi: 10.1093/nar/gkq1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlson DF, Fahrenkrug SC, Hackett PB. Targeting DNA With Fingers and TALENs. Mol Ther Nucleic Acids. 2012;1:e3. doi: 10.1038/mtna.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo M, Pang CWM, Gerken AE, Brock TG. Multiple nuclear localization sequences allow modulation of 5-lipoxygenase nuclear import. Traffic. 2004;5:847–854. doi: 10.1111/j.1600-0854.2004.00227.x. [DOI] [PubMed] [Google Scholar]
- 41.Gassman NR, Clodfelter JE, McCauley AK, Bonin K, Salsbury FR, Scarpinato KD. Cooperative nuclear localization sequences lend a novel role to the N-terminal region of MSH6. PLoS One. 2011;6:e17907. doi: 10.1371/journal.pone.0017907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell O, Jain B, Karr JR, Covert MW, Lu TK. Towards a whole-cell modeling approach for synthetic biology. CHAOS. 2013;23:25112. doi: 10.1063/1.4811182. [DOI] [PMC free article] [PubMed] [Google Scholar]