Abstract
Attempts to characterize cellular behaviors with static, univariate measurements cannot fully capture biological complexity and lead to an inadequate interpretation of cellular processes. Significant biological insight can be gleaned by considering the contribution of dynamic protein post-translational modifications (PTMs) utilizing systems-level quantitative analysis. High-resolution mass spectrometry coupled with computational modeling of dynamic signal-response relationships is a powerful tool to reveal PTM-mediated regulatory networks. Recent advances using this approach have defined network kinetics of growth factor signaling pathways, identified systems level responses to cytotoxic perturbations, elucidated kinase-substrate relationships, and unraveled the dynamics of PTM cross-talk. Innovations in multiplex measurement capacity, PTM annotation accuracy, and computational integration of datasets promises enhanced resolution of dynamic PTM networks and further insight into biological intricacies.
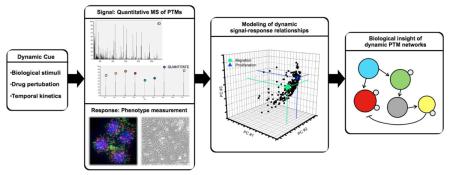
Introduction
Systems biology aims to identify emergent properties: behaviors, such as cell phenotype, defined by the interaction of many components in the network that are not predictable from the analysis of any single component. The classic dogma of molecular biology, in which information flows from DNA to RNA to proteins to coordinate the development and function of a cell, does not adequately explain these emergent properties, and also fails to account for the rapid response of biological systems to altered intra- or extra-cellular conditions. A variety of epigenetic phenomena, including protein-protein interactions, chromatin alterations, non-coding RNAs, and post-translational modifications (PTMs), among others, have been implicated in governing cellular responses and phenotypes. Importantly, none of these epigenetic regulatory events can be accurately inferred from genomic information alone [1–4]. The importance of these additional layers of non-genomic regulation cannot be understated; dynamic epigenetic regulatory networks must be considered to fully appreciate the complete nature of a biological system.
One well-studied mechanism by which cells acutely respond and coordinate activities is through post-translational modification (PTM) based cellular signaling networks. The assortment of PTMs in a eukaryotic cell is staggering, with over 600 different protein modification types annotated in the RESID database (September 2013 release; URL http://pir.georgetown.edu/resid/). Protein PTMs can rapidly modulate complex formation, stability, activity, and spatial localization[4]. Integration of this vast array of PTMs ultimately governs cellular information processing and the corresponding cellular behaviors such as migration, apoptosis, and proliferation that are elicited [5]. Of the routinely studied PTMs, phosphorylation, arguably the most abundant PTM in eukaryotic cells, has been shown to drive signal transduction cascades connecting cell surface receptors to resultant cell phenotypes [6–8]. As such, throughout this review special attention will be paid to the role of dynamic phosphorylation in coordinating information flow within the cell and regulating cellular response to cellular perturbations.
Identification of the altered networks underlying emergent properties typically requires systems-level analysis entailing the collection of multivariate data which can yield hypotheses and predictions that are beyond a scientist's intuition [9,10]. Systems level analyses can also clarify paradoxical findings. For example, the dissection of signaling pathways by traditional reductionist approaches can lead to apparent contradictions in the activity or role of individual proteins. The literature is filled with examples of seemingly conflicting results for a number of highly studied signaling components such as Notch and MAPK [11,12]. One landmark paper from the Yaffe group demonstrated the power of systems-level analysis by considering apoptotic regulation by c-Jun N-terminal kinase (JNK) [13]. Through a series of multivariate sampling measurements and data-driven computational modeling, they concluded that phospho-JNK levels could appear anti-apoptotic, proapoptotic, or uninvolved in apoptosis depending on the `signaling state' of the cell. Therefore, a signaling component can have a multivariate nature and response that can only be fully understood by taking into account the context of the network in which it is functioning. Thus, to fully comprehend the multidirectional intracellular interconnections that exist, techniques must be employed to comprehensively detect and quantify multiple components of a network.
The analytical tools available to unravel these details are expanding but ultimately must satisfy minimal requirements to accurately identify and quantify many components within a single analysis, thereby enabling quantitative analysis at a systems level. Mass spectrometry (MS) based proteomic tools fulfill these requirements and have become a mainstay technology for monitoring system level dynamics of PTMs [14,15]. As such, MS-based strategies have helped to reveal PTM signaling networks by mapping hundreds to thousands of PTM sites in many different cell or tissue types while simultaneously providing quantitative abundances across multiple conditions [16].
To accurately capture information flow and regulatory networks, it is critical to consider the dynamic nature of biological systems. A simple enumeration of the components within a network neglects the relationships and coordination that exists as the system adapts and responds. Cellular states are in constant flux, responding to environmental cues and genetic changes in time and space, so techniques to measure networks in functioning cells must strive to account for these dynamics. One way to achieve this annotation is by meticulous experimental mapping of epigenetic changes across multiple conditions and multiple time points following cell perturbation to gain an understanding of specific systems level responses. This approach switches the emphasis from simply identifying the parts of the system to examining the components that are altered between conditions of interest [17]. This information can be experimentally uncovered by grossly perturbing the state of the cell at the molecular level with some sort of input stimuli (cue), thereby activating diverse PTM conduits (signals) responsible for cellular decision processes and phenotypes (response). These cue-signal-response type measurements, if monitored at the network-level and across multiple systematic conditions, can reveal the dynamic molecular nodes and processes on which further investigation should focus and highlight the relationship of these nodes to quantitative phenotype data if available. As an example, we have analyzed the effects of increased HER2 expression on the tyrosine phosphorylation network response to cell stimulation with epidermal growth factor (EGF) or heregulin [18]. Cellular phenotype response to stimulation was also monitored through quantitative analysis of cell migration and proliferation. Several hundred tyrosine phosphorylation sites were quantified at 4 time points following stimulation with each growth factor in each cell line. The cue-signal-response data was integrated with partial least squares regression (PLSR), enabling the identification of the phosphorylation sites that most strongly correlated with each cell response at each time point. This approach provides putative functional assignments for poorly characterized phosphorylation sites, and the PLSR-based model can be used to predict response to loss of a given phosphorylation site in the network. Given the importance of dynamic signaling networks in regulating the cellular response to perturbation, the remainder of this article will focus on recent reports, tools and approaches relevant to understanding system dynamics of biological processes with a particular emphasis on MS-enabling strategies.
Dynamics of phosphorylation
Mass spectrometric analysis of protein phosphorylation is an immense analytical challenge. Thousands of proteins are expressed in each cell, with most proteins present in multiple different isoforms, and with each isoform potentially modified at multiple phosphorylation sites [19,20]. Due to the transient nature and varied stoichiometry of each phosphorylation site, it has been very difficult to generate a comprehensive catalog of all protein phosphorylation sites in any given biological sample. It is worth mentioning that, in our opinion, the goal of using systems-level tools to probe phosphorylation must move beyond a superficial `cataloging' of phosphorylation sites and phosphorylated proteins to understanding and determining the putative phosphorylation marks that are responsive and dynamic in nature. Thoughtful design and execution of experimental studies to understand phospho-dynamics can provide rich insights into the underlying mechanics of a functioning network, reveal unanticipated biological features and profoundly guide future ventures in a way that simple indexing cannot. Here we will highlight a few recent studies have derived biological insight to cellular signaling mechanisms through MS-based network analysis. For instance, a recent report from the Ferrara group utilized a phospho-proteomic strategy to dissect vascular endothelial growth factor (VEGF)-regulated phosphorylation dynamics in a human endothelial cell model [21]. VEGF receptor signaling had been previously shown to trigger the PI3KAKT, Raf-MEK, and Src-FAK pathways. Using a selective enrichment schema with five immunoaffinity motif antibodies combined with quantitative mass tagging, they selectively investigated temporal phosphorylation profiles of these three pathways upon VEGF stimulation. This kinetic sampling allowed identification of a discrete sequence of phosphorylation dynamics revealing membrane-proximal events, followed by nuclear and transcription factor phosphorylation, and terminal events implying establishment of an integrated signaling network. Furthermore, the acquisition of this network-level phosphorylation dataset allowed resolution of molecular conduits by which VEGF signaling may lead to RTK reprogramming in endothelial cells upon pharmacological intervention. Another recent report used MS-based phospho-proteomics, combined with stable isotope labeling, to survey the network changes to protein phosphorylation during cell-cycle recovery from DNA-damaging agents [22]. Their analysis revealed 154 proteins that were quantitatively dynamic during recovery from DNA damage–induced G2 arrest across multiple time points. Interestingly, 84 of these proteins were previously identified in a screen of targets phosphorylated upon DNA-damage, but only two phosphosites were shared between the induction of DNA repair and the recovery from DNA repair. This result suggests that PTMs governing DNA repair machinery are astonishing complex with multisite phosphorylation often regulating diametrically opposed functions on the same protein.
Each of the above examples used mass spectrometry to query changes in signaling networks at the systems level. In many cases, selected kinases within the network have previously been implicated in regulating a given cellular response. To utilize this a priori knowledge, analog-sensitive kinases (AS-kinases) and bio-orthogonal ATP analogs can be combined with MS-based phospho-proteomics to identify and quantify direct substrates of given kinases [23,24]. To date, quantification of substrate phosphorylation dynamics has been challenging due to the inability of the ATP analog to penetrate the cell membrane. Semi-permeabilizing the cell membrane allows for addition of the ATP analog to the cell, but can significantly alter cell signaling networks and response to stimulation. Continued development of new methods (e.g. microinjection platforms) should enable more high throughput analysis of kinase substrates across different time points and conditions, thereby providing a much more comprehensive map of the signaling networks. This information would facilitate our understanding of how signaling networks re-wire in the context of different disease states, potentially enabling more directed therapeutic strategies to reset the network.
Dynamics of other PTMs and PTM crosstalk
Deciphering the regulation of other protein PTMs has proven immensely challenging due to a combination of poor affinity capture reagents and the low-abundance/low stoichiometry of many PTMs. Despite these challenges, several studies have now documented proteome-wide lysine acetylation, yet none of these studies have quantified acetylation temporal dynamics [25–27]. Novel affinity capture approaches and advancements in MS technologies have allowed network-wide interrogation of several other PTMs. For instance, protein palmitoylation on cysteine residues has recently been analyzed using bioorthogonal labeling with palmitic acid analog 17-octadecynoic acid (17-ODYA) into the endogenous sites of protein palmitoylation, and quantified with isotopic labeling strategies. This novel approach utilized biotin-azide click chemistry and avidin enrichment for LC-MS analysis [28]. Highly dynamic global protein palmitoylation events were defined on proteins implicated in migration, proliferation and cancer, among others. While sites of protein ubiquitination have recently been described using ubiquitin remnant profiling [29], a recent study has leveraged this technique to screen for ubiquitylated sites that are dynamically regulated in response to ultraviolet irradiation. This study uncovered a vital role for dynamic ubiquitination of PCNA associated factor PAF15 during DNA-damage signaling [30]. Although quantification of ubiquitin remnants provided insight into this system, further improvements to this approach may enable selective profiling of specific monoubiquitination or polyubiquitination linkages to illuminate how these differences regulate system-level signaling. Lastly, a recent study by the Bonaldi group combined heavy methyl isotopic labeling, extensive immunoaffinity enrichment of arginine/lysine methylation, and distinct separation schemes to comprehensively investigate non-histone protein methylation in HeLa cells [31].
Another dimension of PTM regulation applies to the coexistence of multiple modifications on the same proteins and the possibility of functional PTM crosstalk. A combinatorial PTM code has been elegantly deciphered to show how acetylation, phosphorylation, and methylation of histone tails leads to chromatin remodeling and modulation of gene expression [32]. The scope and interplay of co-occurring PTMs on other proteins and pathways is less understood, although several studies have described the interaction of proximal acetylation and phosphorylation sites on selected proteins. Traditionally, probing multiple PTMs simultaneously has been restricted due to compatibility of co-enrichment methods and also by difficulties in detecting peptides concurrently modified with unrelated PTMs. Recently, a novel platform was published to permit i) identification of proteins comodified by ubiquitination and phosphorylation, and ii) identification of proteins where these dual PTM marks were found in close sequence proximity [33]. Using this strategy the extent of ubiquitylation-phosphorylation cross-talk in the context of protein degradation was examined. Intriguingly, spatial constraints appear to be at play for some phosphorylation-ubiquitination modification cross-talk and in some cases there can be a preferential directionality to the regulation of PTMs occurring proximally. Other studies have demonstrated the usefulness of systematic in silico integration of phosphorylation, acetylation, and ubiquitination proteomics datasets to reveal PTM interplay or the importance of PTM conservation as an indication of modifications that are more likely to exhibit cross-talk [34,35].
Novel tools and future challenges
The development of innovative technological advances may enhance our experimental dissection of PTM dynamics on a network scale. For example, the ability to expand the multiplex capacity of MS platform analysis would provide further sampling opportunities and overall depth of quantitative coverage. The recently described neutron encoded chemical and metabolic labeling approaches exploit subtle mass defects that occur due to nuclear binding energy variance of stable isotopes and can extend to 12-plex quantification in the same experiment [36,37].
As the breadth of quantitative data increases, it is worthwhile to call attention to the manner in which this data is annotated and interpreted [38,39]. Although systems level studies serve as a rich community resource, ambiguities in peptide identification or PTM site assignments may exist in these datasets, often to detrimental or misleading effects [40]. Given the intent of using proteomics PTM datasets to derive biological insight, it is imperative that the dynamically regulated PTMs be accurately annotated and sites of modification localized precisely. This task relies on automated database matching and scoring of identifications but can benefit from user-assisted validation strategies [41].
With high quality large-scale databases of dynamic temporal phosphorylation profiles, a large number of computational tools can be applied to the data to imply network structure and gain biological insight. In one interesting approach, the Lauffenburger lab applied a combinatorial bioinformatics algorithm (MCAM) to quantitative tyrosine phosphorylation data describing signaling network response to EGF stimulation [42]. Intriguingly, the most frequently co-clustered phosphorylation sites in this computational approach described site-specific protein-protein interactions, including a novel interaction between EGFR and PDLIM1 [43]. In another approach, the prize-collecting steiner tree algorithm utilizes quantitative phosphorylation data and the protein-protein interactome to infer missing nodes and connect co-regulated phosphorylated proteins into pathways and networks [44]. Additional computational tools to infer kinase-substrate relationships, such as NetworKIN, will also benefit from high quality data describing phosphorylation dynamics [45].
In conclusion, measuring system level dynamics of PTMs has the potential to inform our understanding of everything from fundamental biological processes to decoding the complexity involved in disease networks. Network medicine is an emerging area of interest and has recently witnessed an example of a potential temporal network drug (ie. administration of combination therapy in an order- and time-dependent manner) demonstrating the utility of system level studies [46–48]. Leveraging MS technologies to quantify the dynamic interactions of multiple PTM networks simultaneously will make unique contributions to how systems biology is explored.
Highlights
Post-translational modifications (PTMs) modulate diverse cellular functions
PTM signaling networks are dynamic, extensive and interconnect cellular responses
Mass spectrometry PTM analysis can identify and quantify dynamic network components
Integration of dynamic PTM signal-response relationships reveals biological insight
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ivanov AA, Khuri FR, Fu H. Targeting protein-protein interactions as an anticancer strategy. Trends Pharmacol Sci. 2013;34:393–400. doi: 10.1016/j.tips.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rivera CM, Ren B. Mapping Human Epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Federica Calore FLMG. Non-Coding RNAs and Cancer. International Journal of Molecular Sciences. 2013;14:17085. doi: 10.3390/ijms140817085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seet BT, Dikic I, Zhou M-M, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 5.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 6.Krüger R, Kübler D, Pallissé R, Burkovski A, Lehmann WD. Protein and proteome phosphorylation stoichiometry analysis by element mass spectrometry. Anal. Chem. 2006;78:1987–1994. doi: 10.1021/ac051896z. [DOI] [PubMed] [Google Scholar]
- 7.Derouiche A, Cousin C, Mijakovic I. Protein phosphorylation from the perspective of systems biology. Curr Opin Biotechnol. 2012;23:585–590. doi: 10.1016/j.copbio.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 9.Huang PH, Miraldi ER, Xu AM, Kundukulam VA, del Rosario AM, Flynn RA, Cavenee WK, Furnari FB, White FM. Phosphotyrosine signaling analysis of site-specific mutations on EGFRvIII identifies determinants governing glioblastoma cell growth. Mol Biosyst. 2010;6:1227–1237. doi: 10.1039/c001196g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci USA. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwanbeck R, Martini S, Bernoth K, Just U. The Notch signaling pathway: molecular basis of cell context dependency. Eur. J. Cell Biol. 2011;90:572–581. doi: 10.1016/j.ejcb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Fey D, Croucher DR, Kolch W, Kholodenko BN. Crosstalk and signaling switches in mitogen-activated protein kinase cascades. Front Physiol. 2012;3:355. doi: 10.3389/fphys.2012.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 14.Altelaar AFM, Munoz J, Heck AJR. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;14:35–48. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]; •• An excellent introductory overview of MS-proteomics technology platforms highlighting recent technical advancements and discussion of quantitative mass spec applications for PTM and protein network studies.
- 15.del Rosario AM, White FM. Quantifying oncogenic phosphotyrosine signaling networks through systems biology. Curr. Opin. Genet. Dev. 2010;20:23–30. doi: 10.1016/j.gde.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bensimon A, Heck AJR, Aebersold R. Mass spectrometry-based proteomics and network biology. Annu Rev Biochem. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]; •• In this review, an extensive discussion of applying MS-proteomics to understand protein interaction and signaling networks, dynamic modulation of these networks and applications to correlate proteomic data with phenotypic readouts.
- 17.Ideker T, Krogan NJ. Differential network biology. Mol. Syst. Biol. 2012;8:565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf-Yadlin A, Kumar N, Zhang Y, Hautaniemi S, Zaman M, Kim H-D, Grantcharova V, Lauffenburger DA, White FM. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol. Syst. Biol. 2006;2:54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen P. The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem. Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 20.Stasyk T, Huber LA. Mapping in vivo signal transduction defects by phosphoproteomics. Trends Mol Med. 2012;18:43–51. doi: 10.1016/j.molmed.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Zhuang G, Yu K, Jiang Z, Chung A, Yao J, Ha C, Toy K, Soriano R, Haley B, Blackwood E, et al. Phosphoproteomic analysis implicates the mTORC2-FoxO1 axis in VEGF signaling and feedback activation of receptor tyrosine kinases. Sci Signal. 2013;6:ra25. doi: 10.1126/scisignal.2003572. [DOI] [PubMed] [Google Scholar]; •• Demonstrates the use of MS-based phospho-proteomics to define temporal dynamics of the VEGF regulated pathways and functional characterization of network nodes susceptible to pharmacological intervention.
- 22.Halim VA, Alvarez-Fernández M, Xu YJ, Aprelia M, van den Toorn HWP, Heck AJR, Mohammed S, Medema RH. Comparative phosphoproteomic analysis of checkpoint recovery identifies new regulators of the DNA damage response. Sci Signal. 2013;6:rs9. doi: 10.1126/scisignal.2003664. [DOI] [PubMed] [Google Scholar]; • This report defines 154 proteins of the phosphorylation network that are modulated upon recovery from DNA damage and describes methods for functional validation of dynamically phosphorylated proteins.
- 23.Carlson SM, Chouinard CR, Labadorf A, Lam CJ, Schmelzle K, Fraenkel E, White FM. Large-scale discovery of ERK2 substrates identifies ERK-mediated transcriptional regulation by ETV3. Sci Signal. 2011;4:rs11. doi: 10.1126/scisignal.2002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berchowitz LE, Gajadhar AS, van Werven FJ, De Rosa AA, Samoylova ML, Brar GA, Xu Y, Xiao C, Futcher B, Weissman JS, et al. A developmentally regulated translational control pathway establishes the meiotic chromosome segregation pattern. Genes Dev. 2013;27:2147–2163. doi: 10.1101/gad.224253.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2012;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Povlsen LK, Beli P, Wagner SA, Poulsen SL, Sylvestersen KB, Poulsen JW, Nielsen ML, Bekker-Jensen S, Mailand N, Choudhary C. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat Cell Biol. 2012;14:1089–1098. doi: 10.1038/ncb2579. [DOI] [PubMed] [Google Scholar]
- 31.Bremang M, Cuomo A, Agresta AM, Stugiewicz M, Spadotto V, Bonaldi T. Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Mol Biosyst. 2013;9:2231–2247. doi: 10.1039/c3mb00009e. [DOI] [PubMed] [Google Scholar]
- 32.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, part I: covalent histone modifications. Trends Mol Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villén J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A comprehensive, network-level analysis of PTM cross-talk using MS-proteomic methods. Novel methodologies for enrichment of co-modified peptides.
- 34.Woodsmith J, Kamburov A, Stelzl U. Dual coordination of post translational modifications in human protein networks. PLoS Comput. Biol. 2013;9:e1002933. doi: 10.1371/journal.pcbi.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beltrao P, Albanèse V, Kenner LR, Swaney DL, Burlingame A, Villén J, Lim WA, Fraser JS, Frydman J, Krogan NJ. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–425. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hebert AS, Merrill AE, Stefely JA, Bailey DJ, Wenger CD, Westphall MS, Pagliarini DJ, Coon JJ. Amine-reactive neutron-encoded labels for highly-plexed proteomic quantitation. Molecular & Cellular Proteomics. 2013 doi: 10.1074/mcp.M113.032011. doi:10.1074/mcp.M113.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hebert AS, Merrill AE, Bailey DJ, Still AJ, Westphall MS, Strieter ER, Pagliarini DJ, Coon JJ. Neutron-encoded mass signatures for multiplexed proteome quantification. Nat Methods. 2013;10:332–334. doi: 10.1038/nmeth.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A novel quantitation approach exploiting high mass resolution capabilities of state-of-the art mass spectrometers to extend multiplexing capacity and provide higher-content datasets.
- 38.Wu R, Haas W, Dephoure N, Huttlin EL, Zhai B, Sowa ME, Gygi SP. A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat Methods. 2011;8:677–683. doi: 10.1038/nmeth.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salek M, Acuto O. Quantitative dynamics of phosphoproteome: the devil is in the details. Anal. Chem. 2012;84:8431–8436. doi: 10.1021/ac301833k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White FM. The potential cost of high-throughput proteomics. Sci Signal. 2011;4:e8. doi: 10.1126/scisignal.2001813. [DOI] [PubMed] [Google Scholar]; • Provocative discussion of the repercussions from low-quality, minimally validated datasets that are available and often serve as a basis for further studies and biological follow-up experiments. Argues for improved data validation strategies and change of emphasis for the objectives of proteomic-based analyses.
- 41.Curran TG, Bryson BD, Reigelhaupt M, Johnson H, White FM. Computer aided manual validation of mass spectrometry-based proteomic data. Methods. 2013 doi: 10.1016/j.ymeth.2013.03.004. doi:10.1016/j.ymeth.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• A simple, streamlined, freely available software tool developed for expediting the validation of peptide identification and phosphorylation site localization.
- 42.Naegle KM, Welsch RE, Yaffe MB, White FM, Lauffenburger DA. MCAM: Multiple Clustering Analysis Methodology for Deriving Hypotheses and Insights from High-Throughput Proteomic Datasets. PLoS Comput. Biol. 2011;7:e1002119. doi: 10.1371/journal.pcbi.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naegle KM, White FM, Lauffenburger DA, Yaffe MB. Robust co-regulation of tyrosine phosphorylation sites on proteins reveals novel protein interactions. Mol Biosyst. 2012;8:2771–2782. doi: 10.1039/c2mb25200g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang S-SC, Fraenkel E. Integrating proteomic, transcriptional, and interactome data reveals hidden components of signaling and regulatory networks. Sci Signal. 2009;2:ra40. doi: 10.1126/scisignal.2000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MATM, Jørgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pawson T, Linding R. Network medicine. FEBS Lett. 2008;582:1266–1270. doi: 10.1016/j.febslet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 47.Behar M, Barken D, Werner SL, Hoffmann A. The Dynamics of Signaling as a Pharmacological Target. Cell. 2013;155:448–461. doi: 10.1016/j.cell.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149:780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• A landmark study demonstrating the ability to 'rewire' signaling networks and thereby modulate therapeutic response in cancer cells. Implies that network drugs may require optimization of the specific order and timing of co-administered therapies to arrive at optimal efficacy.


