Abstract
Introduction
We investigated the TOMM40-APOE genomic region that has been associated with the risk and age of onset of late-onset Alzheimer's disease (LOAD) to determine if a highly polymorphic, intronic polyT within this region (rs10524523, hereafter 523) affects expression of the APOE and TOMM40 genes. Alleles of this locus are classified: short-S, long-L, very long-VL based on the number of T-residues.
Methods
We evaluated differences in APOE-mRNA and TOMM40-mRNA levels as a function of 523 genotype in two brain regions from APOEε3/3 Caucasian autopsy-confirmed LOAD cases and normal controls. We further investigated the effect of the 523 locus in its native genomic context using a luciferase expression system.
Results
The expression of both genes was significantly increased with disease. Mean expression of APOE and TOMM40-mRNA levels were higher in VL-homozygotes compared to S-homozygotes in temporal and occipital cortexes from Normal and LOAD subjects. Results of a luciferase reporter system were consistent with the human brain mRNA analysis: the 523-VL polyT resulted in significantly higher expression than the S-polyT. While the effect of polyT length on reporter expression was the same in HepG2 hepatoma and SH-SY5Y neuroblastoma cells, the magnitude of the effect was greater in the neuroblastoma than in the hepatoma cells, which implies tissue-specific modulation of the 523-polyT.
Conclusions
These results suggest that the 523 locus may contribute to LOAD susceptibility by modulating the expression of TOMM40 and/or APOE transcription.
Keywords: TOMM40, APOE, polyT polymorphism, mRNA expression, transcription regulation, Alzheimer's disease
1. BACKGROUND
A polymorphism in TOMM40 gene, which is adjacent to and in linkage disequilibrium with APOE, is associated with late onset Alzheimer's disease (LOAD) risk and age of disease onset [1, 2]. The putative risk locus in TOMM40 is an intronic, polyT tract (rs10524523), referred to as 523, that is polymorphic with respect to the number of T residues. The ‘long’ (L) or ‘Very Long’ (VL) alleles refer to homopolymer lengths equal or greater than 20 and 30 T residues respectively, and are associated with earlier age of disease onset in APOE ε3/4[1, 2] (~7 years) Caucasian LOAD patients. This locus has been examined in a number of additional studies. Caselli et al. replicated the association between the longer 523 alleles and earlier onset of LOAD (~9 years) in an independent group of APOE ε3/3 Caucasian subjects drawn from a longitudinal study[3]. Furthermore, in a cognitively healthy, late middle-aged cohort of APOE ε3/3 subjects drawn from a population enriched for family history of LOAD[4], Johnson et al. discovered significant association of 523 with worse performance on primacy retrieval from a verbal list learning task and with reduced gray matter volume in ventral posterior cingulate and medial ventral precuneus, which are brain regions known to be affected in early AD. In a cross sectional study of cognitively healthy elderly, we have also observed APOE-independent associations between the 523 polymorphism and specific cognitive domains of memory and executive control that are preferentially affected in early-stage AD[5]. Together with the original findings, these new studies establish, in Caucasians, the association of 523 with LOAD pathogenesis, particularly for APOE ε3 carriers. However, there is some disagreement in the literature. Two studies do not replicate the association between 523 and age of AD onset[6] [7]. Based on finding that APOE ε3/4 subjects who carried VL/L had earlier disease onset than those who carried S/L[1, 2], the authors assumed that VL would be associated with earlier onset than S in APOE ε3/3 carriers (e.g. S/S<VL/VL). However, while Cruchaga et al. replicated the association between the 523 and AD in APOE ε3/3 subjects, they found that the S allele, rather than the VL allele, was associated with risk of earlier age of onset AD. That is, APOE ε3/3 subjects with the S/S genotype showed a trend towards earlier disease onset [8]. Similarly, Maruszak et. al. also observed a significant association between 523 and LOAD risk in APOE ε3 homozygotes, but reported that the ε3-VL haplotype was significantly more frequent among patients with a later age of onset (≥79 years) contrary to what might have been anticipated based on the original results from APOE ε3/4 heterozygotes [9]. The association between later age of onset of cognitive impairment has now been confirmed for a cohort of Caucasians with carefully ascertained age of onset of cognitive impairment or probable AD[10, 11] (Figure 1, adapted from Crenshaw et al.[11]). Not only do these curves provide age of onset risk information for >97% of Caucasians, and confirm that VL is associated with later age of disease onset in APOE ε3 homozygotes [8] [9], but the study confirms that VL is associated with earlier age of disease onset in APOE ε3/4 subjects, as originally reported[1, 2]. There is, however, a small sub-set of very early onset AD patients with the VL/VL genotype and preliminary data suggests the presence of another genetic variant linked to the VL DNA strand that may result in an onset before age 59 years (Roses et al., unpublished data). These patients are very uncommon and additional series are currently being studied to confirm this finding. Studies of age of onset of this progressive disease are challenging and replication of results is complicated by differences in methods used to ascertain the age of onset (MCI, AD, or some other standardized definition), issues associated with study design, e.g. prospective versus retrospective, longitudinal versus cross-sectional[1, 3, 8, 9, 12, 13] and/or technical limitations and quality control of the 523 genotyping assay[10, 13].
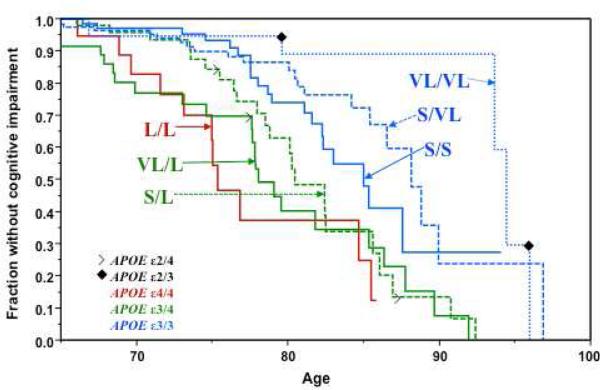
Figure 1. Alzheimer's disease age of onset curves by TOMM40-523 genotypes.
The figure is adapted from Crenshaw et al.[11] Kaplan-Meyer survival curves, where the Y axis shows the percent survival without cognitive impairment, and the X axis represents age. Data was obtained from the Duke Bryan ADRC cohort N=438 subjects: 106 diagnosed with dementia, 332 cognitively normal. N for each genotype: L,L:23; VL,L:54; S,L:72; S,S:100; S,VL:138; VL,VL:51. TOMM40 genotypes and the corresponding APOE genotypes are indicated on the figure. The red line corresponds to APOE ε4/4; the two green lines correspond to APOE ε3/4, and the three blue lines correspond to APOE ε3/3. The data for individuals who carried an APOE ε2 allele are indicated as points on the appropriate TOMM40 genotype curve; open arrowheads and filled diamonds indicate the age at onset of symptoms in the APOE ε2/4 and APOE ε2/3 groups, respectively.
Functional analyses will assist with assessing the contribution of the 523 locus to LOAD risk. Bekris et al. recently showed that a synthetic construct containing the 523 locus acts as an enhancer/silencer of TOMM40 promoter activity in cultured neuronal, but not hepatocyte, cell lines [14]. Our study was designed to determine whether the 523 polyT tract regulates transcription in vivo. We evaluated the association of the 523 genotypes with mRNA expression of TOMM40 and APOE in two different brain regions affected in LOAD from AD cases and controls. In addition, we tested the effect of the 523 locus in its native genomic environment on transcription regulation using a cell based reporter system.
2. MATERIALS AND METHODS
2.1. Brain Samples
Temporal (TC) and occipital (OCC) cortexes, from APOE ε3/3 neurologically healthy controls (n=42) were obtained through the Kathleen Price Bryan Brain Bank (KPBBB) at Duke University, the Brain and Tissue Bank for Developmental Disorders at the University of Maryland, and the Layton Aging & Alzheimer's Disease Center at Oregon Health and Science University. The healthy control brain samples were obtained from postmortem tissues of clinically normal subjects examined in most instances one year prior to death and found to have no cognitive disorder and no neuropathological evidence of PD, AD or other neurodegenerative disorders. TC and OCC from APOE ε3/3 LOAD patients (Braak&Braak stage III-VI; n=69) were obtained through the KPBBB at Duke University. All post mortem intervals (PMI) were <24 hours. All brain tissues donors were Caucasians. Demographics for these samples are summarized in Table 1.
Table 1.
Demographic description of the brain samples
| LOAD | Normal | |
|---|---|---|
| Total subjects (N) | 69 | 42 |
| †TC (N) | 66 | 42 |
| ‡OCC (N) | 69 | 34 |
| Male % | 40 | 50 |
| Age (yr) mean±SD | 76.9±13.3 | 78.2±15.1 |
| §PMI (hr) mean±SD | 12.3±12.1 | 11.4±7.7 |
| Caucasians % | 100 | 100 |
TC- temporal cortex
OCC- occipital cortex
PMI- post mortem interval
2.2. DNA Extraction and Genotyping
Genomic DNA was extracted from brain tissues using the standard Qiagen protocol. DNA concentration and the quality of purification were determined spectrophotometrically. APOE genotypes were determined using a TaqMan-based allelic discrimination assay (Applied Biosystems). Briefly, APOE genotypes were established using two separate SNPs, rs429358334T/C (ABI assay ID: C_3084793_20), and rs7412 472T/C (ABI assay ID: C_904973_10). The assay was conducted using the ABI 7900HT and genotype analysis was performed by the SNP auto-caller feature of SDS software. APOE genotype assignments were as described previously[15].
TOMM40 523 polyT genotypes were determined using a method described previously39,40. In summary, the 523 region of each genomic DNA sample was PCR-amplified using a fluorescently labeled primer. Size of the PCR fragment was determined using an ABI 3730 DNA Analyzer and GeneMapper, version 4.0 software (Applied Biosystems, Foster City, CA). The 523 allele was assigned according to the length of the PCR product and the convention established by Roses et al.: Short (S), ≤19; Long (L) −20-29; Very Long (VL) ≥30[16].
2.3. RNA extraction and cDNA synthesis
Total RNA was extracted from brain samples (100 mg) using TRIzol reagent (Invitrogen, Carlsbad, CA) followed by purification with an RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. RNA concentration was determined spectrophotometrically at 260nm, while the quality of the purified RNA was determined by 260nm/280nm ratio. All of the RNA samples were of acceptable quality having ratios between 1.9 and 2.1. Sample quality and the absence of significant degradation products were confirmed by establishing that every sample had a RNA Integrity Number (RIN), measured on an Agilent Bioanalyzer, of greater than 7. cDNA was synthesized using MultiScribe RT enzyme (Applied Biosystems, Foster City, CA) under the following conditions; 10 min at 25°C and 120 min at 37°C.
2.4. Real time PCR
Real-time PCR was used to quantify the levels of human TOMM40 mRNA and APOE mRNA. Duplicates of each sample were assayed by relative quantitative real-time PCR using the ABI 7900HT to determine the level of TOMM40 and APOE messages relative to the mRNAs for the housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and cyclophilin A (PPIA). ABI MGB probe and primer set assays were used to amplify TOMM40 cDNA (ID Hs01587378_mH, 146bp), APOE cDNA (ID Hs00171168_m1, 108bp); and the two RNA reference controls, GAPDH (ID Hs99999905_m1, 122bp) and PPIA (ID Hs99999904_m1, 98bp)(Applied Biosystems, Foster City, CA). Each cDNA (10 ng) was amplified in duplicate in at least two independent runs (overall ≥ 4 repeats), using TaqMan Universal PCR master mix reagent (Applied Biosystems, Foster City, CA) and the following conditions: 2 min at 50 °C, 10 min at 95 °C, 40 cycles; 15 sec at 95 °C; and 1 min at 60°C. As a negative control for the specificity of the amplification, we used RNA control samples that were not converted to cDNA (no-RT) and no-cDNA/RNA samples (no-template) in each plate. No amplification product was detected in control reactions. Data were analyzed with a threshold set in the linear range of amplification. The cycle number at which any particular sample crossed that threshold (Ct) was then used to determine fold difference, whereas the geometric mean of the two control genes served as a reference for normalization. Fold difference was calculated as 2−ΔΔCt; where ΔCt=[Ct(target)-Ct (reference)] and ΔΔCt =[ΔCt(sample)]-[ΔCt(calibrator)]. The calibrator was a particular brain RNA sample used in every plate for normalization within and across runs. The variation of the ΔCt values among the calibrator replicates was less than 10%. For assay validation, we generated standard curves for TOMM40, APOE and each reference assay, GAPDH and PPIA using different amounts of human brain total RNA (0.1-100 ng). In addition, the slope of the relative efficiency plot for TOMM40 and APOE with each internal control (GAPDH and PPIA) was determined to validate the assays. The slope in the relative efficiency plot for TOMM40 and APOE and the reference genes were <0.1, showing a standard value required for the validation of the relative quantitative method.
2.5. Luciferase Reporter Constructs
The 523 polyT locus and surrounding genomic regions was contained within a 6.874-kb DNA fragment upstream of the APOE translation start site which was amplified from human BAC RP11-47010 (positions 45402974-45409848) using the forward 5’ TGTAACGCGTTGCTGACCTCAAGCTGTCCT 3’ and the reverse 5’ AGCTCGTACGAGAAACTGTCAATCAACCGCCAG 3’ primers that include the MluI and BsiWI restriction sites, respectively. The PCR product was cloned into the pCR-XL-TOPO vector. pGL4.10 (Promega Corporation, Madison, WI), which contains the firefly luciferase coding sequence but lacks eukaryotic promoter or enhancer elements, was modified to include the MluI-BsiWI restriction sites by ligation of synthetic oligos (idtDNA). The ~7-kb DNA, containing the 523 locus, excised by restriction digestion at the MluI- BsiWI sites of pCR-XL-TOPO and cloned into the MluI- BsiWI engineered sites of the pGL4.10 vector. The original BAC used to generate the insert for the first plasmid includes the short allele, T=15, at the 523locus. This pGL4.10 plasmid containing the short allele was designated as p523S. A second plasmid that includes a 523 VL allele (T=34) was constructed using a synthetic vector which contained the 231 bp fragment complimentary to the 5’ sequence of the BAC with T=34 at the 523 locus and the MluI-PmlI restriction sites (idtSMART). The insert was cut at the MluI-PmlI sites of the synthetic vector and was cloned into these sites in p523S, thus replacing the complimentary DNA fragment containing the 523 S allele and generating a new plasmid with the 523 VL allele designated p523VL. Sequencing confirmed the p523S and p523L constructs and the length of their polyT.
2.6. Cell culture and Transfection
The human liver hepatocellular carcinoma cell line, HepG2and the human neuroblastoma cell line,SH-SY5Y, were from American type tissue culture collection. HepG2 cells were grown in Minimum Essential Medium (MEM) (glucose at 4.5 g/liter) / and SH-SY5Y cells were grown in high glucose DMEM/F-12 (1:1). Both media were supplemented with 10% fetal bovine serum, 2 mM glutamine, 1 mM pyruvate, 0.1 mM Non-essential Amino Acids, 100 units/ml penicillin and 100 mg/ml streptomycin. Cells were maintained at 37°C in a humidified, 5% CO2 incubator. 2 ×105 Hep G2 cells or 2×105 SH-SY5Y cells were plated onto each well of a 24-well dish the day prior to transfection. We conducted co-transfection experiments to test expression of each construct. One μg of the empty pGL4.10 vector or 2.5 μg (the molar equivalent to the empty vector) of one of the pGL4.10 derivatives, p523S or p523VL, were mixed with 5 ng of the reference plasmid, pGL4.74, harboring the HSV thymidine kinase promoter upstream of Renilla luciferase, and added to the cells in the presence of Lipofectamine™ 2000 (Life Technologies), according to the manufacturer's instructions. Cells were incubated for 24 hr at 37°C, washed once with phosphate-buffered saline, and then incubated in fresh medium for an additional 24 hr.
For each plasmid (pGL4.10, p523S and p523VL) in each cell line (HepG2, SH-SY5Y), one experiment consisted of performing the transfection and expression assay in triplicate using three wells of cultured cells that were independently transfected in parallel with three individually prepared aliquots of transfection reaction. Each triplicate experiment was repeated four times on separate days.
2.7. Luciferase Assay
Forty-eight hours after transfection the Hep G2 and SH-SY5Y cells were washed with a physiological salts solution and lysed in 100 μl of Passive Lysis Buffer (Promega Corporation, Madison, WI). Firefly luciferase and Renilla luciferase activities were measured in 20 μl of Hep G2 or SH-SY5Y cell lysate using the Dual-Luciferase Reporter assay system (Promega Corporation, Madison, WI) in a luminometer (Turner Biosystems Veritas Microplate Luminometer). “Relative activity” was defined as the ratio of firefly luciferase activity to Renilla luciferase activity and was calculated by dividing luminescence intensities for firefly luciferase by that for Renilla luciferase. “Fold expression” was calculated by dividing the average value of relative activity of each construct with a 523 allele to the relative activity of the pGL4.10 without any insert.
2.8. Statistical analysis
Statistical analyses of brain mRNA were carried out using SAS statistical software, Version 9.1 (SAS Institutes, Cary, NC). Relative TOMM40 mRNA and APOE mRNA expression levels of each sample were measured in replicates and the results of all replicates were averaged. All average values were expressed as mean±SE. Associations of 523 genotypes with TOMM40 and APOE genes expression were assessed using the Generalized Linear Models procedure (PROC GLM). A log transformation (log2) was performed on all mRNA levels to assure normal distribution [17]. Genotypes were coded in the additive model, and also as a dominant model pooling homozygous for VL allele and heterozygous genotypes (S/VL). All models were adjusted for gender, age, PMI, Braak&Braak stage and tissue source. To further control for RNA integrity, analyses were repeated including RIN as a covariate in the models. Correction for multiple testing employed the Bonferroni method.
For the luciferase reporter experiments, statistical significance of differences in expression between the two different 523 allele constructs were analyzed by pairwise comparisons using the Tukey-Kramer HSD test.
3. RESULTS
3.1. The effect of disease status on TOMM40 and APOE mRNAs
TOMM40 and APOE mRNA fold levels [TOMM40/(GAPDH, PPIA); APOE/(GAPDH, PPIA)] were measured in 137 brain tissue samples [temporal cortex (TC), n=66; occipital cortex (OCC), n=69] obtained from 69 AD cases; and in 76 brain tissues (TC, n=42; OCC, n=34) from 42 normal subjects. All brain tissues donors were Caucasians and homozygous for APOE ε3. Table 1 summarizes the demographics of the tissue donors.
We first tested for associations with confounding factors that might affect RNA levels. Expression levels for TOMM40 and APOE mRNAs decreased with age, but this effect was not statistically significant and we did not detect significant associations of TOMM40 or APOE mRNA levels with sex or postmortem interval (PMI) in TC and OCC (data not shown). All of the subsequent analyses were adjusted for age, sex, and PMI.
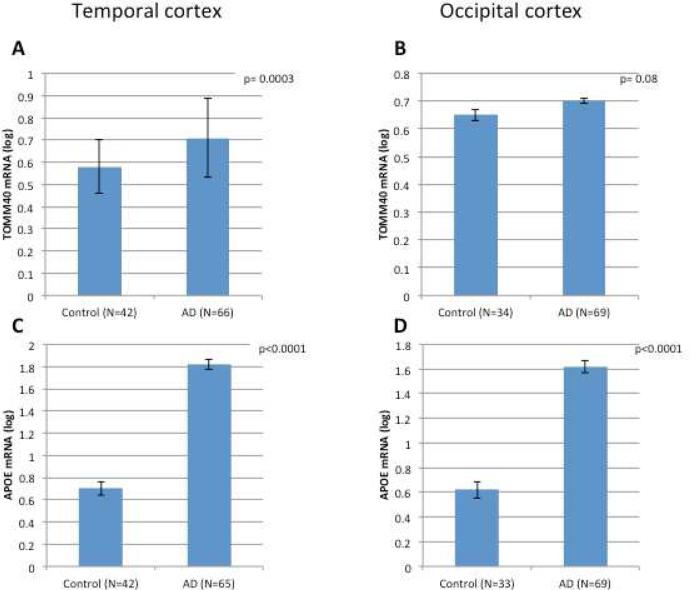
Next, we assessed the effect of disease state on TOMM40 and APOE genes expression. TOMM40 mRNA levels were significantly increased in TC of AD-affected compared to healthy control (p=0.0003; Figure 2A). In OCC there was a small average increase in TOMM40 mRNA in samples from AD cases, however this effect was not significant (p=0.08; Figure 2B). The effect of disease status was much greater for APOE mRNA than TOMM40 mRNA levels. APOE mRNA levels were nearly threefold higher in TC and OCC from AD brains compared to healthy controls (p<0.0001) (Figure 2 C and D). These findings were replicated when adjusting also for RIN.
Figure 2. The effect of disease state on TOMM40 and APOE mRNAs expression levels in human brain tissues from APOE ε3/3 donors.
The study cohort consisted of LOAD and control brain tissues from Caucasian donors with APOE ε3/3 genotype. Fold levels of human TOMM40 mRNA (A and B) and APOE mRNA (C and D) in the temporal (A and C) and occipital (B and D) cortexes were assayed by real-time RT-PCR using TaqMan technology and calculated relative the geometric mean of GAPDH- and PPIA mRNAs reference control using the 2−ΔΔCt method (i.e. results presented are relative to a specific brain RNA sample). The values presented here are means levels± SE adjusted for age, gender, PMI, and source. TC, temporal cortex; OCC, occipital cortex; Normal, clinically and neuropathologically healthy; LOAD, late onset Alzheimer's disease.
3.2. The effect of 523 on TOMM40 and APOE mRNA levels
To rule out the possibility that the effect of the 523 genotype on TOMM40 and APOE mRNA levels was associated with APOE genotype, due to the high LD between these genes, we restricted the analysis of TOMM40 and APOE mRNA levels to APOE ε3/3 subjects.
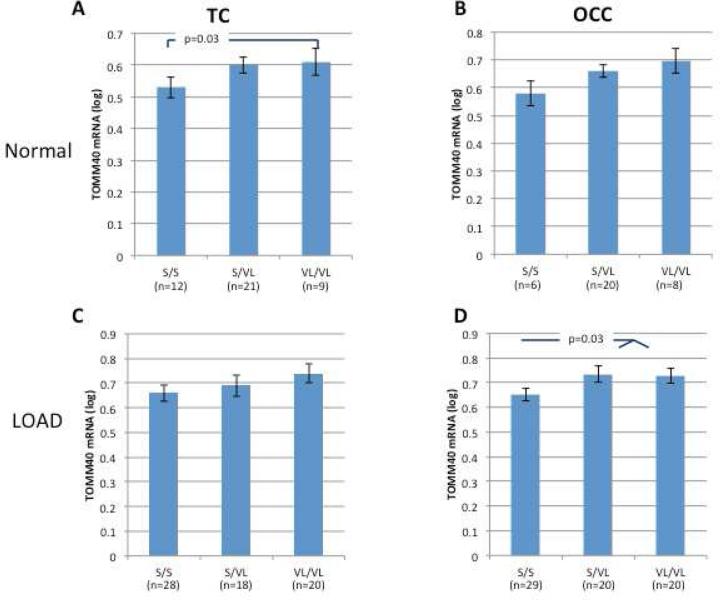
When APOE ε3/3 AD cases and controls are pooled, length of the TOMM40523 poly-T locus was associated with TOMM40 mRNA expression levels in the TC and OCC (n=108 p=0.03; n=103 p=0.05, respectively). Of these subjects, the VL homozygotes had the highest mean TOMM40 mRNA levels. When the APOE ε3/3 group is stratified by disease status, the effect of 523 on TOMM40 mRNA expression was still detectable. In normal TC (n=42), carriers of the VL allele (VL/VL and S/VL) had higher average expression levels of TOMM40 mRNA than the non-carriers (S/S), and significant differences were observed between the S/S and the VL/VL groups (p=0.03; Figure 3A). A similar trend of effect on expression, VL>S, was observed in the Normal OCC, though it did not reach statistical significance (p=0.2, n=34; Figure 3B). In LOAD samples, the 523 VL allele was associated with higher TOMM40 mRNA levels in the OCC (n= 69, S/S vs. S/VL, VL/VL p=0.03); a similar trend, that did not reach statistical significance, was also observed in the TC (n=66, p=0.34) (Figure 3C and D).
Figure 3. The effect of 523 genotypes on TOMM40 mRNA expression levels in human brain tissues from APOE ε3/3 donors.
The study cohort consisted of brain tissues from Caucasian donors with APOE ε3/3 genotype. Cases and control subjects were genotyped for 523. Fold levels of human TOMM40 mRNA in the temporal (A and C) and occipital (B and D) cortexes from normal (A and B) and LOAD (C and D) donors were assayed by real-time RT-PCR using TaqMan technology and calculated relative the geometric mean of GAPDH- and PPIA mRNAs reference control using the 2−ΔΔCt method (i.e. results presented are relative to a specific brain RNA sample). The values presented here are means levels± SE adjusted for age, gender, PMI, source, and Braak&Braak stage. TC, temporal cortex; OCC, occipital cortex; Normal, clinically and neuropathologically healthy; LOAD, late onset Alzheimer's disease.
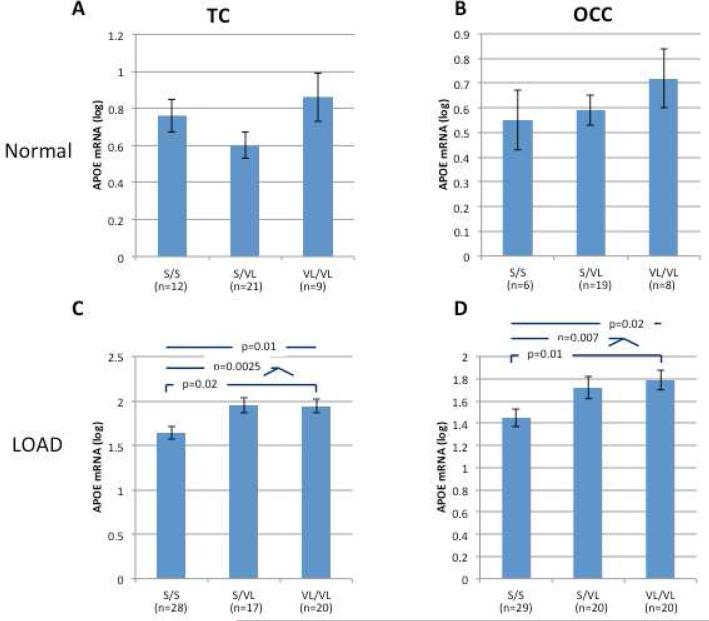
Regardless of disease states, 523 genotype influenced APOE mRNA expression in APOE ε3/3 homozygotes. Homozygotes for the VL allele demonstrated significantly higher levels of APOE mRNA compared to S allele carriers (S/S and S/VL) in the TC (n=107, p=0.01) and OCC (n=102, p=0.06). Analysis of the Normal group showed increased expression in the TC and OCC (n=42 and 33, respectively) for VL/VL homozygotes, but these results were not statistically significant (p=0.19 and 0.6, respectively, Figure 4A and B). The strongest effect of the 523 on APOE mRNA levels was observed in the LOAD samples (Figure 4 C and D). Carriers of the VL allele (S/VL, VL/VL) showed significantly higher levels of APOE mRNA compared to non-carriers (S/S) in the TC (n=65, 20% increase, p=0.0025; Figure 4C) and OCC (n=69, 25% increase p=0.007; Figure 4D) regions. These results remain when RIN was also included in the analyses as covariate.
Figure 4. The effect of 523 genotypes on APOE mRNA expression levels in human brain tissues from APOE ε3/3 donors.
The study cohort consisted of brain tissues from Caucasian donors with APOE ε3/3 genotype. Cases and control subjects were genotyped for 523. Fold levels of human APOE mRNA in the temporal (A and C) and occipital (B and D) cortexes from normal (A and B) and LOAD (C and D) donors were assayed by real-time RT-PCR using TaqMan technology and calculated relative the geometric mean of GAPDH- and PPIA- mRNAs reference control using the 2−ΔΔCt method (i.e. results presented are relative to a specific brain RNA sample). The values presented here are means levels± SE adjusted for age, gender, PMI, source, and Braak&Braak stage. TC, temporal cortex; OCC, occipital cortex; Normal, clinically and neuropathologically healthy; LOAD, late onset Alzheimer's disease.
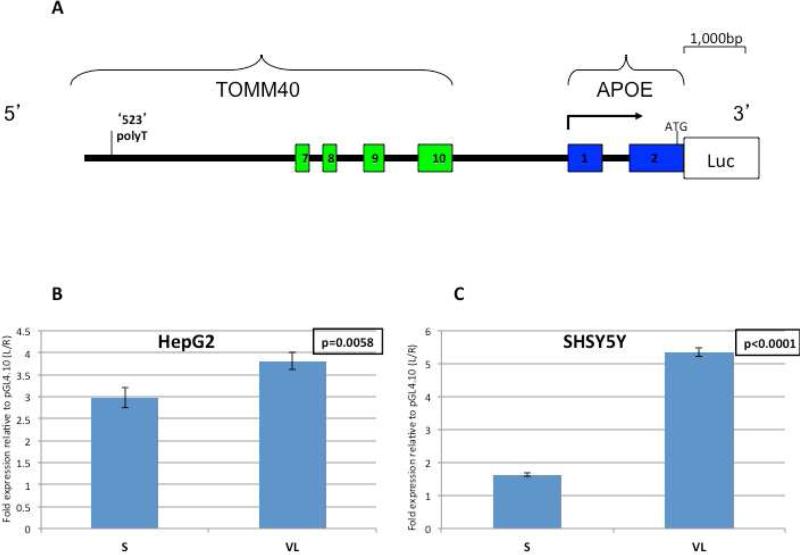
3.3. The effect of 523 on luciferase expression in HepG2 and SH-SY5Y cells
These results suggest the TOMM40 523 region plays a regulatory role in TOMM40 and APOE gene expression in human brain. To further test this hypothesis, we investigated the effect of the 523 locus in its native genomic context using a luciferase expression system. A ~7 kb fragment that includes the 523 locus was amplified from a human BAC and cloned into a luciferase reporter construct as shown in Figure 5A. The poly-T in the native BAC contained 15 T residues and therefore represents the S allele; and the VL allele was introduced by replacing the original sequence of the S allele with a stretch of 34 T residues and the constructs were verified by sequencing. We designated the respective constructs as p523S and p523VL. Luciferase activities as a result of expression from the p523S and p523VL reporter constructs were measured relative to empty vector. We employed two human cell lines, HepG2 cells and the neuronal SH-SY5Y cells, to learn if tissue origin modulated the 523 effect on gene expression. In both cell lines, expression of the p523S and the p523VL constructs, as measured by fold expression of luciferase activity, differed significantly. In HepG2 cells, fold expression of the VL construct was more than 30% greater than expression of the S construct (p=0.0058, Figure 5B). The VL effect was even more pronounced in the SH-SY5Y cells where the p523VL reporter construct yielded ~ 3-fold higher luciferase fold expression compared to the p523S construct (p<0.0001; Figure 5C). The direction of the effect on gene expression, i.e. VL>S, is similar to the effect observed in the analysis of the human brain tissues described above. That the effect was greater in the SH-SY5Y cells than in the hepatoma cells suggests modulation of TOMM40 and APOE expression by the 523 polyT enhancer may be tissue specific.
Figure 5. The Luciferase reporter system for the TOMM40 523 locus.
(A) Schematic representation of the luciferase reporter construct map. The constructs include a ~7 kb human region upstream of the APOE gene translational start site and extending 5’ of the 523 locus. The relative position of the 523 locus is marked in vertical line. The green boxes represent TOMM40 exons (7-10), the blue boxes represent APOE exons (1-2), the solid black line represents introns and intergenic region. The 5’ and 3’ indicate the human genes’ orientation. The arrow above indicates the transcription start site and direction for APOE. The translational start site is marked by ATG. The open box indicates the position of the luciferase reporter gene.
The fold expression of luciferase activity derived by the constructs harboring different 523 alleles in (B) HepG2, and(C) SH-SY5Y cells. Cells were cotransfected with each of the 2 constructs harboring the different 523 alleles, S (p523S) or VL (p523VL), or pGL-4.10 and the Renilla reference control, pGL4.74. For each construct four experiments were performed each in triplicate. The relative activity with each construct was calculated by dividing the luminescence intensity of the Firefly luciferase by that of the cotransfected Renilla luciferase in each independent aliquot of cells and then averaging the three relative luciferase activities seen in each experiment. The fold expression for each construct, p523S and p523VL, was then determined by dividing the average relative activity of each construct to that of the average obtained with pGL-4.10. The average of the ‘fold expression’ of the four independent experiments performed on separate days was calculated. The data represented here are the ‘Fold Expression’ mean± SE. Tukey-Kramer HSD test comparing the ‘fold expression’ of each of the p523S and p523VL constructs revealed p= 0.0058 and <0.0001, in HepG2 (B) and SH-SY5Y (C) cells, respectively.
We investigated the TOMM40-APOE genomic region that has been associated with the risk and age of onset of late-onset Alzheimer's disease (LOAD) to determine the functional effect of a polymorphic, intronic polyT within this region (rs10524523, hereafter 523). Differences in APOE-mRNA and TOMM40-mRNA levels as a function of 523 genotype were evaluated in two brain regions from APOEε3/3 Caucasian autopsy-confirmed LOAD cases and normal controls. The expression of both genes was significantly increased with disease. Mean expression of APOE and TOMM40-mRNA levels were significantly higher in VL-homozygotes compared to S-homozygotes in temporal and occipital cortexes from Normal and LOAD subjects. Results of a luciferase reporter system, in both in HepG2 hepatoma and SH-SY5Y neuroblastoma cells, were consistent with the human brain mRNA analysis: the 523-VL polyT resulted in significantly higher expression than the S-polyT. These results suggest that the 523 locus may contribute to LOAD susceptibility by modulating the expression of TOMM40 and/or APOE transcription.
4. DISCUSSION
Our data demonstrated elevated levels of TOMM40 and APOE transcripts in the brains of LOAD-affected individuals compared to unaffected control brains. Further, the TOMM40 523 genotype was associated with TOMM40 and APOE mRNA levels in two different brain regions that are affected by AD.
Genetic studies on autopsy-confirmed LOAD patients originally reported that the 523 VL allele of TOMM40 is associated with earlier age of LOAD onset relative to the 523 S allele in APOE ε3/4 subjects, i.e. in the context of haplotypes where the 523 L allele is linked to an APOE ε4 allele and the 523 VL or S allele is linked to an APOE ε3 allele[1-3]. This finding was anticipated to generalize to an earlier onset of LOAD onset for VL/VL carriers relative to S/VL and S/S carriers in APOE ε3/3 subjects. However, subsequent studies in longitudinal late-life cohorts showed that 523 VL/VL genotype is associated with later age of LOAD onset relative to 523 S/VL and S/S [10, 11]. Other studies have investigated the association between the 523 VL allele and LOAD-related phenotypes. In a sample of cognitively normal middle-aged APOE ε3/3 subjects, significant association was reported between 523 VL and a reduction in the primacy effect on word list learning task, consistent with impaired consolidation in episodic memory[4]. In these same subjects there was also a reduction in gray matter volume in the ventral posterior cingulate gyrus and the medial ventral precuneus, both areas known to be affected in early AD[4]. We also reported, in a sample of cognitively normal aging subjects, APOE-independent associations between the 523 VL and decline in specific cognitive domains of memory and executive control that are preferentially affected in early stage AD[5]. Furthermore, Bruno et al. showed an increase in CSF cortisol concentration, which suggests hippocampal damage, in individuals who carry two copies of the longer 523 alleles[18]. In another study, they also showed that the CSF levels of neurofilament light proteins, markers of neuronal damage that are elevated in LOAD patients, were significantly higher in subjects with two copies of the longer 523 alleles[19]. Recently, Crenshaw and colleagues thoroughly discussed the observation of variable effects of the 523 VL allele [11]. The authors proposed that the VL/VL genotype (i.e., in APOE ε3/3 subjects) may be associated with pre-clinical stages of AD that are most evident at younger ages (<60 year old) when subtle signs of cognitive decline are not masked by later pathological symptoms and that, in the presence of APOE ε4 (i.e., in APOE ε3/4 subjects), this early effect of VL is worsened. The studies that reported non-replication of the early discovery of the association between 523 genotype and age of LOAD onset are discussed previously[10, 11] and in the introduction.
Here we report data showing that 523 genotype affects expression of the APOE and TOMM40 genes, which provides a possible explanation for the genetic association of the locus with age of LOAD onset and other disease related phenotypes. We demonstrate that the LOAD risk allele, VL, is associated with increased levels of both APOE and TOMM40 transcripts in Normal and AD-affected human brain tissues and a cell-based luciferase reporter system. These results suggest that the increased expression of these genes, driven by the longer 523 allele, may underlay the observed associations between the 523 VL and risk for LOAD-related phenotypes. Furthermore, molecular changes in gene expression occur years before the onset of the disease. Thus, we speculate that the effect of 523 genotype on APOE and TOMM40 expression may reflect presymptomatic AD molecular events and that genetic influence on gene expression remains throughout the development of the disease.
The association trend of the VL allele and higher expression of TOMM40 and APOE mRNAs is evident in two AD relevant cortical areas examined, temporal and occipital neocortex. In particular, the VL/VL genotype group was consistently associated with higher mRNA expression levels compared to the S/S genotype group. Although this is a small sample, differences in mean expression for both TOMM40 and APOE mRNAs approached statistical significance in a number of our experiments. However, only the associations between 523 and APOE mRNA levels in AD-affected temporal and occipital cortexes remain significant after Bonferroni correction (p=0.005 and 0.028, respectively). These observations warrant further investigation in a larger number of samples. A cell-based luciferase reporter system that mimics the effect of the 523 alleles, in their native genomic context, recapitulates the gene expression trends that we observe in the brain tissues, i.e. the VL construct drove significantly higher expression compared to the S construct. This provides additional support for an effect of this locus on transcription of the TOMM40 and APOE genes.
Our results confirm and extent a recent study that revealed a complex transcriptional regulatory region for TOMM40 and APOE expression that extends throughout both genes and is influenced by multiple polymorphisms including the 523 locus[14]. In that study, Bekris et al. fused a promoter DNA fragment with a relative short putative enhancer sequence that contained the 523 locus and demonstrated that 523 length influenced TOMM40 promoter activity. The current study extends their observations in three ways: 1) It demonstrates the transcriptional enhancer activity of 523 in vivo using human brain tissues relevant to disease pathogenesis. 2) A reporter system, with the intact native genomic context of the 523 preserved, mimics the in vivo effect of 523 on gene expression. 3) It shows that the TOMM40-523 effect is regional, that it also influences the transcription of the neighbor gene, APOE.
Two other studies have investigated the association between 523 and TOMM40 mRNA expression in human tissues. The first study used human fibroblast cell lines derived from cognitively-healthy, APOE ε3/4 donors and found no significant differences in TOMM40 mRNA expression[20]. A second group analyzed the expression of TOMM40 and APOE in parietal cortex from subjects chosen without regard to APOE genotype and also did not detect an association between 523 and TOMM40 or APOE mRNA levels[8]. That, these experiments did not detect an association may be explained by the different tissues types assayed, very small sample size (especially when the analysis was repeated using a specific APOE genotype), and differences in the RNA analysis methodologies and study designs. Thus, our study showed for the first time the functional significance of the 523 locus in vivo in human tissues.
mRNA levels are regulated through transcription and posttranscriptional pathways. The 523 polymorphism is a long deoxythymidine homopolymer. It has been proposed that genomic poly-T sequences act as transcriptional enhancer elements[21] [22]. Poly-T sequences may cause nucleosome depletion, which increases the accessibility of the DNA in the region to the transcription machinery or regulatory factors [22]. The results that we report here suggest that the 523 locus does function as an enhancer to regulate the transcription of theTOMM40 and APOE genes.
It has been suggested that alteration of the expression of specific genes may be an important mechanism in the etiology of neurodegenerative disorders including AD[23]. Our data indicate that APOE mRNA levels are increased in LOAD-affected brain tissues compared to controls. Our observation is consistent with other reports of elevated levels of APOE mRNA in AD brains. For example, Zarow et al. report increased APOE mRNA levels in the hippocampus of AD cases compared to controls[24] and Matsui et al. report increased APOE mRNA levels in temporal cortex of AD donors compared to controls[25]. Furthermore, Akram et al. have recently demonstrated that APOE mRNA and protein levels in the inferior temporal gyrus and the hippocampus are strongly, positively correlated with the progression of cognitive dysfunction[26].
We also provide evidence that TOMM40 levels are increased in AD versus control brain. However, there are inconsistent reports in the literature regarding the relationship between LOAD status and TOMM40 expression. TOMM40 mRNA is reduced in whole blood samples from AD subjects compared to matched controls[27]. Another study detected a correlation between TOMM40 mRNA levels and LOAD progression, but was inconclusive regarding the direction of the change, i.e. TOMM40 mRNA levels were higher in the frontal cortex of the majority of LOAD patients, however in the rest of the LOAD samples TOMM40 mRNA was down-regulated[28]. We would like to note here that gene expression comparisons between AD versus control using whole brain tissues might have a limitation due to differences in the cell type composition resulted from neuronal cell loss in AD brains. Thus, disease related changes in gene expression may reflect, in part, differences in the numbers of each cell type (i.e. neurons and glia).
Mitochondrial dysfunction is widely considered to be a key component of the pathophysiology of AD[29, 30]. TOMM40 encodes the central, pore-forming subunit of the Translocation of the Outer Mitochondrial Membrane (TOM) complex, which is the mitochondrial protein import machinery in the outer membrane. The gene product, Tom40, is essential for mitochondrial function and reducing Tom40 elicits cell death[31]. Therefore, an association between increased TOMM40 mRNA expression and LOAD seems paradoxical. However, both the amyloid precursor protein, APP, and its proteolytic, Aβ, products, disrupt mitochondrial function and this is mediated, at least in part, through their respective interactions with Tom40[32, 33]. It is conceivable that by mass action elevated levels of Tom40 accelerate APP and/or Aβ-mediated mitochondrial damage. Alternatively, uncoupled expression of TOMM40 from expression of the genes encoding other members of the TOM complex could either disrupt de novo assembly of new TOM complexes, or the function of pre-existing complexes.
In this study we elucidated the mechanism of action of TOMM40 523, a genetic risk factor for LOAD. Our data provides functional support for the role of the 523 genetic locus in the pathogenesis of LOAD
ACKNOWLEDGMENTS
This work was supported in part by the Ellison Medical Foundation New Scholar award AG-NS-0441-08 (O.C.), the National Institute on Aging (NIA) grant R01 AG040370 (A.R.) and a grant from Zinfandel Pharmaceuticals, Inc., to the Bryan ADRC. We thank the Kathleen Price Bryan Brain Bank (KPBBB) at Duke University funded by NIA AG028377, Layton Aging & Alzheimer's Disease Center at Oregon Health and Science University funded by NIA AG008017, and the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore (NICHD contract no. HHSN275200900011C, Ref. No. NO1-HD-9-0011), for providing us with the brain tissues. We would like also to thanks Dr. Randy Woltjer, and John Ervin for their assistance in obtaining the required brain samples for the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
Dr. Allen D. Roses is the CEO of Zinfandel Pharmaceuticals, Inc., Research Triangle Park, NC. Michael W. Lutz, Donna G. Crenshaw, and William K. Gottschalk are consultants to Zinfandel. Zinfandel Pharmaceuticals, Inc. and Takeda Pharmaceutical Company Ltd. have entered into a worldwide licensing agreement regarding Zinfandel's TOMM40 assay as a biomarker for the risk of Alzheimer's disease, including potential use of the assay in combination with pioglitazone in high-risk older adults with normal cognition.
REFRENCES
- 1.Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, et al. A TOMM40 variable length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J. 2009 doi: 10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lutz MW, Crenshaw DG, Saunders AM, Roses AD. Genetic variation at a single locus and age of onset for Alzheimer's disease. Alzheimers Dement. 6(2):125–31. doi: 10.1016/j.jalz.2010.01.011. PMCID: 2874876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caselli RJ, et al. TOMM40, APOE and age of onset of Alzheimer's disease. Alzheimer's and Dementia. 2010;6:S202. [Google Scholar]
- 4.Johnson S, La Rue A, Hermann B, Xu G, Koscik R, Jonaitis E, et al. The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOE?3/?3 genotype. Alzheimers Dement. 2011;7(4):456–65. doi: 10.1016/j.jalz.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayden KM, McEvoy JM, Linnertz C, Attix D, Kuchibhatla M, Saunders AM, et al. A homopolymer polymorphism in the TOMM40 gene contributes to cognitive performance in aging. Alzheimers Dement. 2012 doi: 10.1016/j.jalz.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu SH, Roeder K, Ferrell RE, Devlin B, DeMichele-Sweet MAA, Kamboh MI, et al. TOMM40 poly-T repeat lengths, age of onset and psychosis risk in Alzheimer disease. Neurobiology of Aging. doi: 10.1016/j.neurobiolaging.2011.06.016. In Press, Corrected Proof. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jun G, Vardarajan BN, Buros J, Yu CE, Hawk MV, Dombroski BA, et al. Comprehensive Search for Alzheimer Disease Susceptibility Loci in the APOE Region. Archives of neurology. 2012:1–10. doi: 10.1001/archneurol.2012.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruchaga C, Nowotny P, Kauwe JSK, Ridge PG, Mayo K, Bertelsen S, et al. Association and Expression Analyses With Single-Nucleotide Polymorphisms in TOMM40 in Alzheimer Disease. Arch Neurol. 2011;68(8):1013–9. doi: 10.1001/archneurol.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maruszak A, Pepłońska B, Safranow K, Chodakowska-Żebrowska M, Barcikowska M, Żekanowski C. TOMM40 rs10524523 Polymorphism's Role in Late-Onset Alzheimer's Disease and in Longevity. Journal of Alzheimer's Disease. 2012;28(2):309–22. doi: 10.3233/JAD-2011-110743. [DOI] [PubMed] [Google Scholar]
- 10.Roses AD, Lutz MW, Crenshaw DG, Grossman I, Saunders AM, Gottschalk WK. TOMM40 and APOE: Requirements for replication studies of association with age of disease onset and enrichment of a clinical trial. Alzheimers Dement. 2013 doi: 10.1016/j.jalz.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Crenshaw DG, Gottschalk WK, Lutz MW, Grossman I, Saunders AM, Burke JR, et al. Using genetics to enable studies on the prevention of Alzheimer's disease. Clin Pharmacol Ther. 2013;93(2):177–85. doi: 10.1038/clpt.2012.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutz MW, Crenshaw DG, Saunders AM, Roses AD. Genetic variation as a single locus and age of onset for Alzheimer's disease. Alzheimer's and Dementia. 2010;6:125–31. doi: 10.1016/j.jalz.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roses AD, Cruchaga C, Nowotny P, Kauwe JS, et al. Association and expression analhyses with single-nucleotide polymorphisms in TOMM40 in Alzheimer disease. [February 21, 2012];Arch Neurol. 2011 Aug;68(8):1013–1019. doi: 10.1001/archneurol.2011.155. Comment on, Available at: http://www.alzforum.org/pap/annotation.asp?powID=120717. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bekris LM, Lutz F, Yu CE. Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. Journal of human genetics. 2011 doi: 10.1038/jhg.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch W, Ehrenhaft A, Griesser K, Pfeufer A, Muller J, Schomig A, et al. TaqMan systems for genotyping of disease-related polymorphisms present in the gene encoding apolipoprotein E. Clin Chem Lab Med. 2002;40(11):1123–31. doi: 10.1515/CCLM.2002.197. [DOI] [PubMed] [Google Scholar]
- 16.Linnertz C, Saucier L, Ge D, Cronin KD, Burke JR, Browndyke JN, et al. Genetic regulation of alpha-synuclein mRNA expression in various human brain tissues. PloS one. 2009;4(10):e7480. doi: 10.1371/journal.pone.0007480. PMCID: 2759540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15(10):1388–92. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruno D, Nierenberg JJ, Ritchie JC, Lutz MW, Pomara N. Cerebrospinal fluid cortisol concentrations in healthy elderly are affected by both APOE and TOMM40 variants. Psychoneuroendocrinology. 2012;37(3):366–71. doi: 10.1016/j.psyneuen.2011.07.006. PMCID: 3207029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruno D, Pomara N, Nierenberg J, Ritchie JC, Lutz MW, Zetterberg H, et al. Levels of cerebrospinal fluid neurofilament light protein in healthy elderly vary as a function of TOMM40 variants. Experimental gerontology. 2012;47(5):347–52. doi: 10.1016/j.exger.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hedskog L, Brohede J, Wiehager B, Pinho CM, Revathikumar P, Lilius L, et al. Biochemical Studies of Poly-T Variants in the Alzheimer's Disease Associated TOMM40 Gene. J Alzheimers Dis. 2012;31(3):527–36. doi: 10.3233/JAD-2012-120580. [DOI] [PubMed] [Google Scholar]
- 21.Anderson JD, Widom J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Molecular and cellular biology. 2001;21(11):3830–9. doi: 10.1128/MCB.21.11.3830-3839.2001. PMCID: 87046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19(1):65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singleton A, Myers A, Hardy J. The law of mass action applied to neurodegenerative disease: a hypothesis concerning the etiology and pathogenesis of complex diseases. Human molecular genetics. 2004:13. doi: 10.1093/hmg/ddh093. Spec No 1:R123-6. [DOI] [PubMed] [Google Scholar]
- 24.Zarow C, Victoroff J. Increased apolipoprotein E mRNA in the hippocampus in Alzheimer disease and in rats after entorhinal cortex lesioning. Experimental neurology. 1998;149(1):79–86. doi: 10.1006/exnr.1997.6709. [DOI] [PubMed] [Google Scholar]
- 25.Matsui T, Ingelsson M, Fukumoto H, Ramasamy K, Kowa H, Frosch MP, et al. Expression of APP pathway mRNAs and proteins in Alzheimer's disease. Brain Res. 2007;1161:116–23. doi: 10.1016/j.brainres.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 26.Akram A, Schmeidler J, Katsel P, Hof PR, Haroutunian V. Association of ApoE and LRP mRNA levels with dementia and AD neuropathology. Neurobiology of Aging. 2012;33(3):628, e1–e14. doi: 10.1016/j.neurobiolaging.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee TS, Goh L, Chong MS, Chua SM, Chen GB, Feng L, et al. Downregulation of TOMM40 expression in the blood of Alzheimer disease subjects compared with matched controls. Journal of psychiatric research. 2012;46(6):828–30. doi: 10.1016/j.jpsychires.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Manczak M, Calkins MJ, Reddy PH. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer's disease: implications for neuronal damage. Human molecular genetics. 2011;20(13):2495–509. doi: 10.1093/hmg/ddr139. PMCID: 3109997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swerdlow RH, Burns JM, Khan SM. The Alzheimer's Disease Mitochondrial Cascade Hypothesis. Journal of Alzheimer's Disease. 2010;20(0):265–79. doi: 10.3233/JAD-2010-100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swerdlow R. Mitochondria and Cell Bioenergetics: Increasingly Recognized Components and a Possible Etiologic Cause of Alzheimer's Disease. Antioxidants & Redox Signaling. 2012;16(12):1434–55. doi: 10.1089/ars.2011.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozjak-Pavlovic V, Ross K, Benlasfer N, Kimmig S, Karlas A, Rudel T. Conserved roles of Sam50 and metaxins in VDAC biogenesis. EMBO Rep. 2007;8(6):576–82. doi: 10.1038/sj.embor.7400982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devi L, Anandatheerthavarada HK. Mitochondrial trafficking of APP and alpha synuclein: Relevance to mitochondrial dysfunction in Alzheimer's and Parkinson's diseases. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2010;1802(1):11–9. doi: 10.1016/j.bbadis.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansson Petersen CA, Alikhani N, Behbahani H, Wiehager B, Pavlov PF, Alafuzoff I, et al. The amyloid β-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proceedings of the National Academy of Sciences. 2008;105(35):13145–50. doi: 10.1073/pnas.0806192105. [DOI] [PMC free article] [PubMed] [Google Scholar]