Abstract
Previously, we demonstrated that primary cultures of rat hepatocytes evidence higher levels of differentiated function when cultured in the presence of a dilute overlay of extracellular matrix (Matrigel). In this investigation, we used DNA microarrays, quantitative RT-PCR, immunoblotting, and cell morphology analyses to evaluate the biological responses imparted by Matrigel overlays on primary cultures of human hepatocytes from five independent donors. Although interindividual variability in responses was evident, our results demonstrated that Matrigel additions typically improved hepatocyte morphology and differentiation character. Results from RNA-profiling experiments indicated that Matrigel additions enhanced hepatocyte RNA expression levels associated with a battery of differentiated features, to levels comparable to those seen in vivo, for genes such as the cytochrome P450s, solute carrier family members, sulfotransferases, certain nuclear transcription factors, and other liver-specific markers, such as albumin, transferrin, and response to the inducer, phenobarbital. In contrast, Matrigel additions were generally associated with reduced RNA expression levels for several cytokeratins, integrins, and a number of stress-related pathways. Decreases in integrin protein expression were similarly detected, although enhanced levels of the gap junction–associated protein, connexin 32, were detected in Matrigel-treated cultures. These data support the concept that ECM functions mechanistically to augment the differentiation character of primary human hepatocytes in culture by mediating a reduction in cellular stress response signaling and by enhancing gap junctional cell-cell communication.
Keywords: extracellular matrix, primary hepatocytes, human, cell culture, microarray, phenobarbital, integrins, connexins
In biological tissues, the importance of microenvironment on cell function is well established. The interactions between cells and the extracellular matrix (ECM) together with cell-cell interactions can have profound effects on cell morphology, function, proliferation, differentiation, and responses to stimuli (Bissell et al., 1990; Caron, 1990; Kocarek et al., 1993; Kono et al., 1995; Lee and Streuli, 1999; Luttringer et al., 2002; Moghe et al., 1996; Musat et al., 1993; Sidhu et al., 2004). ECM is comprised of a complex mixture of biomaterials, including collagens, proteins, proteoglycans, and glycosoaminoglycans, and supports the growth, attachment, and migration of cells in their tissue microenvironments (Rojkind and Ponce-Noyola, 1982).
Several studies have demonstrated that ECM additions to cultures of hepatocellular carcinoma (HCC) cell lines and primary hepatocytes largely enhance cell functional criteria (Kocarek et al., 1993; Kono et al., 1995; Luttringer et al., 2002; Moghe et al., 1996; Musat et al., 1993; Sidhu et al., 2004). The importance of an ECM component in maintenance of cell polarity and morphology has been reported in rat hepatocytes (Bissell et al., 1990; Caron, 1990; Lee and Streuli, 1999; Luttringer et al., 2002; Moghe et al., 1996), as has a modulatory role of ECM in the regulation of rat hepatocyte responses to signaling ligands (Lee and Streuli, 1999). Rat primary hepatocytes cultured in the presence of an ECM appear to display an actin filament organization similar to that of intact liver (Musat et al., 1993), and rat hepatoyctes cultured in collagen or Matrigel (a registered trademark of InVitrogen, Inc., Carlsbad, CA) sandwich configurations demonstrate improved morphology and enhanced levels of albumin gene expression (Bissell et al., 1990; Caron, 1990; Luttringer et al., 2002; Moghe et al., 1996). Similarly, improved levels of albumin, transferrin, and transthyretin—markers of differentiated phenotype—were associated with ECM- and dexamethasone-treated hepatocytes, together with the concomitant suppression of dedifferentiation markers, such as alpha-fetoprotein (AFP) and glutathione transferase π (GSTπ) (Sidhu et al., 2004). It has been reported that cell-ECM contacts in culture enhance signaling and transcription factor binding within the albumin gene promoter (Liu et al., 1991). Whereas primary rat hepatocytes typically lose the expression of the cytochrome P450 genes gradually in culture, the presence of Matrigel was noted to reestablish and maintain P450 expression levels (Kocarek et al., 1993; Omiecinski et al., 1999). Presence of Matrigel also greatly facilitates the gene induction response to phenobarbital (PB), a feature that manifests only in highly differentiated hepatocytes (Ben Ze’ev et al., 1988; LeCluyse et al., 1999; Schuetz et al., 1988; Sidhu et al., 1993, 2004).
Human hepatocytes are more difficult to obtain than those of rodents and are often more difficult to attach in two-dimensional cultures; however, human hepatocytes offer a more accurate reflection of species-specific responses to stimuli, such as pharmaceutical exposures, and provide important insight regarding population variability in chemical response (Hawksworth, 1994; Ulrich et al., 1995; Waring et al., 2003). Due to the scarcity of liver donors for primary human hepatocyte cultures, resulting in small sample sizes, it is essential that optimal and consistent culture conditions be defined. Gene expression profiles and response to drug challenge have been investigated in both attached and suspension-cultured human hepatocytes. Although both cell types were viable and offer at least limited responses to challenge, the gene expression profiles of the two types of culture can be quite different and illustrate the importance of determining optimal culture conditions for human predictive modeling (Waring et al., 2003). Hamilton et al.(2001) examined responses of human hepatocytes to various conditions of culture including ECM additions and cellular plating densities. They reported that although presence of ECM resulted in phenotypic differences in the cells, maintenance of the drug induction response was more dependent on plating density than ECM. In contrast, others have evaluated the effects of Matrigel overlay on human hepatocyte culture and concluded that the presence of ECM markedly facilitated xenobiotic responsiveness (Gross-Steinmeyer et al., 2005).
Despite these observations, the underlying mechanisms of ECM effects on cellular function are largely unknown. Experimental evidence suggests that cues received from the ECM are important in maintaining hepatocyte cell morphology, function, and differentiation status (Bissell et al., 1990; Caron, 1990; Kocarek et al., 1993; Kono et al., 1995; Lee and Streuli, 1999; Luttringer et al., 2002; Moghe et al., 1996; Musat et al., 1993; Sidhu et al., 2004). The integrins play a prominent role in mediating cell-ECM interactions and modulate the signal transduction of extracellular cues. Integrins are transmembrane proteins composed of noncovalently linked alpha and beta subunits. Each alpha and beta subunit contains an extracellular ligand-binding domain, a transmembrane domain, and a short cytoplasmic tail. Although integrins possess no intrinsic enzymatic activity, they function to transduce extracellular signals through direct contact with the cellular cytoskeleton, facilitating the redistribution of intracellular proteins and activation of associated signaling cascades (Clark and Brugge, 1995; Giancotti, 1997; Juliano and Haskill, 1993). Aggregations of integrins cause accumulation and activation of signaling molecules (Milliano and Luxon, 2003), activating Rac, Rho, Cdc42 (Ridley and Hall, 1992; Rottner et al., 1999; Small et al., 1999), and the mitogen activated protein kinase-signaling module (Chen et al., 1994; Short et al., 1998). Overexpression of the beta 1 and beta 3 integrin subunits enhances Rho and Rac activity, respectively, and these activation events are often accompanied by morphological alterations as well as changes in stress fiber formation (Miao et al., 2002).
We conducted this study in order to better assess the biological impact of ECM on primary human hepatocytes in culture and to examine the underlying mechanisms associated with these effects. We hypothesized that ECM additions enhance the differentiation patterning of hepatocytes in culture and their resulting responsiveness to environmental cues through the modulation of cellular integrin-signaling networks and associated formation of focal adhesions.
MATERIALS AND METHODS
Cell culture
Enriched primary human hepatocyte cultures plated on collagen were obtained through the Liver Tissue Procurement and Distribution System, Pittsburgh, which was funded by National Institutes of Health contract #NO1-DK-9-2310. Cells were photographed upon arrival, and the cells were placed in fresh William’s E media containing 1% penicillin/streptomycin, 1% N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), 20µM glutamine, 25nM dexamethasone, 10nM insulin, 1% linoleic acid/bovine serum albumin, 5 ng/ml selenious acid, and 5 µg/ml transferrin. Maintenance medium was changed once per 2 days. Selected cultures were treated with 500µM PB, on day 4 of culture. HepG2 cells were obtained from American Type Culture Collection (Manassas, VA) and cultured in Dulbucco’s Modified Eagle’s Medium with 2mM l-glutamine, 0.1mM nonessential amino acids, 1.5 g/l sodium bicarbonate, 1mM sodium pyruvate, 10% fetal bovine serum, 1% HEPES, and 1% penicillin/streptomycin. Selected cultures were overlayed with 225 µg/ml BD Matrigel Basement Membrane Matrix (BD Biosciences; San Jose, CA), as described previously (Sidhu et al., 2004). All other culturing materials were purchased from Invitrogen. The liver tissue from liver HL#154 was generously provided by Dr Kenneth Thummel (University of Washington, Seattle, WA).
RNA isolation and purification
One milliliter of Trizol reagent (Invitrogen) was pipetted onto hepatocytes following media aspiration. The cell monolayer was scraped after a 2-min incubation at room temperature (RT). Trizol/cell mix was pipetted 10 times and then transferred to a microcentrifuge tube and vortexed for 10 s. Lysates were incubated at RT for 5 min. Chloroform, 200 µl, was added and tubes were shaken for 15 s followed by a 3-min incubation at RT. Samples were centrifuged for 15 min at 12,000 × g at 4°C. The aqueous phase was transferred to a clean tube, and organic phase was stored at −80°C for DNA and protein isolation. Isopropyl alcohol, 500 µl, was added to each sample and incubated at RT for 10 min followed by centrifugation at 4°C for 10 min at 12,000 × g. The RNA pellet was washed with 1ml of 75% ethanol, vortexed, and centrifuged for 5 min at 4°C at 7500×g. Pellets were air dried and then resuspended in nuclease-free water. Samples were incubated at 60°C for 10 min. RNA integrity was confirmed by ethidium bromide visualization following formaldehyde gel electrophoresis.
RNA was further purified using DNA-free (Ambion). Briefly, 3 µl of DNA-free DNase and 10 µl of DNase-free DNase buffer were added to each sample. Reactions were incubated at 37°C for 20 min. Inactivator, 20 µl, was added, and samples were incubated at RT for 2 min followed by centrifugation. Supernatants were transferred to fresh tubes. Ammonium acetate (7.5M) and 100% ethanol were added at 0.1 volumes and 2.5 volumes, respectively. Samples were mixed and incubated at −20°C for 90 min followed by centrifugation at 12,500 rpm for 10 min at 4°C. The resulting pellet was washed in 75% ethanol and then air dried. Pellets were dissolved in nuclease-free water and incubated at 60°C for 10 min. Concentrations were determined by spectrophotometry using a SmartSpec 3000 spectrophotometer (BioRad, Hercules, CA).
cDNA preparation
cDNA was prepared using the High Capacity cDNA Archive Kit (cat# 4322171, Applied Biosystems, Foster City, CA). Briefly, 10 µl of 10× Reverse Transcription Buffer, 4 µl 25× dNTPs, 10 µl of 10× random primers, 5 µl of Multiscribe Reverse Transcriptase (50 U/µl), and 21 µl of nuclease-free water were combined in a 0.5-mlmicrocentrifuge tube. RNA (2 µg) and water were added for a total reaction volume of 100 µl. Reverse transcription was carried out at 25°C for 10 min followed by incubation at 37°C for 2 h.
RT-PCR
Reaction components were prepared for duplicate 25-µl reactions in 96-well plates: 2× TaqMan Universal PCR Master Mix (ABI, Foster City, CA, P/N 4304437), 25 µl; 20× Assays-on-Demand Gene Expression Assay Mix, 2.5 µl; and cDNA diluted in RNase-free water, 22.5 µl. Samples were mixed and then split into individual wells. Using the Applied Biosystems 7300 Real-Time PCR System, plate documents were configured with the appropriate assay and sample information. Thermocycling conditions were as follows: UNG activation, 2 min at 50°C, hold 10 min at 95°C, and for each of 40 cycles, 15 s at 95°C and 1 min at 60°C. Sequence Detection System software (ABI) was used to collect and organize the fluorescence data for analysis.
RT-PCR data analysis
Data were analyzed using the ΔΔCT method (Livak and Schmittgen, 2001). Briefly, CT values for genes of interest were normalized to 18S by generating a ΔCT for each gene ΔCTalb(mg) = (averageCTalb(mg) − averageCT18S(mg)). The ΔΔCT was calculated by normalizing the ΔCT values for treatments to control (ΔΔCTalb = ΔCTalb(mg) − ΔCTalbc). Results were then expressed as fold change over control samples by raising 2−ΔΔCT.
Microarray analysis
Five micrograms of purified RNA was processed for microarray analysis by Paradigm Array Labs (Icoria, Research Triangle Park, NC). After all samples were subjected to and passed quality control measures, RNA was hybridized to Affymetrix Human Genome U133 Plus 2.0 arrays, and results were analyzed by GeneChip Operating Software (Affymetrix, Santa Clara, CA). Raw cell intensity values were computed by the Cell Analysis algorithm, and present, absent, and marginal calls were generated. All individual Matrigel samples were then baselined back to their corresponding controls generating change calls and change call p values. Outputs were transferred to Microsoft Access and filtered for presence in both Matrigel and control samples. Surviving probe sets were used for further study. Complete microarray data sets may be viewed at http://www.vetsci.psu.edu/Omiecinski/index.html.
Cluster analysis
Microarray results were analyzed by ArrayAssist 4.0 (Stratagene, La Jolla, CA) with hierarchical clustering. Raw cell intensities were clustered based on expression in donor samples. Results were then filtered for twofold changes across all donors and clustered again by sample.
GOstat analysis
Lists of changed probe sets were batch analyzed by GOstat (Beissbarth and Speed, 2004) for overrepresented categories of genes. The entire list of probe sets from the Affymetrix Human Genome U133 Plus 2.0 array was used as a reference list to determine overrepresentation. The maximum p value cutoff was set at 0.05, and correction for multiple testing was performed using the Benjamini false discovery rate.
Protein isolation
Isopropanol, 1.5 ml per 1 ml Trizol used, was added to the leftover organic phase after RNA and DNA isolation by Trizol. Samples were incubated at RT for 10 min. Protein was sedimented by centrifugation at 12,000 × g at 4°C for 10 min. After removal of the supernatant, protein pellet was washed three times in 2 ml of 0.3M guanidine hydrochloride in 95% ethanol. The pellets were centrifuged at 7500 × g for 5 min at 4°C following each wash. After the final wash, protein pellets were vortexed in 2 ml of 100% ethanol per 1 ml Trizol used followed by incubation at RT for 20 min and centrifugation at 7500×g for 5 min at 4°C. After removing the supernatant, the pellets were air dried for 15 min and resuspended in 50 µl of 2× buffer (8M urea, 2M thiourea, 0.05M Tris, pH 6.8, 75mM dithiothreitol, 3% sodium dodecyl sulfate, 0.05% Bromphenol blue). After the protein samples were completely redissolved, 50 µl of water was added and protein concentrations were quantified by a modified Bradford assay (BioRad, cat#500-0006).
Western immunoblotting
Twenty micrograms of total protein was heated at 95°C prior to loading onto a precast 10% Tris-HCl sodium dodecyl sulfate– polyacrylamide gel electrophoresis (SDS-PAGE) gel (BioRad). Proteins were separated by denaturing SDS-PAGE (100 v for 1.5 h in 0.03MTris, 0.2Mglycine, 0.025% SDS) and electrophoretically transferred to polyvinylidene fluoride (PVDF) membranes (120 v for 1 h in 0.03M Tris, 0.2M glycine, 20% methanol). Membranes were blocked in 5% nonfat dry milk and TBS-T (0.1% Tween) for 1 h prior to the addition of primary antibody diluted in blocking buffer. Membranes were incubated with primary antibody overnight at 4°C with rocking and then washed thrice for 5 min in TBS-T. Membranes were then incubated with the appropriate secondary horseradish peroxidase–conjugated antibody diluted 1:5000 in blocking buffer for 1 h at RT with rocking. After three 5-min washes in TBS-T, protein-antibody complexes were visualized by chemiluminescence (Lumilight, Roche, Indianapolis, IN) and exposed to x-ray film.
Primary antibodies and dilutions were as follows: α-Keratin 8 (MS-997, Neomarkers, Fremont, CA) 1:200; α-Keratin 18 (MS-142, Neomarkers) 1:200; α-phosphoKeratin 8 (MS-1241, Neomarkers) 1:200; α-phosphoKeratin 18 (MS-1242, Neomarkers) 1:200; α-integrin α5 (AF1864, R&D Systems, Minneapolis, MN) 1:250; and α-connexin 32 (13–8200, Zymed Laboratories Inc, San Francisco, CA) 1:500
RESULTS
Matrigel Overlay Elicits Phenotypic Change in Individual Hepatocyte Samples
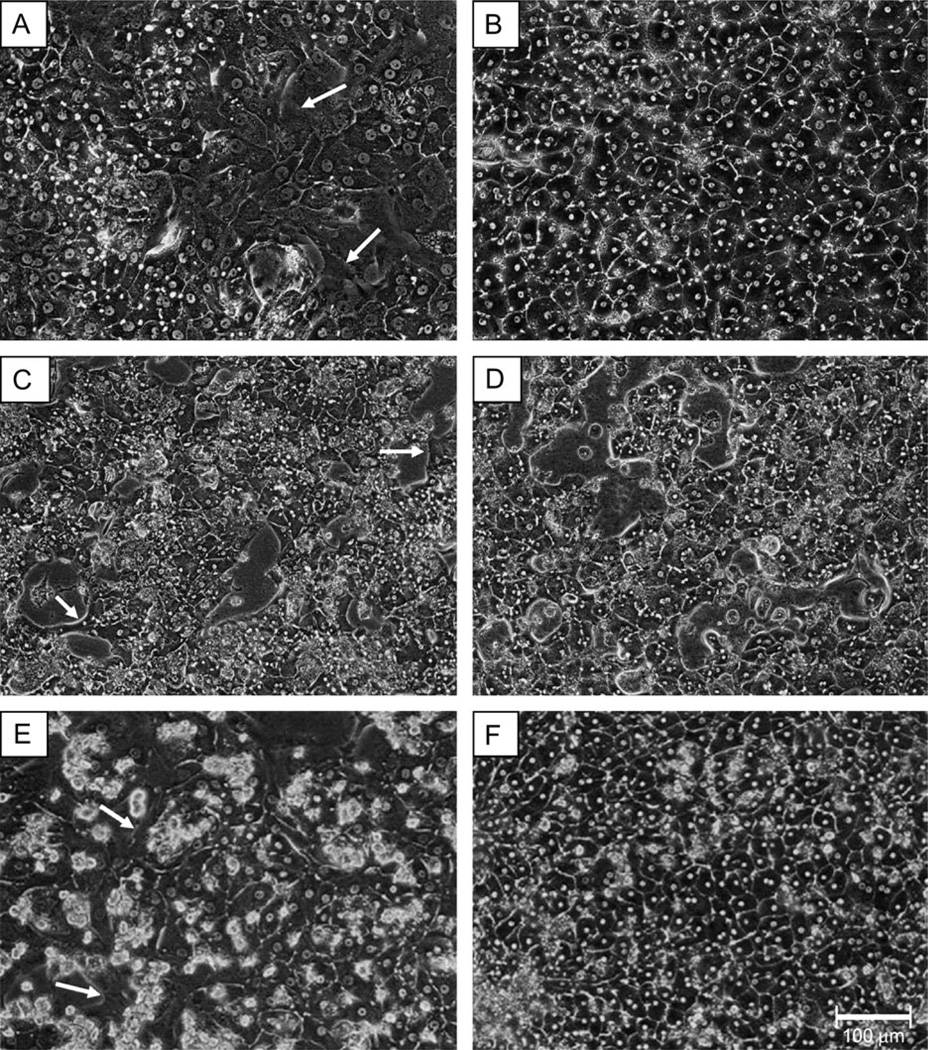
To assess the morphological impact of ECM addition, photomicrographs were obtained from control (Figs. 1A, 1C, and 1E) and Matrigel-treated (Figs. 1B, 1D, and 1F) samples each day over the course of culturing. Photomicrographic results from donors A, B, and C illustrate that the addition of a Matrigel overlay improved hepatocyte morphology, as the cells exhibited more defined cuboidal shape and improved definition of cell borders. In contrast, cells without a Matrigel overlay exhibited a flattened appearance, poorly defined borders, and elaborated spinous processes resembling a fibroblastic character. These cells also failed to form the highly organized networks of cells cultured in the presence of an ECM.
FIG. 1.
Matrigel enhances cellular morphology of primary human hepatocyte cultures for donor A, donor B, and donor C. Primary human hepatocytes from donor A (A and B), donor B (C and D), and donor C (E and F) were cultured in the presence (B, D, F) and absence (A, C, E) of a Matrigel overlay. Photomicrographs were taken under ×20 magnification using phase contrast imaging. Arrows indicate compromised morphology in the absence of a Matrigel overlay.
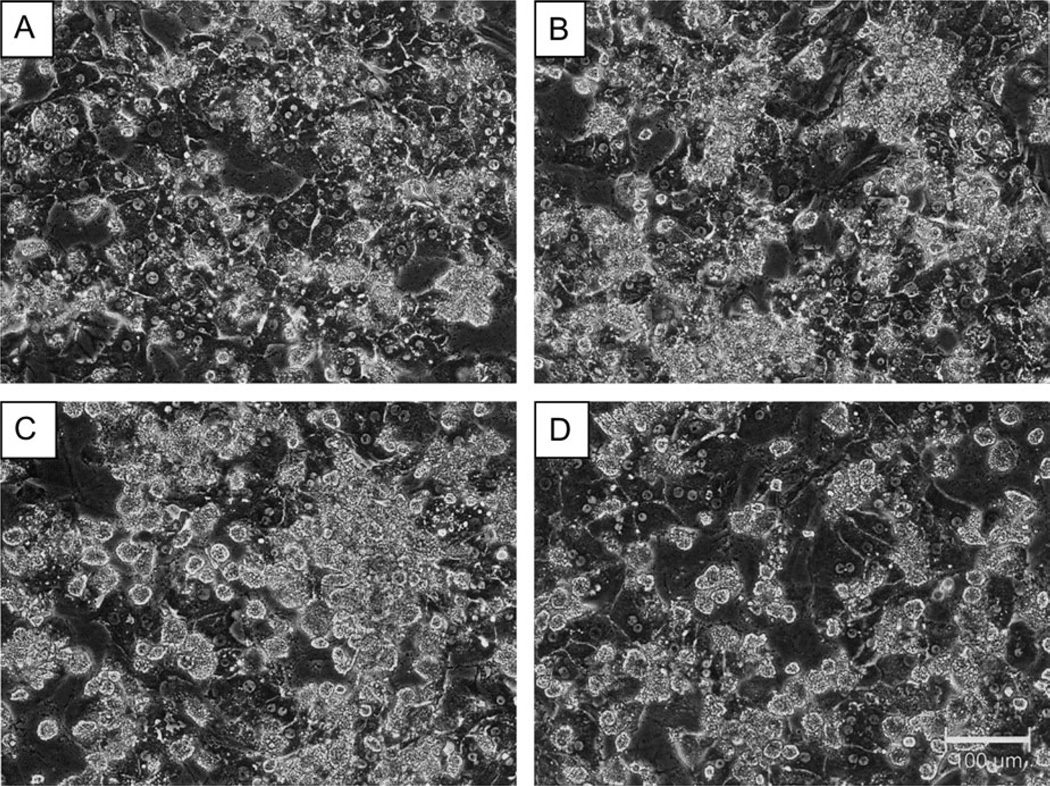
Figure 2 displays photomicrographs taken from donor D and E hepatocytes cultured in the presence (B and D) and absence of Matrigel (A and C). Matrigel additions to these cultures appeared to result in little distinguishable morphological change. However, even from their initial culture, these samples exhibited numerous aggregations of necrotic and apoptotic hepatocytes, both in the presence and absence of ECM. These cells demonstrated disorganized cellular architecture, flattened morphology, and poorly defined cell borders. Therefore, the compromised initial quality of hepatocytes obtained from these two donors was not rescued by Matrigel additions.
FIG. 2.
Matrigel overlay exhibits minimal impact on cellular morphology for primary human hepatocyte cultures obtained from donor D or donor E. Primary human hepatocytes from donor D (A and B) and donor E (C and D) were cultured in the presence (B and D) and absence (A and C) of a Matrigel overlay. Photomicrographs were taken under ×20 magnification using phase contrast imaging.
Overall, our observations indicated that morphological changes in primary human hepatocytes demonstrating low-quality morphology were not phenotypically responsive to ECM, whereas higher quality cultures exhibited further defined improvement in morphology when cultured in the presence of a Matrigel overlay. After reviewing the donor descriptions, no correlation between donor characteristics and culture quality or response to Matrigel could be established (Table 1).
TABLE 1.
Human Hepatocyte Donor Characteristics
| Donor identification |
Age | Gender | Ethnicity | Cause of death |
Smoker/alcohol use |
|---|---|---|---|---|---|
| HH-A | 0.75 | M | C | Anoxia | No/no |
| HH-B | 61 | M | C | Gunshot wound | No/yes (21 drinks/week) |
| HH-C | 3 | F | C | Anoxia | No/no |
| HH-D | 29 | F | C | n/a | No/no |
| HH-E | 46 | F | C | Head injury | Yes (13 years)/yes (3 drinks/week) |
Note. Human hepatocytes were obtained from Dr Steven Strom at the University of Pittsburgh, following the regulations of the Liver Tissue Procurement and Distribution System. C, Caucasian; n/a, not available.
Matrigel Enhances the Differentiation Status of Primary Human Hepatocytes
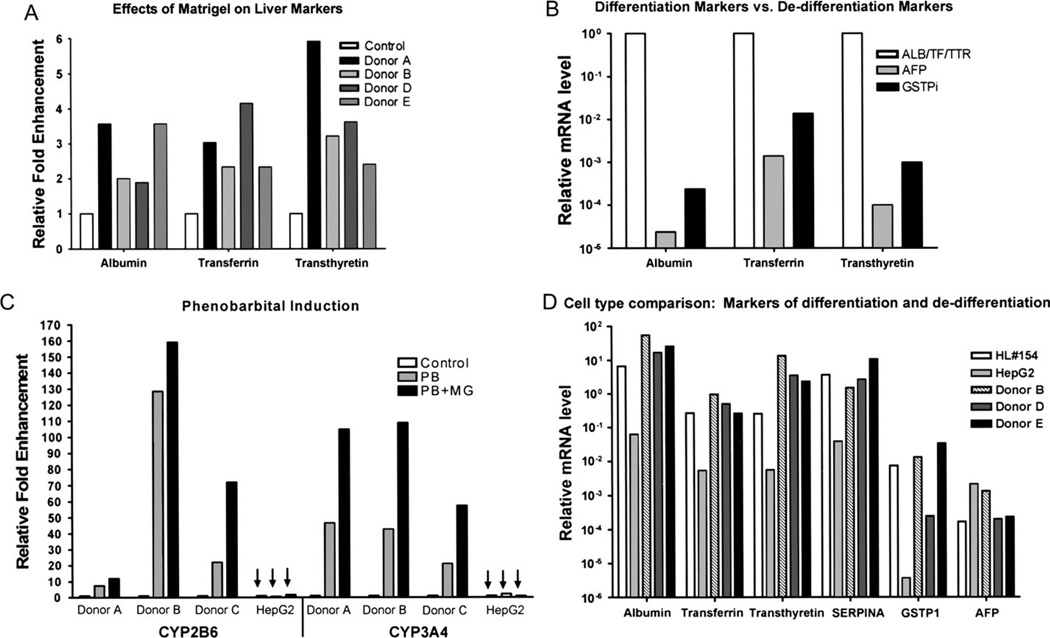
Previous reports have established a supportive role for ECM/Matrigel in hepatocyte culture (Kocarek et al., 1993; Kono et al., 1995; Luttringer et al., 2002; Moghe et al., 1996; Musat et al., 1993; Sidhu et al., 2004). Studies in rat showed that addition of Matrigel to primary hepatocyte cultures increased the expression of differentiation markers (Sidhu et al., 2004). To examine the effects of ECM on primary cultures of human hepatocytes, cells were grown in the presence or absence of a Matrigel overlay. Cultures were monitored over the course of several days, and total RNA was collected. Real-time quantitative PCR was performed on reverse transcription reactions from purified RNA. Results were compared to untreated controls using the ΔΔCt method (Livak and Schmittgen, 2001). The addition of ECM improved the expression of three markers of differentiation: albumin, transferrin, and transthyretin in all four hepatocyte samples tested (Fig. 3A). In addition to enhanced expression of differentiation markers, culturing the hepatocytes in the presence of Matrigel appropriately repressed expression of the dedifferentiation markers, GSTπ and AFP (Fig. 3B).
FIG. 3.
Effects of Matrigel addition on differentiation status of primary human hepatocyte cultures. Total RNA was isolated from primary human hepatocytes cultured for 5 days in the presence or absence of a Matrigel overlay. (A) Transcript levels for the hepatocyte differentiation markers, albumin, transferrin, and transthyretin, were assessed −/+ Matrigel additions using quantitative RT-PCR and the ΔΔCT method. (B) Transcript levels for the hepatocyte dedifferentiation markers, AFP and GSTPi, were similarly ascertained −/+ Matrigel additions and were normalized to each differentiation marker. (C) Primary human hepatocytes were cultured in the absence (control) or presence of Matrigel (MG). Cultures of primary human hepatocytes and HepG2 cells (indicated by arrows) were treated on day 4 with phenobarbital (PB and PB + MG) or left untreated (control) for 24 h prior to RNA isolation. Relative fold changes in transcript levels for the PB-inducible marker genes, CYP2B6 and CYP3A4, are indicated. (D) Total RNA was isolated from HepG2 cells, a section of human liver #154, as well as three different donor samples of primary human hepatocytes that were cultured with a Matrigel overlay. Relative expression analyses for a panel of differentiation and dedifferentiation markers were determined by quantitative RT-PCR analysis, and the results are graphically depicted.
The induction of drug-metabolizing enzymes in response to an external stimulus, in particular PB, represents a complex biological response indicative of a highly differentiated hepatic phenotype. When primary human hepatocytes cultured in the presence or absence of Matrigel (MG and control, respectively) were treated with PB for 24 h prior to isolation of total RNA, induction of the PB-responsive genes, CYP2B6 and CYP3A4, was enhanced in cells cultured in the presence of Matrigel (PB + MG). HepG2 cells, a human HCC cell line, treated in the same fashion exhibited no induction response and no improvement in response in the presence of Matrigel (Fig. 3C). Total RNA was collected from a section of whole human liver, and the expression profile of differentiation and dedifferentiation markers was determined through quantitative RT-PCR. A comparison of human liver, human hepatocytes cultured with Matrigel, and HCC cells was performed. The results demonstrated that primary human hepatocytes cultured in the presence of a Matrigel overlay most closely resemble the expression profile of the human liver while HepG2 cells, although expressing certain markers, differ from the expression levels of the liver by at least 10-fold and as much as 200-fold (Fig. 3D). This result was apparent in both donors D and E, despite the lack of morphological impact of Matrigel on these cultures (Fig. 2). In studies to be reported elsewhere, further comparisons to additional human liver tissues, from six different donors, were also conducted, with similar conclusions derived as that for the representative HL#154 liver presented here. Therefore, the cumulative evidence indicated that a Matrigel overlay was a positive regulator of differentiation status of primary human hepatocytes, facilitating the upregulation of differentiation makers, downregulation of dedifferentiation markers, and enhancement of the response to PB.
Interindividual Variability Contributes to Expression Profile of Donors
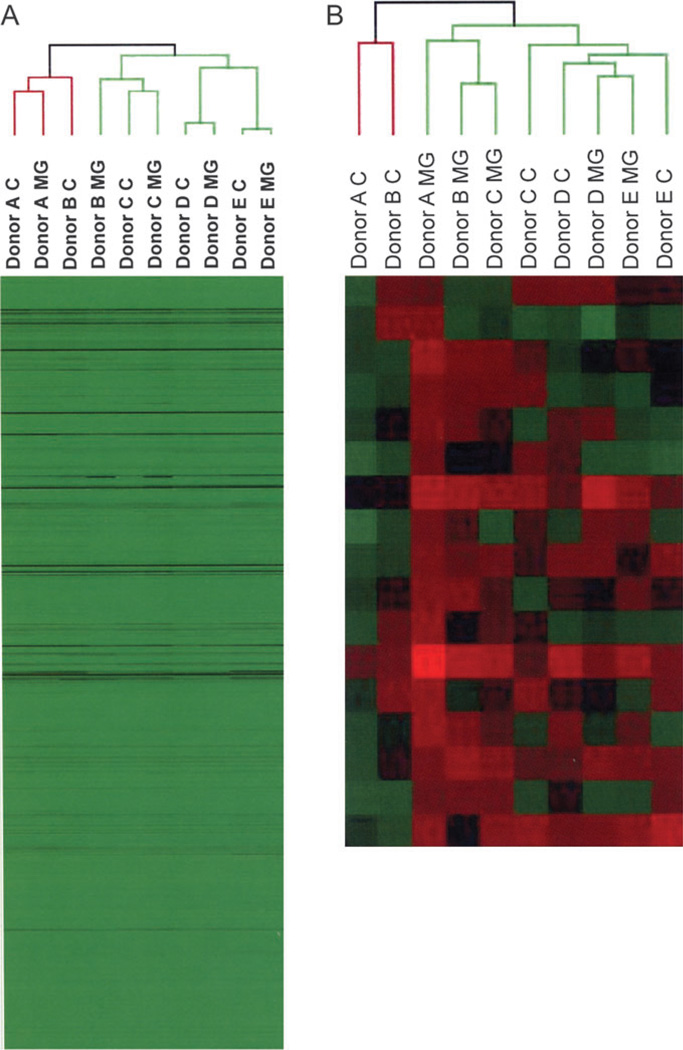
After analysis by microarray sample sets, with and without Matrigel, from all donors were subjected to hierarchical clustering. In Figure 4A, the raw intensity values of all present probe sets in all samples are shown in a heat map. Donors A, C, D, and E cluster together regardless of treatment, indicating the dominance of interindividual variability. When the probe set list was filtered for dramatic changes (twofold or more, either increased or decreased), all the samples tended to cluster by treatment. Matrigel-treated cultures from donors A, B, and C shared similar expression patterns and Matrigel cultures from donors D and E clustered together (Fig. 4B). These clustering patterns reflect the similarity in morphological assessments of the individual cultures (Figs. 1 and 2). In contrast, the clustering of cultures lacking ECM displayed no clear pattern. This highlights the primary dominance of interindividual variability in the absence of ECM treatment. In addition, the ability of Matrigel to similarly impact gene expression regardless of donor is well apparent.
FIG. 4.
Hierarchical clustering analyses of interindividual differences and treatment regimens of primary human hepatocyte samples. Total RNA was isolated from cultures of primary human hepatocytes from five different donors −/+ Matrigel overlay. RNA transcript profiles were analyzed by microarray hybridizations, as described in “Materials and Methods” section. Results were hierarchically clustered (ArrayAssist, Stratagene) before (A) and after (B) filtering for twofold changes in gene expression level between Matrigel-treated samples and untreated controls.
Addition of Matrigel to Primary Human Hepatocyte Cultures Elicits Changes in Gene Expression
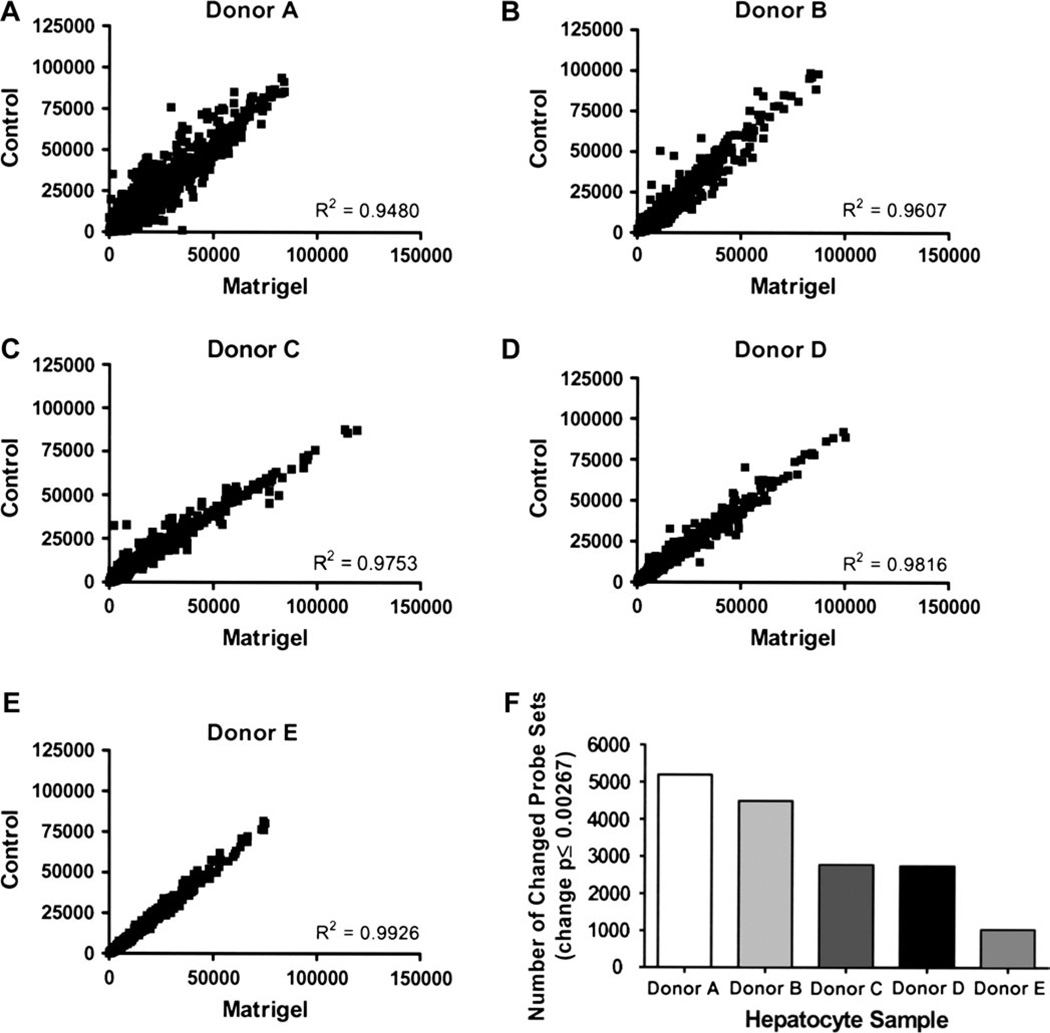
To characterize the changes in gene expression influenced by ECM additions to the primary hepatocyte cultures, individual RNA samples were analyzed by microarray profiling. Gene expression profiles from five donors cultured in the presence or the absence of a Matrigel overlay were examined using the GeneChip Operating Software (Affymetrix). Initially, regression scatterplots were generated by plotting raw intensity values of −/+ ECM data, and correlation coefficients were generated (Figs. 5A–5E). The R2 values obtained −/+ Matrigel were 0.9480, 0.9607, 0.9753, 0.9816, and 0.9926 for donors A, B, C, D, and E, respectively. Although the control and Matrigel samples were highly correlated, measurable gene expression changes were apparent in each donor set. Donor A exhibited 5193 changed probe sets; donor B, 4492 changed probe sets; donor C, 2769 changed probe sets; donor D, 2740 altered probe sets; with donor E exhibiting the fewest numbers of changed probe sets, 1024 (Fig. 5F). In summary for these analyses, although overall levels of gene expression changes consequent to Matrigel overlay additions were subtle, as evidenced by the high R2 values, a minimum of 1000 probe sets were indeed altered as a result of that respective culture condition.
FIG. 5.
Biological impact of Matrigel additions in primary human hepatocyte cultures. Microarray analyses were performed on total RNA collected from primary human hepatocytes cultured in the presence or absence of a Matrigel overlay for either 4 or 5 days. Raw signal intensities for every probe set were plotted for control and Matrigel samples for each donor (A–E). A correlation coefficient was generated for each scatterplot. Total numbers of changed genes (change call p ≤ 0.00267) were reported for each donor (F).
Matrigel Treatment Contributes to Changed Gene Expression Profiles of Primary Human Hepatocytes
Tables 2 and 4 list the probe sets and corresponding genes altered in response to ECM addition among all five hepatocyte donor samples. Table 2 illustrates that several cytochrome P450s, 2C8, 2C9, and 2C19, and epoxide hydrolase 1 (EPHX1) were each increased in all five donor samples treated with Matrigel. Since donors A, B, and C are closely related in expression profiles and demonstrated similar morphological response to Matrigel, the common genes that were upregulated by presence of an ECM in these samples were examined. A total of 193 probe sets, corresponding to 153 genes, were increased by addition of Matrigel to hepatocyte cultures in these three donors. Selected genes are displayed in Table 3. Eleven of the cytochrome P450 gene products were represented by at least one probe set, along with probe sets corresponding to other liver-specific genes like CEBPalpha, FMO5, the nuclear receptors NR1D2 and NR1I3, the solute carrier family proteins, and the sulfotransferases.
TABLE 2.
Universal Gene Expression Increases due to Addition of Matrigel in Five Human Hepatocyte Cultures
| Fold change | |||||||
|---|---|---|---|---|---|---|---|
| Probe set | Description | Gene symbol | Donor A | Donor B | Donor C | Donor D | Donor E |
| 202017_at | Epoxide hydrolase 1, microsomal | EPHX1 | 2.297 | 2.639 | 3.732 | 1.149 | 1.414 |
| 208147_s_at | Cytochrome P450, family 2, subfamily C, polypeptide 8 | CYP2C8 | 3.249 | 3.249 | 2.828 | 2.000 | 2.144 |
| 214421_x_at | Cytochrome P450, family 2, subfamily C, polypeptide 9 | CYP2C9 | 1.625 | 2.144 | 1.866 | 1.414 | 1.231 |
| 216025_x_at | Cytochrome P450, family 2, subfamily C, polypeptide 19 | CYP2C19 | 1.625 | 2.144 | 1.866 | 1.414 | 1.231 |
| cytochrome P450, family 2, subfamily C, polypeptide 9 | CYP2C9 | ||||||
| 220017_x_at | Cytochrome P450, family 2, subfamily C, polypeptide 9 | CYP2C9 | 1.625 | 2.000 | 1.866 | 1.414 | 1.231 |
| 226147_s_at | Polymeric immunoglobulin receptor | PIGR | 1.866 | 3.482 | 1.231 | 1.516 | 1.320 |
Note. Microarray results were filtered for probe sets upregulated among all five donor Matrigel samples. Amount of fold change compared to control for each probe set is reported.
TABLE 4.
Universal Gene Expression Decreases due to Matrigel Addition in Five Human Hepatocyte Primary Cell Cultures
| Fold change | |||||||
|---|---|---|---|---|---|---|---|
| Probe set ID | Description | Gene symbol | Donor A | Donor B | Donor C | Donor D | Donor E |
| 209026_x_at | Beta 5-tubulin | OK/SW-cl.56 | 0.812 | 0.871 | 0.660 | 0.812 | 0.812 |
| 213548_s_at | Hypothetical protein H41 | H41 | 0.758 | 0.379 | 0.660 | 0.758 | 0.616 |
| 217996_at | Pleckstrin homology-like domain, A1 | PHLDA1 | 0.707 | 0.354 | 0.500 | 0.707 | 0.812 |
| 224917_at | Likely ortholog of rat vacuole membrane protein 1 | VMP1 | 0.268 | 0.707 | 0.707 | 0.758 | 0.758 |
Note. Microarray results were filtered for probe sets downregulated among all five donor Matrigel samples. Amount of fold change compared to control for each probe set is reported.
TABLE 3.
Selected Increased Probe Sets in MG-Responsive Human Hepatocyte Cultures
| Fold change | |||||
|---|---|---|---|---|---|
| Probe set | Gene symbol | Description | Donor A | Donor B | Donor C |
| 206262_at | ADH1B | Alcohol dehydrogenase IB (class I), beta polypeptide | 5.657 | 1.625 | 3.031 |
| 204039_at | CEBPA | CCAAT/enhancer-binding protein (C/EBP), alpha | 2.297 | 1.866 | 1.866 |
| 209366_x_at* | CYB5 | Cytochrome b-5 | 1.866 | 1.516 | 1.320 |
| 203979_at | CYP27A1 | Cytochrome P450, family 27, subfamily A, polypeptide 1 | 2.462 | 1.625 | 1.625 |
| 211295_x_at* | CYP2A6 | Cytochrome P450, family 2, subfamily A, polypeptide 6 | 4.925 | 6.063 | 14.929 |
| 207718_x_at | CYP2A7 | Cytochrome P450, family 2, subfamily A, polypeptide 7 | 2.639 | 6.063 | 12.996 |
| 206754_s_at* | CYP2B6 | Cytochrome P450, family 2, subfamily B, polypeptide 6 | 1.414 | 9.849 | 8.574 |
| 216661_x_at* | CYP2C19 | Cytochrome P450, family 2, subfamily C, polypeptide 19 | 1.741 | 2.144 | 2.000 |
| 208147_s_at | CYP2C8 | Cytochrome P450, family 2, subfamily C, polypeptide 8 | 3.249 | 3.249 | 2.828 |
| 220017_x_at* | CYP2C9 | Cytochrome P450, family 2, subfamily C, polypeptide 9 | 1.625 | 2.000 | 1.866 |
| 205999_x_at* | CYP3A4 | Cytochrome P450, family 3, subfamily A, polypeptide 4 | 4.000 | 2.000 | 3.482 |
| 211442_x_at | CYP3A43 | Cytochrome P450, family 3, subfamily A, polypeptide 43 | 1.414 | 2.144 | 2.639 |
| 211843_x_at* | CYP3A7 | Cytochrome P450, family 3, subfamily A, polypeptide 7 | 2.828 | 1.866 | 3.031 |
| 202017_at | EPHX1 | Epoxide hydrolase 1, microsomal | 2.297 | 2.639 | 3.732 |
| 205776_at* | FMO5 | Flavin-containing monooxygenase 5 | 1.741 | 1.866 | 1.625 |
| 225768_at | NR1D2 | Nuclear receptor subfamily 1, group D, member 2 | 1.741 | 2.000 | 1.741 |
| 207007_at | NR1I3 | Nuclear receptor subfamily 1, group I, member 3 | 3.249 | 1.866 | 1.625 |
| 207097_s_at | SLC17A2 | Solute carrier family 17, member 2 | 1.866 | 2.144 | 1.414 |
| 201920_at | SLC20A1 | Solute carrier family 20, member 1 | 1.320 | 2.639 | 1.625 |
| 205972_at | SLC38A3 | Solute carrier family 38, member 3 | 3.249 | 2.144 | 1.866 |
| 225516_at | SLC7A2 | Solute carrier family 7, member 2 | 1.866 | 1.741 | 1.414 |
| 222071_s_at | SLCO4C1 | Solute carrier organic anion transporter family, member 4C1 | 1.866 | 1.625 | 3.031 |
| 203615_x_at | SULT1A1 | Sulfotransferase family, cytosolic, 1A, member 1 | 1.414 | 1.320 | 1.149 |
| 206292_s_at | SULT2A1 | Sulfotransferase family, cytosolic, 2A, member 1 | 1.625 | 1.516 | 1.625 |
Note. Microarray results were filtered for probe sets upregulated in Matrigel samples for donor A, donor B, and donor C. Amount of fold change compared to control for each probe set is reported. An asterisk (*) denotes those genes which had multiple probe sets increased; a single representative probe set with fold changes with respect to control is reported.
The presence of an ECM also resulted in the decrease of four probe sets among all individuals (Table 4), corresponding to beta 5-tubulin (OK/SW-cl.56), hypothetical protein H41 (H41), pleckstrin homology–like domain family A member 1 (PHLDA1), and likely ortholog of rat vacuole membrane protein 1 (VMP1). Among donors A, B, and C, those most responsive to ECM addition, 272 probe sets corresponding to 209 genes exhibited decreased expression in the presence of Matrigel, including actinin, tropomyosin, BMP2, Cdc42, Ras and Ras-related proteins, chemokines, keratins, integrins, JAK1, TGFBR1 and TGFBR2, as well as others. A list of selected genes demonstrating reduced expression levels in the presence of Matrigel is provided in Table 5.
TABLE 5.
Selected Decreased Probe Sets in MG-Responsive Human Hepatocyte Samples
| Fold change | |||||
|---|---|---|---|---|---|
| Probe set | Description | Gene symbol | Donor A | Donor B | Donor C |
| 208636_at* | Actinin, alpha 1 | ACTN1 | 0.660 | 0.758 | 0.574 |
| 205289_at* | Bone morphogenetic protein 2 | BMP2 | 0.500 | 0.406 | 0.574 |
| 211367_s_at* | Caspase 1, apoptosis-related cysteine protease | CASP1 | 0.707 | 0.660 | 0.707 |
| 203065_s_at* | Caveolin 1, caveolae protein, 22 kDa | CAV1 | 0.435 | 0.406 | 0.574 |
| 203324_s_at | caveolin 2 | CAV2 | 0.660 | 0.616 | 0.812 |
| 208727_s_at | Cell division cycle 42 | CDC42 | 0.660 | 0.758 | 0.574 |
| 823_at | Chemokine (C-X3-C motif) ligand 1 | CX3CL1 | 0.707 | 0.758 | 0.536 |
| 204470_at | Chemokine (C-X-C motif) ligand 1 | CXCL1 | 0.406 | 0.660 | 0.500 |
| 206336_at | Chemokine (C-X-C motif) ligand 6 | CXCL6 | 0.406 | 0.574 | 0.435 |
| 200859_x_at* | Filamin A, alpha | FLNA | 0.308 | 0.707 | 0.616 |
| 210338_s_at | Heat shock 70-kDa protein 8 | HSPA8 | 0.871 | 0.812 | 0.812 |
| 201841_s_at | Heat shock 27-kDa protein 1 | HSPB1 | 0.536 | 0.707 | 0.536 |
| 221667_s_at | Heat shock 22-kDa protein 8 | HSPB8 | 0.354 | 0.574 | 0.536 |
| 202727_s_at* | Interferon gamma receptor 1 | IFNGR1 | 0.536 | 0.660 | 0.707 |
| 206295_at | Interleukin 18 | IL18 | 0.287 | 0.287 | 0.379 |
| 202859_x_at* | Interleukin 8 | IL8 | 0.218 | 0.616 | 0.536 |
| 202351_at | Integrin, alpha V | ITGAV | 0.500 | 0.758 | 0.616 |
| 1553530_a_at* | Integrin, beta 1 | ITGB1 | 0.574 | 0.467 | 0.707 |
| 226535_at | Integrin, beta 6 | ITGB6 | 0.354 | 0.467 | 0.500 |
| 1552611_a_at* | Janus kinase 1 | JAK1 | 0.616 | 0.574 | 0.707 |
| 201596_x_at | Keratin 18 | KRT18 | 0.758 | 0.812 | 0.536 |
| 201650_at | Keratin 19 | KRT19 | 0.435 | 0.406 | 0.330 |
| 209016_s_at | Keratin 7 | KRT7 | 0.308 | 0.616 | 0.406 |
| 209008_x_at | Keratin 8 | KRT8 | 0.536 | 0.812 | 0.536 |
| 212119_at | Ras homolog gene family, member Q | RHOQ | 0.812 | 0.812 | 0.758 |
| 224793_s_at | Transforming growth factor, beta receptor I | TGFBR1 | 0.660 | 0.707 | 0.707 |
| 208944_at | Transforming growth factor, beta receptor II (70/80 kDa) | TGFBR2 | 1.000 | 0.707 | 0.707 |
| 218856_at | Tumor necrosis factor receptor superfamily, member 21 | TNFRSF21 | 0.379 | 0.467 | 0.536 |
| 210987_x_at* | Tropomyosin 1 (alpha) | TPM1 | 0.500 | 0.660 | 0.500 |
| 209118_s_at | Tubulin, alpha 3 | TUBA3 | 0.435 | 0.812 | 0.616 |
| 200931_s_at | Vinculin | VCL | 0.574 | 0.660 | 0.574 |
Note. Microarray results were filtered for probe sets downregulated in Matrigel samples for donor A, donor B, and donor C. Amount of fold change compared to control for each probe set is reported. An asterisk (*) denotes those genes which had multiple probe sets increased; a single representative probe set with fold changes with respect to control is reported.
When considering changes occurring in all five individuals, there were certain categories of genes for which expression was either increased or decreased in every donor in response to Matrigel. To investigate common themes of gene expression changes, all the probe sets exhibiting either increased or decreased change in all five donors were analyzed by GOstat (Beissbarth and Speed, 2004). Oxidoreductase-associated genes were the major overrepresented group of genes exhibiting enhanced expression levels (Table 6). Genes in the xenobiotic metabolism ontology category also exhibited increased expression. Overrepresented genes demonstrating reduced expression level in the presence of ECM included actin cytoskeleton, cytoskeleton organization and biogenesis, actin binding, cytoskeletal protein binding, and cellular morphogenesis (Table 7). Overall then, ECM upregulated gene categories within several liver-specific functions, while cytoskeletal- and stress-related categories comprised the downregulated list.
TABLE 6.
Selected GO Terms from Increased Gene List among All Human Hepatocyte Samples
| Best GOs | GO descriptions | Count (1621) |
Total (13,410) |
p Value |
|---|---|---|---|---|
| GO:0016491 | Oxidoreductase activity | 187 | 569 | 6.00 × 10−51 |
| GO:0016614 | Oxidoreductase activity, acting on CH-OH group of donors | 46 | 96 | 4.38 × 10−24 |
| GO:0016616 | Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 43 | 88 | 3.30 × 10−23 |
| GO:0016705 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 40 | 102 | 1.63 × 10−14 |
| GO:0016627 | Oxidoreductase activity, acting on the CH-CH group of donors | 22 | 43 | 2.00 × 10−12 |
| GO:0004497 | Monooxygenase activity | 32 | 85 | 8.96 × 10−11 |
| GO:0016628 | Oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | 12 | 15 | 1.33 × 10−07 |
| GO:0042221 | Response to chemical stimulus | 69 | 314 | 3.34 × 10−06 |
| GO:0016709 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD or NADH as one donor, and incorporation of one atom of oxygen | 12 | 20 | 1.55 × 10−05 |
| GO:0006805 | Xenobiotic metabolism | 13 | 25 | 4.26 × 10−05 |
| GO:0009410 | Response to xenobiotic stimulus | 13 | 27 | 0.000122 |
Note. All increased probe sets for all five donors were combined and batch analyzed by GOstat (Beissbarth and Speed, 2004). Results show the confidence level associated with the GO term overrepresentation.
TABLE 7.
Selected GO Terms from Decreased Gene List among All Human Hepatocyte Samples
| Best GOs | GO Description | Count (2203) |
Total (13,410) |
p Value |
|---|---|---|---|---|
| GO:0003924 | GTPase activity | 48 | 137 | 6.11 × 10−07 |
| GO:0005525 | GTP binding | 85 | 300 | 2.23 × 10−06 |
| GO:0015629 | Actin cytoskeleton | 55 | 183 | 5.39 × 10−05 |
| GO:0007010 | Cytoskeleton organization and biogenesis | 83 | 322 | 0.000294 |
| GO:0003779 | Actin binding | 56 | 214 | 0.00467 |
| GO:0008092 | Cytoskeletal protein binding | 72 | 296 | 0.00771 |
| GO:0000902 | Cellular morphogenesis | 58 | 228 | 0.00804 |
Note. All decreased probe sets for all five donors were combined and batch analyzed by GOstat (Beissbarth and Speed, 2004). Results show the confidence level associated with the GO term overrepresentation.
Imaging analyses indicated the presence of necrotic and apoptotic cellular content in two of the five donor samples, donors D and E (Fig. 2). Strong associations with expression markers for apoptosis and necrosis were not evident for these donors in our GOstat analyses; however, the control versus Matrigel-treated GOstat comparisons performed referenced the initially compromised cellular state, one where these expression markers were already elevated. Although Matrigel supplementation was not able to rescue these cultures from their apoptotic or necrotic phenotype, the addition of an ECM did enhance the expression of other cellular markers, thereby promoting a more closer reflection to that of intact human liver (Fig. 3D).
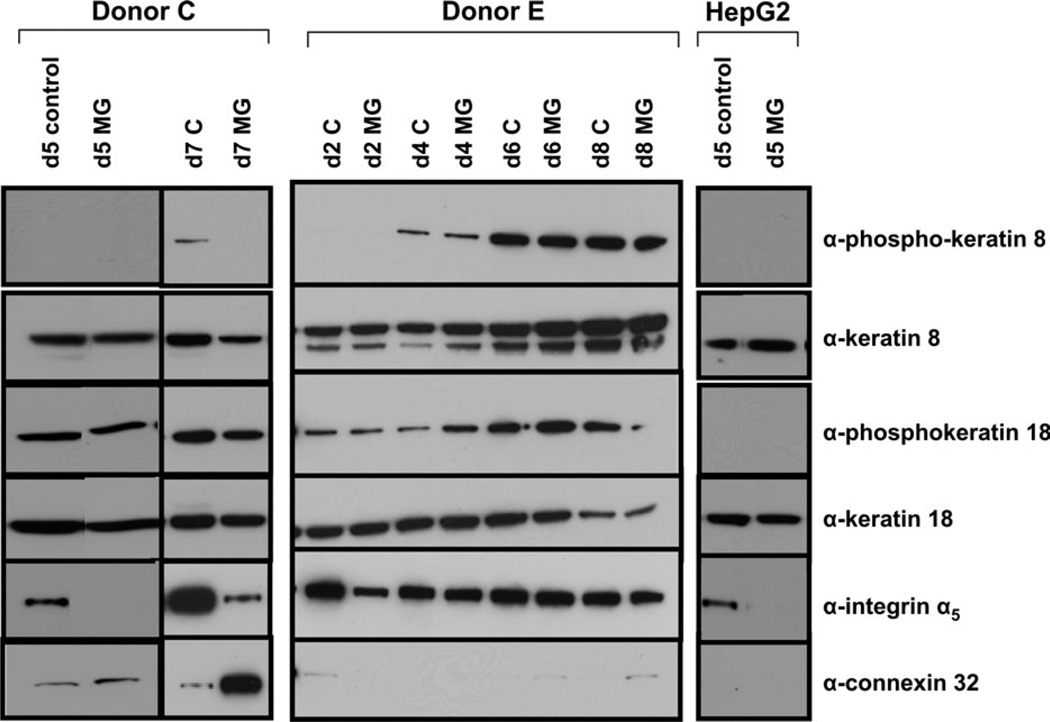
Impact of Matrigel on Protein Expression
To further characterize mechanistic effects of Matrigel as a determinant of phenotypic and genotypic changes in primary human hepatocytes, we performed targeted protein level investigations in the culture system (Fig. 6). In donor C on day 5 (114 h), Matrigel additions appeared to have little effect on the levels of protein expression of keratin 8 or 18, and little effect on the phosphorylation status of these proteins. However, integrin α5 levels were dramatically decreased as a consequence of Matrigel addition; in fact, this protein was undetectable in hepatocytes cultured with Matrigel. Hepatocytes cultured for 7 days (162 h) −/+ Matrigel were also examined for protein expression levels. Integrin α5 was detectable in the Matrigel-treated sample but exhibited an apparent decrease in its respective expression level. Day 5 expression of connexin 32 in control samples was modest but displayed a slight increase in expression in the presence of Matrigel. This increase was more clearly evident in the day 7 samples where Matrigel additions produced a robust increase in the amount of connexin 32 protein. These results reflect trends observed in the microarray data; for example, decreased integrin levels and increased connexin levels were apparent in the mRNA-profiling data as well.
FIG. 6.
Protein expression profiling of primary human hepatocytes and HepG2 cells. Total cellular protein was isolated from primary human hepatocyte cultures and cultures of HepG2 cells, and equal quantities of protein from the respective samples were applied to 10% denaturing gels, separated by SDS-PAGE and then transferred to PVDF membranes and assessed by immunoblotting analyses, as described in “Materials and Methods” section.
Protein expression for donor E also reflected the trends observed in both the microarray data and in the phenotypic assessments, as this individual exhibited only minimal response to ECM. No apparent changes in keratin 8 or 18 phosphorylation status, or in levels of the respective proteins, were noted. The level of integrin α5 appeared mildly decreased in response to Matrigel at day 2 (42 h) but at no other time point. Connexin 32 expression levels were low at all time points, although detectable at day 6 (138 h) and day 8 (186 h). Matrigel-treated samples were increased over controls.
Protein expression profiles were evaluated in two other individuals, donor B and donor D, but were more varied (data not shown). In both these individuals, little change was apparent, −/+ Matrigel, in keratin 8 or 18 protein levels, although the trend was toward decreased expression levels in the presence of ECM. Integrin alpha 5 appeared reduced in the Matrigel-treated samples, whereas, connexin protein levels were largely increased in the presence of Matrigel.
Selected day 5 protein assessments were also performed in HepG2 human hepatoma cells. Although keratin 8 and 18 protein expression was maintained in these cells, no phosphorylation of keratins 8 and 18 was detected. Similarly, there was a complete lack of connexin 32 protein expression in these cells.
Therefore, protein expression data for primary human hepatocytes cultured in the presence of a Matrigel overlay showed the same general trends as microarray data. Decreases in integrin expression and increases in connexin expression were noted in several samples. Conversely, HepG2 cells demonstrated a lack of connexin expression.
DISCUSSION
Addition of an ECM/Matrigel overlay to cultured primary rat hepatocytes has been reported to enhance cellular differentiation status, xenobiotic responsiveness, and hepatocyte morphology (Ben Ze’ev et al., 1988; Moghe et al., 1996; Schuetz et al., 1988; Sidhu et al., 1993, 2004). However, evidence supporting a facilitating role of ECM in primary human hepatocyte culture is mixed, with certain studies indicating a positive role for ECM in human hepatocyte cultures (Gross-Steinmeyer et al., 2005), while others maintain that plating density and not ECM is the primary effector of hepatocyte differentiation status and phenotypic character (Hamilton et al., 2001). In this study, we used gene expression profiling, protein analysis, and morphological characterization to evaluate the biological impact of ECM additions to culture of primary human hepatocytes and demonstrate that ECM indeed enhances the differentiation status of the human hepatocytes and further facilitates the gene responsiveness of these cells to chemical inducer challenge. Our results establish that primary human hepatocytes, when cultured with Matrigel and in a highly defined culture medium, allow cellular expression character that closely resembles that of human liver tissues in vivo and therefore serve as robust model systems for analysis of liver functional biology, toxicological assessment, and biotransformation function.
With respect to cellular morphology, we show that Matrigel additions often facilitate formation of more pronounced cuboidal cell architecture, together with establishment of more highly organized networks of cells possessing clearly defined cell borders and bile canalicular features (Fig. 1). These phenotypic changes are likely the result of changes in cytoskeletal structure and the ability of the cells to form focal adhesions with the ECM. Interindividual differences in responsiveness to ECM were noted, particularly in donors that yielded cells of compromised quality and viability. In these latter cases, it appeared that ECM additions were not sufficient to “rescue” the otherwise poor quality of starting cellular material. Results of hierarchical clustering of gene expression highlight the variability among donors. However, the shift in clustering from individual to treatment supports the conclusion that ECM additions positively impact gene expression character in hepatocytes cultured from donors yielding cells of relatively higher quality (Fig. 4). Reflections of poorer cell quality include the presence of rafts of apoptotic and/or necrotic cells in both control and Matrigel-treated samples. Even hepatocyte samples evidencing poorer cellular morphology demonstrated increases in differentiation marker gene expression, in the presence of Matrigel, and the overall expression profiles of these Matrigel-treated cultures were reasonably well reflective of the intact human liver samples (Figs. 3A and 3D).
Although the overall expression profiles between control and Matrigel samples within a donor were highly correlated, many specific genes demonstrated altered expression character in response to Matrigel (Fig. 5). In particular, the cytochrome P450 genes represented a major group whose expression profile was upregulated by ECM addition in hepatocytes. Epoxide hydrolase 1, another drug-metabolizing enzyme, was upregulated by the presence of ECM in hepatocyte cultures, as were two nuclear receptors, NR1D2 and NR1I3 (Fretland and Omiecinski, 2000). NR1I3, the constitutive androstane receptor, plays an important role mediating PB induction responseness (Sidhu et al., 2004; Wei et al., 2000). Many other genes were increased in the presence of Matrigel, most of them liver specific, including CEBPalpha, FMO5, solute carrier family members, sulfotransferases, and transferrin receptor 2 (Table 3). The enhanced expression of these gene batteries in the presence of Matrigel establishes the utility of this model system for toxicology and biotransformation investigations and demonstrates that the cellular microenvironment is an important determinant for cultured human hepatocytes and that ECM addition enables cellular responses that more accurately reflect those of an intact human liver.
ECM also contributed to the decreased expression of several genes, including certain keratins, integrins, signaling molecules, and chaperones. Consistent with the idea that ECM additions impact the cellular microenvironment, we hypothesized that overrepresented groups demonstrating altered gene expression in the presence of Matrigel would include genes contributing to the cellular cytoskeleton. Indeed, several gene references in this category exhibited decreased expression levels consequent to Matrigel treatment (Table 7). We suggest that Matrigel-induced reductions in cytosketelal-related gene expression likely manifests from the enhanced set of interactions enabled through focal adhesion contacts with cell membrane proteins that are now stimulated by the ECM microenvironment provided. In support of this contention, morphogenesis-related cell surface markers and membrane-associated transcripts were downregulated in response to Matrigel, as were a cadre of stress-related genes. These latter results are consistent with those obtained in primary rat hepatocyte studies that demonstrated the ability of an ECM microenvironment to effectively “quiet” the stress response (Beck et al., 2000; Sidhu and Omiecinski, 1995; Sidhu et al., 2001, 2004). Our microarray data indicate that several stress-related molecules, such as the interleukins, chemokines, and a number of stress-related protein kinases, exhibit reduced transcript expression in primary human hepatocytes cultured in the presence of Matrigel (Table 5), results supporting the concept that the more in vivo–like microenvironment enables a less perturbed cellular state.
Overall, protein expression profiles were reflective of the patterns seen within the microarray data sets. For example, connexin expression levels were increased in the presence of Matrigel while integrin α5 protein levels were consistently reduced in Matrigel-treated samples (Fig. 6). The mRNA expression levels of several integrin pathway–associated genes were decreased in three of five human hepatocyte samples cultured in the presence of Matrigel including integrins, caveolin, various forms of Ras, Rho, Cdc42, Rac, filamen, actinin, actin, vinculin, and Arp2/3 (data not shown). The number of affected components of the integrin-signaling pathway indicates a selective downregulation of this module in response to ECM. We propose that this pathway is specifically affected by the presence of Matrigel since the integrins constitute the main interacting module with the ECM. The downregulation effects of ECM on integrin levels were coupled to reductions in expression levels of their associated downstream signaling cascades. Since integrin-signaling pathways are directly linked with intracellular stress signaling, via activation of molecules such as Rho, Rac, and Cdc42 (Chen et al., 1994; Clark and Brugge, 1995; Giancotti, 1997; Juliano and Haskill, 1993; Juliano et al., 2004; Miao et al., 2002; Short et al., 1998; Stupack and Cheresh, 2002), the downregulation of integrin signaling is likely the major force behind the suppression of cellular stress cascades evident in the Matrigel-cultured hepatocytes.
Levels of connexin mRNA as well as protein were increased in hepatocytes cultured in the presence of Matrigel. Connexins are gap junction proteins that are responsible for cell-cell communication and play important roles in cell growth and differentiation (Bennett et al., 1991; Loewenstein, 1979). Previous reports demonstrated that only hepatocytes cultured in the presence of Matrigel treatment demonstrated connexin 32 expression (Moghe et al., 1996). In addition, an inverse relationship has been established between cell proliferation and gap junction communication (Kojima et al., 1995). Hepatocytes are normally nonproliferative in vivo and remain so under our primary culture conditions (Fausto, 1991; Gomez-Lechon et al., 1990; Lazaro et al., 2003; Strom et al., 1982).We suggest that hepatocytes cultured in the presence of an ECM likely exhibit increased capacity for cell-cell communication, an enhanced state of differentiation and improved responsiveness to complex external stimuli, such as that manifested in the PB induction response.
The results of this investigation demonstrate a positive role for Matrigel in primary cultures of human hepatocytes, in particular in donor samples of higher initial cell quality. In the hepatocyte samples, Matrigel induced both subtle and dramatic changes in mRNA expression in hepatocyte cultures, changes of clear biological impact. ECM additions tended to improve hepatocyte morphology as reflected by enhanced cuboidal shape, more highly defined cell borders, and formation of highly organized cellular networks observed in the presence of matrix. Matrigel additions also resulted in improved hepatocyte differentiation status, as evidenced by enhanced expression of differentiation markers, decreased expression of dedifferentiation markers, and an improved capacity of the cellular response to PB. The mRNA expression of several cytochrome P450s, epoxide hydrolase 1, FMO5, SLCs, CAR, and PB responsiveness was enhanced in hepatocytes cultured in the presence of ECM, supporting the utility of this culture model for both toxicology and drug metabolism research.
Despite some differences in gene expression character, primary human hepatocytes cultured in defined media conditions and in the presence of Matrigel exhibited strikingly similar profiles to those of the intact human liver. In contrast, human HCC cells, often used in liver-based in vitro model studies, exhibited substantially disparate expression profiles when compared to intact human liver (results to be published elsewhere). Mechanistically, we propose that Matrigel facilitates these biological alterations in cultured primary hepatocytes through modification of cell surface marker protein expression, changes that prime the cell for improved in vivo– like behaviors. Gap junction mRNA and protein expression are augmented in Matrigel-treated samples, likely resulting in improved ability of the cultured cells to engage in appropriate cell-cell communication, facilitated as well by the formation of complex cellular networks. Although expression levels of several cell surface markers were downregulated in the presence of matrix, including the integrins and keratins, as were several stress-related gene pathways, these profiles were more highly reflective of those seen in liver tissues themselves. We postulate that provision of a more native configuration of ECM to the cultured cells results in a microenvironment where the cells no longer need to overcompensate for the lack of proper matrix interactions and thereby exhibit reduced stress responses and reduced compensatory need to synthesize cell surface components.
ACKNOWLEDGMENTS
This study was supported by a Toxicogenomics Research Consortium grant from the National Institutes of Environmental Health Sciences, U19 ES11387, and a grant from the National Institute of General Medical Sciences, GM66411.
REFERENCES
- Beck NB, Sidhu JS, Omiecinski CJ. Baculovirus vectors repress phenobarbital-mediated gene induction and stimulate cytokine expression in primary cultures of rat hepatocytes. Gene Ther. 2000;7:1274–1283. doi: 10.1038/sj.gt.3301246. [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Ben Ze’ev A, Robinson GS, Bucher NL, Farmer SR. Cell-cell and cell-matrix interactions differentially regulate the expression of hepatic and cytoskeletal genes in primary cultures of rat hepatocytes. Proc. Natl. Acad. Sci.U.S.A. 1988;85:2161–2165. doi: 10.1073/pnas.85.7.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MV, Barrio LC, Bargiello TA, Spray DC, Hertzberg E, Saez JC. Gap junctions: New tools, new answers, new questions. Neuron. 1991;6:305–320. doi: 10.1016/0896-6273(91)90241-q. [DOI] [PubMed] [Google Scholar]
- Bissell DM, Caron JM, Babiss LE, Friedman JM. Transcriptional regulation of the albumin gene in cultured rat hepatocytes. Role of basement-membrane matrix. Mol. Biol. Med. 1990;7:187–197. [PubMed] [Google Scholar]
- Caron JM. Induction of albumin gene transcription in hepatocytes by extracellular matrix proteins. Mol. Cell Biol. 1990;10:1239–1243. doi: 10.1128/mcb.10.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Kinch MS, Lin TH, Burridge K, Juliano RL. Integrin-mediated cell adhesion activates mitogen-activated protein kinases. J. Biol. Chem. 1994;269:26602–26605. [PubMed] [Google Scholar]
- Clark EA, Brugge JS. Integrins and signal transduction pathways: The road taken. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- Fausto N. Growth factors in liver development, regeneration and carcinogenesis. Prog. Growth Factor Res. 1991;3:219–234. doi: 10.1016/0955-2235(91)90008-r. [DOI] [PubMed] [Google Scholar]
- Fretland AJ, Omiecinski CJ. Epoxide hydrolases: Biochemistry and molecular biology. Chem. Biol. Interact. 2000;129:41–59. doi: 10.1016/s0009-2797(00)00197-6. [DOI] [PubMed] [Google Scholar]
- Giancotti FG. Integrin signaling: Specificity and control of cell survival and cell cycle progression. Curr. Opin. Cell Biol. 1997;9:691–700. doi: 10.1016/s0955-0674(97)80123-8. [DOI] [PubMed] [Google Scholar]
- Gomez-Lechon MJ, Lopez P, Donato T, Montoya A, Larrauri A, Gimenez P, Trullenque R, Fabra R, Castell JV. Culture of human hepatocytes from small surgical liver biopsies. Biochemical characterization and comparison with in vivo. In Vitro Cell. Dev. Biol. 1990;26:67–74. doi: 10.1007/BF02624157. [DOI] [PubMed] [Google Scholar]
- Gross-Steinmeyer K, Stapleton PL, Tracy JH, Bammler TK, Lehman T, Strom SC, Eaton DL. Influence of Matrigel-overlay on constitutive and inducible expression of nine genes encoding drug-metabolizing enzymes in primary human hepatocytes. Xenobiotica. 2005;35:419–438. doi: 10.1080/00498250500137427. [DOI] [PubMed] [Google Scholar]
- Hamilton GA, Jolley SL, Gilbert D, Coon DJ, Barros S, LeCluyse EL. Regulation of cell morphology and cytochrome P450 expression in human hepatocytes by extracellular matrix and cell-cell interactions. Cell Tissue Res. 2001;306:85–99. doi: 10.1007/s004410100429. [DOI] [PubMed] [Google Scholar]
- Hawksworth GM. Advantages and disadvantages of using human cells for pharmacological and toxicological studies. Hum. Exp. Toxicol. 1994;13:568–573. doi: 10.1177/096032719401300811. [DOI] [PubMed] [Google Scholar]
- Juliano RL, Haskill S. Signal transduction from the extracellular matrix. J. Cell Biol. 1993;120:577–585. doi: 10.1083/jcb.120.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano RL, Reddig P, Alahari S, Edin M, Howe A, Aplin A. Integrin regulation of cell signalling and motility. Biochem. Soc. Trans. 2004;32:443–446. doi: 10.1042/BST0320443. [DOI] [PubMed] [Google Scholar]
- Kocarek TA, Schuetz EG, Guzelian PS. Expression of multiple forms of cytochrome P450 mRNAs in primary cultures of rat hepatocytes maintained on matrigel. Mol. Pharmacol. 1993;43:328–334. [PubMed] [Google Scholar]
- Kojima T, Mitaka T, Paul DL, Mori M, Mochizuki Y. Reappearance and long-term maintenance of connexin32 in proliferated adult rat hepatocytes: Use of serum-free L-15 medium supplemented with EGF and DMSO. J. Cell Sci. 1995;108(Pt 4):1347–1357. doi: 10.1242/jcs.108.4.1347. [DOI] [PubMed] [Google Scholar]
- Kono Y, Yang S, Letarte M, Roberts EA. Establishment of a human hepatocyte line derived from primary culture in a collagen gel sandwich culture system. Exp. Cell Res. 1995;221:478–485. doi: 10.1006/excr.1995.1399. [DOI] [PubMed] [Google Scholar]
- Lazaro CA, Croager EJ, Mitchell C, Campbell JS, Yu C, Foraker J, Rhim JA, Yeoh GC, Fausto N. Establishment, characterization, and long-term maintenance of cultures of human fetal hepatocytes. Hepatology. 2003;38:1095–1106. doi: 10.1053/jhep.2003.50448. [DOI] [PubMed] [Google Scholar]
- LeCluyse E, Bullock P, Madan A, Carroll K, Parkinson A. Influence of extracellular matrix overlay and medium formulation on the induction of cytochrome P-450 2B enzymes in primary cultures of rat hepatocytes. Drug Metab. Dispos. 1999;27:909–915. [PubMed] [Google Scholar]
- Lee YJ, Streuli CH. Extracellular matrix selectively modulates the response of mammary epithelial cells to different soluble signaling ligands. J. Biol. Chem. 1999;274:22401–22408. doi: 10.1074/jbc.274.32.22401. [DOI] [PubMed] [Google Scholar]
- Liu JK, DiPersio CM, Zaret KS. Extracellular signals that regulate liver transcription factors during hepatic differentiation in vitro. Mol. Cell Biol. 1991;11:773–784. doi: 10.1128/mcb.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loewenstein WR. Junctional intercellular communication and the control of growth. Biochim. Biophys. Acta. 1979;560:1–65. doi: 10.1016/0304-419x(79)90002-7. [DOI] [PubMed] [Google Scholar]
- Luttringer O, Theil FP, Lave T, Wernli-Kuratli K, Guentert TW, de Saizieu A. Influence of isolation procedure, extracellular matrix and dexamethasone on the regulation of membrane transporters gene expression in rat hepatocytes. Biochem. Pharmacol. 2002;64:1637–1650. doi: 10.1016/s0006-2952(02)01382-5. [DOI] [PubMed] [Google Scholar]
- Miao H, Li S, Hu YL, Yuan S, Zhao Y, Chen BP, Puzon-McLaughlin W, Tarui T, Shyy JY, Takada Y, et al. Differential regulation of Rho GTPases by beta1 and beta3 integrins: The role of an extracellular domain of integrin in intracellular signaling. J. Cell Sci. 2002;115:2199–2206. doi: 10.1242/jcs.115.10.2199. [DOI] [PubMed] [Google Scholar]
- Milliano MT, Luxon BA. Initial signaling of the fibronectin receptor (alpha5beta1 integrin) in hepatic stellate cells is independent of tyrosine phosphorylation. J. Hepatol. 2003;39:32–37. doi: 10.1016/s0168-8278(03)00161-2. [DOI] [PubMed] [Google Scholar]
- Moghe PV, Berthiaume F, Ezzell RM, Toner M, Tompkins RG, Yarmush ML. Culture matrix configuration and composition in the maintenance of hepatocyte polarity and function. Biomaterials. 1996;17:373–385. doi: 10.1016/0142-9612(96)85576-1. [DOI] [PubMed] [Google Scholar]
- Musat AI, Sattler CA, Sattler GL, Pitot HC. Reestablishment of cell polarity of rat hepatocytes in primary culture. Hepatology. 1993;18:198–205. [PubMed] [Google Scholar]
- Omiecinski CJ, Remmel RP, Hosagrahara VP. Concise review of the cytochrome P450s and their roles in toxicology. Toxicol. Sci. 1999;48:151–156. doi: 10.1093/toxsci/48.2.151. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. Distinct patterns of actin organization regulated by the small GTP-binding proteins Rac and Rho. Cold Spring Harb. Symp. Quant. Biol. 1992;57:661–671. doi: 10.1101/sqb.1992.057.01.072. [DOI] [PubMed] [Google Scholar]
- Rojkind M, Ponce-Noyola P. The extracellular matrix of the liver. Coll. Relat. Res. 1982;2:151–175. doi: 10.1016/s0174-173x(82)80031-9. [DOI] [PubMed] [Google Scholar]
- Rottner K, Hall A, Small JV. Interplay between Rac and Rho in the control of substrate contact dynamics. Curr. Biol. 1999;9:640–648. doi: 10.1016/s0960-9822(99)80286-3. [DOI] [PubMed] [Google Scholar]
- Schuetz EG, Li D, Omiecinski CJ, Muller-Eberhard U, Kleinman HK, Elswick B, Guzelian PS. Regulation of gene expression in adult rat hepatocytes cultured on a basement membrane matrix. J. Cell. Physiol. 1988;134:309–323. doi: 10.1002/jcp.1041340302. [DOI] [PubMed] [Google Scholar]
- Short SM, Talbott GA, Juliano RL. Integrin-mediated signaling events in human endothelial cells. Mol. Biol. Cell. 1998;9:1969–1980. doi: 10.1091/mbc.9.8.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu JS, Farin FM, Omiecinski CJ. Influence of extracellular matrix overlay on phenobarbital-mediated induction of CYP2B1, 2B2, and 3A1 genes in primary adult rat hepatocyte culture. Arch. Biochem. Biophys. 1993;301:103–113. doi: 10.1006/abbi.1993.1121. [DOI] [PubMed] [Google Scholar]
- Sidhu JS, Liu F, Boyle SM, Omiecinski CJ. PI3K inhibitors reverse the suppressive actions of insulin on CYP2E1 expression by activating stress-response pathways in primary rat hepatocytes. Mol. Pharmacol. 2001;59:1138–1146. doi: 10.1124/mol.59.5.1138. [DOI] [PubMed] [Google Scholar]
- Sidhu JS, Liu F, andOmiecinski CJ. Phenobarbital responsiveness as a uniquely sensitive indicator of hepatocyte differentiation status: Requirement of dexamethasone and extracellular matrix in establishing the functional integrity of cultured primary rat hepatocytes. Exp. Cell Res. 2004;292:252–264. doi: 10.1016/j.yexcr.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Sidhu JS, Omiecinski CJ. Modulation of xenobiotic-inducible cytochrome P450 gene expression by dexamethasone in primary rat hepatocytes. Pharmacogenetics. 1995;5:24–36. doi: 10.1097/00008571-199502000-00003. [DOI] [PubMed] [Google Scholar]
- Small JV, Kaverina I, Krylyshkina O, Rottner K. Cytoskeleton cross-talk during cell motility. FEBS Lett. 1999;452:96–99. doi: 10.1016/s0014-5793(99)00530-x. [DOI] [PubMed] [Google Scholar]
- Strom SC, Jirtle RL, Jones RS, Novicki DL, Rosenberg MR, Novotny A, Irons G, McLain JR, Michalopoulos G. Isolation, culture, and transplantation of human hepatocytes. J. Natl. Cancer Inst. 1982;68:771–778. [PubMed] [Google Scholar]
- Stupack DG, Cheresh DA. Get a ligand, get a life: Integrins, signaling and cell survival. J. Cell Sci. 2002;115:3729–3738. doi: 10.1242/jcs.00071. [DOI] [PubMed] [Google Scholar]
- Ulrich RG, Bacon JA, Cramer CT, Peng GW, Petrella DK, Stryd RP, Sun EL. Cultured hepatocytes as investigational models for hepatic toxicity: Practical applications in drug discovery and development. Toxicol. Lett. 1995;82–83:107–115. doi: 10.1016/0378-4274(95)03547-8. [DOI] [PubMed] [Google Scholar]
- Waring JF, Ciurlionis R, Jolly RA, Heindel M, Gagne G, Fagerland JA, Ulrich RG. Isolated human hepatocytes in culture display markedly different gene expression patterns depending on attachment status. Toxicol. In Vitro. 2003;17:693–701. doi: 10.1016/s0887-2333(03)00102-4. [DOI] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]