Abstract
Signal transducer and activator of transcription 3 (STAT3) is aberrantly activated in human cancer including lung cancer and has been implicated in transformation, tumorigenicity, and metastasis. One putative downstream gene regulated by Stat3 is MUC1 which also has important roles in tumorigenesis. We determined if Stat3 regulates MUC1 in lung cancer cell lines and what function MUC1 plays in lung cancer cell biology. We examined MUC1 expression in non-small cell lung cancer (NSCLC) cell lines and found high levels of MUC1 protein expression associated with higher levels of tyrosine phosphorylated STAT3. STAT3 knock-down downregulated MUC1 expression whereas constitutive STAT3 expression increased MUC1 expression at mRNA and protein levels. MUC1 knockdown induced cellular apoptosis concomitant with reduced Bcl-XL and sensitized cells to cisplatin treatment. MUC1 knockdown inhibited tumor growth and metastasis in an orthotopic mouse model of lung cancer by activating apoptosis and inhibiting cell proliferation in vivo. These results demonstrate that constitutively activated STAT3 regulates expression of MUC1, which mediates lung cancer cell survival and metastasis in vitro and in vivo. MUC1 appears to be a cooperating oncoprotein with multiple oncogenic tyrosine kinase pathways and could be an effective target for the treatment of lung cancer.
Keywords: MUC1, signal transducer and activator of transcription 3, small interfering RNA, apoptosis, metastasis, lung cancer
Introduction
Signal transducer and activator of transcription (STAT) proteins are persistently activated in a wide variety of human malignancies and can be required for survival of carcinoma cells (1, 2). STAT3 has been found to be a critical mediator of the oncogenic effects in non-small cell lung cancer (NSCLC) (2–4). It is activated by diverse receptor and non-receptor tyrosine kinases and plays a crucial role in oncogenesis by regulating genes important in cellular proliferation, survival, angiogenesis, and invasion/metastasis (5). Critical down-stream genes important for the oncogenic effect of STAT3 include cyclin D, Myc, Bcl-xL, Mcl-1 and VEGF. Other downstream genes have been reported, but their significance in promoting tumor growth remains unclear in lung cancer (6).
One potential gene downstream of STAT3 important in oncogenesis is MUC1. The promoter of the MUC1 gene contains a STAT-responsive element, and STAT3 constitutively binds to promoters of MUC genes (7, 8). Inhibition of STAT3 expression reduced expression of MUC genes and inhibited cellular motility in breast cancer cells (8). MUC1 is a mucin-like glycoprotein shown to be mainly expressed on the apical border of normal secretory epithelial cells (9). There is convincing evidence that the aberrantly overexpressed MUC1 is associated with transformation, tumorigenicity, invasion, and metastasis of carcinomas and thus could be an important downstream target of STAT3 (10–14). MUC1 is synthesized as a single polypeptide which is cleaved in the endoplasmic reticulum into N-terminal (MUC1-N, >200 kDa) and C-terminal (MUC1-C, ~25 kDa) subunits. Previous studies have demonstrated that the MUC1 cytoplasmic tail interacts with a variety of signals involved in cell growth, adhesion, and survival including multiple HER family members, c-Src, protein kinase C, and Grb2/SOS (10, 15–20). MUC1 activates the anti-apoptotic phosphoinositide 3-kinase (PI3K)/Akt and Bcl-xL pathways. MUC1-C also localizes to the mitochondria, attenuating the apoptotic response to DNA damage and conferring resistance to genotoxic anticancer agents (21–23). In addition to these laboratory-based studies, clinical studies have reported that MUC1 expression in lung cancer is associated with nodal metastasis and advanced stage of disease (24).
Based on these studies, we hypothesized that STAT3 might be one of the important regulators of MUC1 transcription and contributes to MUC1 overexpression in these cells. In the present study, we demonstrated that MUC1 is aberrantly overexpressed in a wide variety of NSCLC cell lines, is associated with higher levels of activated STAT3, and is regulated by STAT3 function. The major function of MUC1 is to drive cell survival and invasion/migration of NSCLC cells both in vitro and in vivo. We validated MUC1 as an important regulator of lung cancer cell survival.
Materials and methods
Antibodies
MUC1 Ab-5, a hamster monoclonal antibody which recognizes the MUC1-C of both human and mouse MUC1, and anti-Ki-67 (clone SP6) were purchased from Lab Vision (Fremont, CA) (10, 25). Anti-total STAT3, anti-Bcl-2, anti-Bcl-XL, and goat anti-mouse IgG horseradish peroxidase (HRP)-conjugated secondary antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-phosphorylated STAT3 (Tyr705), anti-total Src, anti-phosphorylated Src (Tyr416), anti-total focal adhesion kinase (FAK), anti-total Akt, anti-phosphorylated Akt (Ser473), and anti-cleaved PARP (C-PARP) were purchased from Cell Signaling Technology (Danvers, MA). Anti-phosphorylated FAK (Tyr397 and Tyr861) was purchased from BioSource (Camarillo, CA). Anti-cytokeratin (clones AE1/AE3) was purchased from Dako (Carpinteria, CA). Alexa Fluor 488 F(ab')2 fragment of goat anti-mouse IgG and Alexa Fluor 633 F(ab')2 fragment of goat anti-rabbit IgG were purchased from Invitrogen (Eugene, OR). Texas red dye-conjugated AffiniPure goat anti-Armenian hamster IgG was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Anti-β-actin and donkey anti-rabbit IgG HRP-conjugated secondary antibody were purchased from Sigma (St. Louis, MO) and Amersham Biosciences (Piscataway, NJ), respectively.
Cell lines and culture
Human NSCLC cells were maintained in RMPI-1640 supplemented with 10% heat-inactivated (56°C, 30 min) newborn calf serum (Invitrogen, Grand Island, NY). HCC827 and H4006 cells were provided by Dr Jon Kurie (University of Texas M.D. Anderson Cancer Center, Houston, TX). H226, H820, and H2279 cells were provided by Dr John D. Minna (University of Texas Southwestern Medical Center, Dallas, TX). PC-9 cells were provided by Dr Matthew Lazzara (Massachusetts Institute of Technology, Boston, MA). The other cell lines were purchased from American Type Culture Collection (Manassas, VA). Monolayer cultures were incubated at 37°C in a 95% humidified atmosphere containing 5% CO2. For generation of the stable luciferase H358 cell line (Luc-H358), H358 cells were transfected with pCMVneo-luciferase plasmid at ~60–70% confluent density using Fugene 6 transfection reagent (Roche, Indianapolis, IN) according to the manufacturer's instructions. Luc-H358 cells were selected with 800 µg/ml G418 sulfate (Invitrogen), beginning 48 h post-transfection, and neomycin-resistant clones were isolated with cloning cylinders by trypsin-EDTA. Stable clones with high-intensity luciferin bioluminescence used for in vivo study were determined with the Xenogen 200 Imaging System 30 min after addition of the 1X D-luciferin solution (Xenogen, Alameda, CA).
Real-time RT-PCR analysis
Cells were harvested at 80% confluent density, and RNA was isolated using Trizol reagent (Invitrogen). cDNA was synthesized from 500 ng of RNA using Protoscript MLV reverse transcriptase (New England Biolabs, Beverly, MA). Expression of GAPDH (internal control) and MUC1 mRNAs was assessed using Taqman FAM-labeled probes (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. A pooled cDNA sample was used in construction of standard curves and calculation of relative expression.
Transfections
A549 cells were transiently transfected with plasmids of pIRES-puro2-vector or pIRES-puro2-MUC1 at ~60–70% confluent density using Fugene 6 transfection reagent as described above. The MUC1 plasmid (17) was kindly provided by Dr Donald Kufe (Dana-Farber Cancer Center, Boston, MA). At 48 h after transfection, cells were harvested and subjected to protein analysis. For adenoviral infections, A549 cells were infected with either a control adenovirus expressing green fluorescent protein (Ad-GFP) or a STAT3 mutant (Ad-STAT3C) as previously described (6). After 13 h of infection, cells were costimulated with IL-6 (20 ng/ml) for an additional 5 h before being collected for RNA analysis. STAT3 antisense and mismatch oligo-nucleotides were kindly provided by ISIS Pharmaceuticals and have been previously described (2).
Small interfering RNA treatment
Cells were reverse transfected with 20 nM small interfering RNA (siRNA) using lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions. The MUC1 siRNA (On-Target plus SMART pool) and the nonspecific control siRNA were supplied by Dharmacon. After a 72-h incubation or at indicated time points, cells were harvested or trypsinized and replated for subsequent experiments.
Cell proliferation assay
Cells were plated in 96-well plates, and siRNA transfection was performed for 120 h as described above. Cellular proliferation was measured by [3-(4,5)-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) analysis (26). Briefly, after cells were washed with PBS, they were incubated in MTT solution for 4 h and then supplemented with 100 µl of dissolving solution (10% SDS in 0.01 M HCl). The absorbance (optical density units) was measured with a microplate spectrophotometer (Bio-Rad Laboratories, Hercules, CA) with Microplate Manager 5.1 software at wavelengths of 590 and 660 nm. Each assay was performed in quadruplicate. Cell viability of siRNA-transfected cells after a 48-h exposure to the chemotherapeutic agent cisplatin (CDDP) was also evaluated by MTT assay.
Flow cytometry analysis
After a 72-h treatment with siRNAs, cells were subjected to flow cytometry analyses of apoptosis and cell cycle alterations. Apoptosis was assayed using Pharmingen (San Diego, CA) PE-conjugated monoclonal active caspase-3 antibody apoptosis kit without modification. We determined the percentage of cells in G1S, and G2/M by propidium iodide staining as described previously (2). A total of 10,000 cells per experimental condition was used for processing and analysis of fluorescence (FACScan; Becton-Dickinson, Franklin Lakes, NJ) using CellQuest software.
Colony formation assay
After a 72-h treatment with siRNAs, cells were collected and mixed (1000/ml) with RPMI-1640 culture medium containing 10% fetal bovine serum (FBS; Invitrogen) and added to 12-well plates (1 ml/well). After cells were plated for 14 days, we counted the number of colonies per well using a Nikon inverted microscope; each assay was performed in triplicate.
Migration and invasion assays
To measure cell migration, we used an 8-µm pore size Boyden chamber (Corning, NY). Equal amounts of control siRNA- and MUC1 siRNA-transfected cells (100,000) in medium plus 0.1% bovine serum albumin were plated on the top chamber, whereas medium with 10% FBS plus 0.1% bovine serum albumin was added to the lower chamber; chambers were plated in duplicate and placed back into the incubator. For the cell invasion assay, we used invasion chambers, which are identical to migration chambers but precoated with 10 µg of growth factor-reduced Matrigel (BD Biosciences, Bedford, MA), as described previously (27). After ~22–24 h, cells remaining on the upper surface of the chamber were removed with wet cotton swabs. Cells on the lower surface of the inserts were fixed and stained with the Hema3 stain set (Fisher Scientific, Middletown, VA). Five fields of migrated or invaded cells were counted under a microscope and averaged.
Western blot analysis
Cells were washed twice with cold PBS and lysed with buffer A [10 mM Tris-HCl (pH 7.4), 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 10 µg/ml leupeptin, and 5 µg/ ml aprotinin] as described previously (26). After incubation for 30 min on ice, the suspensions were centrifuged (15,000 × g for 30 min). The supernatants were removed and stored at -80°C until gel electrophoresis analyses. The protein concentration was determined by the Bio-Rad DC protein estimation assay according to the manufacturer's instructions. For Western blot analysis, ~60–100 µg of whole cell proteins were separated using 10 or 12% SDS-PAGE and transferred to nitrocellulose membranes. After membranes were blocked with 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.1% Tween-20 containing 5% nonfat dry milk at room temperature for 60 min, the membranes were incubated with indicated antibodies at 4°C overnight and then with the HRP-conjugated secondary anti-rabbit or anti-mouse antibodies at room temperature for 60 min. Each protein was detected using the Enhanced Chemiluminescence (Amersham Biosciences) system. β-actin was used as an internal control.
Orthotopic lung tumor model
Female athymic CD-1 nude mice, 8 weeks old, purchased from Charles River Laboratories, were anesthetized in an acrylic chamber using 2.5% isoflurane- air mixture and placed in the right lateral decubitus position. Cells were transfected for 72 h, and then equal amounts of viable control siRNA- and MUC1 siRNA-transfected Luc-H358 cells (1×106 viable cells determined by trypan blue exclusion) in 50 µl of PBS containing 50 µg of growth factor-reduced Matrigel were injected into the left lateral thorax of the mice at the lateral dorsal axillary line as described previously (28). The mice (10 per group) were housed and maintained in specific pathogen-free conditions. The study was approved by and conducted in accordance with the standards established by our Institutional Animal Care and Use Committee. Ten days after injection, three mice per group were randomized to be sacrificed, after which lung tissue specimens were collected and formalin fixed for hematoxylin and eosin (H&E) staining and immunohistochemical analyses. The remaining mice (7 per group) were euthanized and autopsied when the first mouse became moribund, and the relative tumor weight (total tumor-bearing lung weight), pleural effusion, and mediastinal metastasis were evaluated. Orthotopic tumor growth was also evaluated before sacrifice by an in vivo bioluminescence assay using Xenogen Imaging System 200, for which luciferin (150 mg/kg body weight) was injected intraperitoneally 30 min before imaging.
Histology and immunohistochemistry
Formalin-fixed, paraffin-embedded sections (5 µm) were stained with hematoxylin and eosin (H&E). Immunofluorescence combined with quantitative analysis was used to assess in situ expression of MUC1 and Ki-67 as described previously (19, 25). Antigens were retrieved by incubating the tissue in a microwave oven. Optimal concentrations of antibodies were used to detect target molecules, which were ultimately visualized with the use of fluorochrome-labeled secondary antibodies. DNA fragmentation (apoptosis) was detected with the ApopTag red in situ apoptosis detection kit (Chemicon, Temecula, CA) according to the manufacturer's instructions. We used a fully automated, upright Carl Zeiss Axio-Imager Z.1 microscope with a x20/0.5 numerical aperture objective, and DAPI, FITC, Texas red, and Cy5 filter cubes for immunofluorescence image analysis. Images were produced using an AxioCam MRm CCD camera and Axiovision version 4.6 software suite (Carl Zeiss). Total intensities of the immunofluorescence from absorbance of MUC1- and Ki-67-positive cells or the percentage of cells positive for ApopTag in 10 random fields for each tumor were measured with Image Pro Plus version 6.2 software (Media Cybernetics, Silver Spring, MD).
Statistical analysis
Each experiment was repeated two or three times. Data are presented as means ± SD. Statview 5.0 software was used for statistical analyses. Statistical comparisons between control and experimental groups were performed using the χ2 test (for incidence only) and the Student's t-test. Differences were considered to be statistically significant at P<0.05.
Results
MUC1 is expressed in NSCLC cell lines and is regulated by STAT3
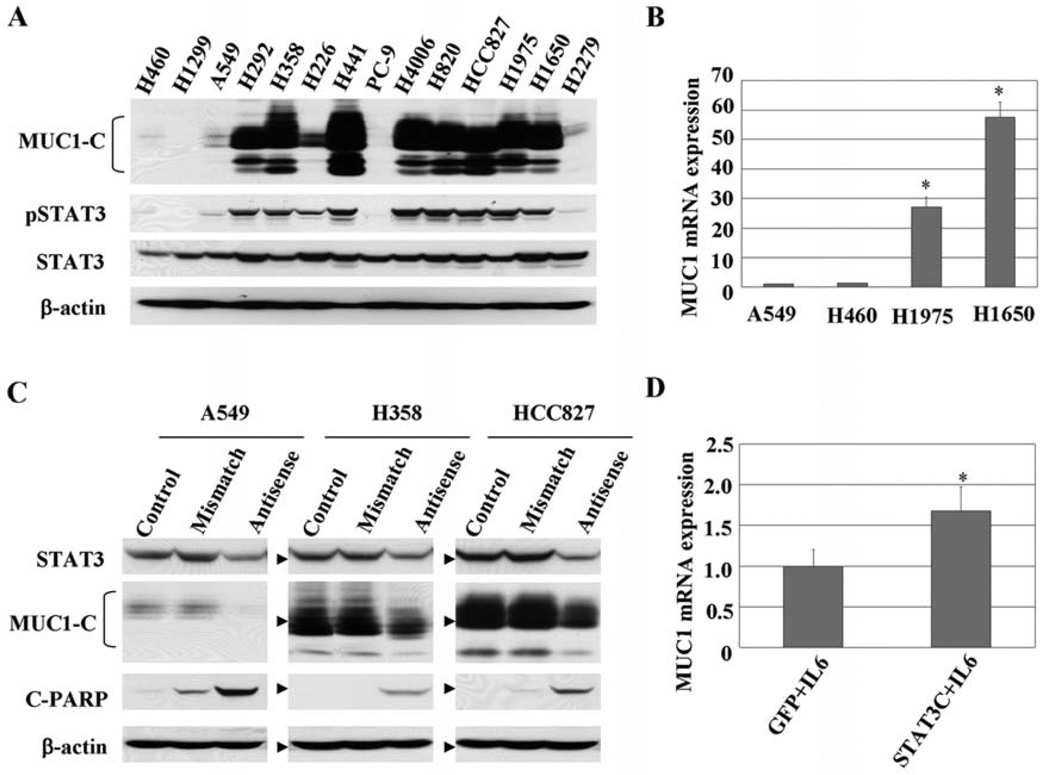
The protein expression levels of MUC1-C and total and tyrosine phosphorylated STAT3 were determined in 14 human NSCLC cell lines (Fig. 1A). We observed high levels of MUC1 protein in 8 cell lines. Consistent with our hypothesis that STAT3 regulates MUC1, cells with high levels of MUC1 generally had enhanced levels of phosphorylated STAT3 (3, 4). Conversely, in H1299 and PC-9 cells, MUC1 was minimally expressed and this corresponded to low levels of phosphorylated STAT3. To determine whether the enhanced levels of protein represent enhanced levels of MUC1 gene transcription, we examined the mRNA levels of MUC1 in a subset of the cells evaluated by real-time quantitative PCR assay. As shown in Fig. 1B, MUC1 mRNA expression levels in both H1975 and H1650 cells with enhanced STAT3 activation were markedly higher than in A549 and H460 cells with lower STAT3 activation. These results demonstrate that enhanced MUC1 gene expression contributes to enhanced levels of MUC1 protein in lung cancer cell lines and suggest that activated STAT3 could be responsible for the expression of MUC1.
Figure 1.
MUC1 is widely overexpressed in NSCLC cells, correlating with STAT3 activation. (A) Protein expression of MUC1-C and total and Tyr705 phosphorylated STAT3 levels were determined in 14 human NSCLC cell lines by Western blot analyses. Equal loading and transfer were shown by repeat probing with β-actin. (B) mRNA levels of MUC1 in a subset of cells were evaluated with real-time RT-PCR. MUC1 mRNA expression levels in each cell line were normalized to GAPDH mRNA, with expression in A549 cells set to an arbitrary value of 1. *P<0.01 compared with A549 or H460 cells. (C) Cells were treated with 700 nM antisense STAT3 oligonucleotides for ~24–48 h, and the effects on MUC1 expression and PARP cleavage were analyzed by Western blot analysis. Equal loading and transfer were shown by repeat probing with β-actin. (D) A549 cells were infected with either control adenovirus (Ad-GFP) or Ad- Stat3C for 13 h along with low levels of IL-6 for an additional 5 h to enhance Stat3C tyrosine phosphorylation. mRNA levels of MUC1 expression were determined by real-time RT-PCR assay and normalized to GAPDH mRNA, with expression in Ad-GFP cells set to an arbitrary value of 1. *P<0.01, **P<0.05 compared with Ad-GFP cells.
To investigate the possibility that activated STAT3 mediates MUC1 expression, STAT3 was knocked down using antisense STAT3 oligonucleotides, and the effect on MUC1 expression was evaluated. As shown in Fig. 1C, STAT3 protein levels of 3 cell lines were reduced by antisense STAT3 oligonucleotide treatment resulting in downregulated levels of MUC1 proteins and cellular apoptosis (increased PARP cleavage). The lack of complete inhibition of MUC1 by antisense STAT3 could be secondary to incomplete STAT3 inhibition or may suggest that other pathways also contribute to MUC1 expression in these lung cancer cells. To provide further evidence linking STAT3 to MUC1, we overexpressed a STAT3 mutant (STAT3C) which constitutively binds DNA and has enhanced transcription with costimulatory tyrosine phosphorylation in lung cancer cells and examined the effects on MUC1 gene expression (29, 30). Cells that received STAT3C had increased mRNA levels of MUC1 expression compared to cells that received no STAT3C (Fig. 1D). Therefore, as shown by two different methods, STAT3 regulates MUC1 expression at both the mRNA and protein levels in lung cancer cells.
MUC1 functions to drive cell survival and colony formation in vitro
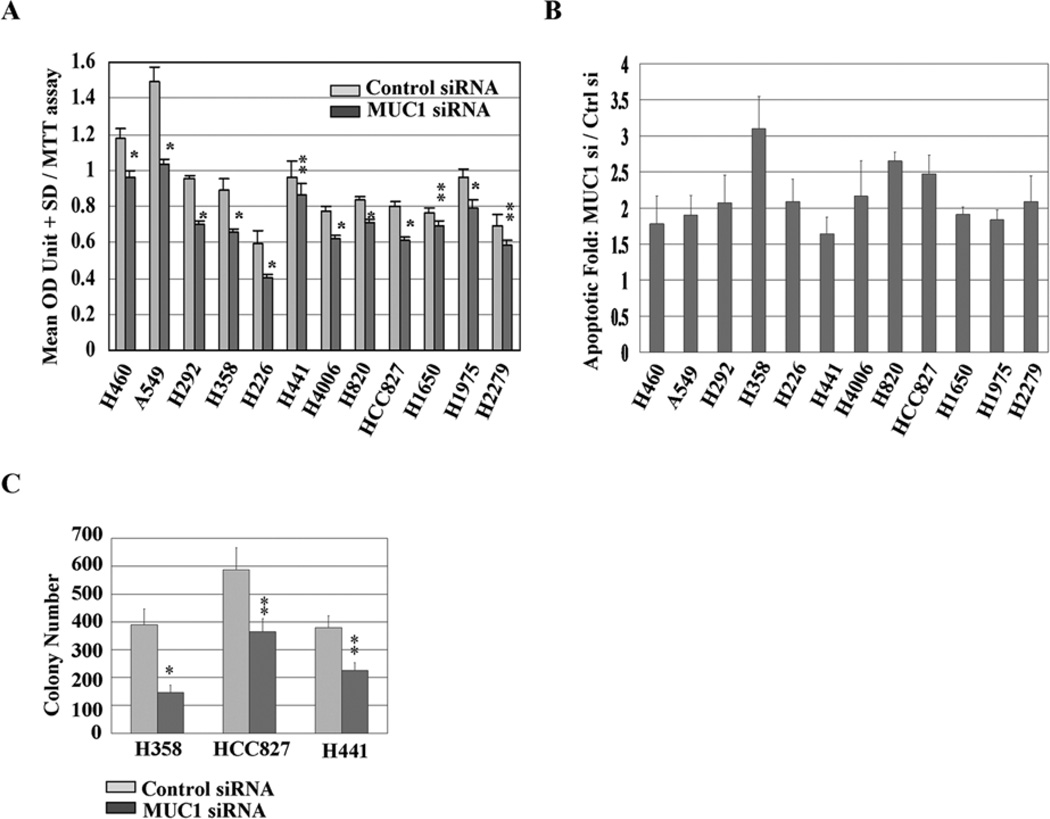
We next exposed NSCLC cell lines to MUC1 siRNA followed by assessment of cellular viabilty. In Fig. 2A, in 12 of 14 cell lines, cellular viability was inhibited by MUC1 siRNA treatment compared to results in the control samples. In these 12 cell lines, MUC1 knockdown by siRNA resulted in modest (~1.6- to 3.1-fold of the control) increases in cellular apoptosis (Fig. 2B). These results indicate that apoptosis contributes to the inhibitory effects of MUC1 siRNA treatment of NSCLC cells. Given the modest effects, we examined the effects of MUC1 knockdown using colony formation assay. MUC1 knockdown cells displayed ~25–63% reduction in colony formation compared to control samples (Fig. 2C). These results suggest that MUC1 may play an important role in the survival of NSCLC cells.
Figure 2.
MUC1 functions to drive cell survival and colony formation. (A) Cells were exposed to siRNA for 120 h, and proliferation rates were measured by MTT assay. *P<0.01, **P<0.05 compared with the control. (B) Cells were exposed to siRNA for 72 h, and cell apoptosis evidenced by positive cells of active caspase-3 in the cells was assessed by flow cytometry analysis. Relative units of apoptosis for the control were set to 1; data for the MUC1 siRNA are shown as fold change relative to control siRNA. (C) Cells treated with siRNA were grown in RPMI-1640 medium supplied with 10% FBS, and colonies were counted after 14 days. *P<0.01, **P<0.05 compared with the control.
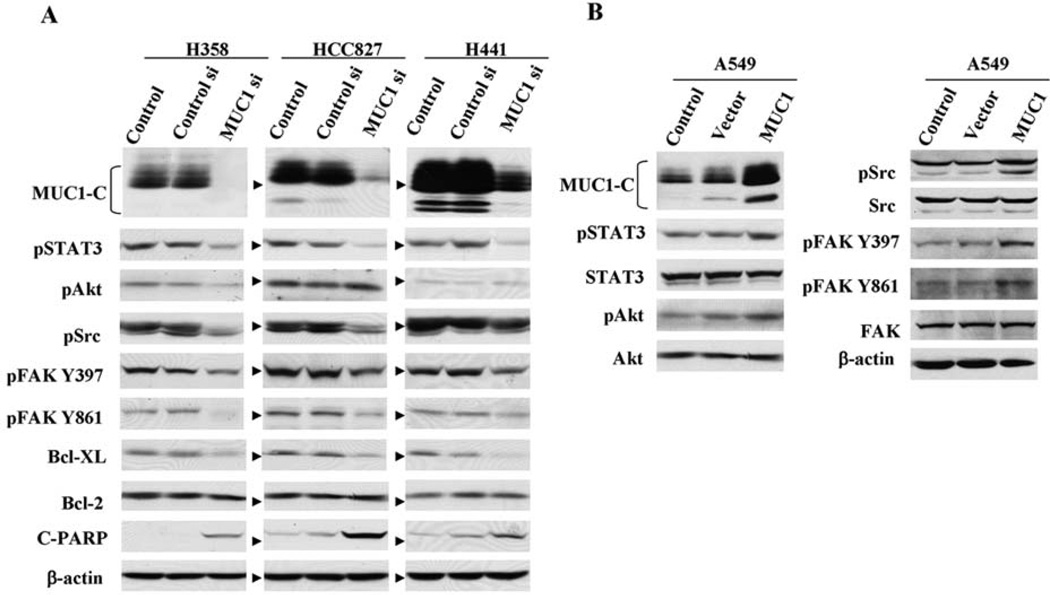
MUC1 knockdown modulates STAT3, SRC, FAK, and Akt signaling pathways in NSCLC cells
To investigate the signaling proteins that are affected by MUC1, we examined the effect of MUC1 knockdown or overexpression on activated STAT3, Akt, Src, and FAK in NSCLC cells. These proteins were chosen for analysis based on earlier studies suggesting they interact with MUC1 or mediate effects of MUC1 signaling (8, 10). H358, HCC827, and H441 cells were exposed to MUC1 siRNA for 72 h, and the effects on phosphorylated signals were detected by Western blot analysis. Exposure of these cell lines to MUC1 siRNA was associated with downregulation of MUC1 protein (Fig. 3A). As shown in Fig. 3A, transfection of cells with MUC1 siRNA resulted in downregulation of phosphorylated tyrosine STAT3, Src, and FAK. The phosphorylated serine Akt, however, was downregulated by MUC1 siRNA only in H358 cells but not in HCC827 and H441 cells, indicating that MUC1 affects various signals in a cell type-dependent manner; whereas, the total proteins of STAT3, Src, FAK, and Akt were not changed by MUC1 siRNA compared to the control (data not shown). The results in HCC827 are consistent with prior studies indicating that mutant EGFR/HER3 signaling is the major regulator of PI3K/Akt in these cells (31). Conversely, in A549 cells transfected with MUC1-C plasmid, levels of the phosphorylated tyrosine proteins STAT3, Src, FAK, and serine phosphorylated Akt were increased (Fig. 3B). These results suggest that in some cell contexts MUC1 regulates activation of STAT3, Src, FAK, and Akt signals in NSCLC cells. We also examined the effect of MUC1 knockdown on anti-apoptotic proteins. Reduction of anti-apoptotic Bcl-XL, but not Bcl-2, was observed with suppressed MUC1 levels along with cleaved PARP in all 3 cell lines (Fig. 3A). These findings indicate that MUC1 knockdown leads to cellular apoptosis via pathways involving Bcl-XL, and MUC1 functions as an oncoprotein to modulate cell survival associated with the Bcl-2 family.
Figure 3.
Effects of MUC1 knockdown or overexpression on multiple cell signals in NSCLC cells. (A) H358, HCC827, and H441 cells were exposed to siRNA for 72 h, and (B) A549 cells were transfected with MUC1-C plasmid for 48 h followed by measurement of phosphorylated tyrosine STAT3, Akt, Src, FAK, and apoptotic-related proteins by Western blot analysis. Equal loading and transfer were shown by repeat probing with β-actin.
MUC1 knockdown sensitizes NSCLC cells to cisplatin treatment
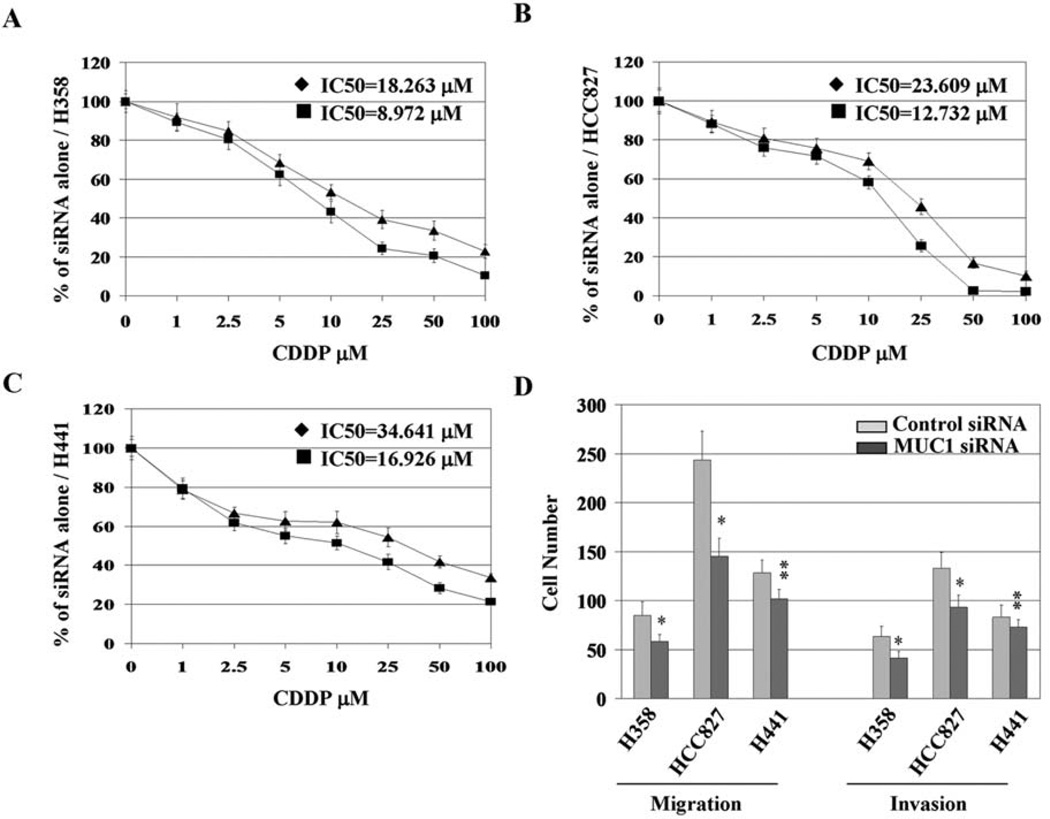
The reduced levels of Bcl-xL in response to MUC1 knockdown suggested that MUC1 knockdown could further sensitize NSCLC cells to chemotherapy-induced apoptosis. To investigate the effects of constitutively over-expressed MUC1 on sensitivity of NSCLC cells to conventional chemotherapeutics, MUC1 siRNA-treated cells were exposed to increasing concentrations of cisplatin, and cell viability was assessed after 48 h and compared with control siRNA-treated cells. As shown in Fig. 4A–C, MUC1 siRNA treatment and exposure to cisplatin resulted in a significantly decreased number of viable cells and a leftward shift of the IC50 of cisplatin compared with cells treated with control siRNA. These results along with previous findings indicate that MUC1 knockdown sensitizes NSCLC cells to chemotherapeutics (22).
Figure 4.
MUC1 knockdown sensitizes NSCLC cells to cisplatin treatment and inhibits metastasis potential. H358 (A), HCC827 (B), and H441 (C) cells were treated with 20 nM siRNA for 72 h, collected and plated in 96-well plates at a density of 10,000 cells/well, and allowed to grow for ~18–24 h before a 48-h exposure to cisplatin in dose gradient. Percentage of cell viability was qualified by MTT assay. IC50 of cisplatin was calculated using Matlab software. ♦, control siRNA + CDDP; ■, MUC1 siRNA + CDDP. (D) MUC1 overexpressing cell lines (H358, HCC827, and H441) were transfected with 20 nM siRNA for 72 h and then assessed for migration and invasion as described in Materials and methods. *P<0.01, **P<0.05 compared with the control.
MUC1 siRNA treatment inhibits the cellular metastasis potential in vitro
We subsequently determined the effects of MUC1 knockdown on the in vitro migration and invasive ability of NSCLC cells. Our data and those of others suggest that Src and its substrate FAK, two important mediators of cell invasion and metastasis, are possible downstream targets of MUC1 (10). As shown in Fig. 4D, cells exposed to siRNA against MUC1 demonstrated statistically significant reductions in both migration and invasion, although MUC1 siRNA treatment resulted in a relative lower inhibitory effect on invasive properties of H441 cells than in the other two cell lines. These results indicate that aberrantly overexpressed MUC1 protein plays a role in the invasion and migration of NSCLC cells in addition to its role in cell survival.
MUC1 knockdown inhibits orthotopic tumor growth and metastasis
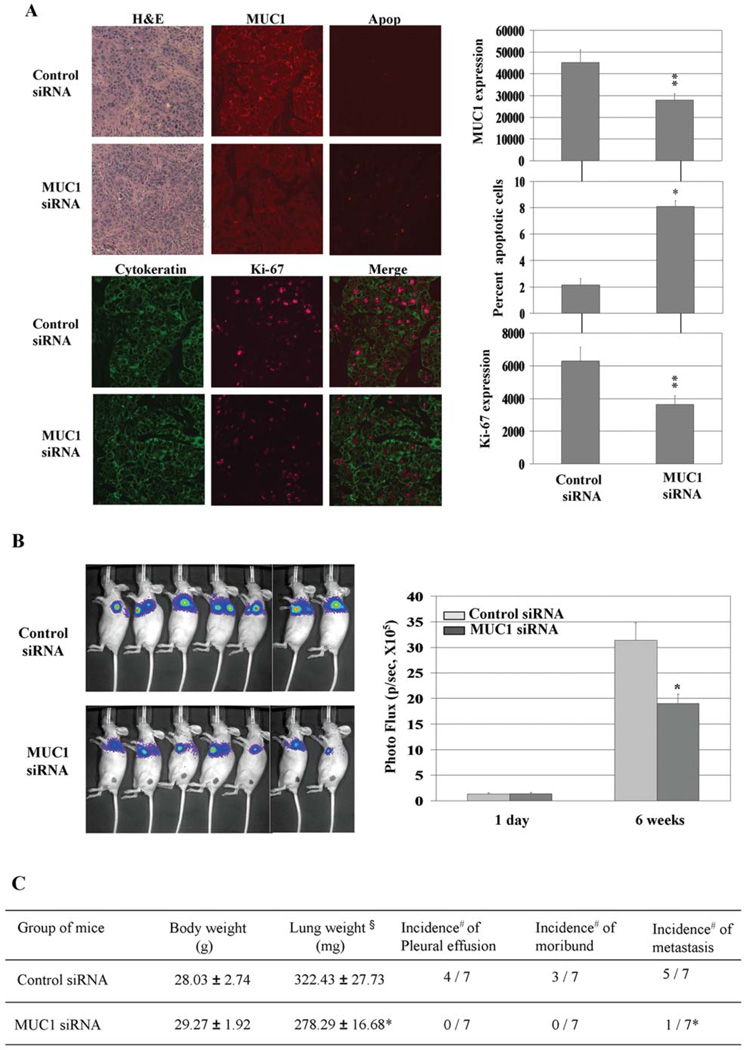
To further investigate the effect of MUC1 on tumor growth and metastasis in mice, H358 cells stably expressing luciferase were exposed to control siRNA or MUC1 siRNA, and equal amounts of viable cells were injected directly into mouse lungs to create orthotopic mouse lung cancer models. These orthotopic models of human lung cancer have been suggested to be more clinically relevant since the tumors originate in the correct microenvironment, can produce pleural and lymph node metastasis which mimics the human syndrome of lung cancer, and recapitulate drug sensitivity better than standard subcutaneous tumor models (28, 32). Within 10 days of injection, microscopic lung tumors were established in both groups and were subjected to immunohistochemical analyses of MUC1, Ki-67 to examine cellular proliferation, and ApopTag to examine for apoptosis 10 days after injection. MUC1 expression was reduced in MUC1- knockdown lung tumors indicating a relative durable effect of the siRNA in this tumor model despite the transient transfection (Fig. 5A). Measurement of the percentage of cells positive for ApopTag showed that MUC1 knockdown resulted in a significant increase in apoptosis compared with the control (Fig. 5A). MUC1 siRNA-transfected cells also expressed less Ki-67 indicative of reduced proliferation of these cells in vivo. Orthotopic tumor growth in the MUC1-knockdown group was significantly delayed compared with the control group, as detected by bioluminescence assay after six weeks of in vivo tumor growth (Fig. 5B). Analysis of lung weight was performed as a surrogate of tumor mass, and in agreement with the bioluminescence data we found significant decreases in lung weight in MUC1 siRNA-treated cells compared to the control. In addition, we found evidence for reduced metastatic potential as the mice with MUC1 siRNA-treated H358 cells had a reduced incidence of mediastinal metastasis and pleural effusions (Fig. 5C). These results support the hypothesis that MUC1 regulates in vivo growth, survival, and metastasis of lung cancer cells.
Figure 5.
MUC1 knockdown induces apoptosis in vivo and inhibits orthotopic tumor growth in CD-1 nude mice. (A) Lung tissue specimens subjected to H&E staining and immunohistochemical analysis of MUC1 and Ki-67 expression and apoptosis (Apop). *P<0.01, **P<0.05 compared with the control. (B) Results of orthotopic tumor growth evaluation using Xenogen 200 imaging system. *P<0.01 compared with the control. (C) Seven mice for each group were sacrificed 6 weeks after tumor injection when control mice became moribund. §, weight designates lung and tumor weight; #, number of positive mice/number of treated mice; *P<0.05 compared with the control siRNA group.
Discussion
In our study, we demonstrated that MUC1 protein and mRNA are constitutively overexpressed in NSCLC cell lines with high levels of activated STAT3. These results further support evidence that overexpression of MUC1 in human carcinoma is caused by increased mRNA expression and that the MUC1 promoter can be stimulated by STAT transcription factors via a STAT-binding site in its promoter (7). In addition, our results strongly suggest that STAT3 plays a role in regulating MUC1 expression at the mRNA and protein levels. Our results add MUC1 as a downstream target of STAT3 which can contribute to the STAT3-dependent phenotype in lung cancer cells (2, 4, 27, 33). These results are consistent with clinical studies in lung cancer showing that MUC1 marks tumors for aggressiveness and suggests that STAT3-MUC1 could be a good molecular pathway target to inhibit tumor growth and metastasis as well as sensitize tumor cells to cytotoxic chemotherapy.
In our studies we found evidence that MUC1 contributed to the activation of STAT3, Akt, Src, and FAK in NSCLC cells although the exact mechanism is unclear. Many of the oncogenic effects of MUC1 stem from its cytoplasmic tail, which binds to several proteins implicated in cancer, including c-Src, EGFR, and downstream signals (10, 16, 17, 19). MUC1 has been shown to affect c-Src signaling in mouse mammary tumor virus-driven polyoma middle T-antigen (PyV MT)-induced mammary tumorigenesis (10). Phosphorylation of MUC1 Tyr46 functions as a binding site for the c-Src SH2 domain via which EGFR and c-Src regulate downstream signals such as β-catenin and GSK3β (17). A large scale analysis of tyrosine phosphorylated proteins in NSCLC did identify peptides corresponding to Tyr46 in cell lines and tumors (34). Thus, the cytoplasmic tail could be important in allowing signaling events to be controlled by MUC1 in lung cancer cells.
It is likely that other signals besides STAT3 cooperate to regulate MUC1 in lung cancer cells. Previous studies in thyroid cells demonstrated that both JAK1/STAT6 and PI3K/Akt pathways differentially affect MUC1 expression at mRNA and protein levels (35). JAK/STAT signaling is mainly thought to modulate MUC1 transcription while PI3K/ Akt signaling is thought to affect MUC1 protein half-life. Overall these results suggest that MUC1, as a downstream target of STAT3, interacts or co-regulates with multiple signals to mediate cell survival and metastasis in NSCLC cells.
Acknowledgements
We thank Dr Donald Kufe (Dana-Farber Cancer Center, Boston, MA) for providing the MUC1 plasmid; Rasa G. Hamilton for the editorial assistance; and Patricia A. Johnston for the administrative assistance. This research was supported, in part, by NIH R01 the Molecular Imaging Core, Mouse Models Core, and the Flow Cytometry Core at the H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL.
References
- 1.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 2.Frank DA. STAT signaling in the pathogenesis and treatment of cancer. Mol Med. 1999;5:432–456. [PMC free article] [PubMed] [Google Scholar]
- 3.Haura EB, Zheng Z, Song L, Cantor A, Bepler G. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clin Cancer Res. 2005;11:8288–8894. doi: 10.1158/1078-0432.CCR-05-0827. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez JV, Greulich H, Sellers WR, Meyerson M, Frank DA. Signal transducer and activator of transcription 3 is required for the oncogenic effects of non-small-cell lung mutations of the epidermal growth factor receptor. Cancer Res. 2006;66:3162–3168. doi: 10.1158/0008-5472.CAN-05-3757. [DOI] [PubMed] [Google Scholar]
- 5.Yu H, Jove R. The STATs of cancer - new molecular targets come of age. Nat Rev. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 6.Dauer DJ, Ferraro B, Song L, et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24:3397–3408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 7.Gaemers IC, Vos HL, Volders HH, van der Valk SW, Hilkens J. A stat-responsive element in the promoter of the episialin/MUC1 gene is involved in its overexpression in carcinoma cells. J Biol Chem. 2001;276:6191–6199. doi: 10.1074/jbc.M009449200. [DOI] [PubMed] [Google Scholar]
- 8.Yuan ZL, Guan YJ, Wang L, Wei W, Kane AB, Chin YE. Central role of the threonine residue within the p+1 loop of receptor tyrosine kinase in STAT3 constitutive phosphorylation in metastatic cancer cells. Mol Cell Biol. 2004;24:9390–9400. doi: 10.1128/MCB.24.21.9390-9400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kufe D, Inghirami G, Abe M, Hayes D, Justi-Wheeler H, Schlom J. Differential reactivity of a novel monoclonal antibody (DF3) with human malignant versus benign breast tumors. Hybridoma. 1984;3:223–232. doi: 10.1089/hyb.1984.3.223. [DOI] [PubMed] [Google Scholar]
- 10.Al Masri A, Gendler SJ. Muc1 affects c-Src signaling in PyV MT-induced mammary tumorigenesis. Oncogene. 2005;24:5799–5808. doi: 10.1038/sj.onc.1208738. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/ MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroeder JA, Masri AA, Adriance MC, et al. MUC1 overexpression results in mammary gland tumorigenesis and prolonged alveolar differentiation. Oncogene. 2004;23:5739–5747. doi: 10.1038/sj.onc.1207713. [DOI] [PubMed] [Google Scholar]
- 13.Spicer AP, Duhig T, Chilton BS, Gendler SJ. Analysis of mammalian MUC1 genes reveals potential functionally important domains. Mamm Genome. 1995;6:885–888. doi: 10.1007/BF00292441. [DOI] [PubMed] [Google Scholar]
- 14.Suwa T, Hinoda Y, Makiguchi Y, et al. Increased invasiveness of MUC1 and cDNA-transfected human gastric cancer MKN74 cells. Int J Cancer. 1998;76:377–382. doi: 10.1002/(sici)1097-0215(19980504)76:3<377::aid-ijc15>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Bharti A, Chen D, Gong J, Kufe D. Interaction of glycogen synthase kinase 3beta with the DF3/MUC1 carcinoma-associated antigen and beta-catenin. Mol Cell Biol. 1998;18:7216–7224. doi: 10.1128/mcb.18.12.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Kuwahara H, Ren J, Wen G, Kufe D. The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1 carcinoma-associated antigen with GSK3 beta and beta-catenin. J Biol Chem. 2001;276:6061–6064. doi: 10.1074/jbc.C000754200. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Ren J, Yu W, et al. The epidermal growth factor receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and beta-catenin. J Biol Chem. 2001;276:35239–35242. doi: 10.1074/jbc.C100359200. [DOI] [PubMed] [Google Scholar]
- 18.Pandey P, Kharbanda S, Kufe D. Association of the DF3/ MUC1 breast cancer antigen with Grb2 and the Sos/Ras exchange protein. Cancer Res. 1995;55:4000–4003. [PubMed] [Google Scholar]
- 19.Schroeder JA, Thompson MC, Gardner MM, Gendler SJ. Transgenic MUC1 interacts with epidermal growth factor receptor and correlates with mitogen-activated protein kinase activation in the mouse mammary gland. J Biol Chem. 2001;276:13057–13064. doi: 10.1074/jbc.M011248200. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto M, Bharti A, Li Y, Kufe D. Interaction of the DF3/MUC1 breast carcinoma-associated antigen and beta-catenin in cell adhesion. J Biol Chem. 1997;272:12492–12494. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- 21.Raina D, Kharbanda S, Kufe D. The MUC1 oncoprotein activates the anti-apoptotic phosphoinositide 3-kinase/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J Biol Chem. 2004;279:20607–20612. doi: 10.1074/jbc.M310538200. [DOI] [PubMed] [Google Scholar]
- 22.Ren J, Agata N, Chen D, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren J, Bharti A, Raina D, Chen W, Ahmad R, Kufe D. MUC1 oncoprotein is targeted to mitochondria by heregulin-induced activation of c-Src and the molecular chaperone HSP90. Oncogene. 2006;25:20–31. doi: 10.1038/sj.onc.1209012. [DOI] [PubMed] [Google Scholar]
- 24.Guddo F, Giatromanolaki A, Patriarca C, et al. Depolarized expression of episialin (EMA, MUC1) in lung adenocarcinoma is associated with tumor progression. Anticancer Res. 1998;18:1915–1920. [PubMed] [Google Scholar]
- 25.Kunnumakkara AB, Guha S, Krishnan S, Diagaradjane P, Gelovani J, Aggarwal BB. Curcumin potentiates antitumor activity of gemcitabine in an orthotopic model of pancreatic cancer through suppression of proliferation, angiogenesis, and inhibition of nuclear factor-kappaB-regulated gene products. Cancer Res. 2007;67:3853–3861. doi: 10.1158/0008-5472.CAN-06-4257. [DOI] [PubMed] [Google Scholar]
- 26.Gao J, Niwa K, Sun W, et al. Non-steroidal anti-inflammatory drugs inhibit cellular proliferation and upregulate cyclo-oxygenase-2 protein expression in endometrial cancer cells. Cancer Sci. 2004;95:901–907. doi: 10.1111/j.1349-7006.2004.tb02200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song L, Morris M, Bagui T, Lee FY, Jove R, Haura EB. Dasatinib (BMS-354825) selectively induces apoptosis in lung cancer cells dependent on epidermal growth factor receptor signaling for survival. Cancer Res. 2006;66:5542–5548. doi: 10.1158/0008-5472.CAN-05-4620. [DOI] [PubMed] [Google Scholar]
- 28.Wu W, O'Reilly MS, Langley RR, et al. Expression of epidermal growth factor (EGF)/transforming growth factor-alpha by human lung cancer cells determines their response to EGF receptor tyrosine kinase inhibition in the lungs of mice. Mol Cancer Ther. 2007;6:2652–2663. doi: 10.1158/1535-7163.MCT-06-0759. [DOI] [PubMed] [Google Scholar]
- 29.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 30.Liddle FJ, Alvarez JV, Poli V, Frank DA. Tyrosine phosphorylation is required for functional activation of disulfidecontaining constitutively active STAT mutants. Biochemistry. 2006;45:5599–5605. doi: 10.1021/bi0525674. [DOI] [PubMed] [Google Scholar]
- 31.Engelman JA, Janne PA, Mermel C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci USA. 2005;2:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onn A, Isobe T, Itasaka S, et al. Development of an orthotopic model to study the biology and therapy of primary human lung cancer in nude mice. Clin Cancer Res. 2003;9:5532–5539. [PubMed] [Google Scholar]
- 33.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 34.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 35.Siragusa M, Zerilli M, Iovino F, et al. MUC1 oncoprotein promotes refractoriness to chemotherapy in thyroid cancer cells. Cancer Res. 2007;67:5522–5530. doi: 10.1158/0008-5472.CAN-06-4197. [DOI] [PubMed] [Google Scholar]