Abstract
Magnetic biosensors, based on nanomaterials and miniature electronics, have emerged as a powerful diagnostic platform. Benefiting from the inherently negligible magnetic background of biological objects, magnetic detection is highly selective even in complex biological media. The sensing thus requires minimal sample purification, and yet achieves high signal-to-background contrast. Moreover, magnetic sensors are also well-suited for miniaturization to match the size of biological targets, which enables sensitive detection of rare cells and small amounts of molecular markers. We herein summarize recent advances in magnetic sensing technologies, with an emphasis on clinical applications in point-of-care settings. Key components of sensors, including magnetic nanomaterials, labeling strategies and magnetometry, are reviewed.
I. INTRODUCTION
Isolating and detecting sparse molecular targets, such as circulating tumor cells, DNAs, microvesicles, and soluble proteins, is of great importance for disease monitoring and diagnostics.1-3 With recent advances of nanomaterials and microfabrication, various new biosensor platforms have been introduced, promising highly sensitive and selective molecular detection.4-7 In these platforms, nanomaterials typically bind to molecular targets and generate distinctive analytical signatures; microfabricated devices could then detect or manipulate labeled targets with spatial and temporal resolution.
With the advent of magnetic nanoparticles (MNPs) which can be effectively rendered molecular-specific, magnetism has become an attractive mechanism for bio-separation and bio-detection.8-12 With recognition ligands conjugated onto their surfaces, MNPs can selectively bind to biological entities of interest, including nucleic acids, proteins, viruses, bacteria, and cells. The binding efficiency is considerably enhanced compared to that of individual recognition ligands, as MNPs provide multiple binding sites (high binding valency).13 Such magnetically labeled targets can be readily distinguished from the remaining sample constituents, because of the intrinsically low magnetic susceptibility of biological objects. This principle has been extensively applied to MNP-based manipulation and separation.14 Advances in magnetic sensing technologies (e.g., NMR, various magnetometers) have further allowed for highly sensitive and yet direct detection of MNP-labeled targets even in native biological samples. Magnetic sensing and sorting is particularly well suited to miniaturization and integration into monolithic chips, for use as an integrated point-of-care diagnostic.12,15-17
This article reviews biological applications of magnetic technology. Considering the breadth of the topic, we will focus on magnetic sensing; for magnetic separation technology, we direct the readers to other comprehensive review articles.18-20. We will first examine the syntheses and properties of MNPs optimized for biosensing, followed by their labeling methodologies. We will then describe different types of magnetic sensors with their representative biological and clinical applications.
II. MAGNETIC NANOPARTICLES FOR BIOSENSING
MNPs are an attractive material for sensing applications because of their unique magnetic properties, facile surface modification, and biocompatibility.21,22 Dextran-coated iron oxide nanoparticles, that were synthesized for clinical uses (i.e., contrast agents for magnetic resonance imagine), were initially adopted as a sensing agent. These particles, however, display low magnetization compared to bulk material, which limits the detection sensitivity. Significant efforts thus have been made to devise new synthetic routes to increase the particle magnetization. One of the representative methods is the thermal decomposition of metal precursors at high temperature in the presence of organic surfactants.23,24 Recent studies further showed enhancement of the particle magnetization through the modulation of particle composition (hybrid structure), size and shape.25-28
Ferrite MNPs
Several different methods have been established to prepare highly crystalline and monodisperse ferrite MNPs. For example, maghemite (γ-Fe2O3) MNPs haven been made from thermal decomposition of iron pentacarbonyl and controlled oxidation.29 Monodisperse magnetite (Fe3O4) particles have been synthesized by thermal decomposition of iron acetylacetonate with 1,2-hexadecanediol used as a reducing agent.30 Iron oxide MNPs can also be synthesized from a simple process using iron oleate as a precursor.24 In this process, iron oleate is prepared by reacting iron chloride with sodium oleate, and uniform MNPs are produced by thermal decomposition of the resulting iron oleate complex. This approach allows large scale synthesis of monodisperse MNPs (up to 40 g scale in a single batch and without a size-sorting process).
The magnetic properties of ferrite particles can be fine-tuned through metal-doping. Among various ferrite types, manganese (Mn)-doped ferrite (MnFe2O4) is known to exhibit high magnetization, due to the high spin quantum number (5/2) of Mn2+ (Fig. 1a).31 It has also been reported that zinc (Zn)-doping to the ferrite host can further increase the saturation magnetization (ms).32 Ferrite [Fe2+(Fe3+)2 (O2−)4] has an inverse spinel crystal structure with 8 tetrahedral (Td) and 16 octahedra (Oh) sites. By replacing Fe3+ with Zn2+ ions in the Td site, the antiparallel spin interactions between the magnetic Fe3+ ions in the Td and the Oh sites are reduced, which results in high magnetization. When 40% of Fe2+ ions are substituted by Zn2+ ions, the resultant Zn-doped ferrite (Zn0.4Fe2.6O4) exhibits the highest ms values. Higher Zn-doping level (> 50%), however, results in a decrease of magnetization, as zinc ferrite (ZnFe2O4) is inherently nonmagnetic. 15 nm Zn-doped ferrite (Zn0.4Fe2.6O4) MNPs exhibited higher magnetization (161 emu/g) than MnFe2O4 MNPs of the same size (125 emu/g), and 15 nm Zn-Mn dual-doped ferrite (Zn0.4Mn0.6Fe2O4) MNPs showed the highest magnetization (175 emu/g).
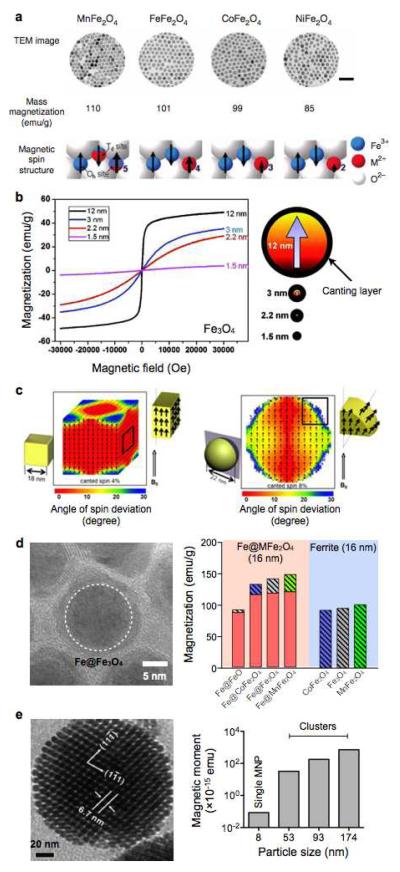
Figure 1. Different types of MNPs developed for magnetic sensing.
(a) Metal-doped ferrite MNPs. The top row shows transmission electron micrograph (TEM) images of 12-nm sized MnFe2O4, Fe3O4, CoFe2O4 and NiFe2O4 MNPs.31 Scale bar, 50 nm. The middle row lists the mass magnetization values of the corresponding MNPs. The bottom row is schematics of spin alignments of magnetic ions in spinel structure. MnFe2O4 exhibits high magnetization due to the high spin quantum number (5/2) of Mn2+. (b) Size-dependent magnetization of Fe3O4 MNPs. As the particle size increases, the relative amount of canted spins decreases, which results in the increase of net magnetization.33 (c) Effect of particle shape on its magnetic property. Cubic MNPs have higher saturation magnetization than spherical particles, since the cubic geometry allows more spins to be aligned in the same direction of applied magnetic fields.28 (d) Fe/Fe3O4 core/shell MNPs. These particle have a hybrid structure, and assume higher magnetization than ferrite particles.40 (e) TEM image of multicore MNPs (left) and magnetization of MNP clusters (right). Clustering MNPs can significantly increase the net magnetic moment of overall particles.41,42 (Reproduced with permission from ref. 31. Copyright 2007 Nature Publishing Group. Reproduced with permission from ref. 33. Copyright 2011 American Chemical Society. Reproduced with permission from ref. 28. Copyright 2012 American Chemical Society. Reproduced with permission from ref. 40, 41 and 42. Copyright 2011, 2007, 2008 John Wiley and Sons, Inc.)
Magnetization of MNPs increases with particle size, since the surface effect (e.g., spin-canting) is reduced in larger particles (Fig. 1b).33 For spherical MNPs, the particle ms can be expressed as ms=Ms•[(r-d)/r]3, where Ms is the saturation magnetization of bulk materials, r is the particle size, and d is the thickness of magnetically disordered (spin-canting) surface layer.9 By fitting the experimental data, Jun et al. reported that d ~ 0.9 nm, which also agreed with theoretical prediction. The spin-canting effect can be reduced by changing the particle shape. For example, cubic ferrite MNPs have been shown to have higher magnetization than spherical ones with the same magnetic volume, as the cubic geometry allows more spins align in the same direction on the particle surface (Fig. 1c).25,28
Fe-core MNPs
The magnetization of ferrite MNPs is eventually bound by ferrite’s bulk magnetization. To overcome this limitation, elemental iron (Fe) has been suggested as an alternative constituent material for MNPs.34,35 Among ferromagnetic crystals, elemental (i.e. non oxidized) Fe assumes the highest ms and relatively low magnetocrystalline anisotropy. Large Fe MNPs can thus possess high magnetic moments while remaining superparamagnetic. Fe MNPs, however, are rapidly oxidized in ambient condition, which necessitates the synthesis of protective shells.35,36 Many different core/shell strategies have been demonstrated, including artificially oxidizing the core27,37 and coating the core with non-magnetic materials.38,39 These methods, however, produce a large fraction of magnetically ‘dead’ volume, leading to smaller magnetic moments than that of similarly-sized ferrite MNPs.
To achieve optimal magnetization of Fe core/shell MNPs, a new synthetic method for Fe/ferrite MNPs was recently developed.40 To preserve the existing elemental Fe-core, the process grew a crystalline ferrite shell around the core. Moreover, the ferrite shell was metal-doped to further improve the particle magnetization.40 The resulting particles showed negligible oxidation of the Fe-core. More importantly, the magnetization values of Fe/MnFe2O4 MNPs were higher (>150%) than those of magnetite-based MNPs (Fig. 1d).
Multicore MNPs
Although magnetic moment can be enhanced by increasing the size of individual particles, this approach often poses technical challenges; large, single-core MNPs tend to be polydisperse and can spontaneously aggregate in suspension. Controlled clustering of superparamagnetic MNPs was thus developed as an alternative approach to increase the net magnetic moment of particles (Fig. 1e).41-43 For example, MNP-loaded polymer micelles have been synthesized by the self-assembly of MNPs and amphiphilic polymers; in this structure, as-synthesized hydrophobic MNPs resided in the hydrophobic core of the amphiphilic polymer micelles. Silica is also a widely used matrix to embed MNP clusters.44,45 While effectively enhancing the overall magnetic moments of particles, these methods also render them high soluble in aqueous solution and amenable for surface chemistry.
III. LABELING STRATEGIES FOR MOLECULAR SENSING
MNP labeling is a critical part of magnetic sensing, which converts molecular information into measurable magnetic signals. For sensitive detection, labeling methods aim to maximally load MNPs onto target molecules. Significant efforts thus have been made to establish 1) optimal coupling chemistries between affinity ligands and MNPs, 2) effective assay configurations for different sensing modalities, and 3) amplification schemes to bind multiple MNPs per target. With such developments, magnetic sensing could be successfully applied to detect a broad range of biological targets, including small molecules, DNAs, viruses, bacteria and mammalian cells.
Conjugation of affinity ligand to MNPs
The targeting specificity of MNPs is conferred by affinity ligands, which include antibodies,46 nucleic acids,47,48 and small molecules.49 Several different chemical strategies have been developed to attach affinity ligands to MNPs.13,50 One promising approach is bioorthogonal chemistry termed BOND (bioorthogonal nanoparticle detection).51 The BOND method uses a rapid, catalyst-free cycloaddition as the coupling mechanism. Antibodies (or other affinity ligands) are modified with trans-cyclooctene and used as scaffolds to couple tetrazine-modified MNPs. This technique is fast, chemoselective, adaptable to other types of nanomaterials, and scalable for biomedical use. Recently, alternative schemes based on complementary oligonucleotide hybridization48 and cyclodextrin/adamantine supramolecular chemistry52 have also been developed. Irrespective of the attachment strategies, functionalized MNPs can be used in different assay configurations, as described below.
Clustering assay
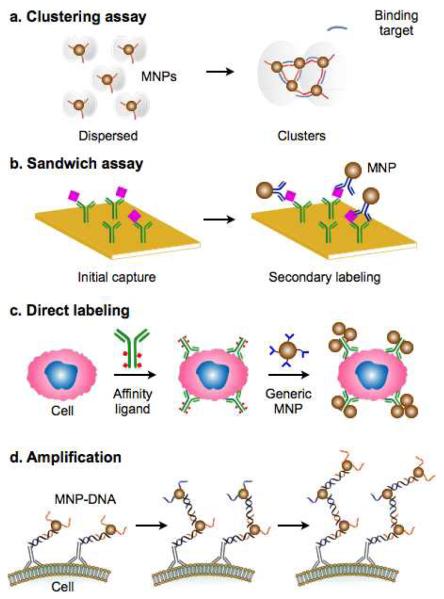
In this assay type, MNPs with multivalent affinity ligands bind simultaneously to a single target molecule, which results in particle aggregation (or disaggregation, if the target can disintegrate preformed MNP clusters; Fig. 2a). Such changes in MNP organization can be detected by either NMR relaxometry53 or Brownian relaxation measurements54. The NMR detection measures changes in the transverse relaxation rate (R2) of water protons, caused by MNP clustering55,56; the Brownian relaxation measurements effectively detect changes of the particles’ hydrodynamic size. The clustering assay is well-suited for rapid analyses. It does not require washing steps to separate bound MNPs from free ones; the binding kinetics can be faster than surface-based detection, as the reaction takes place in the entire sample volume. However, since the extent of MNP clustering depends on the quantitative ratio between MNPs and target molecules, the clustering assay often requires extensive optimization to maximize its detection sensitivity.56 Drawing on its wash-free assay procedure, the clustering assay has been applied to detect small molecules, nucleic acids, and proteins in suspension.53
Figure 2. Molecular targeting and labeling strategies.
(a) Clustering assay. In the presence of binding targets, multivalent MNPs aggregate by crossing-linking.53 Such changes in the organizational state of MNPs can be measured by NMR relaxometry or Brownian relaxation measurements, without the need for additional washing steps. (b) Sandwich labeling. By functionalizing a solid substrate with affinity ligands, small molecular targets can be effectively enriched against a complex biological background. Secondary MNP labeling after the initial capture further brings MNPs close to the sensor surface. (c) Direct labeling. For large biological entities, such as mammalian cells, the whole target can be labelled with affinity ligands and subsequently with MNPs.51 (d) Magnetic amplification. By grafting multiple layers of MNPs onto a target, through the sequential applications of MNPs modified with orthogonal binding partners, magnetic signal can be amplified to detect rare molecular targets.48 (Adopted with permission from ref. 53 and 51. Copyright 2002, 2010 Nature Publishing Group. Reproduced with permission from ref. 48. Copyright 2011 John Wiley and Sons, Inc.)
Sandwich magnetic labeling
To detect molecular targets using surface-based sensors, sandwich magnetic labeling is widely used (Fig. 2b). In this approach, a solid substrate/sensor surface, functionalized with affinity ligands, is used to capture target molecules. This step also effectively increases the local concentration of target molecules and helps to improve the overall assay sensitivity. Captured targets are subsequently coupled with MNPs via secondary affinity ligands. This coupling brings MNPs close to the sensor surface and enables unbound MNPs to be easily washed out. The bound MNPs can be detected using various magnetometers, including magnetoresistance sensors,57,58 Hall elements59, and microcoils.60 The method has been successfully applied to detect a range of biological targets, including proteins, nucleic acids and small molecules.61-64
Direct magnetic labeling
Mammalian cells and bacteria are significantly larger (> several μm) than commonly used MNPs (often < 500 nm). These targets rarely cause clustering of MNPs, and often are not amenable to surface immobilization. Instead, the whole organisms can be directly labeled with MNPs (Fig. 2c)26,27,65. The size difference between target cells and MNPs also allows for easy removal of unbound particles, which significantly increases the overall detection sensitivity. To improve the performance of this targeting assay, and to provide a modular approach for magnetic labeling, the BOND strategy has been adopted for cellular targeting.51 In this method, cells are first targeted with affinity ligands modified with a molecular tag, and subsequently coupled with MNPs. Compared to using ligand-MNP direct conjugates, the two-step labeling generally reduces the required amount of affinity ligands (e.g., antibodies) by more than 10-fold, allows for the use of generic MNPs for a range of different molecular targets, and importantly, improves MNP-loading to target molecules through multiple attachment of MNPs per affinity ligand.66
Magnetic amplification
Following the initial labeling with MNPs, multiple magnetic layers can be formed by sequentially applying MNPs conjugated with orthogonal binding partners.48,67 This scheme amplifies the magnetic signal with each round of labeling, thereby improving the overall detection sensitivity. In one example, Liong et al. employed sequence-specific DNA hybridization to increase the number of MNPs loaded onto cells (Fig. 2d).48 The method was repeated up to 20 rounds of MNP layering, leading to >15-fold enhancement of the signal from the initial targeting.
IV. MAGNETOMETERS FOR SIGNAL DETECTION
Various types of magnetometers can be used to measure the magnetic signal from the labeled biological objects. The detection method could be classified into two groups, namely volumetric and surface-based sensing. In volumetric sensing, the analytical signal is generated from the entire sample volume, which often makes the detection assays simple and fast. The sensing resolution, however, can be limited by the ensemble average nature of signal acquisition. Nuclear magnetic resonance (NMR), magnetic susceptometry, and conventional SQUID (superconducting-quantum-interference-device) magnetometry are representative examples of the volumetric sensing. Surface-based sensors, on the other hand, are complementary to the volumetric ones. By directly detecting individual magnetic objects, surface sensors can achieve higher sensitivity and resolution. A drawback of this sensing mechanism is the requirement to bring or bind target samples to the sensing elements; this could make assays time-consuming and more involved in the device structure. Several solid state magnetometers (e.g., magnetoresistance sensors, Hall effect sensors) have been adopted as a surface-based biosensor.
Miniaturized NMR system
NMR indirectly detects magnetically-labeled objects through the measurement of 1H proton signal. When placed in polarizing NMR magnetic fields, MNPs generate local dipole fields, which efficiently destroy the coherence in the spin-spin relaxation of water protons. Samples containing MNP-labeled objects consequently display higher transverse relaxation rate (R2) than non-targeted ones (Fig. 3a).68 This detection scheme benefits from a built-in signal amplification, since each MNP affects more than millions of surrounding water molecules.69
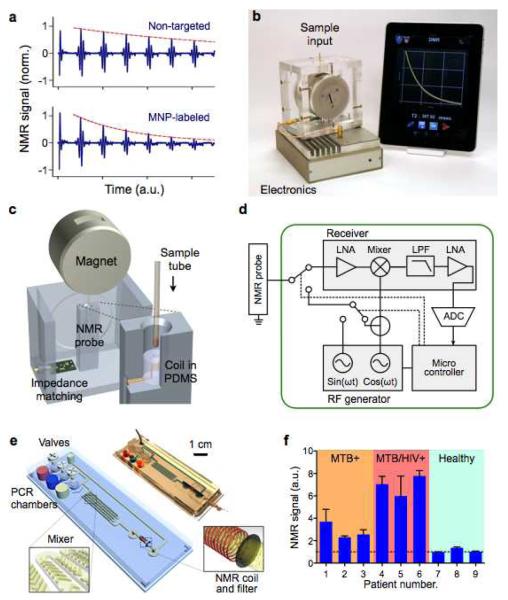
Figure 3. NMR-based magnetic detection.
(a) Assay principle. Samples containing magnetically-labeled biological objects display faster relaxation of proton NMR signal. (b) A new miniaturized NMR (μNMR) system was developed for point-of-care operations. The system features automatic system tuning and user-friendly interface. (c) Schematic of the magnet assembly and the NMR probe. The microcoil is embedded in a polymer (polydimethylsiloxane/PDMS) block with the entire coil-bore accessible, and a thin-walled tube is used for sample-loading.16 (d) The NMR electronics is designed for standalone operation and high programability. (e) A multifunctional fluidic cartridge was developed for bacterial detection.81 The device integrates polymerase-chain-reaction (PCR) chambers, torque-assisted valves, mixing channels and a microcoil. Bacterial samples, PCR reagents, microbeads, and MNPs are loaded onto the chip. After on-chip PCR, magnetic labeling of the microbeads takes place along the mixing channel. The magnetically-labeled beads are then purified and concentrated into the μNMR probe (microcoil) by the membrane filter. (f) The fluidic device in (e) was used to detect Mycobacterium tuberculosis (MTB) in clinical sputum specimens. Compared with samples collected from MTB-positive patients, samples collected from MTB/HIV-positive patients showed higher bacterial burden. Data is represented as mean ± s.d. from triplicate measurements. Reproduced with permission from ref. 16. Copyright 2011 RSC Publishing. Reproduced with permission from ref. 81. Copyright 2013 Nature Publishing Group.)
Significant advances have been made to facilitate NMR-based magnetic detection. These efforts include designing new permanent magnet assembly for NMR field generation,70 integrating NMR transceiver into a custom-designed chip,71 and miniaturizing NMR probes.27,72,73 Such miniaturized NMR (μNMR) systems not only allow for portable operation, but also have higher mass sensitivity. The most recent μNMR system (Fig. 3b) has been further optimized for user-friendly operation in clinical settings. Disposable thin-walled (thickness 25 μm) tubes were used to minimize system contamination as well as to simplify the sample-loading process (Fig. 3c). The designed electronics were programmable for versatile NMR measurements (Fig. 3d). Importantly, it automatically compensated for drifts in the NMR frequency caused by environmental temperature fluctuation, thereby ensuring reliable measurements in clinical settings. The system also interfaced with a mobile device for easy use by medical personnel and for data logging/sharing via a wireless connection. The developed system has been applied in numerous clinical trials, including circulating tumor cell detection74-76, bacterial profiling77-79, and blood-quality monitoring.80
The capacity of the μNMR device can also be improved through integration with microfluidics. For example, Liong et al. demonstrated a μNMR-fluidic cartridge for bacterial detection (Fig. 3e).81 The device performed multifunctions on-chip: target DNA was amplified via polymerase chain reaction (PCR); amplicons were captured onto microbeads and subsequently labeled with MNPs; and the labeled microbeads were concentrated into the NMR probe region. Such integration enabled fast, streamlined assays with the entire reaction completed within 2.5 hours, and also prevented cross-contamination of PCR-amplified products. This platform was used to detect Mycobacterium tuberculosis (MTB) in patient samples (Fig. 3f), and to identify drug-resistant MTB strains.81
Magnetic susceptometry
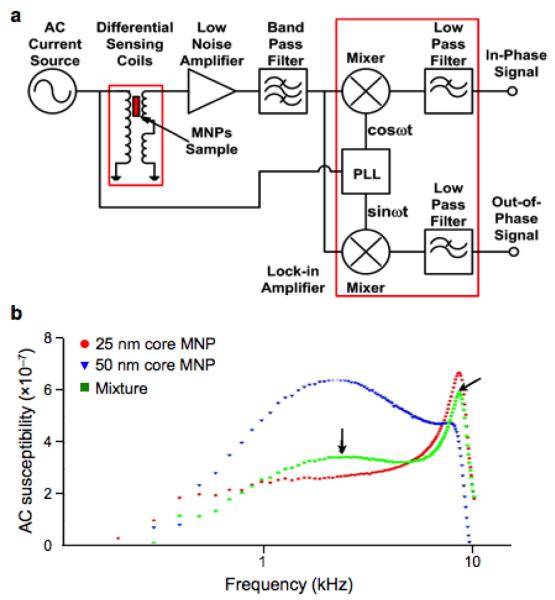
Magnetic susceptometry measures the volumetric responses of MNPs under external magnetic field excitation. Compared to NMR measurement, this approach has a simpler setup, particularly because it does not require a highly uniform magnetic field. Furthermore, direct measurement of the MNPs can permit simultaneous detection of different types of MNPs.82 Susceptometry, however, has a lower sensitivity than NMR-based detection, due to its lack of inherent signal amplification mechanism. A widely used detection scheme is based on Brownian relaxation.82,83 Each MNP in suspension undergoes thermal rotation with a timescale (τB) proportional to the particle’s hydrodynamic volume. Such thermal rotation affects the frequency-dependent magnetic susceptibility (χ) of the MNPs; the quadrant component of χ has its peak at the excitation frequency of 1/τB. Binding of target molecules to MNPs changes the hydrodynamic diameter (and hence τB), and induces a shift in the peak position. Magnetic susceptometry has been used to detect soluble proteins,84 DNA,82 and bacteria,85 with signals measured via induction coils or SQUID. Recently, Park et al. demonstrated a compact susceptometer for Brownian relaxation measurement (Fig. 4a).54 Multiple types of MNPs could be differentiated in a mixture according to their hydrodynamic size (Fig. 4b), wherein the susceptibility spectrum was employed as a unique Brownian relaxation signature. As a model biological application, biotinylated horseradish peroxidase was detected through the clustering assay with streptavidin-coated MNPs.
Figure 4. Multiplexed Brownian detection of differently sized MNPs.
(a) The alternating current (AC) magnetic susceptibility is measured using a quadrature detector. The signals both in-phase and 90 degree out-of-phase with respect to the AC current source are measured, which correspond to the real and imaginary component of the magnetic susceptibility, respectively. PLL, phase-locked loop. (b) The out-of-phase (imaginary) component of the susceptibility has its maximum when the excitation frequency is close to the Brownian relaxation time of the particle. The peak position shifts for differently sized particles, enabling 25 nm core (red) and 50 nm core (blue) MNPs to be measured simultaneously (green). (Reproduced with permission from ref. 54. Copyright 2011 IOP Publishing Ltd.)
Magnetoresistance sensors
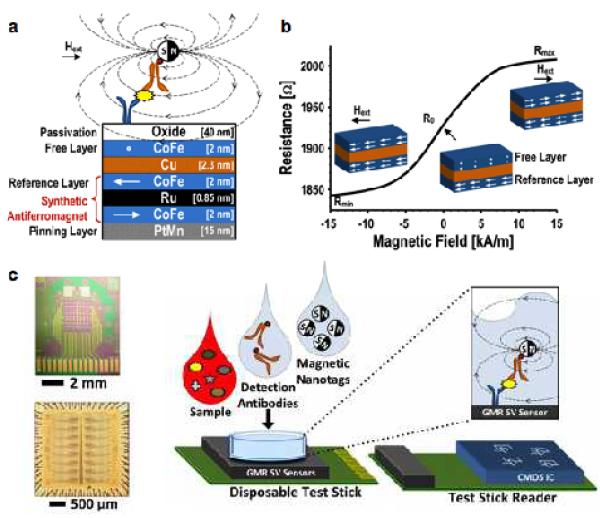
Magnetoresistance (MR) sensors are electrical resistors whose resistance changes under the influence of external magnetic fields. This MR effect is caused by the spin-orbit coupling between conduction electrons and magnetic layers. A representative example is giant magnetoresistance (GMR). The phenomenon takes place in a magnetic structure consisting of ferromagnetic layers separated by non-magnetic metallic layers (Fig. 5a); the relative changes between the layer’s magnetization significantly affects the overall electrical resistance (Fig. 5b). For biosensing applications, GMR sensors operate in conjunction with the magnetic sandwich assay, wherein target molecules are captured onto the ligand-functionalized sensor surface, and subsequently tagged with magnetic probes.
Figure 5. Giant magnetoresistance (GMR) detection of biomarkers.
(a) GMR sensors consist of alternating layers of ferromagnetic and non-magnetic materials. The magnetization of a reference layer is pinned and the magnetization of a free layer is able to change with an applied field. The presence of MNP in close proximity to the sensor creates a local field which changes the magnetization of the free layer. Hext, external magnetic field. (b) As the magnetization of the free layer changes under varying external magnetic fields, the overall electrical resistance of a GMR sensor changes as well. (c) An array of 256 GMR sensors (top) and its interface chip (bottom). The GMR sensor is mounted on a disposable test stick; the interface chip is on the reader stick. This approach has been applied to detect soluble proteins in clinical samples. A sandwich assay is used to bind MNPs close to the GMR sensor surface. CMOS, complementary metal-oxide-semiconductor. (Reproduced with permission from ref. 90. Copyright 2013 IEEE.)
One of the first GMR-based sensors used micrometer-scale magnetic beads for labeling.86 The large size difference between the magnetic probes and the detection targets, however, limited the assay speed and sensitivity.62 By employing MNPs, Osterfeld et al. overcame such challenges and demonstrated sensitive and real-time protein sensing.62 The system was further improved to detect sparse molecular targets in complex biological samples.64 Multiplexed protein detection was also demonstrated with an 8 × 8 sensor array, wherein each GMR sensor was functionalized with different antibodies. These sensors showed a linear dynamic range of over six orders of magnitude, with a limit of detection of ~ 50 aM.64 Beyond protein detection, the MR-based sensors have been further applied to detect specific DNA sequences87 as well as magnetically labeled cells.88,89
More recently, a hybrid GMR system was implemented by combining two separate chips, namely a 256 GMR array and a custom-designed integrate circuit (IC) chip for signal processing (Fig. 5c).90 With such a large array of sensors, this system could function as a protein microarray with the capability to monitor real-time kinetics. The performance of the chip was characterized by detecting an ovarian cancer biomarker, with signal detected at concentrations down to 10 fM.
Miniaturized Hall sensors
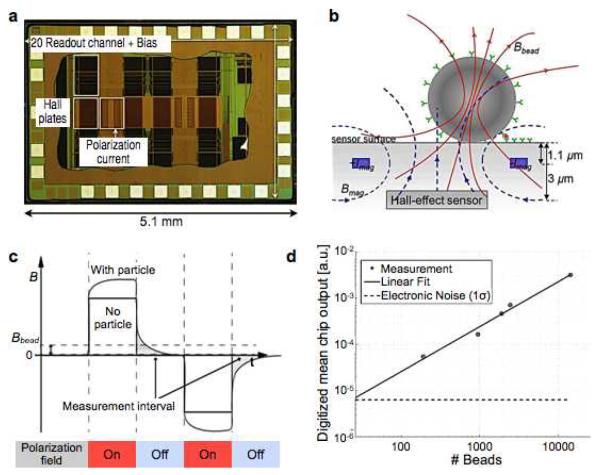
The classical Hall effect refers to the generation of a voltage difference in an electrical conductor subject to a magnetic field. The phenomenon arises as moving charge carriers, deflected by Lorenz force, accumulate on one side of the conductor. While Hall sensors generally have a lower field sensitivity than MR-based sensors, they can assume signal linearity even at high magnetic fields (> 2 T). This attribute makes it possible to use large magnetic fields that fully magnetize MNPs. Furthermore, the sensor fabrication is compatible with the IC process, enabling monolithic integration of large sensor arrays with control electronics. Micrometer-scale Hall (μHall) sensors have been used to detect magnetically-labeled molecular targets.61,91-93. Recently, Gambini et al. significantly advanced the platform by implementing large arrays of μHall sensors (64 × 160 Hall elements) using complementary metal-oxide-semiconductor (CMOS) technology (Fig. 6a). The chip also integrated parallel readout circuitry for high throughput detection, and local micro-electromagnets for controlled polarization of magnetic beads (Fig. 6b). In its operation, the system measured the decaying remnant field of magnetic beads immediately after the magnetization field was turned off (Fig. 6c).94 This approach enabled the weak field that arises from the magnetized bead to be separated from much stronger applied field (102 to 105 times larger), thereby improving the detection sensitivity. In a titration measurement with magnetic beads (Dynabead M280, Invitrogen), the sensor could determine the bead concentration with an uncertainty (1σ) of 25 particles per array, or equivalently a 0.1% coverage of the sensing area (0.64 mm2) (Fig. 6d).95
Figure 6. Integrated Hall sensor for magnetic bead detection.
(a) Die photograph of an integrated Hall sensor integrate circuit (IC). The chip contains 10240 Hall-effect sensors, evaluation electronics, and electromagnets for polarizing field generation. (b) Cross section of a single Hall sensing element. A pair of metal wires on both sides of the Hall-effect sensor are used to generate the polarizing field to magnetize the bead. (c) Magnetic beads are detected via relaxation measurement. The polarization magnetic field is applied to magnetic beads. Subsequently, the field is turned off, and the remnant decaying magnetic field from the bead is measured. The measurement is free from the large offset coming from the polarizing field. (d) Magnetic beads (2.8 μm) were detected in the entire sensing area (0.64 mm2). The sensitivity was down to 0.1% coverage of the sensing area. (Reproduced with permission from ref. 94 and 95. Copyright 2012, 2013 IEEE.)
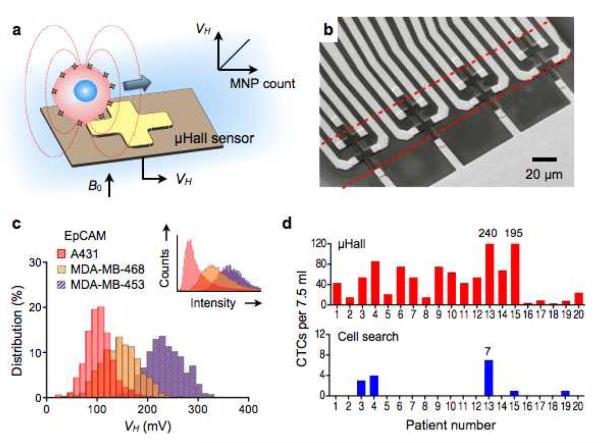
Introducing a new application of the μHall technology, Issadore et al. used the sensor to detect magnetically labeled cells in flow (magnetocytometry).96 Target cells were labeled with MNPs, and the resulting magnetic fields from these cells were measured by a μHall element that reported an electrical voltage (VH) proportional to the MNP counts per cell (Fig. 7a). Because the sensor measures transient signal from moving cells, it could operate in an alternate-current (AC)-coupling mode, unaffected by the static magnetic field (B0) used to magnetize MNPs. The sensor chip had an eight 8 × 8 μm2 μHall elements arranged into an overlapping 2 × 4 array (Fig. 7b), which ensured that individual cells pass directly over at least two μHall elements. Hydrodynamic focusing was also used to stream cells onto the sensing area.96 When compared with conventional flow cytometry, the μHall platform showed good agreement in molecularly screening cells (Fig. 7c). Importantly, the sensor was able to detect individual cells even in the presence of vast numbers of blood cells and unbound reactants. This capability made the platform well-suited for rare cell detection in complex biological media. For example, in a clinical trial with cancer patient samples, the μHall displayed much higher sensitivity in detecting circulating tumor cells than the clinical standard method (Fig. 7d).
Figure 7. MicroHall (μHall) sensor for single cell detection.
(a) Each cell, targeted with MNPs, generates magnetic fields that are detected by the μHall sensor. The Hall voltage (VH) is proportional to the MNP counts. B0, external magnetic field. (b) The sensing area has a 2 × 4 array of μHall elements. The dotted lines indicate the location of fluidic channel. The sensors are arranged into an overlapping array across the fluidic channel width. (c) The μHall system accurately measured the expression levels of epithelial cell adhesion molecule (EpCAM) in different cell lines; the inset shows the same measurements by flow cytometry. (d) Clinical applications of the μHall system. Circulating tumor cells (CTCs) in patient blood samples (n = 20) were detected using either the μHall system (top) or the clinical gold-standard system, CellSearch (bottom). The μHall enumerated a higher number of CTCs across all patient samples. (Reproduced with permission from ref. 96. Copyright 2012 American Association for the Advancement of Science AAAS.)
Diamond magnetometer
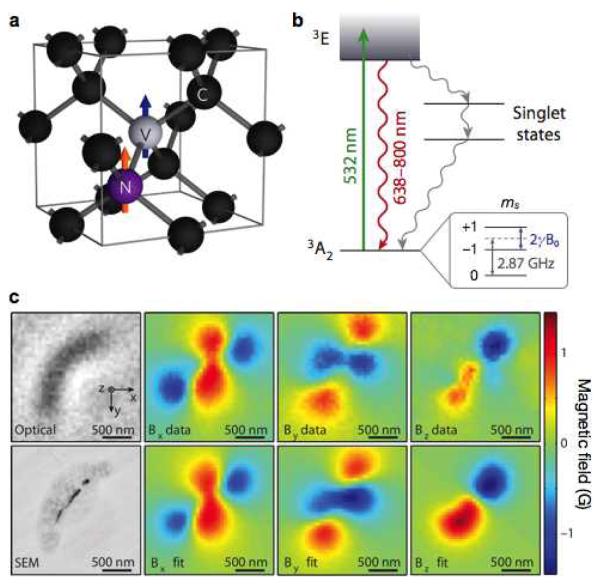
Nitrogen-vacancy (NV) center diamonds have recently emerged as a promising new technology for highly sensitive magnetic detection.97-100 The NV center is formed when a nitrogen atom replaces a carbon atom in the diamond crystal lattice and a vacancy adjoins in one of the four neighboring sites (Fig. 8a). The center has a spin-triplet ground state with sublevels, mS = 0 and ±1. In the absence of external field, the mS = 0 state is spontaneously split from mS = ±1 (zero-field splitting), with an energy gap of 2.87 GHz. In the presence of external field (B0), the energy levels of mS = ±1 further split due to Zeeman effects (Fig. 8b). These energy levels can be detected by applying microwave excitation to the system and concurrently performing fluorescence measurements. By spectroscopically probing the energy levels of mS, the field strength (B0) can be determined with high precision.
Figure 8. Diamond-based magnetic sensing.
(a) Structure of the nitrogen (N) and vacancy (V) inside a diamond lattice. C, carbon. (b) Energy state diagram. The NV center has a spin-triplet ground state (3A2) with a 2.87 GHz zero-field splitting between the ms = 0 and ms = ±1 spin states. Optical excitation (532 nm) pumps spins to the excited state (3E), which leads to the emission of a photon (638–800 nm). The ms = 0 spin state has a stronger fluorescence than the ms = ±1 states. When an external field (B0) is applied, the ms = ±1 states split by 2γB0, where γ is the gyromagnetic ratio of the NV electronic spin. This field (B0) can thus be measured by monitoring the fluorescence emission, while applying a continuous microwave. When the applied microwave frequency is on resonance with either of the ms = ±1 state transitions from ms = 0, the fluorescence rate decreases. (c) Detection of magnetotactic bacteria with a NV-diamond sensor. Top and bottom left images are from optical and scanning electron microscopy (SEM), respectively. Measured magnetic field projections along the x axis (Bx), y axis (By) and z axis (Bz) within the same field-of-view are shown in the top row. The bottom row shows simulated magnetic field projections, assuming that magnetic nanoparticle locations match those in the SEM image. (Reproduced with permission from ref. 100 and 101. Copyright 2012, 2013 Nature Publishing Group.)
NV diamond sensors have a significant potential for biosensing. They can achieve excellent detection sensitivity (3nT/√Hz),98, and operate at room temperature. Standard microscopy can be used to generate wide-field images of magnetic fields with nanoscale resolution. For example, Sage et al. demonstrated magnetic imaging of magnetotactic bacteria with a NV diamond sensor (Fig. 8c).101. Magnetic fields, created by chains of magnetosomes inside the bacteria, were measured, and their full vector map was reconstructed at a sub-cellular spatial resolution (400 nm) in a field of view of 100 μm2. More recently, NV diamond sensor has also been used as a NMR reader.102,103 The exquisite sensitivity of NV diamond detected NMR signal from 104 nuclear spins, enabling NMR measurements on nanometer scale samples.103
V. CONCLUSIONS AND FUTURE PERSPECTIVES
Magnetic biosensor is a promising technology for fast and sensitive molecular diagnostics. A key advantage of magnetic sensing is its intrinsically low background in biological media. Robust assays can thus be performed with minimal purification steps, thereby reducing sample loss and potential degradation of molecular markers. As a synergistic platform, magnetic sensing draws on complementary advantages from multiple disciplines to expand its target versatility and detection functionality. Advances in nanomaterials and sensing technologies, as described in this review, have enabled significant strides towards expanding the reach of magnetic biosensor for a wide range of biological targets and implementing platforms for practical uses in clinical settings.
A major challenge in magnetic detection is to develop a technique for simultaneous detection of multiple (i.e. dozens to hundreds) of biomarkers. Magnetic sensors have a limited capacity for multiplexed detection, because the signal arises from a single parameter (e.g., magnetic moment, relaxation time). Different approaches have been explored to address this limitation. For example, Liong et al. sequentially labeled different molecular markers with MNPs and measured accumulative magnetic signals; the signal difference between consecutive labeling steps correlated with the level of the target marker expression.48 Microfabricated magnetic structures with distinct spectral signatures have been demonstrated as multispectral contrast agents for magnetic resonance imaging.104 The nonlinear magnetization of MNPs also has been exploited to distinguish differently-sized particles by measuring magnetic moments at varying static magnetic field.96 These methods, albeit promising, still need to be optimized for robustness, and extended beyond their current 3-4 biomarker limits. Another appealing direction is to further integrate multiple sensors and magnetic actuators into a single device, so as to empower the platform for more complex, multi-step tasks. Incorporating a large number of sensors and their control circuits into a single device is one promising approach. Such devices will effectively function as a magnetic “imager” with a large field-of-view. This scheme will render the system simple-to-operate without the need for complex microfluidics, and significantly enhance the assay speed through parallel measurements. Such developments will facilitate advances towards the “sample-to-answer” platform, wherein native clinical specimen is the device input and electronic data, presented in a useful format for clinicians, is the device output.105
ACKNOWLEDGEMENT
The authors thank R. M. Westervelt (Harvard) for helpful discussion. H. Shao acknowledges financial support from the B.S.-Ph.D. National Science Scholarship awarded by the Agency for Science, Technology and Research, Singapore. This work was supported in part by NIH Grants R01HL113156, R01EB004626, R01EB010011, HHSN268201000044C, U54-CA119349, K12CA087723-09, and T32-CA79443.
References
- [1].Alix-Panabieres C, Schwarzenbach H, Pantel K. Annu. Rev. Med. 2012;63:199–215. doi: 10.1146/annurev-med-062310-094219. [DOI] [PubMed] [Google Scholar]
- [2].Fan R, Vermesh O, Srivastava A, Yen BKH, Qin L, Ahmad H, Kwong GA, Liu C-C, Gould J, Hood L, Heath JR. Nat. Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Simpson RJ, Lim JWE, Moritz RL, Mathivanan S. Expert Rev. Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- [4].Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cheng MM-C, Cuda G, Bunimovich YL, Gaspari M, Heath JR, Hill HD, Mirkin CA, Nijdam AJ, Terracciano R, Thundat T, Ferrari M. Curr. Opin. Chem. Biol. 2006;10:11–19. doi: 10.1016/j.cbpa.2006.01.006. [DOI] [PubMed] [Google Scholar]
- [6].Giljohann DA, Mirkin CA. Nature. 2009;462:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sorger PK. Nat. Biotechnol. 2008;26:1345–1346. doi: 10.1038/nbt1208-1345. [DOI] [PubMed] [Google Scholar]
- [8].Pankhurst QA, Connolly J, Jones SK, Dobson J. J. Phys. D: Appl. Phys. 2003;36:R167–R181. [Google Scholar]
- [9].Jun Y-W, Seo J-W, Cheon J. Acc. Chem. Res. 2008;41:179–189. doi: 10.1021/ar700121f. [DOI] [PubMed] [Google Scholar]
- [10].Laurent S, Forge D, Port M, Roch A, Robic C, Elst LV, Muller RN. Chem. Rev. 2008;108:2064–2110. doi: 10.1021/cr068445e. [DOI] [PubMed] [Google Scholar]
- [11].Cheon J, Lee J-H. Acc. Chem. Res. 2008;41:1630–1640. doi: 10.1021/ar800045c. [DOI] [PubMed] [Google Scholar]
- [12].Gijs MAM, Lacharme F, Lehmann U. Chem. Rev. 2010;110:1518–1563. doi: 10.1021/cr9001929. [DOI] [PubMed] [Google Scholar]
- [13].Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Nat. Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- [14].Zborowski M, Chalmers JJ. Magnetic cell separation. Elsevier; Amsterdam; Boston: 2008. p. xviii. 454 p., [14] p. of plates. [Google Scholar]
- [15].Haun JB, Castro CM, Wang R, Peterson VM, Marinelli BS, Lee H, Weissleder R. Sci. Transl. Med. 2011;3:71ra16. doi: 10.1126/scitranslmed.3002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Issadore D, Min C, Liong M, Chung J, Weissleder R, Lee H. Lab Chip. 2011;11:2282–2287. doi: 10.1039/c1lc20177h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Muluneh M, Shang W, Issadore D. Adv Healthcare Mater. 2014 doi: 10.1002/adhm.201300502. DOI: 10.1002/adhm.201300502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bhagat AAS, Bow H, Hou HW, Tan SJ, Han J, Lim CT. Med. Biol. Eng. Comput. 2010;48:999–1014. doi: 10.1007/s11517-010-0611-4. [DOI] [PubMed] [Google Scholar]
- [19].Pamme N. Curr. Opin. Chem. Biol. 2012;16:436–443. doi: 10.1016/j.cbpa.2012.05.181. [DOI] [PubMed] [Google Scholar]
- [20].Sajeesh P, Sen AK. Microfluid. Nanofluid. 2013:1–52. [Google Scholar]
- [21].Yoo D, Lee J-H, Shin T-H, Cheon J. Acc. Chem. Res. 2011;44:863–874. doi: 10.1021/ar200085c. [DOI] [PubMed] [Google Scholar]
- [22].Xie J, Liu G, Eden HS, Ai H, Chen X. Acc. Chem. Res. 2011;44:883–892. doi: 10.1021/ar200044b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sun S, Zeng H, Robinson DB, Raoux S, Rice PM, Wang SX, Li G. J. Am. Chem. Soc. 2004;126:273–279. doi: 10.1021/ja0380852. [DOI] [PubMed] [Google Scholar]
- [24].Park J, An K, Hwang Y, Park J-G, Noh H-J, Kim J-Y, Park J-H, Hwang N-M, Hyeon T. Nat. Mater. 2004;3:891–895. doi: 10.1038/nmat1251. [DOI] [PubMed] [Google Scholar]
- [25].Kim D, Lee N, Park M, Kim BH, An K, Hyeon T. J. Am. Chem. Soc. 2009;131:454–455. doi: 10.1021/ja8086906. [DOI] [PubMed] [Google Scholar]
- [26].Lee H, Yoon T-J, Figueiredo J-L, Swirski FK, Weissleder R. Proc. Natl. Acad. Sci. USA. 2009;106:12459–12464. doi: 10.1073/pnas.0902365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lee H, Yoon T-J, Weissleder R. Angew. Chem. Int. Ed. 2009;48:5657–5660. doi: 10.1002/anie.200901791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Noh S.-h., Na W, Jang J.-t., Lee J-H, Lee EJ, Moon SH, Lim Y, Shin J-S, Cheon J. Nano Letters. 2012;12:3716–3721. doi: 10.1021/nl301499u. [DOI] [PubMed] [Google Scholar]
- [29].Hyeon T, Lee SS, Park J, Chung Y, Na HB. J. Am. Chem. Soc. 2001;123:12798–12801. doi: 10.1021/ja016812s. [DOI] [PubMed] [Google Scholar]
- [30].Sun S, Zeng H. J. Am. Chem. Soc. 2002;124:8204–8205. doi: 10.1021/ja026501x. [DOI] [PubMed] [Google Scholar]
- [31].Lee J-H, Huh Y-M, Jun Y.-w., Seo J.-w., Jang J.-t., Song H-T, Kim S, Cho E-J, Yoon H-G, Suh J-S, Cheon J. Nat. Med. 2007;13:95–99. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- [32].Jang J.-t., Nah H, Lee J-H, Moon SH, Kim MG, Cheon J. Angew. Chem. Int. Ed. 2009;48:1234–1238. doi: 10.1002/anie.200805149. [DOI] [PubMed] [Google Scholar]
- [33].Kim BH, Lee N, Kim H, An K, Park YI, Choi Y, Shin K, Lee Y, Kwon SG, Na HB, Park J-G, Ahn T-Y, Kim Y-W, Moon WK, Choi SH, Hyeon T. J. Am. Chem. Soc. 2011;133:12624–12631. doi: 10.1021/ja203340u. [DOI] [PubMed] [Google Scholar]
- [34].Huber DL. Small. 2005;1:482–501. doi: 10.1002/smll.200500006. [DOI] [PubMed] [Google Scholar]
- [35].Qiang Y, Antony J, Sharma A, Nutting J, Sikes D, Meyer D. J. Nanopart. Res. 2006;8:489–496. [Google Scholar]
- [36].Seo WS, Lee JH, Sun X, Suzuki Y, Mann D, Liu Z, Terashima M, Yang PC, McConnell MV, Nishimura DG, Dai H. Nat. Mater. 2006;5:971–976. doi: 10.1038/nmat1775. [DOI] [PubMed] [Google Scholar]
- [37].Peng S, Wang C, Xie J, Sun S. J. Am. Chem. Soc. 2006;128:10676–10677. doi: 10.1021/ja063969h. [DOI] [PubMed] [Google Scholar]
- [38].Ban Z, Barnakov YA, Li F, Golub VO, O’Connor CJ. J. Mater. Chem. 2005;15:4660–4662. [Google Scholar]
- [39].Cheng J, Ni X, Zheng H, Li B, Zhang X, Zhang D. Mater. Res. Bull. 2006;41:1424–1429. [Google Scholar]
- [40].Yoon T-J, Lee H, Shao H, Weissleder R. Angew. Chem. Int. Ed. 2011;50:4663–4666. doi: 10.1002/anie.201100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ge J, Hu Y, Biasini M, Beyermann WP, Yin Y. Angew. Chem. Int. Ed. 2007;46:4342–4345. doi: 10.1002/anie.200700197. [DOI] [PubMed] [Google Scholar]
- [42].Zhuang J, Wu H, Yang Y, Cao YC. Angew. Chem. Int. Ed. 2008;47:2208–2212. doi: 10.1002/anie.200705049. [DOI] [PubMed] [Google Scholar]
- [43].Hickey RJ, Meng X, Zhang P, Park S-J. ACS Nano. 2013;7:5824–5833. doi: 10.1021/nn400824b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lee JE, Lee N, Kim H, Kim J, Choi SH, Kim JH, Kim T, Song IC, Park SP, Moon WK, Hyeon T. J. Am. Chem. Soc. 2010;132:552–557. doi: 10.1021/ja905793q. [DOI] [PubMed] [Google Scholar]
- [45].Yoon T-J, Lee H, Shao H, Hilderbrand SA, Weissleder R. Adv. Mater. 2011;23:4793–4797. doi: 10.1002/adma.201102948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Molday RS, Mackenzie D. J. Immunol. Methods. 1982;52:353–367. doi: 10.1016/0022-1759(82)90007-2. [DOI] [PubMed] [Google Scholar]
- [47].Patolsky F, Weizmann Y, Katz E, Willner I. Angew. Chem. Int. Ed. 2003;42:2372–2376. doi: 10.1002/anie.200250379. [DOI] [PubMed] [Google Scholar]
- [48].Liong M, Tassa C, Shaw SY, Lee H, Weissleder R. Adv. Mater. 2011;23:H254–H257. doi: 10.1002/adma.201101401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lin P-C, Tseng M-C, Su A-K, Chen Y-J, Lin C-C. Anal. Chem. 2007;79:3401–3408. doi: 10.1021/ac070195u. [DOI] [PubMed] [Google Scholar]
- [50].Sun EY, Josephson L, Kelly KA, Weissleder R. Bioconjugate Chem. 2006;17:109–113. doi: 10.1021/bc050290e. [DOI] [PubMed] [Google Scholar]
- [51].Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Nat. Nanotechnol. 2010;5:660–665. doi: 10.1038/nnano.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Agasti SS, Liong M, Tassa C, Chung HJ, Shaw SY, Lee H, Weissleder R. Angew. Chem. Int. Ed. 2012;51:450–454. doi: 10.1002/anie.201105670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Perez JM, Josephson L, O’Loughlin T, Hogemann D, Weissleder R. Nat. Biotechnol. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- [54].Park K, Harrah T, Goldberg EB, Guertin RP, Sonkusale S. Nanotechnology. 2011;22:085501. doi: 10.1088/0957-4484/22/8/085501. [DOI] [PubMed] [Google Scholar]
- [55].Brown KA, Vassiliou CC, Issadore D, Berezovsky J, Cima MJ, Westervelt RM. J. Magn. Magn. Mater. 2010;322:3122–3126. doi: 10.1016/j.jmmm.2010.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Min C, Shao H, Liong M, Yoon T-J, Weissleder R, Lee H. ACS Nano. 2012;6:6821–6828. doi: 10.1021/nn301615b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Xu L, Yu H, Akhras MS, Han S-J, Osterfeld S, White RL, Pourmand N, Wang SX. Biosens. Bioelectron. 2008;24:99–103. doi: 10.1016/j.bios.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li G, Sun S, Wilson RJ, White RL, Pourmand N, Wang SX. Sens. Actuators A. 2006;126:98–106. doi: 10.1016/j.sna.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Manandhar P, Chen K-S, Aledealat K, Mihajlovic G, Yun CS, Field M, Sullivan GJ, Strouse GF, Chase PB, Molnar S. von, Xiong P. Nanotechnology. 2009;20:355501. doi: 10.1088/0957-4484/20/35/355501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wang H, Mahdavi A, Tirrell DA, Hajimiri A. Lab Chip. 2012;12:4465–4471. doi: 10.1039/c2lc40392g. [DOI] [PubMed] [Google Scholar]
- [61].Aytur T, Foley J, Anwar M, Boser B, Harris E, Beatty PR. J. Immunol. Methods. 2006;314:21–29. doi: 10.1016/j.jim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- [62].Osterfeld SJ, Yu H, Gaster RS, Caramuta S, Xu L, Han S-J, Hall DA, Wilson RJ, Sun S, White RL, Davis RW, Pourmand N, Wang SX. Proc. Natl. Acad. Sci. USA. 2008;105:20637–20640. doi: 10.1073/pnas.0810822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Koets M, Wijk T. van der, van Eemeren JTWM, Amerongen A. van, Prins MWJ. Biosens. Bioelectron. 2009;24:1893–1898. doi: 10.1016/j.bios.2008.09.023. [DOI] [PubMed] [Google Scholar]
- [64].Gaster RS, Xu L, Han S-J, Wilson RJ, Hall DA, Osterfeld SJ, Yu H, Wang SX. Nat. Nanotechnol. 2011;6:314–320. doi: 10.1038/nnano.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lewin M, Carlesso N, Tung C-H, Tang X-W, Cory D, Scadden DT, Weissleder R. Nat. Biotechnol. 2000;18:410–414. doi: 10.1038/74464. [DOI] [PubMed] [Google Scholar]
- [66].Shao H, Min C, Issadore D, Liong M, Yoon T-J, Weissleder R, Lee H. Theranostics. 2012;2:55–65. doi: 10.7150/thno.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Peterson VM, Castro CM, Lee H, Weissleder R. ACS Nano. 2012;6:3506–3513. doi: 10.1021/nn300536y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gillis P, Koenig SH. Magn. Reson. Med. 1987;5:323–345. doi: 10.1002/mrm.1910050404. [DOI] [PubMed] [Google Scholar]
- [69].Lee H, Sun E, Ham D, Weissleder R. Nat. Med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Danieli E, Perlo J, Blumich B, Casanova F. Angew. Chem. Int. Ed. 2010;49:4133–4135. doi: 10.1002/anie.201000221. [DOI] [PubMed] [Google Scholar]
- [71].Sun N, Liu Y, Lee H, Weissleder R, Ham D. IEEE J. Solid-State Circuits. 2009;44:1629–1643. [Google Scholar]
- [72].Massin C, Vincent F, Homsy A, Ehrmann K, Boero G, Besse P-A, Daridon A, Verpoorte E, de Rooij NF, Popovic RS. J. Magn. Reson. 2003;164:242–255. doi: 10.1016/s1090-7807(03)00151-4. [DOI] [PubMed] [Google Scholar]
- [73].Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Nat. Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ghazani AA, Castro CM, Gorbatov R, Lee H, Weissleder R. Neoplasia. 2012;14:388–395. doi: 10.1596/neo.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ghazani AA, Pectasides M, Sharma A, Castro CM, Mino-Kenudson M, Lee H, Shepard J-AO, Weissleder R. Nanomedicine. 2014;10:661–668. doi: 10.1016/j.nano.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Castro CM, Ghazani AA, Chung J, Shao H, Issadore D, Yoon T-J, Weissleder R, Lee H. Lab Chip. 2014;14:14–23. doi: 10.1039/c3lc50621e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chung HJ, Reiner T, Budin G, Min C, Liong M, Issadore D, Lee H, Weissleder R. ACS Nano. 2011;5:8834–8841. doi: 10.1021/nn2029692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Budin G, Chung HJ, Lee H, Weissleder R. Angew. Chem. Int. Ed. 2012;51:7752–7755. doi: 10.1002/anie.201202982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Chung HJ, Castro CM, Im H, Lee H, Weissleder R. Nat. Nanotechnol. 2013;8:369–375. doi: 10.1038/nnano.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Rho J, Chung J, Im H, Liong M, Shao H, Castro CM, Weissleder R, Lee H. ACS Nano. 2013;7:11227–11233. doi: 10.1021/nn405016y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Liong M, Hoang AN, Chung J, Gural N, Ford CB, Min C, Shah RR, Ahmad R, Fernandez-Suarez M, Fortune SM, Toner M, Lee H, Weissleder R. Nat. Commun. 2013;4:1752. doi: 10.1038/ncomms2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Stromberg M, de la Torre TZG, Goransson J, Gunnarsson K, Nilsson M, Svedlindh P, Stromme M. Anal. Chem. 2009;81:3398–3406. doi: 10.1021/ac900561r. [DOI] [PubMed] [Google Scholar]
- [83].Connolly J, St Pierre TG. J. Magn. Magn. Mater. 2001;225:156–160. [Google Scholar]
- [84].Astalan AP, Ahrentorp F, Johansson C, Larsson K, Krozer A. Biosens. Bioelectron. 2004;19:945–951. doi: 10.1016/j.bios.2003.09.005. [DOI] [PubMed] [Google Scholar]
- [85].Grossman HL, Myers WR, Vreeland VJ, Bruehl R, Alper MD, Bertozzi CR, Clarke J. Proc. Natl. Acad. Sci. USA. 2004;101:129–134. doi: 10.1073/pnas.0307128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Baselt DR, Lee GU, Natesan M, Metzger SW, Sheehan PE, Colton RJ. Biosens. Bioelectron. 1998;13:731–739. doi: 10.1016/s0956-5663(98)00037-2. [DOI] [PubMed] [Google Scholar]
- [87].Graham DL, Ferreira HA, Feliciano N, Freitas PP, Clarke LA, Amaral MD. Sensor Actuat. B-Chem. 2005;107:936–944. [Google Scholar]
- [88].Loureiro J, Andrade PZ, Cardoso S, da Silva CL, Cabral JM, Freitas PP. Lab Chip. 2011;11:2255–2261. doi: 10.1039/c0lc00324g. [DOI] [PubMed] [Google Scholar]
- [89].Freitas PP, Cardoso FA, Martins VC, Martins SAM, Loureiro J, Amaral J, Chaves RC, Cardoso S, Fonseca LP, Sebastião AM, Pannetier-Lecoeur M, Fermon C. Lab Chip. 2012;12:546–557. doi: 10.1039/c1lc20791a. [DOI] [PubMed] [Google Scholar]
- [90].Hall DA, Gaster RS, Makinwa KAA, Wang SX, Murmann B. IEEE J. Solid-State Circuits. 2013;48:1290–1301. doi: 10.1109/JSSC.2013.2245058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Besse P-A, Boero G, Demierre M, Pott V, Popovic R. Appl. Phys. Lett. 2002;80:4199–4201. [Google Scholar]
- [92].Mihajlovic G, Xiong P, Molnar S. von, Ohtani K, Ohno H, Field M, Sullivan GJ. Appl. Phys. Lett. 2005;87:112502. [Google Scholar]
- [93].Togawa K, Sanbonsugi H, Lapicki A, Abe M, Handa H, Sandhu A. IEEE Trans. Magn. 2005;41:3661–3663. [Google Scholar]
- [94].Liu PP, Skucha K, Duan Y, Megens M, Kim J, Izyumin II, Gambini S, Boser B. IEEE J. Solid-State Circuits. 2012;47:1056–1064. doi: 10.1109/jssc.2012.2185339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gambini S, Skucha K, Liu PP, Kim J, Krigel R. IEEE J. Solid-State Circuits. 2013;48:302–317. doi: 10.1109/jssc.2012.2185339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Issadore D, Chung J, Shao H, Liong M, Ghazani AA, Castro CM, Weissleder R, Lee H. Sci. Transl. Med. 2012;4:141ra92. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Balasubramanian G, Chan IY, Kolesov R, Al-Hmoud M, Tisler J, Shin C, Kim C, Wojcik A, Hemmer PR, Krueger A, Hanke T, Leitenstorfer A, Bratschitsch R, Jelezko F, Wrachtrup J. Nature. 2008;455:648–651. doi: 10.1038/nature07278. [DOI] [PubMed] [Google Scholar]
- [98].Maze JR, Stanwix PL, Hodges JS, Hong S, Taylor JM, Cappellaro P, Jiang L, Dutt MVG, Togan E, Zibrov AS, Yacoby A, Walsworth RL, Lukin MD. Nature. 2008;455:644–647. doi: 10.1038/nature07279. [DOI] [PubMed] [Google Scholar]
- [99].Taylor JM, Cappellaro P, Childress L, Jiang L, Budker D, Hemmer PR, Yacoby A, Walsworth R, Lukin MD. Nat. Phys. 2008;4:810–816. [Google Scholar]
- [100].Waldherr G, Beck J, Neumann P, Said RS, Nitsche M, Markham ML, Twitchen DJ, Twamley J, Jelezko F, Wrachtrup J. Nat. Nanotechnol. 2012;7:105–108. doi: 10.1038/nnano.2011.224. [DOI] [PubMed] [Google Scholar]
- [101].Sage D. Le, Arai K, Glenn DR, DeVience SJ, Pham LM, Rahn-Lee L, Lukin MD, Yacoby A, Komeili A, Walsworth RL. Nature. 2013;496:486–489. doi: 10.1038/nature12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Mamin HJ, Kim M, Sherwood MH, Rettner CT, Ohno K, Awschalom DD, Rugar D. Science. 2013;339:557–560. doi: 10.1126/science.1231540. [DOI] [PubMed] [Google Scholar]
- [103].Staudacher T, Shi F, Pezzagna S, Meijer J, Du J, Meriles CA, Reinhard F, Wrachtrup J. Science. 2013;339:561–563. doi: 10.1126/science.1231675. [DOI] [PubMed] [Google Scholar]
- [104].Zabow G, Dodd S, Moreland J, Koretsky A. Nature. 2008;453:1058–1063. doi: 10.1038/nature07048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Issadore D, Westervelt R. Point-of-Care Diagnostics on a Chip. Springer; [Google Scholar]