Abstract
Type 2 diabetes mellitus is a common disabling disease with onset in middle-aged individuals, caused by an imbalance between insulin production and action. Genetic studies point to major genetic components, but, with the exception of maturity-onset diabetes of the young (MODY), specific diabetes susceptibility genes remain to be identified. Recent studies showed that a dominant negative mutation in the insulin promoter factor-1 (IPF-1), a pancreatic β-cell specific transcription factor, causes pancreatic agenesis and MODY. Thus, we investigated 192 French, non-MODY type 2 diabetic families for mutations in IPF-1. We identified 3 novel IPF-1 mutations, including 2 substitutions (Q59L and D76N) and an in-frame proline insertion (InsCCG243). Functional transactivation assays of these IPF-1 mutant isoforms in a β-pancreatic tumor cell line transfected with a transcriptional reporter and IPF-1 expression plasmids demonstrate a significant inhibition of basal insulin promoter activity (stronger with the InsCCG243 mutant). We find that the InsCCG243 mutation is linked, in 2 families, to an autosomal dominant-like late-onset form of type 2 diabetes, in which insulin secretion becomes progressively impaired. The lower penetrance D76N and Q59L mutations were more prevalent and were associated with a relative risk of 12.6 for diabetes and with decreased glucose-stimulated insulin-secretion in nondiabetic subjects. We propose that IPF-1 mutations can cause MODY or apparently monogenic late-onset diabetes and that they represent a significant risk factor for type 2 diabetes in humans.
Introduction
Type 2 diabetes is a common disorder resulting from a polygenic and heterogeneous etiology and characterized by chronic hyperglycemia due to an imbalance between insulin production and action (1). Genetic studies have now provided some clues for the complex inheritance of type 2 diabetes, owing to the recent mapping of several type 2 diabetes major susceptibility genes (2–4). Similar approaches have been even more successful in the identification of specific genes that cause maturity-onset diabetes of the young (MODY), an autosomal dominant form of type 2 diabetes characterized by an early onset (usually ≤25 years) and a primary defect in insulin secretion (reviewed in ref. 5). Proper transcriptional regulation of glucose homeostasis-specific genes appears to be crucial for maintaining normoglycemia, as 4 of the 5 MODY clinical subtypes described to date result from mutations affecting transcription factors (6–10).
The homeodomain-containing protein, IPF-1 (also known as STF1, IDX1, and PDX1), is a transcription factor critically required for the embryonic development of the pancreas and for the transcriptional regulation of endocrine pancreas-specific genes in adults, such as insulin, glucose transporter-2 (GLUT2) and glucokinase in β-cells, and somatostatin in δ-cells (reviewed in ref. 11). IPF-1 is normally expressed in all cells of the pancreatic embryonic bud, and its absence in mice arrests pancreas development at the bud stage, leading to pancreatic agenesis (12, 13). Recently, a homozygosity for a base deletion mutation in IPF-1 codon 63 (Pro63fsdelC), resulting in a premature stop codon, was reported to cause pancreatic agenesis in a child (14). Individuals who are heterozygous carriers of the Pro63fsdelC mutation develop MODY (MODY4; ref. 9). IPF-1 deficiency in this MODY4 subtype results from the expression, due to the Pro63fsdelC, of an internally translated dominant negative isoform of IPF-1 that inhibits the wild-type expressed IPF-1 isoform (15). Therefore, less-severe IPF-1 mutations may be candidates for inheritable defects in the common late-onset forms of type 2 diabetes. To address this question, we examined the protein encoding and regulatory sequences of IPF-1 gene for mutations in a group of 64 randomly selected French families with late-onset type 2 diabetes.
Here we describe 3 novel defective IPF-1 mutations affecting up to 6.2% of French families with late-onset type 2 diabetes. We also provide statistical, physiological, and functional evidence that, depending on the mutation impairing effect, IPF-1 mutations are associated with diabetes either through a monogenic-like mode of inheritance or in a polygenic context of the disease susceptibility.
Methods
Subjects.
Sixty-four unrelated type 2 diabetic patients from French ancestry were screened by PCR and direct sequencing for mutations in the IPF-1 gene and its regulatory sequences. These families were randomly selected from a large collection of multiplex type 2 diabetic families recruited throughout France (16). These families had no known etiology for diabetes. These probands, 34 men and 30 women, were aged 62.5 ± 11.6 years (mean ± SD), had a body mass index (BMI) of 25.3 ± 3.8 kg/m2, presented fasting plasma glucose and insulin levels of 9.9 ± 3.9 mmol/L and 12.5 ± 5.0 mU/L, respectively. They were treated by oral hypoglycemic agents (50.8%), insulin (36.0%), or both (13.2%) and had an average age at diagnosis of diabetes of 41.7 ± 7.1 years. Two normoglycemic subjects with no familial history of diabetes were considered as controls for gene screening. Genomic DNA was extracted from peripheral blood lymphocytes using standard procedures. For prevalence studies, cohorts of 192 unrelated type 2 diabetic probands (selected from the same dataset of French Caucasian families and including the 64 probands selected for gene screening) and of 231 normoglycemic control subjects were studied. The control group was composed of normoglycemic spouses of patients from type 2 diabetic families, all of whom had no familial history of diabetes and underwent an oral glucose tolerance testing. The characteristics of type 2 diabetic and control cohorts have been described previously (17).
IPF-1 gene screening.
Sixty-four type 2 diabetic patients were screened for mutations in the IPF-1 gene and its regulatory sequences. The upstream enhancer and proximal promoter segments and the 2 exons with their adjacent intronic sequences of IPF-1 gene were amplified by PCR using specific oligonucleotides as described recently (18): (forward) 5′—GCCGCAGACAATGGACTC—3′ and (reverse) 5′—AGATGCCCTTGCTGTCACC—3′ for the upstream enhancer segment; (forward) 5′—GCCTAGCCTTTCAGTGCG—3′ and (reverse) 5′—TGGGTCCTTGTAAAGCTG—3′ for the proximal promoter; (forward) 5′—CCATGAACGGCGAGGAGC—3′ and (reverse) 5′—CAGGCTTACCTGCCCACT—3′ for exon 1 and (forward) 5′—GCCCTGTGTCGCCCGCAG—3′ and (reverse) 5′—TTGAAGCCCCTCA-GCCAG—3′ for exon 2. For direct DNA sequencing protocols, all these forward and reverse primers were extended at their 5′ ends with the universal M13 forward and reverse primers, respectively. The enhancer segment, located approximately 1.8 kb upstream the transcriptional start site of IPF-1, was investigated because it is highly homologous to its murine counterpart and contains regulatory elements that may be essential for islet specific IPF-1 expression (D.A. Stoffers, unpublished study).
PCR was carried out on 100 ng of genomic DNA in a 50-μL volume, 0.25 mM of each primer, and 1.5 U of AmpliTaqGold (Perkin-Elmer Applied Biosystems, Forster City, California, USA). For all fragments, PCR cycling consisted in an initial denaturation at 94°C for 12 minutes followed by 30 cycles of a denaturation step at 94°C for 30 seconds, an annealing step at 57°C for 30 seconds, and an extension step at 72°C for 45 seconds, with a final extension step at 72°C for 10 minutes. Because IPF-1 gene sequences are highly G-C rich, we used DMSO in PCR and sequencing reactions at a 5% vol/vol final concentration. The PCR products were sequenced on both strands using M13 primers.
Genotyping of IPF-1 identified sequence nucleotide variations.
The presence of the Q59L variation results in the loss of an Alu I enzymatic restriction site and thus was analyzed using a simple PCR-RFLP procedure. Genotyping of the D76N substitution was examined by a restriction cleavage assay using a nested forward oligonucleotide with an introduced mismatch at the second nucleotide from the 3′ end (5′—TCCCCGTACGAGGTGCCCCCCCTCGCCGTC—3′); the reverse primer corresponded to the wild type sequence (5′—CGGTTGGGCTCCTCCAGGAC—3′). The introduced mismatch in the forward primer resulted in a Sal I polymorphic restriction site, depending on the presence or absence of the codon 76 G→A transition, and allowed a simple screening for this variation. The InsCCG243 mutation in IPF-1 exon 2 presents like a trinucleotide repeat expansion. Thus, analysis for its presence or absence was performed using standard fluorescent-based genotyping protocols, with the flanking oligonucleotides: 5′—AAGGAGGAGGACAAGAAG—3′ and 5′—CAGCGGGCGCA-CAGC-AC—3′, and genotyping was performed as for simple tandem repeats on an ABI-Prism 377 DNA sequencer (Perkin-Elmer Applied Biosystems, Forster City, California, USA). Using these oligonucleotides, the wild-type allele corresponded to a fragment of 148 bp, whereas the presence of the InsCCG243 mutation yielded a 151-bp fragment. Specificity of the D76N and the InsCCG243 variations genotyping protocols was checked by direct DNA sequencing on both strands of random samples of the corresponding PCR products (data not shown).
Functional evaluation of mutant IPF-1 proteins.
A transactivation assay was used to test the function of the mutant IPF-1 proteins Q59L, D76N, and InsCCG243 compared with wild-type IPF-1 (15). The Q59L, D76N, and InsCCG243 mutations were introduced into the expression vector pCMV5 containing the human IPF-1 coding sequences using site-directed mutagenesis (QuickChange kit; Stratagene, La Jolla, California, USA). Mutant IPF-1 expression plasmids were confirmed by DNA sequencing. HIT-T15 cells that express high levels of endogenous IPF-1 and insulin were transiently cotransfected with 2.5 μg of a reporter plasmid containing the rat insulin I promoter linked to the transcriptional reporter chloramphenicol acetyl transferase (–410 rINSI CAT) and 0.3–33 ng of expression vector pCMV5 containing the wild-type or mutant IPF-1 coding sequences. The relative abilities of the different IPF-1 proteins to activate the reporter were assessed by measuring CAT activity in HIT-T15 cell extracts prepared 48 hours after transfection as described previously (15). The relative levels of expression of the wild-type IPF-1 and the InsCCG243, Q59L, and D76N mutant forms were determined by SDS-PAGE and Western immunoblot assays using antiserum α253 in extracts of the HIT-T15 cells used for transactivation assays (14, 15). For these Western immunoblot assays, 500 ng of the same IPF-1 expression plasmids was transfected to the HIT-T15 cells to allow assay of plasmid expressed IPF-1 above a background of endogenous IPF-1. Statistical significance was determined by both Student’s t test and Wilcoxon rank sum test.
Results
One diabetic proband from each of the 64 randomly selected French type 2 diabetic families was screened for mutations of IPF-1 gene. In the coding sequence of IPF-1, we identified 3 novel nucleotide sequence variations: CAG→CTG and GAC→AAC variations in exon 1 predicted to result, respectively, in a glutamine-to-leucine substitution at codon 59 (Q59L) and in an aspartic acid-to-asparagine substitution at codon 76 (D76N). Of the 64 families screened, the Q59L and the D76N were found in 1 and 2 families, respectively. The third sequence variation was a CCG in-frame insertion at codon 243 (InsCCG243), predicted to result in the addition of an extra-proline in a hexa-proline tract encoded by exon 2 of IPF-1 (amino acids 239–244). This insertion mutation was identified in 1 family of the 64 families investigated. The Q59L, D76N, and InsCCG243 variations were typed in an extended cohort of 192 unrelated type 2 diabetic probands (including the initial group of 64 probands subjected to gene screening) and in a group of 231 unrelated normoglycemic control subjects. The genotype frequencies of these 3 mutations are shown in Table 1.
Table 1.
IPF-1 gene sequence variations identified in French families with late-onset type 2 diabetes
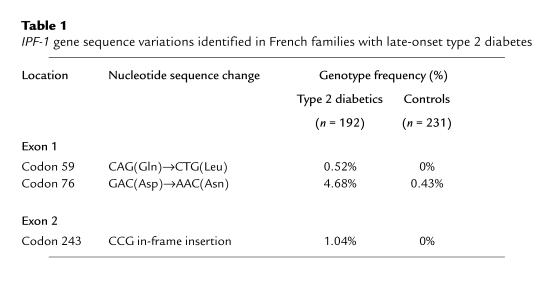
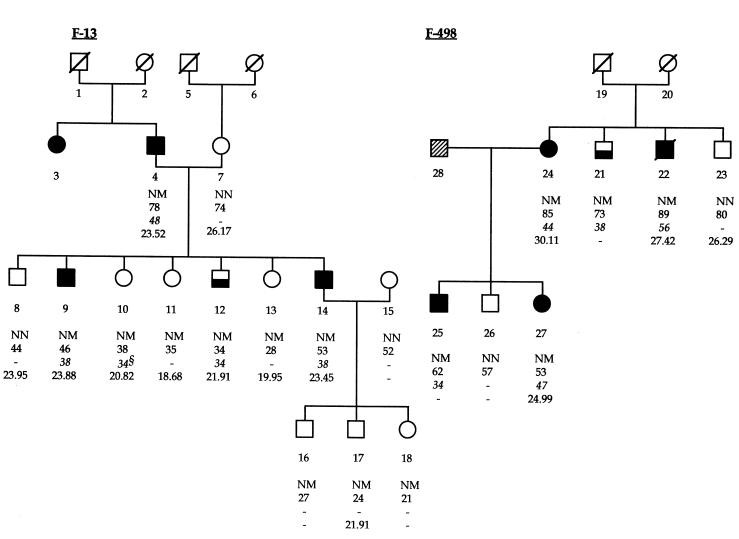
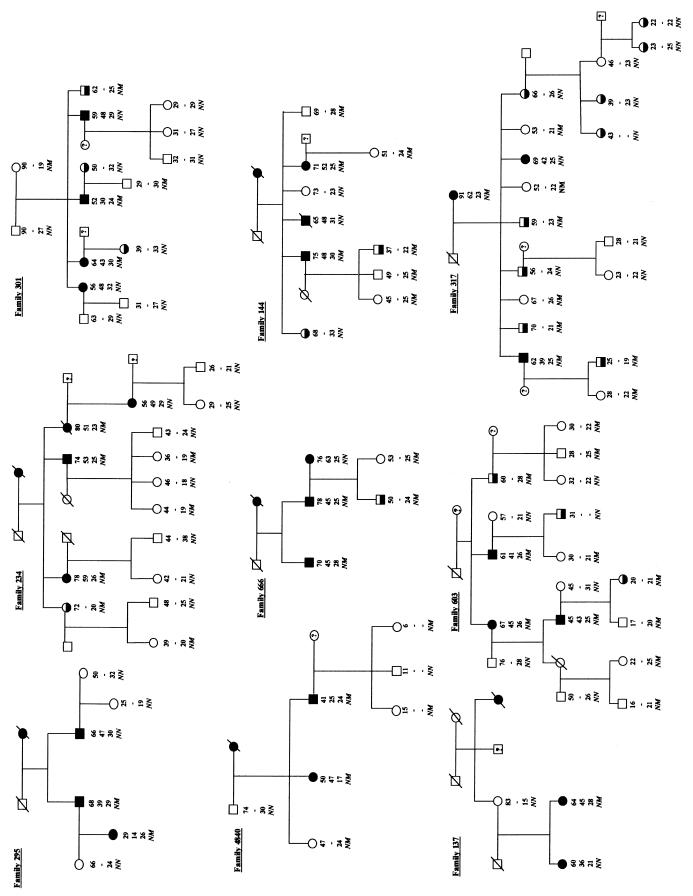
The InsCCG243 mutation was found in 2 families in the cohort of 192 diabetic families, but was not detected on screening of 462 normal chromosomes from 231 unrelated healthy controls (Table 1). Intrafamilial transmission analysis showed that this InsCCG243 variation cosegregated with type 2 diabetes in the 2 families (F-13 and F-498; Figure 1). The lod scores for this mutation in families F-13 and F-498 were 0.77 and 0.17, respectively. All affected members of these 2 families were heterozygous for this insertion mutation, with a mean age at diagnosis of 43.5 ± 7.5 years (mean ± SD). Analysis of 4 middle-aged nondiabetic subjects carrying the IPF-1 InsCCG243 mutation (Figure 1, individuals 10, 11, 12, and 13), for which insulin-secretion profile records were available in 1992 and again in 1997 showed a progressive decline in both basal and 2-hour insulin/glucose ratios during oral glucose tolerance test (OGTT) examination (P < 0.02; Figure 2).
Figure 1.
Segregation of the IPF-1 InsCCG243 variation in pedigrees F-13 and F-498. Filled, half-filled, and open symbols represent subjects with overt type 2 diabetes mellitus, impaired glucose tolerance, and normal glucose tolerance, respectively. Hatched symbols represent untested subjects. The first line under the symbols corresponds to the identification number. The second line shows the genotype at codon 243 of IPF-1 (italicized): N, Normal allele; M, Mutant allele (InsCCG243). The third and fourth lines show, respectively, the current age of subjects and the age of diagnosis of diabetes (or glucose intolerance, italicized), when appropriate. The fifth line shows the BMI (kg/m2). §: Subject 10 presented with impaired glucose tolerance at 34 years of age.
Figure 2.
Five-year evolution of insulin-secretion profiles in 4 middle-aged carriers of the IPF-1 InsCCG243 mutation. The plots show comparisons of plasma glucose levels (Figure 3a), plasma insulin levels (Figure 3b), and insulin/glucose ratios (Figure 3c), between 1992 (filled bars) and 1997 (hatched bars), with the paired Student’s t test. Basal (0 minutes) and 2 hours (120 minutes) mean values (± SEM) during oral glucose tolerance tests are presented. *P = 0.02; **P = 0.011.
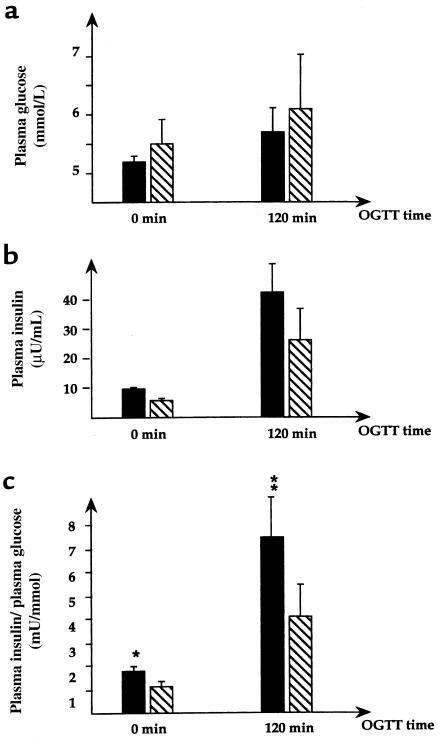
To assess the IPF-1 mutant isoforms for a possible loss of function in the transactivation of gene transcription, their ability to activate the insulin promoter was evaluated in the HIT-T15 insulinoma cell line. HIT-T15 cells express IPF-1 and, similar to many β-cell tumor cell lines, are subject to squelching of cotransfected insulin promoters when high amounts of exogenous IPF-1 expression vectors are used in transfection studies. Therefore, we used small quantities of expression vectors (300 pg to 33 ng) and found that whereas a small increase or no change in insulin promoter activity was observed in response to cotransfected wild-type IPF-1, the InsCCG243 inhibited the basal activity of the insulin promoter by 60% (Figure 3a). This result was not due to differences in the efficiencies of expression of IPF-1 wild-type and InsCCG243 mutant isoforms, as ascertained by Western blot analysis of nuclear extracts prepared from transfected cells (Figure 3b). This attenuation (>50%) of the function of endogenous IPF-1 to activate the insulin gene promoter assessed in HIT-T15 cells is consistent with the linkage of IPF-1 InsCCG243 mutation with type 2 diabetes in the 2 families and its possible involvement in the observed impairment of insulin secretion.
Figure 3.
Functional and Western immunoblot analyses of mutant IPF-1 proteins. (a) Transactivation assays were done in HIT-T15 insulinoma cells that express endogenous IPF-1. Cells were transiently transfected with a rat insulin-promoter-CAT reporter plasmid and 0.3–33 ng of expression plasmids for wild-type IPF-1 (WT) or the mutant IPF-1 isoforms. The number of independent experiments is 13–14, done on 2–3 different occasions. Basal CAT activities ranged from 6% to 15% conversion to acetylated forms of chloramphenicol. Mean values ± SEM are given compared to basal values (transfection with empty vector) normalized for the independent experiments. P values, shown above the bars (± SEM), were determined by the Wilcoxon rank sum test comparing the effect of each mutation to wild-type IPF-1 (WT). For the same comparisons, P values using the unpaired 2-sided Student’s t test were 0.052, 0.28, and < 0.001 for D76N, Q59L, and InsCCG243, respectively. (b) Determination of IPF-1 expression levels by Western immunoblot using antiserum α253 (14), specific for the detection of the COOH-terminal region of IPF-1: 500 ng of IPF-1 expression plasmids was transfected to the HIT-T15 cells. The relative expression levels of the wild-type (lane 2), Q59L (lane 4), and InsCCG243 (lane 5) IPF-1 forms were the same. The D76N form (lane 3) was expressed at a lower level than that of the others. After correction for expression efficiency by Western immunoblot analysis, the D76N mutant function was approximately 60% that of the wild-type IPF-1. Lane 1 corresponds to extracts from cells transfected with the empty vector.
Like the InsCCG243 mutation, the Q59L mutation was not detected upon the screening of 462 normal chromosomes of nondiabetic controls (Table 1). However, this variant was present in several but not all diabetic members of a single family (lod score = 0.7; data not shown). A diabetes-causing mutation was also identified in the β-pancreatic transcription factor IB1 (islet Brain 1) in this particular pedigree, and family members who inherited both IB1 S59N and IPF-1 Q59L mutations have a more severe form of diabetes compared with the inheritance of IB1 mutation only (G. Waeber et al., unpublished study), suggesting an additive deleterious effect of the IPF-1 Q59L variant.
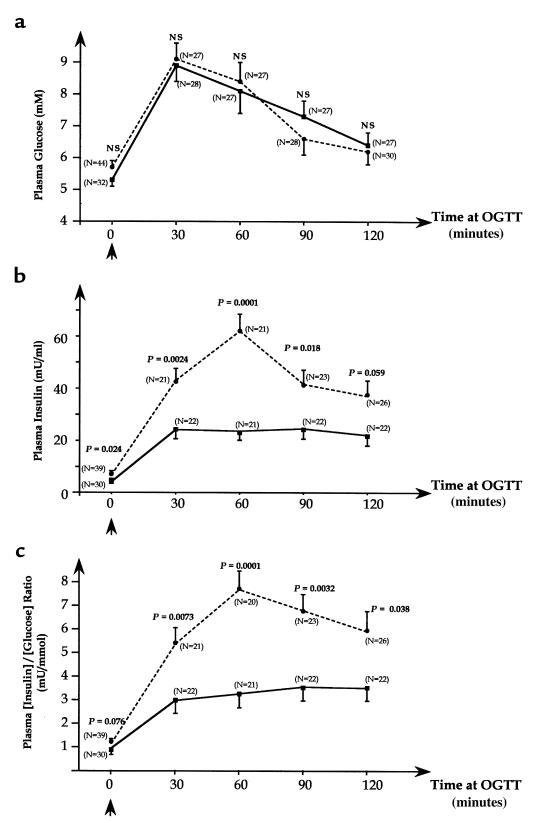
The third IPF-1 variant (D76N) was identified in 9 families in whom it did not perfectly cosegregate with diabetes. This variation was seen in several but not all overtly diabetic subjects from these 9 families (Figure 4). Transmission disequilibrium could not be tested in these pedigrees because most parents were not available. However, the genotyping of 192 unrelated diabetic individuals and 231 control subjects showed a significant association between the IPF-1 D76N variant and diabetes with a relative risk of 11.3 (4.68% in diabetic probands versus 0.43% in controls; P = 0.0041; Table 1). These findings strongly suggest that the IPF-1 D76N variant may contribute to the genetic susceptibility for type 2 diabetes although it is insufficient to cause diabetes by itself. To test this hypothesis, we analyzed insulin-secretion profiles in young normoglycemic and glucose intolerant subjects of these 9 families. Glucose levels during the OGTT were not different between carriers and noncarriers of the D76N variant (Figure 5a). However, plasma insulin levels in response to the glucose load were significantly decreased in subjects carrying the heterozygous D76N variant (0.0001 < P < 0.024; Figure 5b). Plasma insulin/glucose ratio were also significantly reduced in subjects with the IPF-1 D76N mutation compared with IPF-1 wild-type carriers during the OGTT, particularly at 60 minutes (P = 0.0001; Figure 5c). Furthermore, the first phase of the insulin-secretion response (expressed as the insulin increment at 30 or 60 minutes) was markedly decreased in subjects with the D76N mutation (0.0001 < P < 0.0031; data not shown). Because these carriers of the D76N mutation maintain normoglycemia despite lower insulin response to glucose, they should exhibit increased insulin sensitivity. Indeed, these subjects were significantly leaner than wild-type IPF-1 carriers (BMI ± SEM: 22.4 ± 0.4 vs. 25.8 ± 1.0 kg/m2; P = 0.0036) and therefore may be less insulin resistant. These observations suggest that the IPF-1 D76N may be a significant risk factor for an impairment of insulin secretion and late-onset diabetes.
Figure 4.
Segregation of the IPF-1 D76N variation in the 9 type 2 diabetic families in which it was detected. These families belong to a large collection, ascertained in France for the presence of at least 2 sibs presenting with overt type 2 diabetes, and additional family members were recruited, when available (ref. 16). Filled, half-filled, and open symbols represent subjects with overt type 2 diabetes mellitus, impaired glucose tolerance, and normal glucose tolerance, respectively. Under each symbol, the first line corresponds the actual age; the second line shows the age at diagnosis of overt type 2 diabetes (when appropriate). The third line shows individual BMI values (kg/m2), and the fourth line shows the genotype at IPF-1 codon 76 (italicized): N, Normal allele; M, mutant allele.
Figure 5.
Glucose levels and insulin-secretion profiles in young normoglycemic and glucose intolerant subjects from the 9 families carrying the IPF-1 D76N mutation. Only nonovertly diabetic subjects were included in the analysis. The maximal number glucose and insulin values for the studied subjects were included as possible, and the numbers of individual data used in the comparisons are shown above each line-point (N). Carriers of the IPF-1 D76N mutation (continuous line) compared with IPF-1 wild-type carriers (dotted line) for the mean (± SEM) of plasma glucose levels (Figure 5a), insulin levels (Figure 5b), and insulin/glucose ratios (Figure 5c) during standard oral glucose tolerance tests. For each OGTT time point (0, 30, 60, 90, and 120 minutes), the P values are shown.
Functional expression assays of the IPF-1 Q59L and D76N mutants showed a significant inhibitory effect, less than that observed with the InsCCG243 mutation (20–25% inhibition), on the function of endogenous IPF-1 to activate the insulin gene promoter in HIT-T15 cells (Figure 3a). Western immunoblot analysis showed that the Q59L mutant allele was expressed with the same efficiency as the wild-type allele (Figure 3b). In contrast, the expression of D76N was less than that of the wild-type allele (Figure 3b), indicating that this mutation may alter expression and/or reduce stability of IPF-1. When corrected for expression efficiency by Western immunoblot analyses, the D76N mutant function was considerably lower than that of the wild-type IPF-1 (Figure 3b). Our findings suggest that both IPF-1 Q59L and D76N variations, by affecting IPF-1 activity in a subtle way, can be considered as significant risk factors for the development of type 2 diabetes in a polygenic context, i.e., in the presence of other genetic variations.
Discussion
In the present study, we identified 3 novel mutations in IPF-1 associated with the common (non-MODY) form of type 2 diabetes. These mutations occurred in structurally conserved domains of IPF-1 protein, and all the 3 mutants, when expressed in vitro, showed a significant decrease in IPF-1 transactivation function. These observations not only confirm the role for IPF-1 in monogenic subtypes of type 2 diabetes (9) but provide the first evidence, to our knowledge, that IPF-1 may represent a diabetes-predisposing gene in a non-negligible proportion (∼6%) of the common forms of type 2 diabetes.
One of the IPF-1 mutations described here, the InsCCG243, is linked to an autosomal-dominant form of type 2 diabetes, with progressive insulin-secretion impairment. This insulin secretion defect observed is similar to that observed previously in prediabetic subjects carrying diabetogenic mutations in HNF-4α/MODY1 gene (17, 19). The InsCCG243 mutation nondiabetic carriers showed lower than normal insulin levels at high glucose levels (17, 19). Nevertheless, this observation requires a more extensive clinical confirmation using appropriate glycemic clamp studies in this InsCCG243 mutation young carriers in families F-13 and F-498, compared with matched controls with no familial history of diabetes. As a consequence, the young subjects carrying the IPF-1 InsCCG243 mutation are likely to be at a high risk for the development of type 2 diabetes. However, the dominant-like transmission of diabetes in families F-13 and F-498 suggests undiagnosed MODY, with several subjects being nonpenetrant. It is noteworthy that 7 members of the U.S. pedigree, with the IPF-1 Pro63fsdelC heterozygous mutation (9), are now identified with documented onset of diabetes before the age of 30, 3 of whom had glucose intolerance before the age of 25 years (J.F. Habener, personal communication). These subjects have severe impaired insulin secretion, as determined using a 5-step glucose clamp, indicating that this particular family is a defined MODY by the strictest criteria. In contrast, no family member in the French F-13 and F-498 families was diagnosed for diabetes before the age of 30, and the relatively late age at diagnosis of diabetes associated with the InsCCG243 mutation (43.5 years) is clearly suggestive of a late-onset form of type 2 diabetes rather than a MODY phenotype. The InsCCG243 mutation occurs at the COOH-terminal border of IPF-1 homeodomain for which a transactivation function has been reported previously, using truncation deletions, in the context of the somatostatin reporter (20). Furthermore, polyproline tracts in transcription factors are known to comprise essential transactivation domains and to modulate transactivation activities (21, 22). In this regard, the transactivation function of the IPF-1 InsCCG243 mutant isoform was reduced at least by half, when compared with the IPF-1 wild-type isoprotein in the β-pancreatic HIT-T15 cell line. Collectively, the present findings suggest that the IPF-1 InsCCG243 mutation likely contributes by itself to the development of glucose intolerance and type 2 diabetes.
On the other hand, we identified 2 additional IPF-1 mutations, the Q59L and D76N substitutions, associated with a less severe phenotype. Both mutations are highly associated with type 2 diabetes with an overall relative risk of 12.6 for developing diabetes, although these mutations do not strictly cosegregate with diabetes. The Q59L and D76N variations occur in the NH2-terminal region of IPF-1, close to the subdomain COOH (amino acids 60–73), that is required to synergistically activate the enhancer-mediated transcription of the insulin gene, along with another insulin gene activator(s) (23). Accordingly, both mutations showed a significant but mild effect (∼25% reduction) on transactivation potential of IPF-1 in the context of the pancreatic β-cell (HIT-T15 cell line), consistent with the lower penetrance of these mutations (i.e., absence of tight linkage with diabetes in the families). In young nonovert diabetic subjects, the D76N mutation was found highly associated with a decrease of insulin secretion in response to glucose. Therefore D76N and the Q59L mutations may be significant risk factors for an impairment of insulin-secretion and may be involved in the polygenic basis of late-onset diabetes. These genetic defects affecting β-cell function and/or in β-cell mass may participate to a progressive impairment of glucose homeostasis, when combined with other inherited metabolic abnormalities together with age and obesity-related insulin resistance, which increase the demand for insulin release.
Overall, the present study suggests that IPF-1 mutations may act distinctly in the pathogenesis of human type 2 diabetes, depending on the degree of IPF-1 functional impairment. Mutations that result in a profound alteration of IPF-1 function, such as the previously described dominant negative Pro63fsdelC mutation, cause MODY4 and, when present in a homozygous state, cause pancreatic agenesis (9, 14). The less-severe InsCCG243 mutation, which results in a 50% IPF-1 loss of function, is responsible for a late-onset form of type 2 diabetes with an apparent autosomal-dominant inheritance. It is noteworthy that the IPF-1 Pro63fsdelC mutation causes a frame shift and premature termination of translation, resulting in a reinitiation of translation and expression of a dominant inhibitor of gene transcription (15). In addition to the haplo-insufficiency (present for both mutations), the inhibition of the normal allele may account for the very early onset of diabetes. Although severely impairing IPF-1 alleles, such as the Pro63fsdelC mutation are rare in Caucasians, additional IPF-1 mutations such the D76N and Q59L substitutions seem to be more prevalent in familial type 2 diabetes (combined prevalence ∼5.2%). Such mutations may represent a significant component of the polygenic background predisposing to type 2 diabetes in humans. Nevertheless, considering the difficulties in identifying diabetes genes in humans, it remains to be determined how such “mild” IPF-1 mutations can predispose to an impairment of insulin-secretion and glucose intolerance. Notably, several HNF-1α mutations, responsible for a severe MODY3 phenotype, exhibited in vitro only a 20–30% decrease in transactivation potentials (M. Vaxillaire, personal communication), suggesting that even “modest” functional impairment of transcription factors may have in vivo deleterious effects. Therefore, we believe that the defective IPF-1 mutations described here may contribute to type 2 diabetes by promoting insulin deficiency. It is noteworthy that mice heterozygous for a null mutation in IPF-1 develop glucose intolerance and late-onset type 2 diabetes (24), whereas mice with a pancreatic β-cell–specific conditional knockout develop a MODY-like diabetes, with a profound impairment of Glut2 expression (25). Taken together, these findings strengthen the idea that intact IPF-1 activity is required to maintain normoglycemia and underscores the importance of IPF-1 along with other transcription factors that control pancreatic islet development/function in glucose homeostasis as well as in the pathogenesis of human type 2 diabetes. In this regard, IPF-1 is the first homeoprotein-encoding gene, to our knowledge, in which genetic defects are involved in a human non-monogenic disorder.
Acknowledgments
We thank the diabetic patient volunteers and their families for their participation in the study. This work is supported by the BIOMED II-program No. BMH4-CT950662. These studies were supported in part by U.S. Public Health Service grant DK 30457 (to J.F. Habener). E.H. Hani was supported by The Institut de Recherches Internationales Servier. D.A. Stoffer is a recipient of a Mentored Clinical Scientist Development Award (DK 02456). J.F. Habener is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Doris A. Stoffers’s present address is: University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania 19104, USA.
References
- 1.DeFronzo RA. Pathogenesis of type 2 diabetes: metabolic and molecular implications for identifying diabetes genes. Diabet Rev. 1997;5:177–269. [Google Scholar]
- 2.Hanis CL, et al. A genome-wide search for human non–insulin-dependent (type2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nat Genet. 1996;13:161–166. doi: 10.1038/ng0696-161. [DOI] [PubMed] [Google Scholar]
- 3.Mahtani MM, et al. Mapping of a gene for type 2 diabetes associated with an insulin secretion defect by a genome scan in Finnish families. Nat Genet. 1996;14:90–94. doi: 10.1038/ng0996-90. [DOI] [PubMed] [Google Scholar]
- 4.Pratley RE, et al. An autosomal genomic scan for loci linked to prediabetic phenotypes in Pima Indians. J Clin Invest. 1998;101:1757–1764. doi: 10.1172/JCI1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hattersley AT. Maturity-onset diabetes of the young: clinical heterogeneity explained by genetic heterogeneity. Diabet Med. 1998;15:15–24. doi: 10.1002/(SICI)1096-9136(199801)15:1<15::AID-DIA562>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 6.Froguel P, et al. Familial hyperglycemia due to mutations in glucokinase: definition of a subtype of diabetes mellitus. N Engl J Med. 1993;328:697–702. doi: 10.1056/NEJM199303113281005. [DOI] [PubMed] [Google Scholar]
- 7.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 8.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-1α gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 9.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early onset type II diabetes (MODY4) linked to IPF1. Nat Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 10.Horikawa Y, et al. Mutation in hepatocyte nuclear factor-1 gene (TCF2) associated with MODY. Nat Genet. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 11.Habener JF, Stoffers DA. A newly discovered role of transcription factors involved in pancreas development and the pathogenesis of diabetes mellitus. Proc Assoc Am Physicians. 1998;110:12–21. [PubMed] [Google Scholar]
- 12.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 13.Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 14.Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 15.Stoffers DA, Stanojevic V, Habener JF. Insulin promoter factor-1 gene mutation linked to early-onset type 2 diabetes mellitus directs expression of a dominant negative isoprotein. J Clin Invest. 1998;102:232–241. doi: 10.1172/JCI2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vionnet N, et al. Genetics of NIDDM in France: studies with 19 candidate genes in affected sib pairs. Diabetes. 1997;46:1062–1068. doi: 10.2337/diab.46.6.1062. [DOI] [PubMed] [Google Scholar]
- 17.Hani EH, et al. A missense mutation in hepatocyte nuclear factor-4 alpha, resulting in a reduced transactivation activity, in human late-onset non-insulin-dependent diabetes mellitus. J Clin Invest. 1998;101:521–526. doi: 10.1172/JCI1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevre JC, Hani EH, Stoffers DA, Habener JF, Froguel P. Insulin promoter factor 1 gene is not a major cause of maturity-onset diabetes of the young in French Caucasians. Diabetes. 1998;47:843–844. doi: 10.2337/diabetes.47.5.843. [DOI] [PubMed] [Google Scholar]
- 19.Byrne MM, et al. Altered insulin secretory responses to glucose in subjects with a mutation in the MODY1 gene on chromosome 20. Diabetes. 1995;44:699–704. doi: 10.2337/diab.44.6.699. [DOI] [PubMed] [Google Scholar]
- 20.Lu M, Miller C, Habener JF. Functional regions of the homeodomain protein IDX-1 required for transactivation of the rat somatostatin gene. Endocrinology. 1996;137:2959–2967. doi: 10.1210/endo.137.7.8770920. [DOI] [PubMed] [Google Scholar]
- 21.Mermod N, O’Neill EA, Kelly TJ, Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989;58:741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 22.Gerber HP, et al. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 23.Peshavaria M, Henderson E, Sharma A, Wright CV, Stein R. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol Cell Biol. 1997;17:3987–3996. doi: 10.1128/mcb.17.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dutta S, Bonner-Weir S, Montminy M, Wright CV. Regulatory factor linked to late-onset diabetes? Nature. 1998;392:560. doi: 10.1038/33311. [DOI] [PubMed] [Google Scholar]
- 25.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. Beta-cell–specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]