Abstract
Mothers of children with neonatal lupus erythematosus (NLE) and heart block, as well as patients with Sjögren’s syndrome (SS) and systemic lupus erythematosus, have serum autoantibodies that recognize SS-A/Ro autoantigens including the 60-kDa ribonucleoprotein. By yeast 2-hybrid screening, we identified a novel 75-kDa protein (pp75) that interacts with the carboxyl 70% of 60-kDa SS-A/Ro. The specificity of interaction was confirmed using mammalian 2-hybrid and chemical crosslinking studies. Immunoprecipitation with radiolabeled HeLa cell extracts showed that pp75 was phosphorylated and associated with 2 other phosphoproteins of 64 kDa. In Northern blot analysis, pp75 was expressed in all tissues analyzed; the highest expression was in the human heart. Based on immunofluorescence of transfected HeLa cells, pp75 is localized predominantly in the cytoplasm, an observation confirmed by immunohistochemistry in untransfected cells. Based on Western blot and ELISA assays, sera from 14 of 84 mothers of children with NLE recognized pp75, including 1 mother in whom anti–SS-A/Ro antibodies were not detected. In addition, sera from 5 of 80 patients with SS were positive for anti-pp75 antibody. Identification of molecular partners is a first step toward elucidating the functions and possible involvement in pathogenesis of long-recognized autoantigens such as 60-kDa SS-A/Ro, which are at present poorly understood.
Introduction
The presence of anti–SS-A/Ro antibodies in the sera of mothers whose children have cardiac and cutaneous manifestations of neonatal lupus erythematosus (NLE) is nearly universal (1–3). SS-A/Ro antibodies have been eluted from an affected fetal heart, suggesting that these antibodies may participate in the pathogenic event (4). Maternal antibodies are actively transported across the placenta in the early second trimester. In the fetal circulation, these antibodies may initiate myocardial inflammation that permanently damages the cardiac conduction system. Clinically, mothers of children with NLE can have systemic lupus erythematosus (SLE) or Sjögren’s syndrome (SS), but many are entirely asymptomatic and are only identified by the birth of an affected child (2). SS-A/Ro antibodies are also found in sera from patients with SLE (35%) and SS (60%) (5).
Anti–SS-A/Ro antibodies recognize both 52-kDa and 60-kDa SS-A/Ro proteins (6). The 60-kDa protein is the predominant component of a family of low-abundant ribonucleoprotein particles (RNPs) containing small cytoplasmic RNAs known as hY1-5 (7, 8). These Ro or hY RNPs are biochemically and immunologically heterogeneous (9). The cDNAs for human 60-kDa SS-A/Ro antigens have been analyzed in several laboratories (10, 11). The amino acid sequence of the 60-kDa SS-A/Ro is generally conserved among different species (12–14). It is a member of the RNA-binding protein family with RNA-binding consensus sequence in the NH2-terminal domain. Recent data indicate that the antigen exists in at least 2 forms derived from alternative splicing of mRNA: 60α and 60β. The α and β are alternative spliced products, differing in the usage of the last exon. The predominant form is 60α, which has been shown to bind hY-RNA directly in an in vitro reconstitution assay (11).
The subcellular localization of SS-A/Ro has been controversial, with different studies demonstrating the 60-kDa and 52-kDa protein in cytoplasmic or nuclear compartments (9). These discrepant observations may be secondary to different physiological conditions present in analysis, as well as to different cell types. The hY RNP complexes may dissociate in the cytoplasm and reassemble in the nucleoplasm (15). Non–hY RNA-associated 60-kDa SS-A/Ro is predominantly found in the nucleus (9). It is not clear whether dissociation of 60-kDa protein from hY RNA requires the participation of other proteins.
The cellular functions of the SS-A/Ro antigens have not been characterized. Because there is a direct interaction between 60-kDa SS-A/Ro and hY RNA using immunoprecipitation experiments, it has been postulated that the 60-kDa protein is involved in hY RNA biogenesis. The 60-kDa protein has also been proposed to function as part of a novel quality control or discard pathway for 5S rRNA precursors in Xenopus oocytes (16). However, given that 5S RNA is not known to be associated with the 60-kDa SS-A/Ro in mammalian cells, the function of the SS-A/Ro protein in mammals remains to be elucidated. Accordingly, the identification of known or novel cellular proteins that physically interact with the 60-kDa SS-A/Ro antigen was initiated to gain insight into the cellular function of SS-A/Ro and its potential role in the pathogenesis of NLE. It is possible that SS-A/Ro antigens form complexes with other cellular components that participate in triggering autoantibody formation. This study focused on the 60α, as it is the major form expressed in human cells, and used yeast and mammalian 2-hybrid systems with biochemical confirmation of protein interaction.
Methods
Yeast 2-hybrid cloning.
A human foreskin keratinocyte cDNA library constructed with the yeast 2-hybrid vector pGAD10 was purchased from CLONTECH Laboratories Inc. (catalog no. HL4034AB; Palo Alto, California, USA) and used for screening of target proteins associated with 60-kDa SS-A/Ro. A bait plasmid was constructed with the GAL4 DNA-binding domain (GAL4-DB) conjugated to amino acids 165–538 of the 60-kDa SS-A/Ro protein using the plasmid vector pBD-GAL4 Cam from Stratagene cloning system (La Jolla, California, USA). As the 60-kDa SS-A/Ro protein (538 amino acids) has a RNA-binding domain (aa60–160) in the NH2-terminal region (6, 11), the rationale for the design of the bait construct to identify a protein interaction partner was that the RNA-binding domain should be excluded to reduce background in the yeast 2-hybrid cloning system. An estimated 1.9 million cDNAs were cotransformed with the bait into the yeast strain YRG-2 and plated on triple amino acid–deficient (His/Leu/Trp-deficient) medium according to the library screening protocol provided by CLONTECH. The plasmid DNA from 3 positive yeast clones were extracted and transformed into Escherichia coli XL-1 Blue selected for the antibiotic ampicillin (50 μg/mL).
Isolation of 5′ sequence of pp75 cDNA using 5′ RACE and hybridization with WI38 library.
Because the cDNA obtained from the yeast 2-hybrid system was a partial sequence, RACE methodology was used to obtain overlapping 5′ clones from human placenta cDNAs (5′ RACE-Ready cDNAs; CLONTECH). In addition, a human lung fibroblast WI38 library constructed in the λZap II vector was purchased from Stratagene and screened with an oligonucleotide probe based on 5′ sequence of pAD cDNA insert. Two partially complementary synthetic oligonucleotides:
5′CTAAGTCGGAGCGAGTTGGGTCGCCGAAGCTCTCTGGGGG-3′
5′CCCAGGATGGGACCCCCCTTTTCCCCCAGAGAGCTTCGGC-3′
were designed (overlap underlined), synthesized using Applied Biosystems Inc. (Foster City, California, USA) model 394 synthesizer, mixed in equimolar amounts, labeled with α-[32P]dATP using Klenow polymerase fill-in reaction and purified for use in hybridization screening of the cDNA library using standard methods (17). Positive clones were identified after plaque purification and were subcloned in vivo into pBluescript plasmid using R408 helper phage.
Sequence analysis.
Nucleotide sequences were determined in both strands by using dye terminator sequencing and a Applied Biosystems semiautomated sequencer 377 and were compiled using SeqEd software (Applied Biosystems, Inc.). Both nucleotide and deduced amino acid sequences were analyzed for similarity with known sequences using BLAST search (18) and ExPASy Proteomics tools (19).
Mammalian 2-hybrid system.
The mammalian Matchmaker 2-hybrid assay kit based on the CAT reporter was obtained from CLONTECH. As in the yeast 2-hybrid system, the first mammalian vector pM encodes the GAL4-DB designed to generate a fusion protein with a “bait” polypeptide. The second mammalian vector pVP16 encodes the activation domain (AD) derived from the VP16 protein of herpes simplex virus and is designed to express a fusion protein linked to a “target” polypeptide. The 60-kDa SS-A/Ro and pp75 cDNAs were subcloned from the yeast expression constructs into mammalian expression vectors pM and pVP16 to generate pM-60 (bait) and pVP-75 (target), respectively. The efficiency of the mammalian 2-hybrid assay was improved by substituting the CAT reporter system with a more sensitive luciferase reporter construct pGAL-TK-LUX kindly provided by J. Li (Division of Oncovirology, The Scripps Research Institute). The plasmid pGAL-TK-LUX contains sequence encoding luciferase under the control of a minimal TK promoter and 5 copies of GAL4 DNA-binding site. The plasmids pM-60 and pVP-75 were cotransfected with pGAL-TK-LUX into COS-7 cells using DOTAP transfecting agent (Boehringer Mannheim Corp., Indianapolis, Indiana, USA). After overnight incubation, transfected cells were harvested and cell lysates were analyzed for luciferase activity as described previously (20) in relative light unit (RLU) using the luminometer Monolight 2010 by Analytical Luminescence Laboratory (San Diego, California, USA). Control plasmids from CLONTECH were included separately or combined as appropriate. Positive controls include pM3-VP16 encoding fusion protein of GAL4-DB and VP16 AD, pM-53 encoding GAL4-DB fused to p53, and pVP-T encoding VP16 AD fused to the SV40 large T antigen. Negative controls include empty vector pM and pVP16 and pVP-CP encoding a viral coat protein.
Chemical crosslinking of SS-A/Ro and pp75.
HeLa cells were grown as monolayer in DMEM culture medium containing 10% FCS, detached from culture flask using trypsin/EDTA (GIBCO BRL, Grand Island, New York, USA), washed by serum-free DMEM 2 times and once in a buffer containing 20 mM HEPES (pH 7.2), 10 mM MgCl2, and 250 mM sucrose. Cells were counted, aliquoted in 6.25 × 105 each, and transferred to 1.5-mL microcentrifuge tubes. After centrifugation, cell pellets were resuspended in 50 μL 1X lysis buffer (150 mM NaCl, 30 mM Tris-HCl [pH 7.5], 1% Nonidet P-40, and 1.2% glycerol containing protease inhibitor cocktail without EDTA [Boehringer Mannheim Corp.]). Cell lysates corresponding to 6.25 × 105 cells were used in crosslinking reactions using 1, 3, or 10 μL fresh 25 mM DTSSP stock (3,3′-dithiobis[sulfosuccinimidylpropionate]; Pierce Chemical Co., Rockford, Illinois, USA). Crosslinking reactions were performed on ice for 2 hours before quenching by the addition of 2.5 μL 1 M Tris-HCl (pH 7.5). Crosslinked cell lysates were initially analyzed by immunoblot and subsequently as substrate in immunoprecipitation using human anti–SS-A/Ro and followed by immunoblotting analysis of the immunoprecipitates using rabbit anti-pp75 antibody (see later here).
In vitro translation product and recombinant protein.
cDNAs subcloned into pBluescript (Stratagene) or pET28 (Novagen, Madison, Wisconsin, USA) plasmids were used as DNA templates for in vitro translation with TnT-coupled reticulocyte lysate systems (Promega Corp., Madison, Wisconsin, USA) in the presence of [35S]methionine. The translation products were analyzed by 15% SDS-polyacrylamide gel, viewed after autoradiography and used as antigen substrate in immunoprecipitation as described previously (21, 22).
Cell labeling and immunoprecipitation.
HeLa cells were cultured and radiolabeled overnight with [35S]methionine or [32P]orthophosphate as described previously (23). Cell extracts were prepared using either buffer A (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1.5 mM MgCl2, 0.5% Nonidet P-40) or buffer B containing multiple phosphatase inhibitors (30 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 1.2% glycerol, 1 mM sodium vanadate, 2.5 mM sodium pyrophosphate, 50 mM NaF, 1 mM DTT) containing Complete protease inhibitor cocktail (Boehringer Mannheim, Corp.). A standard immunoprecipitation reaction contained 100 μL 10% protein A-Sepharose, 500 μL buffer NET2 (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS, 0.02% sodium azide), 10 μL 10 mg/mL BSA, 40 μL labeled cell extract, 5–15 μL serum and protease inhibitors. After incubation for 1 hour, beads were washed 5 times in NET2, eluted in an equal volume of Laemmli sample buffer (24), eluate run on a 12.5% polyacrylamide gel, and exposed to x-ray film at –70°C.
Northern blot analysis of mRNA expression.
A multiple tissue Northern blot with 2 μg poly-A selected mRNA on each lane was purchased from CLONTECH. A [32P]UTP1-labeled antisense RNA probe of 231 bases was transcribed from BamHI linearized DNA template and purified using 6% polyacrylamide gel in 7M urea (17). The blot was preincubated in hybridization buffer ExpressHyb (CLONTECH) at 68°C for 30 minutes followed by the addition of RNA probe at 1 × 106 cpm/mL for 60 minutes. The blot was washed and exposed to x-ray film at –70°C. To confirm the accuracy of samples loaded, the same blot was later stripped and reprobed with an actin probe according to the manufacturer’s instructions.
RT-PCR analysis of mRNA expression.
Second to fourth passages of human keratinocytes from neonatal foreskins were obtained from Clonetics (San Diego, California, USA) and cultured as described previously (25). Other human cell lines were obtained for analysis from the American Tissue Culture Collection (Rockville, Maryland, USA), including acute lymphoblastic leukemia MOLT-4, lung carcinoma SK-LU-1 and Calu-3, larynx epidermoid carcinoma HEp-2, breast cancer T47D, bladder carcinoma T24, and cervix epitheloid carcinoma HeLa. For cell lines grown as monolayers, cells were harvested from culture flasks using trypsin, and total cellular RNA was purified using UltrospecRNA isolation reagents (Biotecx, Houston, Texas, USA). Specific mRNA was analyzed using a simplified RT-PCR method described previously (26). Total RNA (0.2 μg) and 0.5 μM primers (0.5 μL each) were heated to 70°C for 10 minutes and then quickly chilled on ice. Remaining components including 1.25U Taq polymerase (GIBCO BRL), 100 U SuperScript II RNase H-RT (GIBCO BRL), 20 U RNase inhibitor (Promega Corp.), 0.25 μL 10 mM dNTPs, and 2.5 μL 10X PCR buffer containing 500 mM KCl, 100 mM Tris-HCl (pH 8.3), 15 mM MgCl2, and 0.1% gelatin were added to a final total volume of 25 μL. For the RT-PCR detection of pp75 mRNA, the sense primer 5′-CTGGGAGCATTCAGCCTGTGA-3′ and antisense primer 5′-ACATTGGAGGTGGTCAAGAG-3′ were designed based on sequence described in Figure 1 with expected PCR products of 1,041 bp. All reaction components were mixed in a single 0.5-mL microcentrifuge tube before thermal cycling. The RT-PCR program consisted of a RT step (50°C for 1 hour) and a denaturing step (94°C for 3 minutes), followed by 30 cycles of PCR (94°C for 5 seconds, 55°C for 5 seconds, and 72°C for 1 minute). RT-PCR products were analyzed by agarose gel electrophoresis. The mRNA of the housekeeping gene coding for GAPDH was analyzed as an internal control (10 ng total RNA per reaction).
Figure 1.
Cloning of human 60-kDa SS-A/Ro–associated protein pp75. (a) Schematic representation of cDNAs pINP1 derived from yeast 2-hybrid cloning, 5′ RACE, WI38 cDNA library screen, and clone KIAA0857. The shaded boxes represent the open reading frame (ORF). (b) Nucleotide and deduced amino acid sequence of pp75/KIAA0857. Nucleotide sequence of 3,017 bases for the WI38 cDNA was submitted to GenBank under accession number AF153085. The nucleotide sequences corresponding to the ORF between sequence submitted under AF153085 and clone KIAA0857 are identical; 4 single base substitutions and insertion/deletion are observed in the 3′ untranslated regions. The methionine start site (Kozak’s sequence CCGCCATGG) and poly-A signal AATAAA are underlined. The GC-rich 5′-untranslated region is highlighted by a thick underline of 5 or more consecutive GC residues. The protein has relatively high percentage of serine residues (13.94%; boldface and italics). A leucine zipper motif is double-underlined, and the 4 leucine residues are circled. RT-PCR primers and antisense RNA probe for the analysis of mRNA expression in different cells and tissues are indicated by arrows in both panels. The BamHI site used to generate antisense RNA probe is boxed in b.
Expression of recombinant pp75 in E. coli and production of rabbit anti-pp75 antibody.
The cDNA in original pINP1 (aa465–653) and pWI38 (aa105–653) were subcloned into the expression vector pET28 (Novagen) and transformed into the E. coli BL-21 (DE3) for recombinant protein production. The pET28 vector produces an NH2-terminal 6x Histidine fusion tag to allow efficient purification of recombinant proteins using Ni2+ affinity chromatography (QIAGEN Inc., Valencia, California, USA). Two New Zealand White female rabbits R2808 and R2809 were each immunized by subcutaneous injections of 1 mg purified recombinant pp75 (aa465–653) in an equal volume of CFA (Difco Laboratories, Detroit, Michigan, USA). After 30 days, the animals were boosted with 1 mg of the same recombinant protein in Freund’s incomplete adjuvant (Difco Laboratories). Antisera were collected 10 days later and assayed for specific antibody. Before the immunization, preimmune sera were collected and used as negative controls. Activities of antisera were monitored by Western blot and immunofluorescence assay.
Western blot.
Cell extracts or recombinant proteins were separated by 15% polyacrylamide gel followed by electrotransfer to nitrocellulose paper (27). Human or rabbit sera diluted 1:100–1,000 were incubated with the nitrocellulose for 1 hour. After buffer washes, the nitrocellulose was probed for 1 hour with horseradish peroxidase–conjugated goat anti-human IgG or goat anti-rabbit IgG diluted 1:2,000–5,000. Reactivities were detected by the chemiluminescence method using ECL reagents (Amersham Life Sciences, Buckinghamshire, United Kingdom) as described previously (27).
Anti-pp75 ELISA.
Standard ELISA was carried out according to established method described previously (28). Briefly, an Immulon 2 high binding microtiter plate (Dynex Technologies, Inc., Chantilly, Virginia, USA) was coated with Ni-column affinity purified recombinant protein (1 μg/mL) diluted in PBS. Human sera were obtained from the Research Registry for Neonatal Lupus (29) and serum bank at the W.M. Keck Autoimmune Disease Center. Sera diluted 1:200 were incubated with antigen, and bound antibodies were detected with horseradish peroxidase–conjugated goat anti-human IgG (Caltag Laboratories, South San Francisco, California, USA) and the substrate 2,2′-azinobis(3-ethylbenzthiazoline sulfonic acid) (Boehringer Mannheim Corp.).
Transient transfection of recombinant pp75.
The coding region of pp75 cDNA was subcloned into pCMV-T7Tag plasmid vector constructed such that the expressed protein is in frame with the upstream 11-amino acids-epitope tag (T7Tag, Ala-Ser-Met-Thr-Gly-Gly-Gln-Gln-Met-Gly-Arg) and transcription is driven by the upstream human cytomegalovirus (CMV) enhancer, promoter, and intron elements. The cDNA was transiently transfected into HeLa cells seeded on 8-chamber slides (Nalge Nunc International Corp., Naperville, Illinois, USA) using DOTAP transfecting agent (Boehringer Mannheim Corp.) for 20 hours. Cells were fixed in ice-cold methanol-acetone (1:3 volume) for 2 minutes, stained with murine mAb to T7Tag (Novagen; 1:1,000) and FITC-conjugated goat anti-mouse IgG. The subcellular location of expressed pp75 was observed with fluorescence microscopy.
Results
Identification of 60-kDa SS-A/Ro interaction protein partner using yeast 2-hybrid cloning.
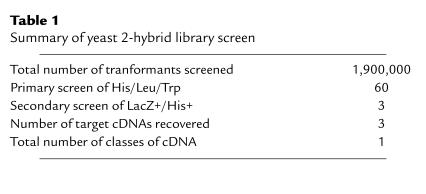
The rationale for selection of a keratinocyte cDNA library as the initial target in which to identify potential protein-protein interaction partners of the 60-kDa SS-A/Ro protein was based on the observation that anti–SS-A/Ro antibodies are highly associated with 2 autoimmune diseases, subacute cutaneous lupus and neonatal lupus, in which skin rashes are major symptom manifestations. Table 1 summarizes the number of clones in each step of the screening protocol. Approximately 1,900,000 cDNA clones were cotransformed with the 60-kDa protein bait construct into the yeast strain YRG-2 plated on triple amino acid–deficient (His/Leu/Trp-deficient) medium. There were 60 viable colonies detected up to 10 days after initial plating. When the 60 clones were analyzed for the production of β-galactosidase (LacZ gene product) by incubation with X-gal substrate, only 3 clones gave the blue color reaction. Plasmid DNA were extracted from these 3 yeast clones and transformed into E. coli XL1-Blue using ampicillin selection to recover only target cDNA clones, as the pBD-GAL4 CAM bait plasmid does not harbor resistance to ampicillin. The recovered plasmid (Interaction protein 1 [pINP1]) in all 3 positive clones contained a common cDNA insert of 2.3 kb, and sequence analysis showed that their inserts were identical. The interaction between the 60-kDa SS-A/Ro bait and candidate target plasmid pINP1 in the yeast system was further confirmed by retransforming the bait and pINP1 plasmid into the same yeast strain and showing their ability to express β-galactosidase. The observed protein-protein interaction was specific, as pINP1 did not interact with the pAD-GAL4 CAM plasmid vector alone or with bait plasmid encoding the 52-kDa SS-A/Ro protein in this yeast 2-hybrid assay.
Table 1.
Summary of yeast 2-hybrid library screen
pINP1 encodes a partial novel cDNA.
The nucleotide sequence for pINP1 was determined, and a deduced open reading frame was identified (Figure 1a). Sequence analysis using BLAST search showed that pINP1 cDNA insert was novel and unreported at the time (see later here). The pINP1 cDNA was subcloned into the pET28 vector for production of recombinant protein in E. coli and used in an in vitro translation system. Both E. coli and the in vitro translation system generated a recombinant polypeptide of ∼29 kDa detected in the SDS-PAGE gel. Rabbit antibodies produced using the 29-kDa recombinant protein as immunogen were shown to recognize a 75-kDa protein in various cell extracts (see later here). These data suggest that the pINP1 was a partial cDNA missing 5′ sequence.
Full-length pp75.
To characterize further the structure and function of this novel 75-kDa protein, we obtained 5′ upstream cDNA using both 5′ RACE methodology and library hybridization screening. Initial attempts using the RACE method were of limited success. The longest cDNA produced only 145 bp extra 5′ sequence compared with pINP1 (Figure 1). A substantially longer cDNA (pWI38) was obtained by screening the WI38 λZAPII cDNA library using 2 partially complementary synthetic oligonucleotides designed based on 5′ sequence of pINP1 cDNA insert. Figure 1a summarizes 3 types of cDNA obtained from yeast 2-hybrid, 5′ RACE, and hybridization cloning. Sequence analysis of pWI38 showed complete identity with original yeast 2-hybrid– and 5′ RACE–derived cDNAs and thus provided an independent verification of the previous clones. Recently, the nucleotide sequence of pWI38 was shown to be essentially identical to that reported for a human brain cDNA clone KIAA0857 (GenBank accession AB020664; ref. 30), which is longer at both ends (Figure 1). KIAA0857 was uncharacterized but estimated to encode a protein of greater than 50 kDa (30). The complete nucleotide and deduced amino acid sequences are shown in Figure 1b. There is now evidence to show that KIAA0857 encodes the full-length cDNA for pp75. First, the first ATG was found at nucleotide position 242 from the 5′ end and is in excellent agreement with Kozak’s consensus sequence (CCGCCATGG) for a methionine initiation codon (31). Second, the upstream sequence of the putative methionine start codon is consistent with 5′-untranslated regions, which are often populated with GC-rich domains; 197 of 241 nucleotides in 5′-untranslated region is GC (81.7% GC content; see Figure 1b, thick underline). Third, the deduced open reading frame contains 653 amino acids with a calculated molecular mass of 70 kDa is in close agreement with the observed electrophoretic mobility of pp75 (see later here).
The putative full-length pp75 protein has a relatively high percentage of serine residues (13.94%) consistent with the observation that it is a phosphorylated polypeptide (see later here). Analysis of the predicted polypeptide showed that there are putative cAMP- and cGMP-dependent protein kinase (n = 3), protein kinase C (n = 8), and casine kinase II phosphorylation sites (n = 14), although there are currently no experimental data to support that pp75 is regulated by these kinases. The protein deduced from the cDNA was designated as 60-kDa SS-A/Ro–associated protein pp75 because a 75-kDa protein in HeLa cell lysate was detected by rabbit antiserum against recombinant novel protein using Western blot (see later here).
pp75 complexed with pp64 doublet.
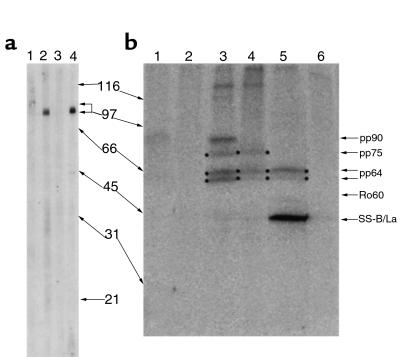
The original cDNA from pINP1 (BglII fragment) was subcloned into the BamHI site of E. coli expression plasmid pET28c to generate the construct pET-INP1. Recombinant protein expressed in BL21 (DE3) bacteria migrated at 29 kDa and was purified by nickel affinity column before immunization of rabbits. Western blot analysis using sera from both rabbit R2808 and R2809 showed that the major band detected in HeLa cell extract was 75 kDa (Figure 2a). In immunoprecipitation assay using extracts from HeLa cells metabolically labeled with [32P]phosphate, postimmune sera from both rabbits gave the 75-kDa protein band and 2 additional bands, a doublet at 64 kDa (pp64; Figure 2b). As hY RNAs are associated with the 60-kDa SS-A/Ro protein, pp75 does not appear to be a stable component of the 60-kDa SS-A/Ro ribonucleoprotein complex because [32P]-labeled hY RNAs were not detected in immunoprecipitation using rabbit anti-pp75 (data not shown). The pp64 bands did not represent RNA species, as RNase digestion of the immunoprecipitate did not affect these bands, whereas RNA species associated with SS-A/Ro and SS-B/La were removed by the same RNase treatment. In sum, these data suggested that pp75 is complexed with pp64 doublet, as pp64 is not directly recognized by the rabbit anti-pp75 antisera in Western blot. Interestingly, 1 of the prototype Sjögren’s syndrome sera used in the laboratory immunoprecipitated pp64 doublet in addition to SS-B/La in repeated experiments (Figure 2b, lane 5), providing evidence that pp64 is also an autoantigen in SS. However, none of the other prototype SS-A/Ro sera analyzed showed reactivity to pp64, although extensive screening was not performed. The association of pp64 doublet and pp75 is not further explored in the present study but remains an area of active investigation in our laboratory.
Figure 2.
Western blot and immunoprecipitation of pp75. (a) Western blot analysis of whole-cell lysate of HeLa cells using preimmune (lanes 1 and 3) and antisera (lanes 2 and 4) from rabbit R2808 (lanes 1 and 2) and R2809 (lanes 3 and 4). Both rabbit antisera recognized predominantly a single protein of 75 kDa (arrow). (b) Immunoprecipitation of cell extract from [32P]phosphate-labeled HeLa cells using preimmune rabbit sera (lanes 1 and 2), rabbit anti-pp75 from R2809 (lane 3) and R2808 (lane 4), SS serum Cas with antibody to SS-A/Ro and SS-B/La (lane 5), and normal human serum control (lane 6). The 60-kDa SS-A/Ro (Ro60) is poorly phosphorylated and is not detected in this assay. In repeated experiments, both rabbit antisera recognized phosphoprotein of 75 kDa (pp75) and a doublet of 64 kDa (pp64), and serum from R2809 also recognized a 90-kDa phosphoprotein (pp90), whereas the higher molecular weight bands seen in lanes 3 and 4 are not consistently observed.
Interaction between 60-kDa SS-A/Ro and pp75 protein in mammalian system.
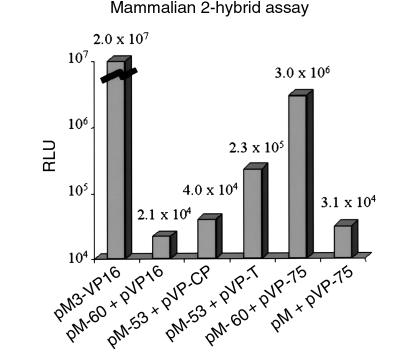
To substantiate further the biologic “authenticity” of the protein-protein interaction detected in the yeast system, the 60-kDa SS-A/Ro bait and the candidate target pp75 cDNA were subcloned in frame into eukaryotic plasmids pM and pVP16, respectively. Figure 3 shows the results when these plasmids plus controls were cotransfected into COS-7 cells. The bait plasmid containing 60-kDa SS-A/Ro (pM-60) plus empty target vector pVP16 gave a background luciferase signal of 2.1 × 104 RLU (pM-60 + pVP16). For simplicity, control individual bait or target plasmid cotransfected with reporter pGAL-TK-LUX gave the same level of background reading and is not shown in Figure 3. Other negative controls shown in Figure 2 include the p53 protein as bait and unrelated control target (pM-53 + pVP-CP: 4 × 104 RLU) and the empty bait vector and candidate target pp75 (pM + pVP-75: 3.1 × 104 RLU). The positive control pM3-VP16 consisting of both the GAL4 DNA-binding and VP16 activation domain fusion protein gave 2.0 × 107 RLU representing a 1,000-fold higher signal than negative controls. The nonfusion positive control representing the interaction of p53 and SV40 large T antigen separately gave a 10-fold higher signal (pM-53 + pVP-T: 2.3 × 105 RLU) than negative controls. Interestingly, the interaction of 60-kDa SS-A/Ro and pp75 gave a 100-fold higher signal than the negative controls and a 13-fold higher signal than the positive control interaction of p53 and SV40 large T (pM-60 + pVP-75: 3 × 106 RLU). Accordingly, the protein-protein interaction of 60-kDa SS-A/Ro and pp75 detected in yeast 2-hybrid cloning could be reproducibly confirmed in the mammalian system.
Figure 3.
Two-hybrid assay confirmed interaction of 60-kDa SS-A/Ro and pp75 in COS-7 cells. The 60-kDa SS-A/Ro and pp75 cDNAs were subcloned into the mammalian expression vector pM and pVP16, respectively, as described in Methods. Bait and target plasmids in different combination with positive and negative controls were cotransfected together with a third reporter plasmid pGAL-TK-LUX into COS-7 cells. After overnight incubation, transfected cells were harvested and cell lysates were analyzed for luciferase activity and expressed as RLU. The interaction between 60-kDa SS-A/Ro and pp75 (pM-60 + pVP-75, 3.0 × 106 RLU) was 13-fold higher than the positive control interaction between p53 and SV40 large T antigen (pM-53 + pVP-T, 2.3 × 105 RLU) and approximately 100-fold higher than background signals ranging from 2.1 × 104 to 4 × 104 RLU from different negative controls. pM3-VP16, bait-target fusion protein control. pM-60, 60 kDa SS-A/Ro bait. pVP16, empty target. pM-53, p53 bait. pVP-CP, viral coat protein negative control target. pVP-T, SV40 large T antigen target. pVP-75, pp75 target. pM, empty bait.
Crosslinking of pp75 and 60-kDa SS-A/Ro.
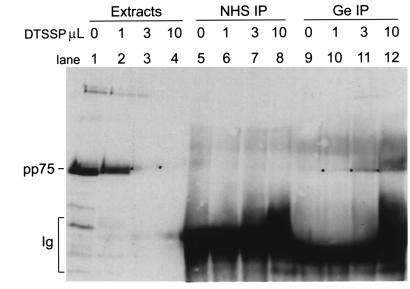
To rule out potential artifact associated with the 2-hybrid system, independent evidence for the interaction of pp75 and 60-kDa SS-A/Ro was sought. It was postulated that the interaction of 60-kDa SS-A/Ro with pp75 is transient but can be stabilized with a chemical crosslinking reaction. DTSSP was chosen as the crosslinking agent because it is a water-soluble compound and the crosslinked product can be cleaved in the presence of β-mercaptoethanol in denaturing SDS-PAGE. Figure 4 shows representative crosslinking results using different amount of DTSSP and resultant effects on antigenicity of pp75 and its association with SS-A/Ro. The rationale for using 3 increasing concentrations of DTSSP was based on preliminary titration experiments of the crosslinking agent showing that excessive crosslinking destroyed epitopes recognized by rabbit anti-pp75 antibodies. This is demonstrated in Figure 4, lanes 1–4, showing that the reactivity of rabbit anti-pp75 was significantly reduced when 3 or 10 μL DTSSP was added (Figure 4, lanes 3 and 4). When the crosslinked extracts were immunoprecipitated with either a normal human serum or the prototype anti–SS-A/Ro serum Ge (21, 32), and the immunoprecipitates were analyzed for the presence of pp75 using the rabbit antibody in Western blot, serum Ge coprecipitated pp75 when DTSSP was added (Figure 4, lanes 10–12). In contrast, there was no reactivity with normal human serum (Figure 4, lanes 6–8) or with serum Ge in the absence of crosslinking (Figure 4, lane 9). Ge antibody did recognize 60-kDa SS-A/Ro and appeared to require high concentration of the crosslinker DTSSP to coimmunoprecipitate pp75.
Figure 4.
Chemical crosslinking of SS-A/Ro and pp75. Fresh HeLa cell lysates were prepared and crosslinked with DTSSP as described in Methods using 0, 1, 3, or 10 μL crosslinking agent. Western blotting analysis using rabbit anti-pp75 antibody was performed on the crosslinked lysate alone (extracts, lanes 1–4) or immunoprecipitates by a normal human serum (NHS IP, lanes 5–8) or prototype anti–SS-A/Ro serum Ge (Ge IP, lanes 9–12). Reactivity to human Ig heavy chain primarily resulted from the cross-reaction of goat anti-rabbit Ig secondary reagent.
Expression of cellular pp75.
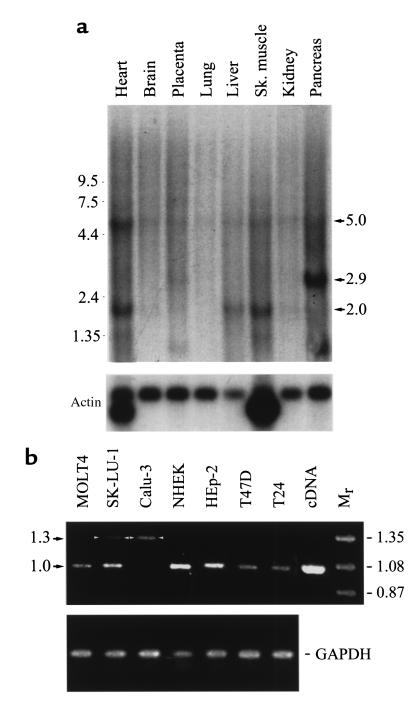
The expression of pp75 was examined in several normal human tissues and cell lines known to express 60-kDa SS-A/Ro. Figure 5a shows the results of Northern blot for the detection of pp75 mRNA using an antisense RNA probe. An approximately 5.0-kb band was detected in all tissues with the highest expression noted in the heart. Additional low–molecular weight bands of 2.9 and 2.0 kb were also observed in some but not all tissues. The strong 2.9-kb species observed in the pancreas and 2.0-kb RNA species in heart and skeletal muscle may represent alternatively spliced products, but this has not been further investigated. These bands were not detected when the same blot was stripped and reprobed for β-actin mRNA.
Figure 5.
Expression of pp75 cellular RNA. (a) Northern blot analysis of pp75 mRNA in normal human tissues. Multiple tissue blot was hybridized with antisense RNA probe for pp75 (top panel) and actin (bottom panel) as described in Methods. (b) RT-PCR analysis of human pp75 mRNA in selected human cell lines and primary keratinocytes (NHEK). Control for pp75 cDNA and molecular weight markers (Mr) are shown on the right. Total RNA samples of 10 ng each were analyzed for internal control GAPDH (bottom panel).
Figure 5b shows the results of RT-PCR analysis for pp75 mRNA in all cell lines examined including lung carcinoma SK-LU-1 and Calu-3, normal human cultured keratinocytes (NHEK), larynx epidermoid carcinoma HEp-2, breast cancer T47D, bladder carcinoma T24, and cervix epitheloid carcinoma HeLa (data not shown). All cell lines gave the expected 1-kb band alone, except Calu-3, which gave primarily a 1.3-kb band, and SK-LU-1, which gave both 1-kb and 1.3-kb bands; the 1.3-kb species may also represent alternatively spliced mRNA for pp75 in these cell types. Using the same RT-PCR assay, the 1-kb RT-PCR product of pp75 was detected in mRNA from human brain, kidney, liver, pancreas, and testis (data not shown). In short, the expression of pp75 appears to be ubiquitous and is similar to the expression observed for 60-kDa SS-A/Ro.
pp75 localized predominantly to the cytoplasm.
The subcellular localization of pp75 in HeLa and HEp-2 cells was examined by indirect immunofluorescence microscopy using rabbit anti-pp75 antisera at 1:50 or 1:100 dilution. Cytoplasmic staining were observed in both cell types compared with control preimmune sera. To obtain an independent determination for the subcellular localization of pp75, the open reading frame of pp75 was subcloned and fused to an NH2-terminal epitope T7Tag in a eukaryotic expression plasmid and transiently transfected into HeLa cells. The recombinant pp75 in transfected cells was detected using mouse anti-T7Tag mAb and immunofluorescence microscopy. Primarily, cytoplasmic staining was observed in all transfected cells, although there was great variation in the level of expression (Figure 6).
Figure 6.
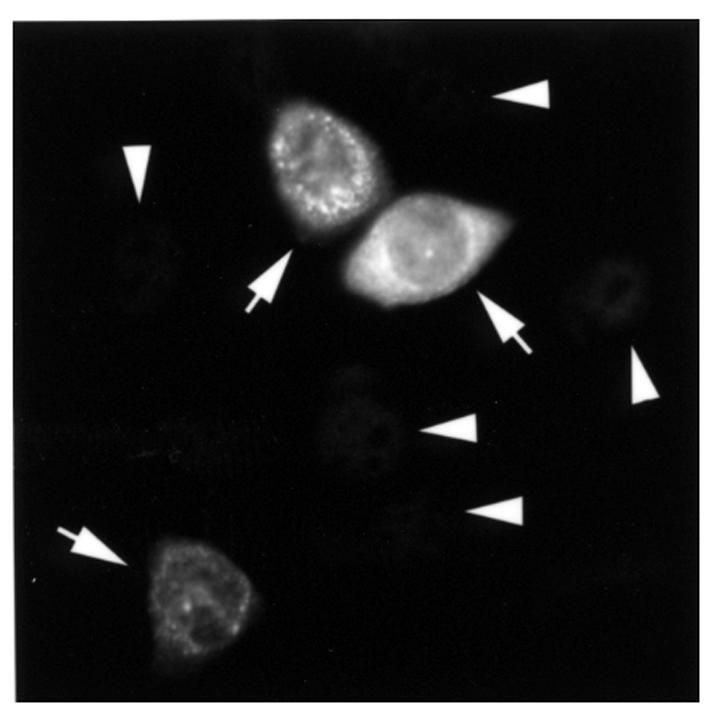
Transient transfection of pp75 cDNA showed cytoplasmic expression in HeLa cells. A CMV promoter–driven expression construct for the epitope-tagged pp75 protein was transfected into HeLa cells and observed after overnight incubation. Recombinant protein was identified predominantly in the cytoplasm of transfected cells (arrows) with weak to strong staining, whereas untransfected cells (arrowheads) gave background nuclear staining from the T7tag mAb.
Autoantibody to pp75 in sera from the mothers of children with NLE and SS.
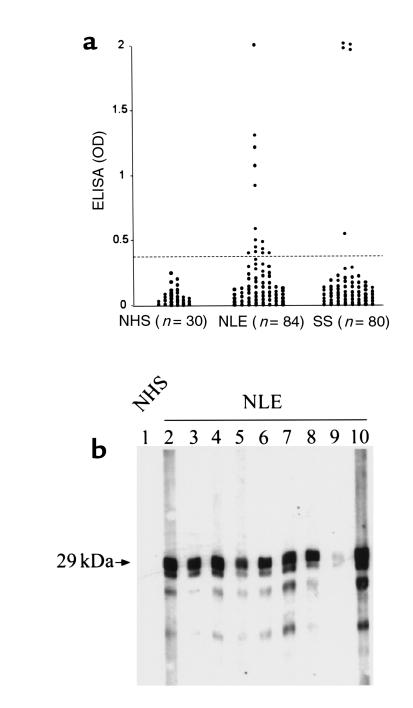
Because autoantibodies are often detected to proteins that form macromolecular complexes (33), recombinant protein of pp75 (aa105–653) was used as substrate to establish an ELISA to determine the frequency of autoantibodies to pp75 in sera from mothers of children with NLE. Fourteen (16.67%) of 84 sera from the mothers of children with NLE, and 5 (6.25%) of 80 sera from patients with SS, had detectable autoantibodies greater than 3 SD above the mean for 30 sera from normals (Figure 7a). The positive sera were confirmed by Western blot using recombinant pp75 as substrate (Figure 7b) and an immunoprecipitation assay using in vitro translation product (data not shown). No reactivities were observed with normal human sera in Western blot and the immunoprecipitation assay.
Figure 7.
Human autoantibody to recombinant pp75 in sera from mothers of children with NLE and from patients with SS. (a) Eighty-four sera from mothers of children with NLE, 80 sera from patients with SS, and 30 normal human sera were analyzed for autoantibody to recombinant pp75 (aa105–653) using ELISA format. Circles show a representative set of data for each of the samples, and the dash line is the cut-off calculated as mean ± 3 SD. At least 3 repeats were performed for the ELISA. (b) Representative Western blot showing reactivity of 9 ELISA positive among the NLE sera to recombinant COOH-terminal 29-kDa fragment of pp75 (aa465–653). OD, optical density.
Of the 14 seropositive mothers, 4 are asymptomatic, 4 were diagnosed with SLE, 2 have SS, 2 have SLE/SS, and 2 have an undifferentiated autoimmune disease. Thirteen mothers had children with cardiac manifestations of NLE alone, and 1, with cutaneous disease alone; however, one of the mothers whose serum was clearly reactive with pp75 did not have anti–SS-A/Ro or SS-B/La antibodies by ELISA or Western blot.
Discussion
Among autoantibody-autoantigen systems described in systemic rheumatic disease, autoantibodies to SS-A/Ro are exceptional in that they are strongly associated with NLE, subacute cutaneous lupus, and SS (32, 34). Although proof of pathogenicity is not definitive, accumulating evidence suggests that anti–SS-A/Ro antibodies contribute to the cardiac and tissue damage in NLE (4, 35–37). A molecular definition of the SS-A/Ro autoantigens has been provided by the cloning of cDNAs, but little is known about their biologic function other than binding to hY RNAs or their role in disease pathogenesis and tissue injury. The initial approach to define further the 60-kDa SS-A/Ro used yeast 2-hybrid cloning with the goal of identifying interaction proteins. A strong interaction between the 60-kDa SS-A/Ro protein and a novel 75-kDa phosphoprotein was identified. In vitro and in vivo association between these 2 proteins was confirmed using the mammalian 2-hybrid system and chemical crosslinking experiments.
The newly identified pp75 is a novel protein showing no significant homology with any well-defined protein in GenBank. However, in the Expressed Sequence Tag database, there are several other unidentified human proteins with sequence similarity (range, 25–50%) to pp75 primarily in the NH2-terminal domain of approximately 100 amino acid residues. Some of these sequences clearly represent partial cDNAs or genomic fragments. One hypothesis is that these related proteins are identical to the pp64 proteins detected to be associated with pp75 in the immunoprecipitation assay. The pp64 proteins were clearly recognized by 1 of the prototype SS-A/Ro/SS-B/La sera, although pp75 was not coprecipitated. The 2 rabbits R2808 and R2809 immunized with the COOH-terminal domain of pp75, which does not shared sequence similarity with the related proteins described here, recognized only pp75 from a whole-cell extract in Western blot.
Sequence analysis of pp75 indicated that it contains many serine residues, some of which may be phosphorylated as supported by immunoprecipitation using extracts of [32P]-labeled HeLa cells. Given that the 60-kDa SS-A/Ro is a weakly phosphorylated protein (ref. 38; D. Wang and E.K.L. Chan, unpublished study), the interaction of pp75 and 60-kDa SS-A/Ro may share a common pathway regulated by protein phosphorylation. The subcellular localization of pp75 is predominantly cytoplasmic, suggesting that the interaction of pp75 and 60-kDa SS/Ro is also restricted to this compartment as originally described for the hY RNPs (7, 8). However, the hY RNAs from HeLa cells were not immunoprecipitated using rabbit anti-pp75 antibodies, indicating that pp75 is not an integral component of the hY RNP complex. Both pp75 and 60-kDa SS-A/Ro are expressed in many cell types and tissues, suggesting that their putative association is not restricted to specific cell types and tissues. As the scan for tissue expression of pp75 showed the highest level in human heart, further work is needed to quantitate carefully the levels of pp75 and the 60-kDa SS-A/Ro in larger number of samples.
Autoantibodies to pp75 in the sera from SS-A/Ro–positive mothers of children with NLE may be yet another example of intermolecular spreading that has been demonstrated by several investigators for the SS-A/Ro-SS-B/La system (39–41). Secondary immune responses are probably explained by the capacity of B cells to process and present a wide array of T-cell epitopes. In the present study, 1 human serum contained antibodies to pp75 in the absence of detectable reactivity to 60-kDa SS-A/Ro, suggesting that pp75 alone is antigenic.
The physical association of pp75 and 60-kDa SS-A/Ro is also reflected in the clinical spectrum of the patients who have anti-pp75 antibodies. With the exception of 1 positive serum from an asymptomatic mother whose child has congenital heart block, all positive pp75 sera contain antibodies to 60-kDa SS-A/Ro. Therefore, it was not surprising to find that anti-pp75 antibodies are present in mothers with both cutaneous and cardiac manifestations of NLE. In addition, using sera from the patients with SS to screen antibody against pp75, we found that 5 of 80 sera also have the autoantibody activities.
The involvement of pp75 in the pathogenesis of NLE is yet undefined, but 2 requisite conditions have been met: approximately 25% of the sera from mothers of affected children do contain these antibodies; and Northern blot analysis indicates that human heart is a tissue with the highest expression of pp75. The placental transport of maternal anti-pp75 antibodies may target this vulnerable tissue and initiate a specific interaction to cause damage.
Acknowledgments
This is publication 11932-MEM from The Scripps Research Institute. We thank M. Lu for providing the pCMV-T7tag plasmid, and J. Li for the GAL4 regulated luciferase reporter plasmid pGAL-TK-LUX. D. Wang is a recipient of an Arthritis Foundation Fellowship Award. This work was supported by grants from the National Institutes of Health (NIH; AR-41803 and AR-42455) and by contract AR-42220 for the Research Registry for Neonatal Lupus. This work was also supported in part by the Sam and Rose Stein Charitable Trust and NIH grant M01RR00833 provided to the General Clinical Research Center of the Scripps Research Institute.
References
- 1.Buyon JP, et al. Identification of mothers at risk for congenital heart block and other neonatal lupus syndromes in their children. Comparison of enzyme-linked immunosorbent assay and immunoblot for measurement of anti-SS-A/Ro and anti–SS-B/La antibodies. Arthritis Rheum. 1993;36:1263–1273. doi: 10.1002/art.1780360911. [DOI] [PubMed] [Google Scholar]
- 2.Buyon JP. Neonatal lupus. Curr Opin Rheumatol. 1996;8:485–490. doi: 10.1097/00002281-199609000-00016. [DOI] [PubMed] [Google Scholar]
- 3.Silverman E, Mamula M, Hardin JA, Laxer R. Importance of the immune response to the Ro/La particle in the development of congenital heart block and neonatal lupus erythematosus. J Rheumatol. 1991;18:120–124. [PubMed] [Google Scholar]
- 4.Reichlin M, et al. Concentration of autoantibodies to native 60-kD Ro/SS-A and denatured 52-kd Ro/SS-A in eluates from the heart of a child who died with congenital complete heart block. Arthritis Rheum. 1994;37:1698–1703. doi: 10.1002/art.1780371120. [DOI] [PubMed] [Google Scholar]
- 5.von Muhlen CA, Tan EM. Autoantibodies in the diagnosis of systemic rheumatic diseases. Semin Arthritis Rheum. 1995;24:323–358. doi: 10.1016/s0049-0172(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Chetrit E, et al. A 52-kD protein is a novel component of the SS-A/Ro antigenic particle. J Exp Med. 1988;167:1560–1571. doi: 10.1084/jem.167.5.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolin SL, Steitz JA. Genes for two small cytoplasmic Ro RNAs are adjacent and appear to be single-copy in the human genome. Cell. 1983;32:735–744. doi: 10.1016/0092-8674(83)90059-4. [DOI] [PubMed] [Google Scholar]
- 8.Wolin SL, Steitz JA. The Ro small cytoplasmic ribonucleoproteins: identification of the antigenic protein and its binding site on the Ro RNAs. Proc Natl Acad Sci USA. 1984;81:1996–2000. doi: 10.1073/pnas.81.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruijn GJ, Simons FH, van Venrooij WJ. Intracellular localization and nucleocytoplasmic transport of Ro RNP components. Eur J Cell Biol. 1997;74:123–132. [PubMed] [Google Scholar]
- 10.Ben-Chetrit E, Gandy BJ, Tan EM, Sullivan KF. Isolation and characterization of a cDNA clone encoding the 60-kD component of the human SS-A/Ro ribonucleoprotein autoantigen. J Clin Invest. 1989;83:1284–1292. doi: 10.1172/JCI114013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutscher SL, Harley JB, Keene JD. Molecular analysis of the 60-kDa human Ro ribonucleoprotein. Proc Natl Acad Sci USA. 1988;85:9479–9483. doi: 10.1073/pnas.85.24.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labbe JC, Jannatipour M, Rokeach LA. The Caenorhabditis elegans rop-1 gene encodes the homologue of the human 60-kDa Ro autoantigen. Gene. 1995;167:227–231. doi: 10.1016/0378-1119(95)00695-8. [DOI] [PubMed] [Google Scholar]
- 13.O’Brien CA, Margelot K, Wolin SL. Xenopus Ro ribonucleoproteins: members of an evolutionarily conserved class of cytoplasmic ribonucleoproteins. Proc Natl Acad Sci USA. 1993;90:7250–7254. doi: 10.1073/pnas.90.15.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, Buyon JP, Chan EKL. Cloning and expression of mouse 60 kDa ribonucleoprotein SS-A/Ro. Mol Biol Rep. 1996;23:205–210. doi: 10.1007/BF00351170. [DOI] [PubMed] [Google Scholar]
- 15.Farris AD, Puvion-Dutilleul F, Puvion E, Harley JB, Lee LA. The ultrastructural localization of 60-kDa Ro protein and human cytoplasmic RNAs: association with novel electron-dense bodies. Proc Natl Acad Sci USA. 1997;94:3040–3045. doi: 10.1073/pnas.94.7.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien CA, Wolin SL. A possible role for the 60-kD Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes Dev. 1994;8:2891–2903. doi: 10.1101/gad.8.23.2891. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY. 2.108–2.107.
- 18.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ExPASy Proteomics. http://www.expasy.ch/www/tools.html. Accessed July, 1999.
- 20.de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan EKL, Hamel JC, Buyon JP, Tan EM. Molecular definition and sequence motifs of the 52-kD component of human SS-A/Ro autoantigen. J Clin Invest. 1991;87:68–76. doi: 10.1172/JCI115003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan EKL, Imai H, Hamel JC, Tan EM. Human autoantibody to RNA polymerase I transcription factor hUBF. Molecular identity of nucleolus organizer region autoantigen NOR-90 and ribosomal RNA transcription upstream binding factor. J Exp Med. 1991;173:1239–1244. doi: 10.1084/jem.174.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan EKL, Francoeur AM, Tan EM. Epitopes, structural domains, and asymmetry of amino acid residues in SS-B/La nuclear protein. J Immunol. 1986;136:3744–3749. [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Wang D, Chan EKL. 17-beta-estradiol increases expression of the 52 and 60-kD SS-A/Ro autoantigens in human keratinocytes and breast cancer cell line MCF-7. J Invest Dermatol. 1996;107:610–614. doi: 10.1111/1523-1747.ep12584194. [DOI] [PubMed] [Google Scholar]
- 26.Goblet C, Prost E, Whalen RG. One-step amplification of transcripts in total RNA using the polymerase chain reaction. Nucleic Acids Res. 1989;17:2144. doi: 10.1093/nar/17.5.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan, E.K.L., and Pollard, K.M. 1997. Detection of autoantibodies to ribonucleoprotein particles by immunoblotting. In Manual of clinical laboratory immunology. N.R. Rose, E.C. de Macario, J.D. Folds, H.C. Lane, and R.M. Nakamura, editors. American Society of Microbiology. Washington, DC. 928–934.
- 28.Rubin, R.L. 1997. Enzyme-linked immunosorbent assay for antibodies to native DNA, histones and (H2A-H2B)-DNA. In Manual of clinical laboratory immunology. N.R. Rose, E.C. de Macario, J.D. Folds, H.C. Lane, and R.M. Nakamura, editors. American Society of Microbiology. Washington, DC. 935–941.
- 29.Buyon JP, et al. Autoimmune-associated congenital heart block: demographics, mortality, morbidity and recurrence rates obtained from a national neonatal lupus registry. J Am Coll Cardiol. 1998;31:1658–1666. doi: 10.1016/s0735-1097(98)00161-2. [DOI] [PubMed] [Google Scholar]
- 30.Nagase T, et al. Prediction of the coding sequences of unidentified human genes. XII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1998;5:355–364. doi: 10.1093/dnares/5.6.355. [DOI] [PubMed] [Google Scholar]
- 31.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan, E.K.L., and Buyon, J.P. 1994. The SS-A/Ro antigen. In Manual of biological markers of disease. W.J. van Venrooij and R.N. Maini, editors. Kluwer Academic Publishers. Dordrecht, the Netherlands. 1–18.
- 33.Tan EM, Chan EKL, Sullivan KF, Rubin RL. Antinuclear antibodies (ANAs): diagnostically specific immune markers and clues toward the understanding of systemic autoimmunity. Clin Immunol Immunopathol. 1988;47:121–141. doi: 10.1016/0090-1229(88)90066-9. [DOI] [PubMed] [Google Scholar]
- 34.Reichlin M. Autoantibodies to the RoRNP particles. Clin Exp Immunol. 1995;99:7–9. doi: 10.1111/j.1365-2249.1995.tb03464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horsfall AC, Venables PJ, Taylor PV, Maini RN. Ro and La antigens and maternal anti-La idiotype on the surface of myocardial fibres in congenital heart block. J Autoimmun. 1991;4:165–176. doi: 10.1016/0896-8411(91)90015-5. [DOI] [PubMed] [Google Scholar]
- 36.Garcia S, et al. Cellular mechanism of the conduction abnormalities induced by serum from anti-Ro/SSA-positive patients in rabbit hearts. J Clin Invest. 1994;93:718–724. doi: 10.1172/JCI117025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boutjdir M, et al. Arrhythmogenicity of IgG and anti–52-kD SSA/Ro affinity-purified antibodies from mothers of children with congenital heart block. Circ Res. 1997;80:354–362. doi: 10.1161/01.res.80.3.354. [DOI] [PubMed] [Google Scholar]
- 38.Ramos-Deleon M, Herrera-Esparza R. Tyrosine kinase participates in phosphorylation of the Ro60 ribonucleoprotein. Rev Rhum Engl Ed. 1998;65:89–93. [PubMed] [Google Scholar]
- 39.Topfer F, Gordon T, McCluskey J. Intra- and intermolecular spreading of autoimmunity involving the nuclear self-antigens La (SS-B) and Ro (SS-A) Proc Natl Acad Sci USA. 1995;92:875–879. doi: 10.1073/pnas.92.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keech CL, Gordon TP, McCluskey J. The immune response to 52-kDa Ro and 60-kDa Ro is linked in experimental autoimmunity. J Immunol. 1996;157:3694–3699. [PubMed] [Google Scholar]
- 41.Tseng CE, et al. The 52-kD protein as a target of intermolecular spreading of the immune response to components of the SS-A/Ro-SS-B/La complex. Arthritis Rheum. 1996;40:936–944. doi: 10.1002/art.1780400523. [DOI] [PubMed] [Google Scholar]