Abstract
The development of nervous system connectivity depends upon the arborization of dendritic fields and the stabilization of dendritic spine synapses. It is well established that neuronal activity and the neurotrophin BDNF modulate these correlated processes. However, the downstream mechanisms by which these extrinsic signals regulate dendritic development and spine stabilization are less well known. Here we report that a substrate of BDNF signaling, the Ankyrin Repeat-rich Membrane Spanning protein (ARMS) or Kidins220, plays a critical role in the branching of cortical and hippocampal dendrites and in the turnover of cortical spines. In the barrel somatosensory cortex and the dentate gyrus, regions where ARMS/Kidins220 is highly expressed, no difference in the complexity of dendritic arbors was observed in 1-month-old adolescent ARMS/Kidins220+/− mice compared to wild-type littermates. However, at 3 months of age, young adult ARMS/Kidins220+/− mice exhibited decreased dendritic complexity. This suggests that ARMS/Kidins220 does not play a significant role in the initial formation of dendrites but, rather, is involved in the refinement or stabilization of the arbors later in development. In addition, at 1 month of age the rate of spine elimination was higher in ARMS/Kidins220+/− mice than in wild-type mice, suggesting that adolescent ARMS/Kidins220+/− mice exhibit reduced spine stability. Taken together, these data suggest that ARMS/Kidins220 is important for the growth of dendritic arbors and spine stability during an activity- and BDNF-dependent period of development.
Keywords: spines, dendritic arbors, barrel cortex, dentate gyrus
INTRODUCTION
During development, individual neurons extend and elaborate their axons and dendrites and form synapses with other neurons to establish neuronal networks. The initial growth of branches and synapses occurs through a coordinated and dynamic process of formation and elimination. Dendritic branches project filopodia, which explore space through extension and retraction until contact is made with an axonal branch (McFarlane, 2000; Scott and Luo, 2001). Stabilization of these contacts is achieved functionally through the recruitment of pre- and postsynaptic components to form synapses and morphologically through the maturation of the filopodia into spines. The stabilization of synapses and spines regulates the growth of both axonal and dendritic arbors by anchoring newly added branches from which further branching can occur (Niell et al., 2004; Meyer and Smith, 2006; Ruthazer et al., 2006).
The initial formation of neuronal morphology relies largely on gene programs intrinsic to neurons (Goldberg, 2004), but extrinsic signals play an important role in the maturation and refinement of network connections well into adolescence. Neuronal activity, often as a result of sensory experience, is especially crucial for proper morphological development. During critical periods of refinement, neuronal activity can sculpt the segregation of inputs into ocular dominance columns in the visual cortex (Wiesel and Hubel, 1974) and regulate axon elimination at neuromuscular junctions (Sanes and Lichtman, 1999). Often neuronal activity mediates structural modifications by initiating changes in molecular mechanisms.
Several extrinsic molecules have been identified in the control of dendrite formation and branching. These include insulin-like growth factor (Niblock et al., 2000), thyroid hormone (Heuer and Mason, 2003), hepatocyte growth factor (Gutierrez et al., 2004), and BDNF (Cohen-Cory and Lom, 2004; Horch, 2004). As a major activity-dependent factor, BDNF has an established role in modulating synaptic plasticity (Korte et al., 1995; Patterson et al., 1996; Gottschalk et al., 1999) and is also directly responsible for the morphogenesis of dendritic spines and dendritic growth (Cohen-Cory and Fraser, 1995; McAllister et al., 1995; Horch and Katz, 2002; Liu et al., 2007). BDNF is in the family of neurotrophins that includes NGF, NT-3, and NT-4 and acts through the TrkB receptor tyrosine kinase and the p75 receptor (Chao, 2003). In the developing visual system, BDNF directly influences the growth of synapses and axonal and dendritic arbors (Hu et al., 2005; Sanchez et al., 2006). Further support for the importance of BDNF in morphological development comes from the deficits in spines and dendritic branching seen in BDNF−/− and TrkB−/− mice (Xu et al., 2000; Gorski et al., 2003; Chen et al., 2006).
Because BDNF uses common signaling pathways through generic receptor tyrosine kinase phosphorylation, it is not known how specificity is generated to account for regulation of spine remodeling and dendritic branching. One unique signaling molecule downstream of the TrkB receptor is the Ankyrin Repeat-rich Membrane Spanning protein (ARMS)(Kong et al., 2001), or Kidins220 (Iglesias et al., 2000), a transmembrane scaffold protein that has been implicated in synaptic functions due to its regulation by neuronal activity (Cortes et al., 2007). In addition to being a substrate of phosphorylation by Trks, ARMS/Kidins220 can also be phosphorylated by Eph receptors, another signaling system important for the development of neuronal morphology (Kong et al., 2001; Klein, 2004).
We hypothesized that ARMS/Kidins220 may be involved in the BDNF signaling that regulates dendritic branching and spine development. To investigate the role of ARMS/Kidins220 in morphological development in vivo, we examined dendritic branching and spine dynamics in ARMS/Kidins220+/− mice. Our data suggest that ARMS/Kidins220 is important for the proper morphological development of dendritic arbors and spines during an activity- and BDNF-dependent developmental period.
METHODS
Brain lysates and Western blot
Whole brain, cortex, and hippocampus were dissected from C57Bl6 mice at various ages and frozen at −70° C. The tissues were homogenized in a lysis buffer containing 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 2 µg/ml leupeptin, 1 mM sodium orthovanadate, 2 µg/ml aprotinin, 1 mM PMSF, and 10 mM sodium fluoride. After cell debris was cleared, 20 µg of P1-P30 lysates and 40 µg of 1- and 3-month lysates were separated on SDS-PAGE gels, transferred to PVDF membranes (Millipore), and probed using an anti-ARMS/Kidins220 rabbit polyclonal antibody (1:4000)(Kong et al., 2001) and an anti-tubulin mouse monoclonal antibody (Sigma). Immunoreactive proteins were visualized by ECL detection (Amersham Biosciences) and film autoradiography.
Immunohistochemistry
The immunohistochemistry was performed as previously described (Bath et al., 2008) with slight modifications. C57Bl6 mice were anesthetized with sodium pentobarbital and transcardially perfused with 0.1% sodium nitrite in phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA) in PBS. Brains were dissected out, post-fixed in 4% PFA for one hour, and incubated overnight at 4° C in PBS containing 30% sucrose. Following complete immersion in the sucrose solution, brains were cut into 40-µm thick serial coronal sections using a freezing microtome.
Immunostaining was performed on free-floating sections as follows. First, sections were blocked for one hour in a buffer containing 10% normal horse serum, 10% normal goat or donkey serum, and 0.1% Triton X-100 in PBS. They were then incubated with anti-ARMS/Kidins220 rabbit polyclonal antibody (2 µg/mL)(Kong et al., 2001) or rabbit IgGs (Santa Cruz) and Hoechst stain (1:1000; Molecular Probes) in block buffer for about 36 hours at 4° C. After 3 × 10-minute washes with PBS, the sections were incubated with Alexa-555 goat anti-rabbit secondary antibody (Invitrogen) for 2 hours in PBS with 0.1% Triton X-100, washed extensively, and mounted on slides.
Low resolution images were taken with a Nikon Eclipse E800 microscope fitted with a Nikon Plan Apo 20X, 0.75 numerical aperture (NA) objective using a Zeiss AxioCam HRc digital camera. High resolution confocal images were taken with a Zeiss LSM510 laser-scanning confocal microscope equipped with a Zeiss Plan Neofluor 40X, 1.3 NA oil immersion objective and processed using LSM510 and NIH ImageJ software. The composite image was assembled using Adobe Photoshop software.
ARMS/Kidins220 mutant mice
To generate a targeting construct for ARMS/Kidins220 knockout mice, a genomic library from 129SV/J mice (Stratagene) was screened using mouse ARMS/Kidins220 cDNA. A neomycin-resistance gene flanked by FRT sites was inserted downstream of exon 14, which encodes the end of the second transmembrane domain and part of the intracellular loop [Figure 2(A)]. Exon 14 and the neomycin cassette were flanked by loxP sites to generate a conditional allele. The truncated diphtheria toxin gene was placed at the end of the long arm for negative selection.
Figure 2. Generation of a mutant ARMS/Kidins220 mouse.
(A) Schematic of the wild-type ARMS allele, the targeting vector, and the targeted allele before and after Cre recombination. Neo, neomycin resistance gene; DT-A, diphtheria toxin. B, A, and N denote the sites for the restriction enzymes BamHI, ApaI, and NotI; red rectangles denote exons (numbered in red); and green triangles denote loxP sites.
(B) Southern blot analysis of genomic DNA from mutant mice after digestion with BamHI. The 5’ probe, as indicated in the schematic in (A), detects a 5 Kb fragment from wild-type alleles (WT), a 6.4 Kb fragment from mutants before actin-Cre recombination (Mut, top panel), and a 4.5 Kb fragment from mutants after actin-Cre recombination (Mut, bottom panel).
(C) Western blot analysis of ARMS protein levels in the whole brain of ARMS+/− mice and wild-type littermates at 4 months of age. ARMS+/− mice showed a 30–40% reduction in ARMS protein compared to wild-type. Tubulin was used as a loading control.
The targeting vector was linearized at a unique upstream NotI site and was electroporated into CJ7 embryonic stem (ES) cells as previously described (Tessarollo, 2001). G418-resistant colonies were screened for homologous recombination by Southern blot analysis with a 5’ 0.4-kb external probe. ES clones were injected into C57BL/6 blastocysts to generate chimeric mice. The chimeras were bred with C57BL/6 mice and the resulting progeny were kept on a 129-C57BL/6 mixed genetic background. Exon 14 was removed by crossing the chimeras with mice that expressed the Cre recombinase driven by the actin promoter. Southern blot analysis confirmed the presence of mutant alleles in mice before and after actin-Cre recombination [Figure 2(B)]. ARMS/Kidins220 genotyping was performed by PCR with tail genomic DNA and the following three primers: ARMS KO1F, 5´ ggtttcacacgccttctgtg; ARMS KpnF, 5´ cacattgggtacctggagc; and ARMS KO3B: 5´ ccagacctctgtctgccag. A 500 bp product is amplified from the WT allele, and a 200 bp product is amplified from the mutant allele. Lysates were made from brains from 4-month-old ARMS/Kidins220+/− mice and wild-type littermates, and Western blot analysis was performed, as described above.
For the in vivo imaging studies, ARMS/Kidins220+/−; YFP mice were generated by crossing ARMS/Kidins220+/− males with C57BL/6 females expressing YFP driven by the Thy1 promoter in a subset of Layer 5 cortical pyramidal cells (Feng et al., 2000).
All animals were housed and bred in the Skirball Institute Animal Facility. All animal care and procedures were done in accordance with protocols approved by the NYU School of Medicine Institutional Animal Care and Use Committee.
Golgi impregnation, neuronal tracing, and Sholl analyses
Golgi impregnation and neuronal tracing were performed as previously described (Chen et al., 2006). Brains from 1- and 3-month-old male ARMS/Kidins220+/− mice and wild-type littermates were dissected out and Golgi impregnated using the FD Rapid GolgiStain Kit (FD NeuroTechnologies) according to the manufacturer’s protocol. The brains were cut into 150-µm thick serial coronal sections using a Leica VT1000S vibratome.
From the outset, genotype information for all slides was coded, and neurons were selected and traced blind to the genotype. Golgi-labeled dentate gyrus granule cells and barrel somatosensory cortex Layer 5 pyramidal cells were identified by morphology and through consultation with an atlas (Franklin and Paxinos, 2004) to determine their location in the proper region. Neurons were selected for tracing and analysis if they were isolated enough from other impregnated cells that dendrite paths were unambiguous, if impregnation clearly filled the dendritic tree, and if no branches were cut off. The entire dendritic arbor of the granule cells and the basal dendritic arbor of the pyramidal cells were traced under 40X magnification using Neurolucida software (MicroBrightField, Inc.).
Sholl analyses using a 10-µm radius, as well as additional measures of dendritic complexity such as the number of primary dendrites, total dendritic length, number of branch points, and number of branch tips, were performed on traced neuronal arbors using Neuroexplorer software (MicroBrightField, Inc.)(Sholl, 1953). Student’s t-test statistical analyses were done using Prism software (GraphPad).
Cell culture and immunocytochemistry
Primary cortical neurons were dissected from E18–19 Sprague Dawley rat embryos in Ca2+- and Mg2+-free HBSS supplemented with 0.37% glucose, digested with 0.05% trypsin, mechanically dissociated with fire-polished Pasteur pipettes, and plated on poly-D-lysine-coated coverslips in a 24-well plate at 40K neurons per well. Cells were grown in Neurobasal media (Gibco) supplemented with B27 supplement (Gibco), 0.37% glucose, 0.5 mM glutamine, 1.2 µg/ml 5-fluoro-2-deoxyuridine, and, in certain cultures, 25 ng/ml BDNF. Fresh media was added to the cells every 3–4 days.
GFP-expressing ARMS/Kidins220 and control shRNA lentivirus were previously described (Cortes et al., 2007). Cells were infected on DIV0–1 and fixed on DIV4 and DIV7 with 4% paraformaldehyde and 20% sucrose in PBS. Cells were permeabilized with 0.1% Triton X-100 in PBS for 3 minutes, blocked with TBS buffer containing 10% normal goat serum, 2% BSA, and 0.25% fish skin gelatin, and incubated in a humidified chamber overnight with rabbit polyclonal anti-MAP2 (1:2000, Abcam), chicken polyclonal anti-GFP (1:2000, Abcam), and mouse monoclonal anti-GAD-65 (1:500, BD Pharmingen) primary antibodies in block buffer. After washing with TBS wash buffer containing 0.25% fish skin gelatin, cells were incubated with anti-rabbit Alexa 555, anti-chicken Alexa 488, and anti-mouse Alexa 647 (1:500, Invitrogen) secondary antibodies in block buffer for one hour, and washed again. Coverslips were mounted onto slides with Mowiol mounting solution.
In vitro neurite analysis
Slides were coded during the acquisition of images and quantification of neurites. Images of GFP-positive infected neurons were taken with a Zeiss confocal microscope as described above. Primary neurites of imaged neurons were quantified by examining the overlap of MAP2 staining with GFP staining and counting the number of double-labeled projections arising from the soma. Any GABAeric neuron displaying GAD-65 expression in the soma was excluded from the analysis. Student’s t-test statistical analyses were done using Prism software (GraphPad).
In vivo imaging of dendritic spines
Transcranial two-photon microscopy and data quantification were performed as previously described (Grutzendler et al., 2002; Xu et al., 2007). One-month-old male ARMS/Kidins220+/−; YFP mice and ARMS/Kidins220+/+; YFP wild-type littermates were analyzed.
RESULTS
ARMS/Kidins220 protein expression is developmentally regulated
The ARMS/Kidins220 protein (hereafter referred to as ARMS) was originally discovered as a substrate for protein kinase D (PKD)(Iglesias et al., 2000) and as an interacting protein of Trk and p75 receptors (Kong et al., 2001). ARMS is a transmembrane protein that is highly expressed in the nervous system. In addition to its presence on the cell surface, biochemical and cell biological analyses indicate that ARMS is localized prominently in intracellular vesicular compartments (Arevalo et al., 2006).
Here we investigated the function of ARMS in neuronal development. We examined the expression of ARMS protein through the early stages of development. ARMS protein expression is high at birth and decreases over the next month [Figure 1(A)]. ARMS protein levels do not change visibly between adolescence at one month and young adulthood at three months [Figure 1(B)]. Thus, the developmental regulation of ARMS is most significant in the early postnatal period.
Figure 1. ARMS/Kidins220 is developmentally regulated in the postnatal brain and is highly expressed in the barrel somatosensory cortex and the dentate gyrus of the hippocampus.
(A) Western blot analysis of ARMS protein levels in whole mouse brain at the indicated postnatal age (days). Tubulin is shown as a loading control.
(B) Western blot analysis of ARMS protein levels in whole brain, cortex, and hippocampus of 1- and 3-month-old mice. Tubulin is shown as a loading control.
(C) Coronal sections from wild-type mice were stained with anti-ARMS antibody. Composite image of ARMS staining in the hippocampus and the surrounding cortex. White boxes mark regions of higher magnification in the somatosensory cortex (SC) and the dentate gyrus (DG). In the cortex, ARMS is prominently expressed in Layer 5 pyramidal neurons. In the hippocampus, ARMS is highly expressed in the cell bodies in all regions but is expressed more strongly in the dendrites of the dentate gyrus (molecular layer, ML) than in the dendrites of CA3 or CA1 (stratum radiatum, SR). Scale bar, 100 µm.
(D) Higher magnification image of ARMS staining in the barrel somatosensory cortex. In Layer 5 pyramidal neurons, ARMS is expressed in cell body as well as in processes extending from the soma (arrows). Scale bar, 100 µm.
(E) Higher magnification image of ARMS staining in the dentate gyrus. Scale bar, 100 µm.
To visualize ARMS protein distribution in the developing brain, we carried out immunohistochemical analysis of brain sections from 1-month-old mice. ARMS stained throughout the brain and prominently in the hippocampus and in the cortex [Figure 1(C)]. In the cortex, there is high expression of ARMS in Layer 5 pyramidal neurons, where it can be detected in cell bodies and in dendritic projections [Figure 1(D), arrows highlight projections]. In the hippocampus, ARMS is found in the somas of the cells of all three major hippocampal regions – the dentate gyrus, CA1, and CA3 – as well as in hilar neurons [Figures 1(C) and 1(E)]. ARMS is also expressed in dendrites, with notably higher expression in the molecular layer of the dentate gyrus than in the stratum radiatum of CA3 or CA1 [Figure 1(C)].
To study the effects of ARMS in vivo, we generated ARMS mutant mice by targeting exon 14 of the ARMS locus, which encodes part of a transmembrane domain [Figure 2(A)]. LoxP sites were inserted into the genome to flank the exon, and the removal of the exon after Cre-mediated recombination was confirmed by Southern blot [Figure 2(B)]. ARMS−/− mice displayed very early embryonic lethality and could not be studied, but ARMS+/− mice are viable and fertile. They exhibit grossly normal brain morphology despite a 30–40% decrease of ARMS protein compared to wild-type mice [Figure 2(C)].
ARMS/Kidins220 mutant mice display decreased dendritic complexity in the barrel somatosensory cortex
The expression of ARMS in dendrites suggests that it may participate in dendritic and spine changes during development. To study the effect of ARMS on neuronal morphology, we first examined the dendritic arbors of neurons from brain slices of ARMS+/− mice and wild-type littermates. Neurons were labeled using the Golgi-Cox staining method at 1 month of age, which corresponds to adolescence, and at 3 months of age, which corresponds to young adulthood.
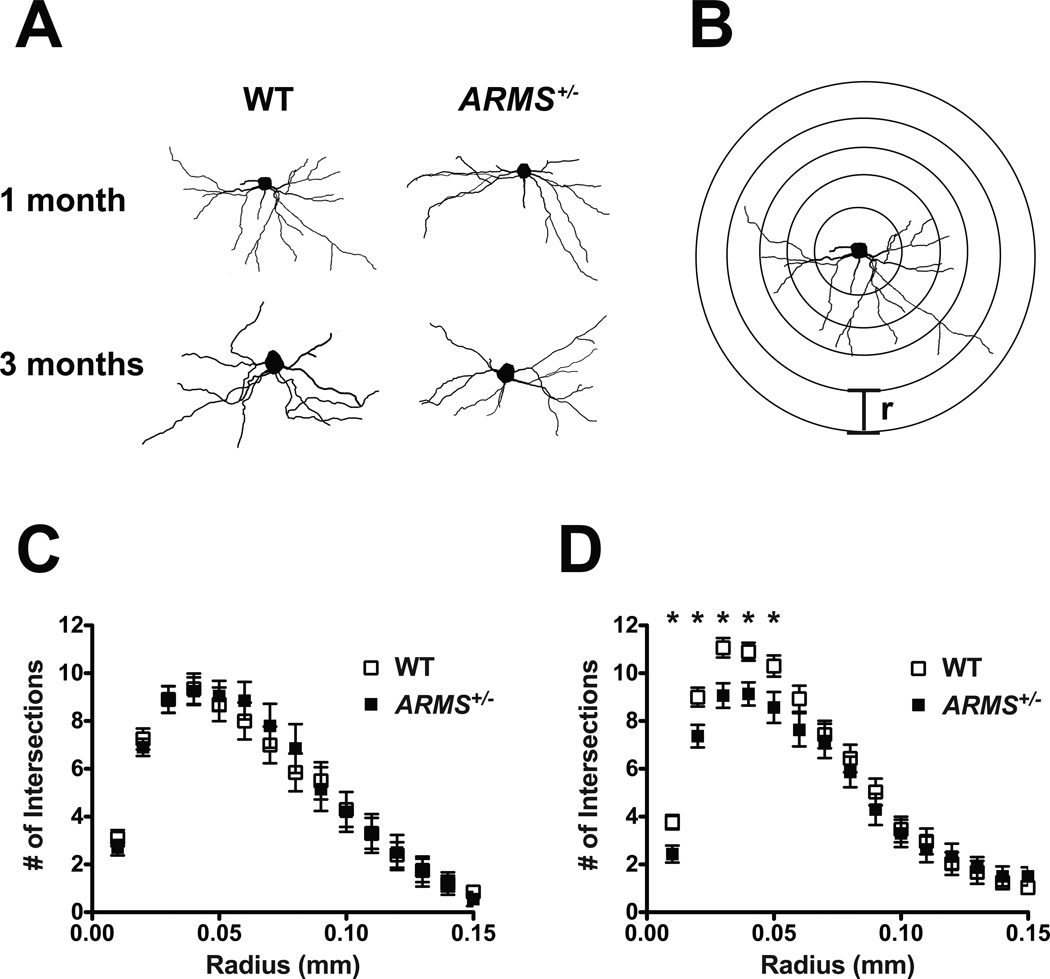
To investigate the role of ARMS in dendritic morphogenesis, the barrel field cortex, a well-studied somatosensory region that responds to whisker stimulation, was first examined. We traced the arbors of the basal dendrites of Layer 5 pyramidal neurons, a cell population that expresses ARMS strongly [Figures 3(A) and 1(D)]. To determine the branching complexity of the dendritic arbors, we analyzed the tracings using the Sholl method, which measures the number of times dendrites intersect each ring in a series of concentric rings radiating from the cell soma a given radius apart [Figure 3(B)](Sholl, 1953).
Figure 3. Barrel cortex pyramidal neurons exhibit decreased dendritic complexity in young adult but not adolescent ARMS/Kidins220+/− mice.
(A) Representative tracings of the basal dendrites of Layer 5 pyramidal neurons in the barrel cortex of 1- and 3-month-old ARMS+/− mice and wild-type littermates.
(B) Schematic of the Sholl method for determining dendritic complexity (not to scale). Radiating concentric rings of a set interval (r) are centered on the cell soma of a neuronal tracing, and the number of dendrites intersecting each ring is quantified. Analyses were performed with a radius interval of 10 µm.
(C) Sholl analyses of the basal dendrites of Layer 5 pyramidal neurons in the barrel cortex of 1-month-old (adolescent) ARMS+/− mice and wild-type littermates (n = 20 neurons from two mice per genotype). There was no difference in dendritic complexity between the two genotypes.
(D) Sholl analyses of the basal dendrites of Layer 5 pyramidal neurons in the barrel cortex of 3-month-old (young adult) ARMS+/− mice and wild-type littermates (n = 30 neurons from two mice per genotype). ARMS+/− mice displayed less complex dendritic arbors than wild-type mice (*p < 0.05, Student’s t-test).
At 1 month of age, there was no significant difference in dendritic complexity between the ARMS+/− mice compared to wild-type littermates [Figure 3(C)]. By 3 months of age, the curve of dendritic complexity for wild-type mice shifted upward, indicating increased arborization over the 2-month period [Figure 3(D)]. However, for ARMS+/− mice, the curve did not shift significantly from 1–3 months of age and was significantly lower than that of wild-type mice at 3 months of age. These data indicate that while dendrites in wild-type animals continued to grow and branch between 1–3 months of age, dendrites in ARMS+/− mice did not increase in complexity during this time. This suggests that ARMS plays a role in dendritic branching in these neurons in the developmental period between adolescence and young adulthood.
ARMS/Kidins220 mutant mice display dendritic complexity in the dentate gyrus
To determine whether these changes were specific for the barrel cortex or if ARMS affected dendritic arborization in other brain regions, we analyzed the dendritic arbors of granule cells in the dentate gyrus where ARMS is also highly expressed [Figures 4(A) and 1(C)]. As in the barrel cortex, at 1 month of age the granule cell arbors of ARMS+/− mice were similar in complexity to those of wild-type mice [Figure 4(B)]. Between 1–3 months of age both ARMS+/− and wild-type mice continued to increase their dendritic complexity, but at 3 months of age the branching in ARMS+/− mice was significantly lower than in wild-type mice [Figure 4(C)]. These data suggest that, like in barrel cortex neurons, granule cells require ARMS for proper dendritic branching between 1–3 months of age. As the requirement for ARMS does not appear until after 1 month, ARMS may be more important for the refinement and stabilization of arbors than the initial patterning.
Figure 4. Dentate gyrus granule cells also exhibit decreased dendritic complexity in young adult but not adolescent ARMS/Kidins220+/− mice.
(A) Representative tracings of dentate gyrus granule cells of 1- and 3-month-old wild-type and ARMS+/− mice.
(B) Sholl analyses of granule cell dendrites of 1-month-old (adolescent) ARMS+/− mice and wild-type littermates (n = 24 neurons from two mice per genotype). There was no difference in dendritic complexity between the two genotypes.
(C) Sholl analyses of granule cell dendrites of 3-month-old (young adult) ARMS+/− mice and wild-type littermates (n = 21 neurons from two mice per genotype). ARMS+/− mice displayed less complex dendritic arbors than wild-type mice (*p < 0.05, Student’s t-test).
In addition to the Sholl analysis, we also examined other measures of dendritic complexity in the barrel cortex and the dentate gyrus at 1 and 3 months. The number of primary dendrites in Layer 5 pyramidal cortical neurons as well as total dendritic length, the number of branch points, and the number of branch tips in both cortical neurons and hippocampal granule cells were quantified [Supplementary Table 1]. No significant differences were seen except for a decrease in the number of primary dendrites in cortical neurons in 3-month-old ARMS+/− mice.
Decreasing ARMS/Kidins220 levels in vitro leads to decreased BDNF-induced primary neurite outgrowth
To see what effect ARMS levels have on the growth of neurons in culture, we decreased the levels of ARMS in primary cortical cultures using shRNA lentivirus and examined primary neurite outgrowth. We infected cells with GFP-expressing lentivirus within 24 hours of plating to knock down ARMS in cultures grown in regular media conditions as well as in media supplemented with BDNF. Cells were fixed at DIV4 and DIV7 and stained with MAP2. GFP-expressing cells were imaged, and primary neurites displaying MAP2 staining were counted [Figure 5].
Figure 5. Decreasing ARMS/Kidins220 levels in primary cortical cultures leads to decreased BDNF-induced primary neurite growth.
Primary cortical cultures grown with and without BDNF supplementation were infected with lentivirus containing GFP-tagged ARMS or control shRNA, and the number of primary neurites of excitatory neurons was counted at DIV4 and DIV7 (n = 20 for all conditions; data is plotted as mean ± s.e.m.; *p < 0.05, Student’s t-test)
Under regular media conditions, no difference in primary neurite outgrowth could be seen at DIV4 between control shRNA- and ARMS shRNA-infected neurons (control shRNA = 10.55 ± 0.51, n = 20; ARMS shRNA = 10.70 ± 0.62, n = 20; p > 0.05); however, at DIV7, neurons infected with ARMS shRNA lentivirus displayed fewer primary dendrites than control-infected neurons (control shRNA = 16.15 ± 0.88, n = 20; ARMS shRNA = 13.65 ± 0.49, n = 20; p > 0.05). ARMS shRNA-infected cells grown in BDNF displayed a reduction in the number of primary neurites at both DIV4 (control shRNA = 14.70 ± 0.65, n = 20; ARMS shRNA = 13.00 ± 0.48, n = 20; p < 0.05) and DIV7 (control shRNA = 19.85 ± 0.56, n = 20; ARMS shRNA = 17.50 ± 0.67, n = 20; p < 0.05). These data support a role for ARMS in BDNF-induced dendritic growth.
ARMS/Kidins220 mutant mice display decreased spine stability in vivo
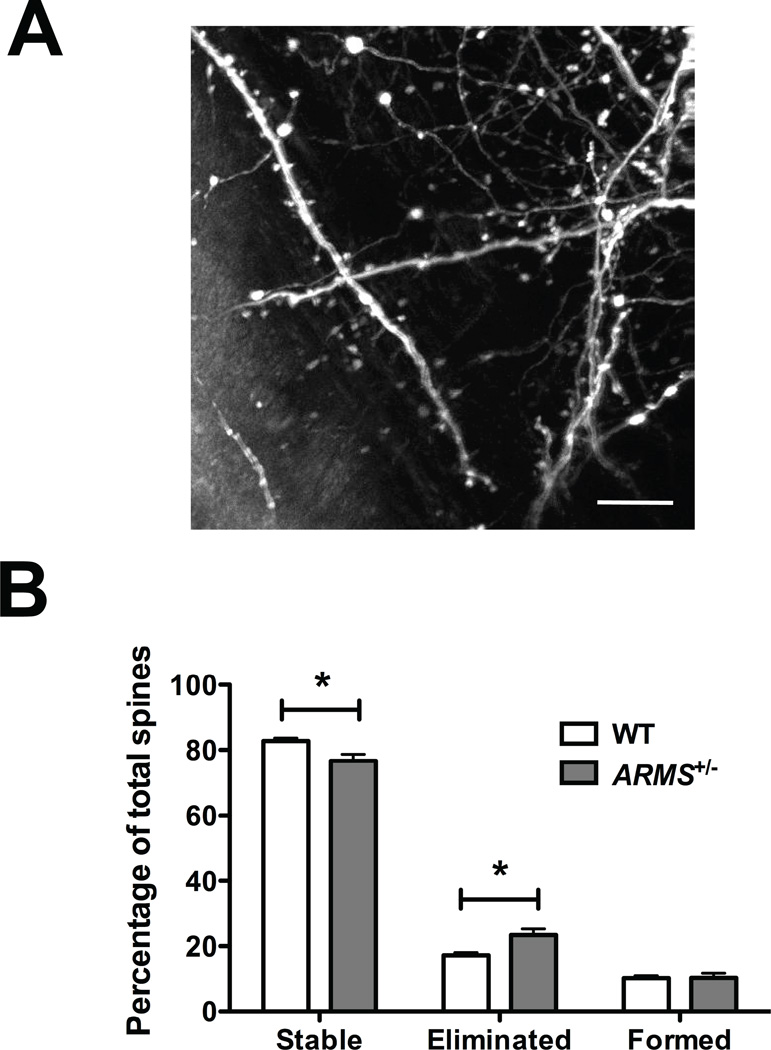
Dendritic branching is highly correlated with synapse stabilization, which can be reflected in the dynamics of dendritic spines. As the molecules involved in this process are not completely defined, ARMS represents a candidate protein to regulate this aspect of neuronal morphology. Spine formation and elimination over time can be studied through transcranial two-photon microscopy in living animals that express yellow fluorescent protein (YFP) in a subset of neurons (Feng et al., 2000; Grutzendler et al., 2002). Using mice that express YFP driven by the Thy-1 promoter in a subset of Layer 5 pyramidal neurons, labeled apical dendrites that project to Layer 1 can be imaged through a thinned-skull window at many time points during development, and the fate of individual spines on those dendrites can be tracked to determine the degree of spine turnover [Figure 6(A)].
Figure 6. ARMS/Kidins220+/−; YFP adolescent mice exhibit decreased spine stability due to increased spine elimination.
(A) A collapsed two-photon confocal image stack taken through a transcranial window over the barrel somatosensory cortex of a YFP-expressing mouse. Apical dendrites projected to Layer 1 from YFP-expressing Layer 5 pyramidal neurons are clearly visible and are studded with spines and filopodia. Scale bar, 10 µm.
(B) Analysis of spines on apical dendrites in Layer 1 projected from YFP-labeled Layer 5 pyramidal neurons of ARMS+/−; YFP mice (ARMS heterozygotes; n = 3) and ARMS+/+; YFP littermates (ARMS wild-type; n = 4). Two sets of transcranial two-photon microscropy images were taken: the first view at 1 month of age and the second view 2 weeks later. Individual dendrites were analyzed for spine content at both time points. Spines present at both time points were considered “stable,” spines no longer present at the second time point were considered “eliminated,” and spines identified at the second time point only were considered newly “formed.” Spine counts were plotted as a percentage of the total spines identified at the first view. ARMS+/−; YFP mice exhibited decreased spine stability as a result of an increased rate of spine elimination (*p < 0.05, Student’s t-test). The rate of spine formation was not significantly different between genotypes. The data is plotted as mean ± s.e.m.
To determine whether ARMS affects spine dynamics in vivo, we generated ARMS+/−; YFP mice and imaged dendritic protrusions in the barrel cortex. Animals were imaged first at 1 month of age and again 2 weeks later, and spines that were stable, eliminated, or newly formed were quantified. During this period, ARMS+/−; YFP mice displayed a lower percentage of stable spines (stable spines: WT = 82.75 ± 0.85%, n = 4; ARMS+/− = 76.66 ± 2.03%, n = 3; p > 0.05) due to an increased rate of spine elimination compared to ARMS+/+; YFP littermates (eliminated spines: WT = 17.25 ± 0.85%, n = 4; ARMS+/− = 23.33 ± 2.03%, n = 3; p < 0.05)[Figure 6(B)]. However, spine formation rate was the same for both groups of mice (newly-formed spines: WT = 10.25 ± 0.75%, n = 4; ARMS+/− = 10.33 ± 1.45%, n = 3; p > 0.05). These data suggest that ARMS influences spine dynamics and may play a role in stabilizing spines.
Taken together, the results of the Sholl analyses of dendritic branching and the two-photon microscopy in vivo analyses of spine turnover in ARMS+/− mice suggest that ARMS regulates spine stability and branch formation during a critical developmental period.
DISCUSSION
Abundant evidence has established BDNF as an activity-dependent molecule that controls synaptic function and morphological plasticity. However, a major unanswered question is how neurotrophic factors are able to regulate changes in neuronal morphology in a temporal and selective manner. Many signaling proteins are associated with and become phosphorylated by Trk receptors, including Shc, Gab1, and phospholipase C-γ, but these proteins interact with other receptor tyrosine kinases and are unlikely to confer specificity to Trk signaling (Chao, 1992). Of all the proteins that are directly phosphorylated by the TrkB receptor, the ARMS/Kidins220 protein represents one of the most unique substrates (Kong et al., 2001; Arevalo et al., 2004).
ARMS/Kidins220 is a transmembrane scaffold protein that is implicated in synaptic functions due to its regulation by neuronal activity. In hippocampal cultures, ARMS protein levels increase when activity is blocked using a sodium channel inhibitor; conversely, ARMS levels decrease when neurons are activated using a GABA receptor inhibitor (Cortes et al., 2007). ARMS is similar in function to several other activity-dependent scaffold proteins in the post-synaptic density, including PSD-95 and Shank, which, in addition to regulating synaptic functions, also regulate spines and dendritic morphology (Vessey and Karra, 2007). These scaffold proteins use many specialized protein-protein interaction domains, such as the SAM domain, PDZ tail motif, and ankyrin repeats that are found in the ARMS molecule, to form macromolecular complexes that link trafficking machinery, glutamate receptors, and cytoskeletal regulators.
There is strong evidence for a trafficking role for the ARMS protein. It was originally identified as the first known substrate of PKD, a protein kinase C-related kinase that is implicated in membrane trafficking events (Iglesias et al., 2000). ARMS also is capable of directly interacting with kinesin subunits (Bracale et al., 2007). The participation of ARMS in intracellular trafficking events may ultimately influence the activities of growth cones or the movement of cytoskeletal or ion channel proteins. In PC12 cells, it has been shown that ARMS is highly expressed at the tips of neurites (Iglesias et al., 2000). During NGF induction of neuronal differentiation, there is a direct engagement of the TrkA receptor with ARMS, which leads to sustained MAPK activation that regulates neurite outgrowth (Arevalo et al., 2004). Notably, it has been found that MAP kinase activation is involved in the ability of BDNF to increase dendritic spine density in hippocampal neurons (Alonso et al., 2004). Hence, the expression of ARMS at specific periods during development provides a mechanism that may couple neurotrophin signaling directly to MAP kinase signaling. The number of potential interactions with ARMS indicates that ARMS may represent a dynamic multi-functional protein responsible for the specialized effects of neurotrophins.
To study the properties of the ARMS protein in vivo, ARMS-deficient mice were generated by targeting an exon encoding part of a transmembrane domain. Homozygous ARMS knockout mice exhibited embryonic lethality, indicating the necessity of this protein in early development. For this reason, we analyzed ARMS+/− mice, which are viable and fertile. Western blot analysis of tissues from the heterozygous mice indicated that ARMS expression was decreased by 30–40% instead of 50%. Since ARMS is normally present at high levels in neuronal cells, this suggests that proper neuronal function requires ARMS protein levels to be tightly regulated and maintained. Nevertheless, the modest decrease of ARMS protein in ARMS+/− produced deficits in neuronal morphology.
ARMS+/− mice displayed dendritic branching in the cortex and hippocampus that was normal at 1 month of age but was altered by 3 months. In wild-type mice, branching continued during this period while growth in ARMS+/− mice was blunted. The timing of the influence of ARMS coincides with the timing of BDNF’s effects on dendritic growth. BDNF levels are very low during development and increase dramatically 2–4 weeks after birth (Baquet et al., 2004). The striking increase suggests that one role of this molecule is to respond to activity-regulated events in the adolescent and adult brain. Consistent with this delayed developmental action of BDNF, dendrites form and branch normally in the cortex of conditional BDNF knockout mice in the early postnatal weeks with reductions in dendritic complexity appearing at 3–5 weeks of age (Gorski et al., 2003). Since TrkB receptors and ARMS protein are co-expressed at this time, it is likely that BDNF exerts its effects through TrkB and ARMS signaling. The evidence that decreasing ARMS levels in cultured cortical neurons blunts the effect BDNF exerts on neurite outgrowth supports this potential role for ARMS in BDNF-induced dendrite growth.
BDNF exerts a strong influence upon synapses during development. For example, in the developing retinotectal system in Xenopus, BDNF applied to the tectum stabilized the synapses formed between retinal ganglion cell axons and tectal dendrites, leading to the subsequent stabilization of axonal arbors (Hu et al., 2005; Sanchez et al., 2006). An examination of spine turnover in ARMS+/− mice showed that ARMS also may influence synapses by promoting the stabilization of spines. In 1-month-old animals, the rate of spine elimination over a 2-week period was higher than that in wild-type littermates. Thus, spine stability was decreased in ARMS+/− mice. Interestingly, the rate of spine formation was unchanged, suggesting that the formation and elimination of spines occurs through distinct mechanisms. The effect of ARMS levels is relevant to the adolescent period since spine turnover rates in older ARMS+/− mice were similar to wild-type (data not shown). These data corroborate the finding in dendrites that ARMS influences morphology changes during the adolescent developmental period, and it is possible that ARMS may therefore participate in the mechanisms downstream of BDNF that regulates synapse stability.
The findings from the ARMS+/− mice support the synaptotropic hypothesis of arborization, which posits that synapse formation and maturation govern the stability of growing arbors (Vaughn, 1989; Meyer and Smith, 2006; Ruthazer et al., 2006; Cline and Haas, 2008). Many groups have examined the role of activity and BDNF in regulating developing synapses and arborization (Lu, 2003; Cohen-Cory and Lom, 2004; Hu et al., 2005; Sanchez et al., 2006). Analysis of the ARMS+/− mice indicates that both spines and dendrites are regulated by this BDNF substrate. It is possible that the greater elimination of spines in these animals leads to the destabilization of newly formed dendrites and subsequent impairment in branching. Because the effect of ARMS on dendritic branching is not seen until after the adolescent period, ARMS may be involved in the refinement of connections mediated by neuronal activity and BDNF signaling during a critical period of morphological development. Understanding the mechanisms by which ARMS acts downstream of BDNF to regulate spine stabilization and dendritic branching will rely on future developmental and signal transduction studies.
Supplementary Material
Dendritic parameters as indicated were measured in the basal dendrites of Layer 5 pyramidal neurons in the barrel cortex and dentate gyrus granule cells of 1- and 3-month-old ARMS+/− mice and wild-type littermates. (n = as indicated for Sholl analyses). The number of primary dendrites in ARMS+/− mice at 3 months of age was significantly fewer than that of wild-type mice (n = 30 neurons from two mice per genotype; data = mean ± s.e.m.; *p < 0.05, Student’s t-test).
ACKNOWLEDGEMENTS
We thank Deqiang Jing, Kevin G. Bath, and Francis S. Lee for their technical guidance and resources; Feng Pan and Jhon Sutachan-Rubio for their technical support; Katrin Deinhardt, Vladimir Camarena, and Kate L. Foster for their critical readings of the manuscript; and members of the Moses V. Chao, Barbara L. Hempstead, and Francis S. Lee laboratories for helpful discussions. This work was supported by the NIH Medical Scientist Training program (SHW), by a Marie Curie International Reintegration Grant within the 7th European Community Framework Programme to JCA, by the NCI Intramural Research Program, Center for Cancer Research of the NIH (LT), and by NIH grants to MVC (NS21072 and HD23315) and to WBG.
REFERENCES
- Alonso M, Medina JH, Pozzo-Miller L. ERK1/2 activation is necessary for BDNF to increase dendritic spine density in hippocampal CA1 pyramidal neurons. Learn Mem. 2004;11:172–178. doi: 10.1101/lm.67804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo JC, Pereira DB, Yano H, Teng KK, Chao MV. Identification of a switch in neurotrophin signaling by selective tyrosine phosphorylation. J Biol Chem. 2006;281:1001–1007. doi: 10.1074/jbc.M504163200. [DOI] [PubMed] [Google Scholar]
- Arevalo JC, Yano H, Teng KK, Chao MV. A unique pathway for sustained neurotrophin signaling through an ankyrin-rich membrane-spanning protein. Embo J. 2004;23:2358–2368. doi: 10.1038/sj.emboj.7600253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J Neurosci. 2004;24:4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath KG, Mandairon N, Jing D, Rajagopal R, Kapoor R, Chen ZY, Khan T, Proenca CC, Kraemer R, Cleland TA, Hempstead BL, Chao MV, Lee FS. Variant brain-derived neurotrophic factor (Val66Met) alters adult olfactory bulb neurogenesis and spontaneous olfactory discrimination. J Neurosci. 2008;28:2383–2393. doi: 10.1523/JNEUROSCI.4387-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracale A, Cesca F, Neubrand VE, Newsome TP, Way M, Schiavo G. Kidins220/ARMS is transported by a kinesin-1-based mechanism likely to be involved in neuronal differentiation. Mol Biol Cell. 2007;18:142–152. doi: 10.1091/mbc.E06-05-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV. Growth factor signaling: where is the specificity? Cell. 1992;68:995–997. doi: 10.1016/0092-8674(92)90068-n. [DOI] [PubMed] [Google Scholar]
- Chao MV. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, Herrera DG, Toth M, Yang C, McEwen BS, Hempstead BL, Lee FS. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline H, Haas K. The regulation of dendritic arbor development and plasticity by glutamatergic synaptic input: a review of the synaptotrophic hypothesis. J Physiol. 2008;586:1509–1517. doi: 10.1113/jphysiol.2007.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Cory S, Fraser SE. Effects of brain-derived neurotrophic factor on optic axon branching and remodelling in vivo. Nature. 1995;378:192–196. doi: 10.1038/378192a0. [DOI] [PubMed] [Google Scholar]
- Cohen-Cory S, Lom B. Neurotrophic regulation of retinal ganglion cell synaptic connectivity: from axons and dendrites to synapses. Int J Dev Biol. 2004;48:947–956. doi: 10.1387/ijdb.041883sc. [DOI] [PubMed] [Google Scholar]
- Cortes RY, Arevalo JC, Magby JP, Chao MV, Plummer MR. Developmental and activity-dependent regulation of ARMS/Kidins220 in cultured rat hippocampal neurons. Dev Neurobiol. 2007;67:1687–1698. doi: 10.1002/dneu.20542. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G. New York: Elsevier Academic Press; 2004. The mouse brain in stereotaxic coordinates; p. 360. [Google Scholar]
- Goldberg JL. Intrinsic neuronal regulation of axon and dendrite growth. Curr Opin Neurobiol. 2004;14:551–557. doi: 10.1016/j.conb.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Zeiler SR, Tamowski S, Jones KR. Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J Neurosci. 2003;23:6856–6865. doi: 10.1523/JNEUROSCI.23-17-06856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk WA, Jiang H, Tartaglia N, Feng L, Figurov A, Lu B. Signaling mechanisms mediating BDNF modulation of synaptic plasticity in the hippocampus. Learn Mem. 1999;6:243–256. [PMC free article] [PubMed] [Google Scholar]
- Grutzendler J, Kasthuri N, Gan WB. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- Gutierrez H, Dolcet X, Tolcos M, Davies A. HGF regulates the development of cortical pyramidal dendrites. Development. 2004;131:3717–3726. doi: 10.1242/dev.01209. [DOI] [PubMed] [Google Scholar]
- Heuer H, Mason CA. Thyroid hormone induces cerebellar Purkinje cell dendritic development via the thyroid hormone receptor alpha1. J Neurosci. 2003;23:10604–10612. doi: 10.1523/JNEUROSCI.23-33-10604.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horch HW. Local effects of BDNF on dendritic growth. Rev Neurosci. 2004;15:117–129. doi: 10.1515/revneuro.2004.15.2.117. [DOI] [PubMed] [Google Scholar]
- Horch HW, Katz LC. BDNF release from single cells elicits local dendritic growth in nearby neurons. Nat Neurosci. 2002;5:1177–1184. doi: 10.1038/nn927. [DOI] [PubMed] [Google Scholar]
- Hu B, Nikolakopoulou AM, Cohen-Cory S. BDNF stabilizes synapses and maintains the structural complexity of optic axons in vivo. Development. 2005;132:4285–4298. doi: 10.1242/dev.02017. [DOI] [PubMed] [Google Scholar]
- Iglesias T, Cabrera-Poch N, Mitchell MP, Naven TJ, Rozengurt E, Schiavo G. Identification and cloning of Kidins220, a novel neuronal substrate of protein kinase D. J Biol Chem. 2000;275:40048–40056. doi: 10.1074/jbc.M005261200. [DOI] [PubMed] [Google Scholar]
- Klein R. Eph/ephrin signaling in morphogenesis, neural development and plasticity. Curr Opin Cell Biol. 2004;16:580–589. doi: 10.1016/j.ceb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Kong H, Boulter J, Weber JL, Lai C, Chao MV. An evolutionarily conserved transmembrane protein that is a novel downstream target of neurotrophin and ephrin receptors. J Neurosci. 2001;21:176–185. doi: 10.1523/JNEUROSCI.21-01-00176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci U S A. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Grishanin RN, Tolwani RJ, Renteria RC, Xu B, Reichardt LF, Copenhagen DR. Brain-derived neurotrophic factor and TrkB modulate visual experience-dependent refinement of neuronal pathways in retina. J Neurosci. 2007;27:7256–7267. doi: 10.1523/JNEUROSCI.0779-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B. BDNF and activity-dependent synaptic modulation. Learn Mem. 2003;10:86–98. doi: 10.1101/lm.54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- McFarlane S. Dendritic morphogenesis: building an arbor. Mol Neurobiol. 2000;22:1–9. doi: 10.1385/MN:22:1-3:001. [DOI] [PubMed] [Google Scholar]
- Meyer MP, Smith SJ. Evidence from in vivo imaging that synaptogenesis guides the growth and branching of axonal arbors by two distinct mechanisms. J Neurosci. 2006;26:3604–3614. doi: 10.1523/JNEUROSCI.0223-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niblock MM, Brunso-Bechtold JK, Riddle DR. Insulin-like growth factor I stimulates dendritic growth in primary somatosensory cortex. J Neurosci. 2000;20:4165–4176. doi: 10.1523/JNEUROSCI.20-11-04165.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niell CM, Meyer MP, Smith SJ. In vivo imaging of synapse formation on a growing dendritic arbor. Nat Neurosci. 2004;7:254–260. doi: 10.1038/nn1191. [DOI] [PubMed] [Google Scholar]
- Patterson SL, Abel T, Deuel TA, Martin KC, Rose JC, Kandel ER. Recombinant BDNF rescues deficits in basal synaptic transmission and hippocampal LTP in BDNF knockout mice. Neuron. 1996;16:1137–1145. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Ruthazer ES, Li J, Cline HT. Stabilization of axon branch dynamics by synaptic maturation. J Neurosci. 2006;26:3594–3603. doi: 10.1523/JNEUROSCI.0069-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AL, Matthews BJ, Meynard MM, Hu B, Javed S, Cohen Cory S. BDNF increases synapse density in dendrites of developing tectal neurons in vivo. Development. 2006;133:2477–2486. doi: 10.1242/dev.02409. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW. Development of the vertebrate neuromuscular junction. Annu Rev Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- Scott EK, Luo L. How do dendrites take their shape? Nat Neurosci. 2001;4:359–365. doi: 10.1038/86006. [DOI] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Tessarollo L. Manipulating mouse embryonic stem cells. Methods Mol Biol. 2001;158:47–63. doi: 10.1385/1-59259-220-1:47. [DOI] [PubMed] [Google Scholar]
- Vaughn JE. Fine structure of synaptogenesis in the vertebrate central nervous system. Synapse. 1989;3:255–285. doi: 10.1002/syn.890030312. [DOI] [PubMed] [Google Scholar]
- Vessey JP, Karra D. More than just synaptic building blocks: scaffolding proteins of the post-synaptic density regulate dendritic patterning. J Neurochem. 2007;102:324–332. doi: 10.1111/j.1471-4159.2007.04662.x. [DOI] [PubMed] [Google Scholar]
- Wiesel TN, Hubel DH. Ordered arrangement of orientation columns in monkeys lacking visual experience. J Comp Neurol. 1974;158:307–318. doi: 10.1002/cne.901580306. [DOI] [PubMed] [Google Scholar]
- Xu B, Gottschalk W, Chow A, Wilson RI, Schnell E, Zang K, Wang D, Nicoll RA, Lu B, Reichardt LF. The role of brain-derived neurotrophic factor receptors in the mature hippocampus: modulation of long-term potentiation through a presynaptic mechanism involving TrkB. J Neurosci. 2000;20:6888–6897. doi: 10.1523/JNEUROSCI.20-18-06888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HT, Pan F, Yang G, Gan WB. Choice of cranial window type for in vivo imaging affects dendritic spine turnover in the cortex. Nat Neurosci. 2007;10:549–551. doi: 10.1038/nn1883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dendritic parameters as indicated were measured in the basal dendrites of Layer 5 pyramidal neurons in the barrel cortex and dentate gyrus granule cells of 1- and 3-month-old ARMS+/− mice and wild-type littermates. (n = as indicated for Sholl analyses). The number of primary dendrites in ARMS+/− mice at 3 months of age was significantly fewer than that of wild-type mice (n = 30 neurons from two mice per genotype; data = mean ± s.e.m.; *p < 0.05, Student’s t-test).