Abstract
Chronic administration of lysergic acid diethylamide (LSD) every other day to rats results in a variety of abnormal behaviors. These build over the 90 day course of treatment and can persist at full strength for at least several months after cessation of treatment. The behaviors are consistent with those observed in animal models of schizophrenia and include hyperactivity, reduced sucrose-preference, and decreased social interaction. In order to elucidate molecular changes that underlie these aberrant behaviors, we chronically treated rats with LSD and performed RNA-Sequencing on the medial prefrontal cortex (mPFC), an area highly associated with both the actions of LSD and the pathophysiology of schizophrenia and other psychiatric illnesses. We observed widespread changes in the neurogenetic state of treated animals four weeks after cessation of LSD treatment. QPCR was used to validate a subset of gene expression changes observed with RNA-Seq, and confirmed a significant correlation between the two methods. Functional clustering analysis indicates differentially expressed genes are enriched in pathways involving neurotransmission (Drd2, Gabrb1), synaptic plasticity (Nr2a, Krox20), energy metabolism (Atp5d, Ndufa1) and neuropeptide signaling (Npy, Bdnf), among others. Many processes identified as altered by chronic LSD are also implicated in the pathogenesis of schizophrenia, and genes affected by LSD are enriched with putative schizophrenia genes. Our results provide a relatively comprehensive analysis of mPFC transcriptional regulation in response to chronic LSD, and indicate that the long-term effects of LSD may bear relevance to psychiatric illnesses, including schizophrenia.
Keywords: LSD, schizophrenia, mPFC, serotonin, rat, RNA-Seq
1. Introduction
Lysergic acid diethylamide (LSD) is a classic hallucinogen that can produce a profound acute intoxication characterized by hallucinations, delusions, detachment from reality, and paranoia (Nichols, 2004). Certain effects of LSD are recognized to be similar to those presenting in schizophrenia, and have led us and others to propose the use of LSD to model aspects of schizophrenia and psychosis (Geyer and Vollenweider, 2008, Marona-Lewicka et al., 2011, Halberstadt and Geyer, 2013, Hanks and Gonzalez-Maeso, 2013). Like other hallucinogens, LSD directly activates 5-HT2A receptors and indirectly modulates glutamatergic neurotransmission to produce its primary effects (Nichols, 2004). LSD also has affinity for several other receptors, including serotonin 5-HT1A, 5-HT2C, dopamine D1, and D2, which all may contribute to its behavioral pharmacology (Halberstadt and Geyer, 2011). LSD is unique among hallucinogens, however, in that it produces two distinct temporal phases, "an acute phase (about 4 hours in man) and a 4- to 6-hr second phase. During the second 6 hr, the ‘TV show in the head’ no longer compels interest; subjects think often that the effect is over but fairly regularly report (at perhaps 10 hr after an initial dose) that they had been at the least self-centered, and usually suspicious, with ideas of reference or even paranoid convictions” (Freedman, 1984). Freedman also noted that acute LSD intoxication disrupts cognitive control and attentional processes, alongside the sensory changes and delusional tendencies characteristic of the first and second phases (Freedman, 1968). In rat drug discrimination experiments, LSD also produces a unique biphasic temporal profile with a second phase mediated by dopaminergic mechanisms (Marona-Lewicka et al., 2005).
Although the effects of acute LSD have been well studied, long-term administration of LSD in humans has not. Tolerance quickly develops to most of the overt effects with daily dosing, but sensitivity returns to baseline after three days of abstinence (Belleville et al., 1956). Experimenters who gave autistic children LSD daily for many months noted less tolerance than had been reported in adults, and relatively few adverse events (Bender et al., 1961). In general, however, no studies have specifically monitored long-term behavioral changes resulting from repeated LSD administration in normal healthy humans, and LSD is not a drug taken on a daily basis over several months by recreational users. Repeated recreational LSD use on a less frequent basis has been associated with psychological, perceptual, and cognitive abnormalities including decreased performance on tests of visual perception and spatial orientation, a high incidence of magical thinking, decreased performance on tests of nonverbal abstract reasoning, and hallucinogen persisting perception disorder (HPPD) (Blacker et al., 1968, McGlothlin et al., 1969, Abraham, 1982, Halpern and Pope, 2003). Prolonged psychotic reactions have been reported following LSD use, however, the incidence of these phenomena is rare and may be related to preexisting vulnerability (Nichols, 2004).
We recently reported that chronic administration of LSD results in a variety of persistent abnormal behaviors in rats (Marona-Lewicka et al., 2011). Altered behaviors include hyperactivity, hyper-reactivity, abolished preference for sucrose solution, and altered social behaviors (Marona-Lewicka et al., 2011). These behaviors are similar to many that present in rodent models of schizophrenia. Interestingly, the aberrant behavioral changes induced by LSD continue for at least many months after discontinuance of the LSD treatment. For example, increased locomotion persists for at least three months following cessation of LSD treatment. Both olanzapine and haloperidol temporarily attenuate this hyperlocomotion in doses that do not affect control activity.
In an effort to understand the neurochemical and molecular basis for the persistent changes in behavior induced by chronic LSD, we performed high-throughput RNA sequencing to profile gene expression changes in the medial prefrontal cortex (mPFC) of rats chronically treated with LSD. We have found that many genes are persistently dysregulated four weeks after discontinuation of LSD treatment. Functional clustering indicates that the processes in which these genes are enriched include neurotransmitter function, synaptic plasticity, metabolism, and endocrine function. Significantly, genes and networks affected by chronic LSD demonstrate substantial overlap with genes and pathways implicated in schizophrenia. Together with our pharmacological and behavioral results, these data support the notion that rats chronically treated with LSD may serve as a useful new model of the sustained, abnormal functioning observed in psychiatric illnesses such as schizophrenia.
2. Methods
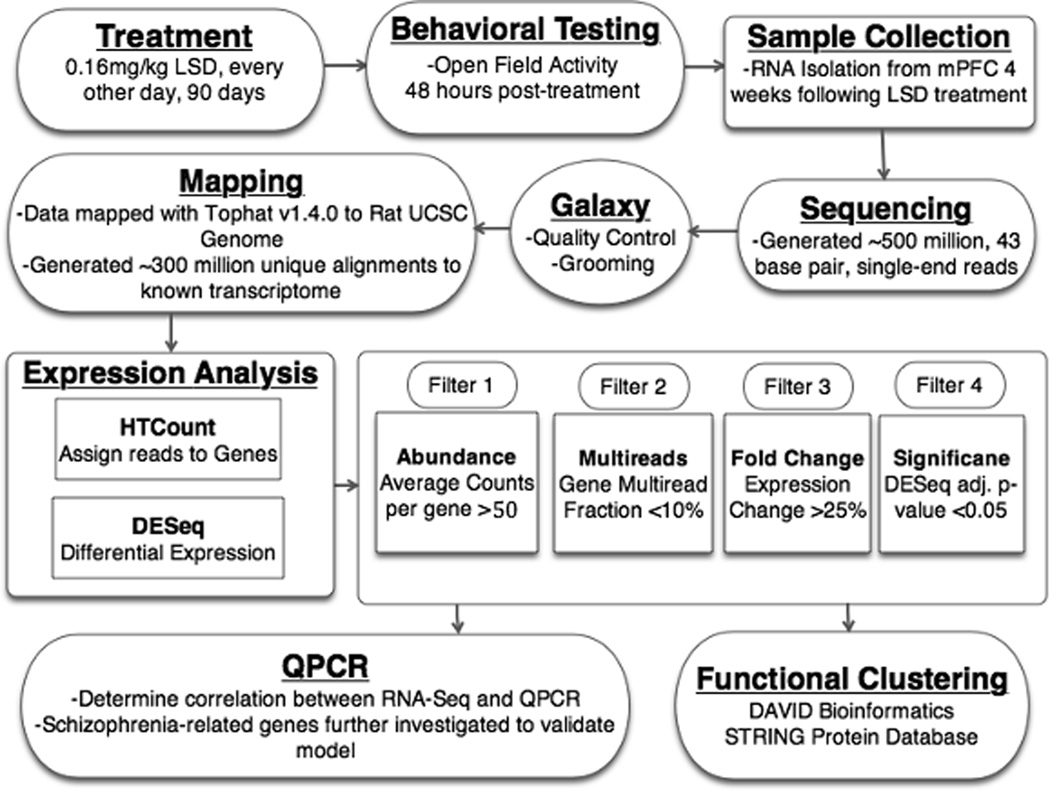
A summary of the methods can be found in Figure 1.
Figure 1.
An outline of experimental procedures is shown. A.) Rats 180–200g (N=10/group) are treated with saline or 0.16 mg/kg LSD, every other day, for 90 days. B.) Locomotor testing at 48 hours post-treatment demonstrating persistent hyperactivity is followed by collection of brains at four weeks post-treatment. C.) Sample preparation consists of isolation of mPFC and extraction of RNA, followed by standard Illumina library preparation protocol. D.) Sequencing performed by Vanderbilt Genome Resources Core, resulting in over 500 million, 43 bp, single-end reads. E.) Raw data uploaded to Galaxy Public Instance for further manipulation and mapping F.) Topaht v 1.4.0 was used to map reads to 2004 UCSC Rat Genome, and generated over 300 million unique alignments to the transcriptome. G.) HTCount was used for counting reads aligning to gene loci, and DE-Seq v1.82 was used for differential expression testing. H.) Increasingly stringent filters applied to data result in a collection of differentially expressed genes for downstream analysis. I.) Subset of differentially expressed genes and schizophrenia-related genes further tested with QPCR. J.) Functional clustering analysis performed to reveal highly represented pathways which are persistently altered by chronic LSD.
2.1 Animals and treatment
Male Sprague-Dawley rats obtained from Harlan (Indianapolis, IN) were aged 6–8 weeks (180–200 g) at the start of treatment. Animals were housed individually for seven days before treatment, during which time they were handled for 15 min each day. During treatment, rats were individually housed in translucent home cages, allowed free access to food and water, and kept on a 12:12 h light/dark cycle. For treatment, rats were injected (i.p.) with either sterile saline, or LSD (0.16 mg/kg) every other day for 90 days. LSD [(+)-lysergic acid diethylamide tartrate], was synthesized in the laboratory of Dr. D.E. Nichols. Animals used in these studies were maintained in accordance with the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals, and all protocols were approved by the Institutional Animal Care and Use Committees of both LSUHSC and Purdue University.
2.2 Sample preparation
After discontinuation of LSD, all animals (N=10/treatment group) were behaviorally tested to confirm the presence of hyperactivity as previously described, (Marona-Lewicka et al., 2011) and at four weeks were euthanized by decapitation. Brains were removed and immediately frozen on dry ice and stored at −80 °C until processing. The mPFC from one hemisphere was dissected and processed for simultaneous isolation of total RNA, protein, and gDNA using the Norgen All-In-One kit (Toronto, CAN) following manufacturer’s directions. RNA quality was assayed on an Agilent Bioanalyzer. All samples sequenced exhibited an RIN > 8.
2.3 RNA Sequencing
Total RNA (4 µg) was provided to the Genome Sciences Resource core at Vanderbilt University School of Medicine for sequencing (https://gtc.vanderbilt.edu/). Sample libraries were prepared with Illumina mRNA sample preparation kits. Briefly, this method requires mRNA enrichment through polyA tail selection, cDNA transcription, fragmentation, fragment size selection, adapter ligation, and cluster generation. Sequencing was performed on an Illumina GA IIX. Read length was 43 base pairs, and sequencing resulted in an average depth of ~26 million reads/sample. Each sample was sequenced on a separate lane of an Illumina flow cell, requiring three flow cells in total (20 independent samples; 8 lanes/flow cell).
2.4 Data Analysis
We developed our own workflow for analyzing RNA-sequencing data using publicly available resources. A Web-based graphical front end for publicly available sequencing packages, including Tophat and Cufflinks, named GALAXY (Giardine et al., 2005) was used for the initial analysis (http://main.g2.bx.psu.edu/).
Data files (Illumina v1.3 FASTQ) were uploaded to GALAXY and converted to Sanger format using the FASTQ Groomer (Blankenberg et al., 2010). Mapping was performed with Tophat (Trapnell et al., 2009) v1.4.0 to map individual reads to the UCSC Rat 2004 Genome (rn4). Segments with a minimum length of 21 bp were mapped independently, with a maximum of two mismatches allowed per segment, with 0 mismatches allowed in anchor regions of spliced reads (eight base pairs on either side of a splice junction). Known junctions were provided through the UCSC RefSeq annotation, and unknown junctions were built de novo with Tophat. A maximum of 10 alignments were allowed per read, and any read exceeding this number of genomic alignments was discarded. For gene expression quantification, we performed initial trial runs with both Cufflinks (Trapnell et al., 2010), available on GALAXY, and an R package DE-Seq (Anders and Huber, 2010). After several initial trials with various iterations and parameter settings of the two algorithms, we chose to perform the final expression analysis with DE-Seq v1.82, based on the robustness, simplicity, and interpretability of the results.
BAM files were converted to SAM using the BAM-to-SAM conversion tool (Li et al., 2009) on GALAXY. Alignments were assigned to gene loci using HT-Seq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html). Gene loci were defined by the UCSC RefSeq annotation, and HTSeq was run with the Union_Intersection parameter. HTSeq by default only counts reads uniquely mapping to a single location in the transcriptome, and these are the count data we used for differential expression testing. We additionally modified the output to count non-uniquely mapped reads to determine which genes had large fractions of discarded multireads. A table of gene counts for 16950 known rat genes and 20 samples provided the input to DE-Seq. DE-Seq uses the negative binomial distribution to model counts distributed across genes (Anders and Huber, 2010), and the output consists of normalized average gene counts for each condition, along with p-values, and p-values adjusted for multiple testing for every gene.
For the final list of candidate genes we employed multiple filters: 1) Defined Genes; 2) Genes with >50 reads aligned; 3) Genes with less than 10% of total reads consisting of multireads; 4) Genes with expression changes ≥ 25% compared to control; 5) Genes with p-values <0.05 (adjusted for multiple comparison), according to DE-Seq v1.82. Functional clustering analysis of the final candidate list was performed using the DAVID Bioinformatics Resource (http://david.abcc.ncifcrf.gov/home.jsp) (Huang da et al., 2009b,a) and the STRING Protein Interaction Database (Franceschini et al., 2013).
2.5 Individual Gene Expression analysis by QPCR
First strand cDNA was synthesized using the ImPromp-II kit from Promega (Madison, WI). For the quantitative RT-PCR (QPCR) experiments, the Universal ProbeLibrary system from Roche (Indianapolis, IN) was used to design primer/probe pairs. All primers were aligned to transcripts as defined by the UCSC RefSeq annotation used in the sequencing analysis, and spanned exon/exon boundaries where possible. Distinction was not made between potential splice isoforms, but primers were aligned to exons in the primary transcript of a gene. Primers were synthesized by IDT (Coralville, IA) (sequences provided in Supplementary Table S1). Triplicate amplification reactions using the first strand cDNA sample from each rat were performed on a Roche 480 LightCycler II using the Roche Light Cycler Master Mix following the manufacturer’s directions. Amplification of Ywhah, a housekeeping gene consistently expressed among all samples, was performed simultaneously on every plate to normalize amplification thresholds. Relative expression was determined using the ΔΔ-Ct method. Significance of expression changes was determined by performing one-tailed Student’s t-tests, in the direction implicated by fold changes measured with RNA-Seq. All genes marked as significant exhibit p<0.05, and a false detection rate (FDR) <10% (Benjamini Y, 1995).
3. RESULTS
3.1 Sequencing Analysis
An overview of the experimental procedures is provided in Figure 1. Treated animals exhibited increased locomotor activity in the open field post-treatment, consistent with treated groups of animals previously described, (Marona-Lewicka et al, 2011) and were sacrificed four weeks after the last treatment with LSD or saline. Sequence data from all 20 samples were of high quality, with well over 99% of the reads in each sample exhibiting an average PHRED quality score of over 35, and over 90% of reads lacking a single base call with a PHRED score <20. Data exhibit the expected characteristics of single-end, random-primed Illumina GAII sequences (Hansen et al., 2010).
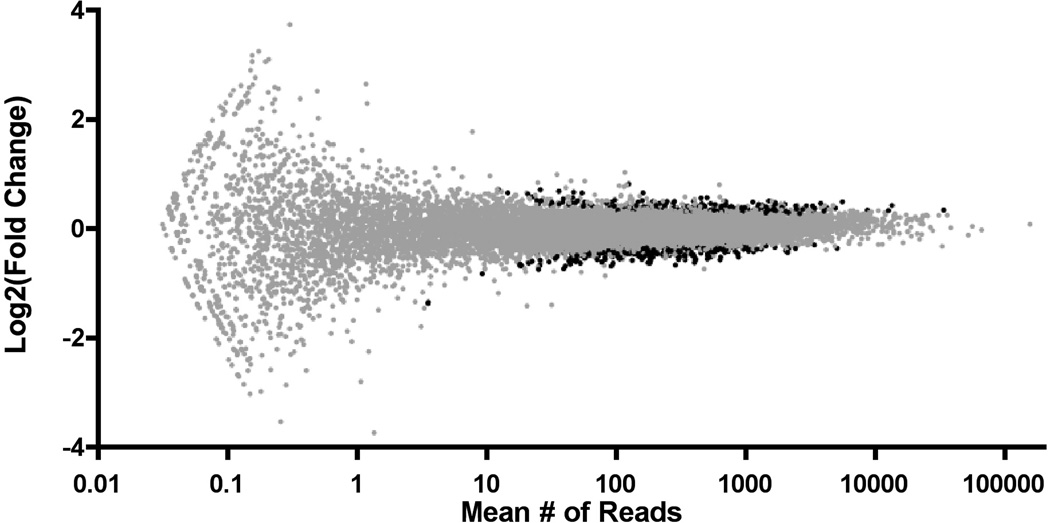
Tophat was used to map reads to the genome, and HTSeq was used to assign reads to individual genes (Supplementary Table S2). 527 million reads were sequenced, with an average of 26.4 million reads per sample resulting in 474 million alignments to the genome, of which 309 million reads mapped uniquely to a known gene. 58% of generated reads mapped uniquely with high confidence to a known gene, and only these reads were used in downstream differential expression analysis. Of the 42% of mapped reads that were not aligned uniquely to known genes, many were multireads that mapped to two or more places on the genome with equal probability. Also, many represent poorly annotated or unknown genes in the rat genome and were ignored. Additionally, a small fraction mapped into ambiguous or overlapping exonic regions. Based on RNA-sequencing results in highly annotated genomes (Daines et al., 2011, Nookaew et al., 2012), a high percentage (>95%) of the reads are likely generated by genuine mRNA transcripts and are not artifacts. The raw, per-gene count data generated in HTSeq provides the input to DE-Seq. The count data for each gene in a given sample is then normalized by a scaling factor calculated in DE-Seq, allowing fold-changes across conditions to be calculated. The distribution of the differential expression data is shown in Figure 2, and indicates that chronic LSD induces widespread but relatively low-magnitude changes in gene expression in the mPFC.
Figure 2.
Global differential gene expression changes are shown above, as calculated with DE-Seq v1.82. Each defined gene with >0 reads is placed on the x-axis according to average number of mapped reads and on the y-axis according to Log2(LSD counts/Control counts). Differentially expressed genes (adj. p<0.05) are shown in black.
Criteria were applied to all quantified genes to produce a list of genes potentially affected by chronic LSD. Of 14740 defined genes with at least one mapped read, 10097 genes in the mPFC had average expression levels reaching 50 counts/gene/sample, given our sequencing depth. Of these genes, 9605 had little or no potentially confounding multiread fraction (<10%). From these highly expressed transcripts, 636 exhibited an absolute expression change of > 25%, and 402 exhibited differential expression according to DE-Seq (p < 0.05, adjusted for multiple testing). There were 283 transcripts that were both >25% changed and had a significance of p < 0.05. A spreadsheet detailing expression data for all genes passing differential expression criteria can be found in Supplementary Table S4.
3.2 QPCR Validation
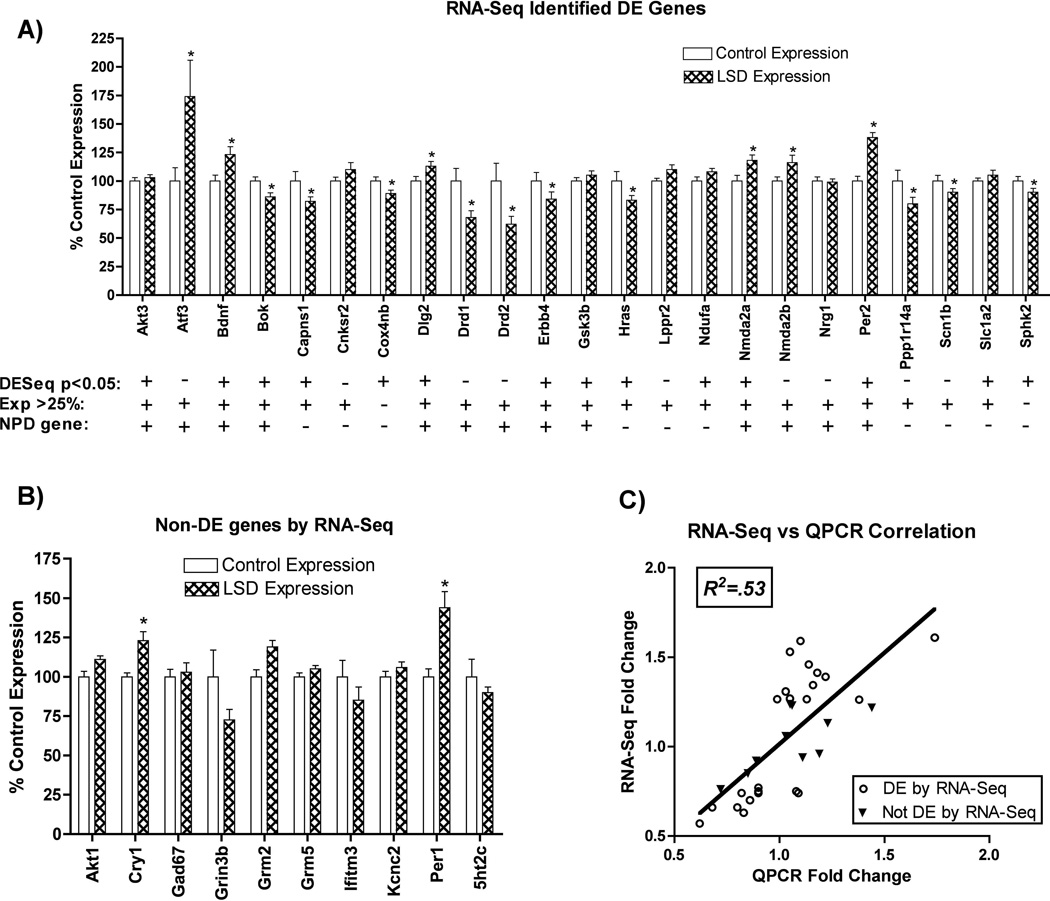
To validate our sequencing analysis as a suitable strategy to measure changes in gene expression, we used QPCR to measure expression levels for a subset of the same transcripts analyzed with RNA sequencing. From our most stringent list, those with >25% expression changes and p < 0.05, we tested 12 genes and found a QPCR validation rate of 66.7% (8/12). We further tested an additional 11 genes from an expanded list that represented 755 transcripts that exhibited either > 25% expression or a DE-Seq significance of p < 0.05, and observed a QPCR validation rate of 72.7% (8/11). Because we observed similar validation rates between our most stringent list and the expanded list, we moved forward with the expanded list for our final pool of candidates that represent genes with > 25% differential expression or a DE-Seq significance of p < 0.05. In total, our methodology has achieved a validation rate of 69.6% (16/23) (Figure 3A).
Figure 3.
QPCR gene expression changes are shown relative to control expression. A) 23 genes found to be significantly different using RNA-Seq (adj. p<0.05 or FoldChange >25%) were chosen to be further tested by QPCR. Genes marked with “*” were found to by QPCR to be significantly differentially expressed (p<0.05, t-test, N=10, FDR<10%) Symbols ("+" or "−") below each gene indicate whether the gene exhibits: 1) DESeq significance (adj. p<0.05) 2) DESeq FoldChange >25% 3) Evidence for involvement in neuropsychiatric disease(NPD). B) 10 genes not found to be differentially expressed by RNA-Seq, but thought to be relevant to psychiatric diseases were tested by QPCR, and results are shown relative to control expression. Genes marked with “*” were found to by QPCR to be significantly differentially expressed (p<0.05, t-test, N=10, FDR<10%). C) QPCR and RNA-Seq Correlation shown above. All 33 genes tested by QPCR are shown; open circles represent genes differentially expressed by RNA-Seq, and triangles represent genes not differentially expressed by RNA-Seq. The horizontal and vertical position of each dot represents QPCR Fold Change and RNA-Seq Fold Change, respectively. Pearson’s correlation coefficient was calculated after linear regression, R2=.53 (p<0.001). Best Fit slope = 1.02 +/− 0.17.
Some genes potentially relevant to psychiatric illness were identified from our candidate pool and tested as part of our validation as described above. To determine whether there are neuropsychiatric disease (NPD) relevant genes altered but not detected by sequencing, we tested 10 genes implicated by others in psychiatric disease that did not meet criteria for inclusion in our final candidate pool. Of these, 2/10 were found to be differentially expressed by QPCR (Figure 3B). To determine the accuracy of differential expression measurements obtained with RNA-Seq, we compared fold changes measured with QPCR to fold changes measured by RNA-Seq for each of the 33 total genes measured by QPCR. We observed a highly significant correlation between expression levels as determined by RNA-Seq and QPCR (Pearson correlation, R2 = .53, p<0.0001) (Figure 3C), indicating a general concurrence between QPCR and RNA-Seq for estimating fold changes.
3.3 Functional Clustering
In order to investigate which processes were most affected by LSD treatment and which changes might underlie the persistent changes in behavior, we performed functional clustering analysis on the 755 genes in our final candidate pool. The DAVID Bioinformatic Database and STRING Protein Interaction Database were mined for our candidate gene lists, and the most significant results of the DAVID Functional Clustering are shown in Table 1. Broad processes such as neurotransmission and synaptic plasticity were among the most affected. The graphical output from the STRING analysis is shown in Supplementary Figure S1. Under stringent interaction settings, about one-third of the genes are represented by multiple interconnected nodes. Several functionally related clusters are present, including neurotransmitter receptor function, and effector signaling pathways. Interestingly, other clusters including those for RNA processing, energy metabolism, and endocrine function are also present.
Table 1.
DAVID Functional Clustering Table. The 20 most significant functional categories as determined by the relative enrichment of these functions with differentially expressed genes are shown. The number of differentially expressed genes in a given category is listed along with the p-value for the category, and the adjusted p-value for multiple testing. Significance is calculated by comparing the fraction of differentially expressed genes that fit the functional category to the fraction of all genes that are part of the category using a modified Fisher’s exact test (EASE score).
| Functional Group | Count | P-Value | Benjamini |
|---|---|---|---|
| transmission of nerve impulse | 36 | 6.25E−10 | 1.67E−06 |
| synaptic transmission | 30 | 4.01E−09 | 5.36E−06 |
| regulation of synaptic plasticity | 17 | 4.95E−08 | 4.41E−05 |
| synapse | 37 | 7.35E−07 | 9.63E−05 |
| regulation of system process | 34 | 1.61E−07 | 1.07E−04 |
| synapse part | 30 | 3.21E−07 | 1.26E−04 |
| behavior | 40 | 2.72E−07 | 1.45E−04 |
| regulation of neurological system process | 24 | 7.04E−07 | 2.69E−04 |
| regulation of transmission of nerve impulse | 23 | 8.51E−07 | 2.84E−04 |
| regulation of synaptic transmission | 22 | 1.12E−06 | 3.33E−04 |
| non-membrane-bounded organelle | 108 | 4.86E−06 | 4.78E−04 |
| learning or memory | 19 | 2.42E−06 | 6.46E−04 |
| cell projection organization | 33 | 7.28E−06 | 0.001766915 |
| cell-cell signaling | 31 | 8.52E−06 | 0.001895161 |
| cytoskeleton | 63 | 2.74E−05 | 0.002149377 |
| regulation of neuronal synaptic plasticity | 11 | 1.22E−05 | 0.002509196 |
| Huntington’s disease | 22 | 2.21E−05 | 0.003179466 |
| phosphorylation | 51 | 1.90E−05 | 0.003611326 |
| plasma membrane | 128 | 7.88E−05 | 0.005148794 |
| phosphorus metabolic process | 57 | 3.42E−05 | 0.005687728 |
To identify potential overlap of our candidate set of genes with those identified as relevant to psychiatric illness, we sought to explicitly compare our gene expression results to comprehensive studies of genetic vulnerability for mental disorders in humans. A variety of methods have been used to associate genetic variation with psychiatric disorders, including genome-wide association studies (GWAS), linkage analysis, and post-mortem gene expression analysis. One technique for meta-analysis that is termed ‘convergent functional genomics’ (CFG) uses human genetic data, biomarkers, genetic animal models, and drug-induced animal models from many studies to rank genes’ potential involvement in schizophrenia, (Ayalew et al., 2012) as well as other psychiatric disorders including bipolar disorder and alcoholism (Patel et al., 2010). The relative enrichment of our identified genes among the top-ranked genes from schizophrenia and bipolar disorder is shown in Table 2. One would expect approximately 4.55% (755/16,950) of the top CFG genes to appear randomly in our differential expression list; we observed 14.3% (6/42, p=0.006, chi-squared, Yate’s correction) in the case of schizophrenia, and 8.9% (5/56, p=0.19, chi-squared, Yate’s correction) in the case of bipolar disorder. This 3.2-fold relative enrichment of genes highly implicated in schizophrenia on our differential expression list suggests a significant degree of overlap between genes involved in schizophrenia and those affected by chronic LSD. Additionally, comparison of LSD-altered gene clusters with pathway analysis generated from top CFG genes (see Ayalew et al., 2012) reveals that three of the top six CFG pathways (glutamate receptor signaling, G protein-coupled receptor signaling, and synaptic long-term potentiation) are also enriched with genes differentially expressed in response to chronic LSD.
Table 2.
Convergent Functional Genomics(CFG) is one method of meta-analysis to identify genes which confer susceptibility to complex genetic disorders, including schizophrenia and bipolar disorder. The comparison of chronic LSD induced, differentially expressed genes to the top ranked genes implicated in schizophrenia and bipolar disorder reveals 6 genes common to both lists for schizophrenia, (Bdnf, Drd2, Grin2b, Prkca, Slc1a2, Tnik) and 5 genes in common for bipolar disorder (Bdnf, Gsk3b, Klf12, Ptprt, Syn3). This represents a significant (p=0.006) enrichment of 3.2 fold for schizophrenia genes and a nonsignificant (p=0.19) enrichment of 2.0 fold for bipolar disorder genes (chi-squared, Yates’ corrected, 2×2 contingency table).
| Top Ranked Schizophrenia Genes (CFG score >4.0)(Ayalew et al., 2012) |
Top Ranked Bipolar Genes (CFG score > 6.5)(Patel et al., 2010) |
|---|---|
|
Expect: 4.45% (# of DE genes/Total genes: 755/16950) |
Expect: 4.45% (# of DE genes/Total genes: 755/16950) |
| Found: 14.3% (6/42 genes) | Found: 8.9% (5/56 genes) |
| Relative Enrichment: 3.2 Fold | Relative Enrichment: 2.0 Fold |
| Odds Ratio: 3.6 | Odds Ratio: 2.1 |
| Significance: p=0.006 | Significance: p=0.19 |
4. Discussion
4.1 Summary of gene expression changes
In this work we have characterized global mRNA changes that occur in the mPFC four weeks after discontinuation of chronic LSD administration. Extended exposure to LSD produces widespread and long-lasting abnormalities in gene expression that accompany persistent abnormal behaviors. Examination of altered genes using two different functional clustering techniques shows that neurotransmission and synaptic plasticity processes are among the most highly affected by chronic LSD. Specifically, genes relevant to GABA, glutamate, and dopamine systems are highly represented.
The receptor interactions of LSD within the cortex are complex, but hallucinogens are known to alter cellular and network activity in the PFC when given acutely (Celada et al., 2008, Wood et al., 2012). The canonical microcircuitry of the cortex includes reciprocal connections between pyramidal neurons and GABA-ergic interneurons that are rich in 5-HT2A and 5-HT1A receptors (Celada et al., 2013). We simultaneously observed significantly higher levels of the GABA-A ion channels (Gabrb1, Gabrb2, and Gabrg3) in chronic LSD-treated animals, together with a >40% decrease in expression of a GABA transporter gene (Slc6a13). These changes may reflect long-term adaptations in mPFC pyramidal neurons to alterations in GABA release from interneurons, or more broadly, may indicate a disruption in baseline excitatory/inhibitory balance amidst the persistent abnormal drive resulting from chronic stimulation of 5-HT receptors in the cortex.
The acute disruption of cortical electrophysiology by classic psychedelics involves glutamatergic mechanisms as well, as previously demonstrated using both LSD and DOI (Lambe and Aghajanian, 2006). We measured changes in several genes related to glutamate signaling following chronic dosing, including increases in the expression of NDMA receptor subunits (NR2a and NR2b). These ion channel subunits are critical for many forms of synaptic plasticity including LTP and LTD, and contribute to a wide variety of cortical synapse development and maintenance functions (Yashiro and Philpot, 2008). Although LSD is known to cause excessive glutamate signaling acutely (Muschamp et al., 2004), the observed alterations in the glutamate system are consistent with long-term changes in synaptic plasticity.
We previously observed that genes involved in synaptic plasticity increased in expression following acute LSD, including transcription factors and cytoskeletal protein genes (Nichols and Sanders-Bush, 2002, 2004). Whereas the expression levels of these genes generally return to baseline several hours after acute treatment, some do not (Nichols et al., 2003). We hypothesize that with repeated stimulation these factors lead to synaptic adaptations in the cortex that eventually stabilize over the course of treatment, reaching a persistent aberrant state. This hypothesis is further supported by the highly significant enrichment of other synaptic plasticity genes among transcripts altered by chronic LSD, such as increases in Bdnf and Krox20 mRNA. These two growth factors are both increased in expression following acute hallucinogen dosing (Vaidya et al., 1997, Nichols and Sanders-Bush, 2002), are involved in a large variety of important synaptic functions, and have been linked to dopamine signaling in the PFC (De Steno and Schmauss, 2009, Xing et al., 2012).
The cortical dopamine system appears to be persistently affected by chronic LSD. We observe about a 40% decrease in the mRNA expression of the dopamine receptor genes Drd1 and Drd2, suggestive of receptor downregulation. That may be due to repeated excess dopaminergic activity in the frontal cortex following chronic LSD. LSD, which is also a dopamine receptor agonist, may be directly acting at the receptors, or it may act through indirect modulation of dopamine release mediated by 5-HT2A receptor activation in the mPFC (Pehek et al., 2006).
Many of the changes we observe after chronic LSD are consistent with the known pharmacology of 5-HT2A ligands and the pathways they couple with acutely, including a decrease in phospholipase C beta (Plcb1) (Sanders-Bush and Conn, 1986), an increase in calcium/calmodulin dependent kinases (Camk4, Cask), and a decrease in a calcium/calmodulin dependent kinase inhibitor (Camk2n2) (Arvanov et al., 1999). Further, we have noted other patterns of gene expression in metabolic and endocrine genes resulting from chronic LSD that may offer insight into the neural mechanisms that underlie the long-term behavioral changes. For example, we found decreases in the expression of four NADH dehydrogenase subunit genes (Ndufs8, Ndufb2, Ndufb7, and Nudufa1), along with decreases in three vacuolar ATPase H+ transport genes (Atp6v0b, Atp6v0e2, Atp6v0c), and a decrease in the mitochondrial ATP synthase subunit gene Atp5d. We also measured significant decreases in two cytochrome c oxidase IV subunit genes (Cox7a2, Cox8a), a related gene (Cox4nb), and in glutathione S-transferases (Gstt2, Gstp2). Our data suggest that altered cortical metabolism, mitochondrial dysfunction, and potentially oxidative stress result from chronic LSD treatment. Interestingly, mitochondrial dysfunction and oxidative stress have been associated with a variety of psychiatric illnesses, and mitochondrial disease can result in psychiatric symptoms (Anglin et al., 2012, Andreazza et al., 2013). Similar patterns of abnormal gene expression and biochemical dysfunction have been found in the PFC of schizophrenia patients (Prabakaran et al., 2004).
Thyrotropin releasing hormone mRNA (Trh) and two thyroid hormone receptor genes (Thra, Thrb) were significantly altered in mPFC gene expression, potentially implicating the hypothalamo-pituitary-thyroid axis in the effects of chronic LSD. Thyrotropin releasing hormone (TRH) has been implicated in a wide range of CNS actions, including modulating acetylcholine, dopamine, and serotonin signaling (Horita, 1998). Additionally, TRH has been shown to modulate a variety of behaviors, including arousal and locomotor activity (Hara et al., 2009). A large cluster of other hormone and neuropeptide-related genes also were significantly regulated by LSD, including those for nuclear orphan receptor-1 (Nor1), orexin receptor (Hcrtr1), tachykinin 2 (Tac2), and Neuropeptide Y (Npy), among others. These systems have the ability to modulate neurotransmission and behavior, and neuropeptide systems have been implicated in psychiatric diseases, including schizophrenia and bipolar disorder (LaCrosse and Olive, 2013, Seifuddin et al., 2013).
4.2 Considerations Regarding Translational Relevance
Psychiatric illness is notoriously difficult to model in animals for several reasons. One fundamental issue for translational research concerns the often nebulous distinctions between psychiatric diseases as they are currently defined. Dimensions of abnormal behavior are often similar between different nosologies, and an overlap in environmental and genetic risk factors for psychiatric illness is being increasingly appreciated (Insel and Cuthbert, 2009). For example, genomic analyses of large patient populations have recently revealed a substantial correlation between single nucleotide polymorphisms (SNPs) and liability for different psychiatric diseases, including bipolar disorder with schizophrenia, and schizophrenia with major depressive disorder (Lee et al., 2013).
In recognizing such issues, the NIMH has adopted the Research Domain Criteria (RDoC) project to organize research around specific functional domains, which reflect fundamental biological mechanisms and are applicable across traditional psychiatric disease boundaries (Insel et al., 2010). These domains are currently broadly defined to include positive valence, negative valence, social functioning, cognitive processing, and arousal/regulatory systems. The domains are further subdivided into more specific constructs, which can be described in a matrix across multiple levels of analysis, including genes, circuits, and behaviors (see NIMH website for details).
The work presented here concerns persistent genetic changes in the mPFC circuit associated with disruptions in behaviors across several functional domains. The behaviors that are known to be altered by chronic LSD fit within fundamental domains currently defined by the RDoC, including arousal (hyperactivity), social functioning (approach and aggression), and positive/negative valence (sucrose preference). Many genes regulated by chronic LSD have been previously implicated as important to these dimensions of behavior within the current RDoC matrix. A summary of perturbed behavioral subdomains and potentially relevant gene changes is shown in Supplementary Table 5 in an abbreviated form of the RDoC matrix. Although patterns of gene expression may covary with behaviors, the evidence from the current study is correlative.
Further work will be required to draw definitive conclusions regarding the relative importance of any specific gene or pathway in generating aspects of abnormal behavior, and to separate primary from secondary phenomena. Studies investigating more specific subdomains of behavior affected by chronic LSD are warranted, particularly concerning the cognitive domain. One caveat to our results is that we have only examined mRNA expression, which does not always parallel protein expression. Therefore, it will be important to measure changes in protein expression in relation to the observed mRNA changes to fully understand functional changes across units of analysis. Additionally, identifying alterations in circuits outside the mPFC will be critical to form a more complete picture of how chronic LSD is causing persistent behavioral changes. However, global mRNA sequencing in the mPFC has provided important clues as to which gene networks might be contributing to the observed behavioral dimensions.
Whereas a multitude of pharmacological and genetic manipulations can lead to similar behaviors in animal models, they often converge on common pathways (Eyles et al., 2012). Psychiatric disorders are both highly heritable and highly polygenic (Ruderfer et al., 2013). Many disparate alterations, such as individually rare copy number variants and combinations of SNPs, converge to produce deficits in common pathways (Kirov et al., 2012, Xu et al., 2012, Gulsuner et al., 2013). Recently, large scale GWAS have identified SNP arrays for bipolar disorder and schizophrenia that can explain a fraction (~1/3) of the estimated total heritability of these diseases (Fernandes et al., 2013, Lee et al., 2013). Other studies indicate little patient-population overlap at the level of individual mutations, but increasing overlap at gene and pathway levels (Ayalew et al., 2012). Comparison of our work to a recent CFG analysis of schizophrenia (Ayalew et al., 2012) at the gene level revealed a substantial enrichment of CFG-prioritized genes in the genes affected by chronic LSD. CFG genes were identified though a variety of methods and studies, however, and were not limited to specific tissues like mPFC. This precludes drawing a precise comparison with our results, moreover, such comparisons are strictly correlative. Despite differences in methodology, comparison of CFG top canonical pathways to our functional clusters reveals similarities at a network interaction level, implicating glutamate signaling and synaptic plasticity as key processes (Ayalew et al., 2012).
No animal model can recapitulate the spectrum of a human psychiatric disorder. However, as the fundamental constructs of biobehavioral function are defined in humans, the validity of animal work can be evaluated with respect to specific functional domains. We have found that chronic LSD results in long-lasting behavioral and genetic changes that are consistent with several biobehavioral dimensions relevant to psychiatric disorders. We propose that further study of the development and maintenance of the stable, aberrant state produced by chronic LSD may be informative to further understanding the etiology of psychiatric disorders, including schizophrenia.
Supplementary Material
Highlights.
-
-
Chronic LSD results in persistent behavioral changes in rats.
-
-
We measured transcriptome changes with RNA-Seq in mPFC in response to chronic LSD.
-
-
QPCR measurements validated the mRNA alterations measured with RNA-Seq.
-
-
Chronic LSD affects synaptic plasticity, neurotransmission, neuroendocrine, and metabolic pathways.
-
-
Molecular and behavioral data indicate chronic LSD is relevant to psychiatric disease.
Acknowledgments
We acknowledge support for this work by NIH grants MH083689 from NIMH to CDN, and DA02189 from NIDA and the Robert C. and Charlotte P. Anderson endowment to DEN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix: Supplementary material
Supplementary information is available at the Neuropharmacology website.
Conflict of Interest:
The authors have no conflicts of interest to report.
Bibliography
- Abraham HD. A chronic impairment of colour vision in users of LSD. Br J Psychiatry. 1982;140:518–520. doi: 10.1192/bjp.140.5.518. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreazza AC, Wang JF, Salmasi F, Shao L, Young LT. Specific subcellular changes in oxidative stress in prefrontal cortex from patients with bipolar disorder. J Neurochem. 2013;127:552–561. doi: 10.1111/jnc.12316. [DOI] [PubMed] [Google Scholar]
- Anglin RE, Garside SL, Tarnopolsky MA, Mazurek MF, Rosebush PI. The psychiatric manifestations of mitochondrial disorders: a case and review of the literature. J Clin Psychiatry. 2012;73:506–512. doi: 10.4088/JCP.11r07237. [DOI] [PubMed] [Google Scholar]
- Arvanov VL, Liang X, Magro P, Roberts R, Wang RY. A pre- and postsynaptic modulatory action of 5-HT and the 5-HT2A, 2C receptor agonist DOB on NMDA-evoked responses in the rat medial prefrontal cortex. Eur J Neurosci. 1999;11:2917–2934. doi: 10.1046/j.1460-9568.1999.00708.x. [DOI] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, Winiger E, Breier A, Shekhar A, Amdur R, Koller D, Nurnberger JI, Corvin A, Geyer M, Tsuang MT, Salomon D, Schork NJ, Fanous AH, O'Donovan MC, Niculescu AB. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleville RE, Fraser HF, Isbell H, Logan CR, Wikler A. Studies on lysergic acid diethylamide (LSD-25). I. Effects in former morphine addicts and development of tolerance during chronic intoxication. AMA Arch Neurol Psychiatry. 1956;76:468–478. [PubMed] [Google Scholar]
- Bender L, Goldschmidt L, Sankar DV. Treatment of autistic schizophrenic children with LSD-25 and UML-491. Recent Adv Biol Psychiatry. 1961;4:170–179. doi: 10.1007/978-1-4684-8306-2_17. [DOI] [PubMed] [Google Scholar]
- Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- Blacker KH, Jones RT, Stone GC, Pfefferbaum D. Chronic users of LSD: the "acidheads". Am J Psychiatry. 1968;125:97–107. [PubMed] [Google Scholar]
- Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A. Manipulation of FASTQ data with Galaxy. Bioinformatics. 2010;26:1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celada P, Puig MV, Artigas F. Serotonin modulation of cortical neurons and networks. Front Integr Neurosci. 2013;7:25. doi: 10.3389/fnint.2013.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celada P, Puig MV, Diaz-Mataix L, Artigas F. The hallucinogen DOI reduces low-frequency oscillations in rat prefrontal cortex: reversal by antipsychotic drugs. Biol Psychiatry. 2008;64:392–400. doi: 10.1016/j.biopsych.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Daines B, Wang H, Wang L, Li Y, Han Y, Emmert D, Gelbart W, Wang X, Li W, Gibbs R, Chen R. The Drosophila melanogaster transcriptome by paired-end RNA sequencing. Genome Res. 2011;21:315–324. doi: 10.1101/gr.107854.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Steno DA, Schmauss C. A role for dopamine D2 receptors in reversal learning. Neuroscience. 2009;162:118–127. doi: 10.1016/j.neuroscience.2009.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyles D, Feldon J, Meyer U. Schizophrenia: do all roads lead to dopamine or is this where they start? Evidence from two epidemiologically informed developmental rodent models. Transl Psychiatry. 2012;2:e81. doi: 10.1038/tp.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes CP, Christoforou A, Giddaluru S, Ersland KM, Djurovic S, Mattheisen M, Lundervold AJ, Reinvang I, Nothen MM, Rietschel M, Ophoff RA, Hofman A, Uitterlinden AG, Werge T, Cichon S, Espeseth T, Andreassen OA, Steen VM, Le Hellard S. A genetic deconstruction of neurocognitive traits in schizophrenia and bipolar disorder. PLoS One. 2013;8:e81052. doi: 10.1371/journal.pone.0081052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman DX. On the Use and Abuse of LSD. Arch Gen Psychiatry. 1968;18:330–347. doi: 10.1001/archpsyc.1968.01740030074008. [DOI] [PubMed] [Google Scholar]
- Freedman DX. LSD: The Bridge from Human to Animal. New York: Raven Press; 1984. [Google Scholar]
- Geyer MA, Vollenweider FX. Serotonin research: contributions to understanding psychoses. Trends Pharmacol Sci. 2008;29:445–453. doi: 10.1016/j.tips.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, Nimgaonkar VL, Go RC, Savage RM, Swerdlow NR, Gur RE, Braff DL, King MC, McClellan JM. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halberstadt AL, Geyer MA. Serotonergic hallucinogens as translational models relevant to schizophrenia. Int J Neuropsychopharmacol. 2013:1–16. doi: 10.1017/S1461145713000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern JH, Pope HG., Jr Hallucinogen persisting perception disorder: what do we know after 50 years? Drug Alcohol Depend. 2003;69:109–119. doi: 10.1016/s0376-8716(02)00306-x. [DOI] [PubMed] [Google Scholar]
- Hanks JB, Gonzalez-Maeso J. Animal models of serotonergic psychedelics. ACS chemical neuroscience. 2013;4:33–42. doi: 10.1021/cn300138m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010;38:e131. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara J, Gerashchenko D, Wisor JP, Sakurai T, Xie X, Kilduff TS. Thyrotropin-releasing hormone increases behavioral arousal through modulation of hypocretin/orexin neurons. J Neurosci. 2009;29:3705–3714. doi: 10.1523/JNEUROSCI.0431-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horita A. An update on the CNS actions of TRH and its analogs. Life Sci. 1998;62:1443–1448. doi: 10.1016/s0024-3205(98)00087-3. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, Sanislow C, Wang P. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- Insel TR, Cuthbert BN. Endophenotypes: bridging genomic complexity and disorder heterogeneity. Biol Psychiatry. 2009;66:988–989. doi: 10.1016/j.biopsych.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, Grozeva D, Fjodorova M, Wollerton R, Rees E, Nikolov I, van de Lagemaat LN, Bayes A, Fernandez E, Olason PI, Bottcher Y, Komiyama NH, Collins MO, Choudhary J, Stefansson K, Stefansson H, Grant SG, Purcell S, Sklar P, O'Donovan MC, Owen MJ. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCrosse AL, Olive MF. Neuropeptide systems and schizophrenia. CNS Neurol Disord Drug Targets. 2013;12:619–632. doi: 10.2174/1871527311312050010. [DOI] [PubMed] [Google Scholar]
- Lambe EK, Aghajanian GK. Hallucinogen-induced UP states in the brain slice of rat prefrontal cortex: role of glutamate spillover and NR2B-NMDA receptors. Neuropsychopharmacology. 2006;31:1682–1689. doi: 10.1038/sj.npp.1300944. [DOI] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, Mowry BJ, Thapar A, Goddard ME, Witte JS, Absher D, Agartz I, Akil H, Amin F, Andreassen OA, Anjorin A, Anney R, Anttila V, Arking DE, Asherson P, Azevedo MH, Backlund L, Badner JA, Bailey AJ, Banaschewski T, Barchas JD, Barnes MR, Barrett TB, Bass N, Battaglia A, Bauer M, Bayes M, Bellivier F, Bergen SE, Berrettini W, Betancur C, Bettecken T, Biederman J, Binder EB, Black DW, Blackwood DH, Bloss CS, Boehnke M, Boomsma DI, Breen G, Breuer R, Bruggeman R, Cormican P, Buccola NG, Buitelaar JK, Bunney WE, Buxbaum JD, Byerley WF, Byrne EM, Caesar S, Cahn W, Cantor RM, Casas M, Chakravarti A, Chambert K, Choudhury K, Cichon S, Cloninger CR, Collier DA, Cook EH, Coon H, Cormand B, Corvin A, Coryell WH, Craig DW, Craig IW, Crosbie J, Cuccaro ML, Curtis D, Czamara D, Datta S, Dawson G, Day R, De Geus EJ, Degenhardt F, Djurovic S, Donohoe GJ, Doyle AE, Duan J, Dudbridge F, Duketis E, Ebstein RP, Edenberg HJ, Elia J, Ennis S, Etain B, Fanous A, Farmer AE, Ferrier IN, Flickinger M, Fombonne E, Foroud T, Frank J, Franke B, Fraser C, Freedman R, Freimer NB, Freitag CM, Friedl M, Frisen L, Gallagher L, Gejman PV, Georgieva L, Gershon ES, Geschwind DH, Giegling I, Gill M, Gordon SD, Gordon-Smith K, Green EK, Greenwood TA, Grice DE, Gross M, Grozeva D, Guan W, Gurling H, De Haan L, Haines JL, Hakonarson H, Hallmayer J, Hamilton SP, Hamshere ML, Hansen TF, Hartmann AM, Hautzinger M, Heath AC, Henders AK, Herms S, Hickie IB, Hipolito M, Hoefels S, Holmans PA, Holsboer F, Hoogendijk WJ, Hottenga JJ, Hultman CM, Hus V, Ingason A, Ising M, Jamain S, Jones EG, Jones I, Jones L, Tzeng JY, Kahler AK, Kahn RS, Kandaswamy R, Keller MC, Kennedy JL, Kenny E, Kent L, Kim Y, Kirov GK, Klauck SM, Klei L, Knowles JA, Kohli MA, Koller DL, Konte B, Korszun A, Krabbendam L, Krasucki R, Kuntsi J, Kwan P, Landen M, Langstrom N, Lathrop M, Lawrence J, Lawson WB, Leboyer M, Ledbetter DH, Lee PH, Lencz T, Lesch KP, Levinson DF, Lewis CM, Li J, Lichtenstein P, Lieberman JA, Lin DY, Linszen DH, Liu C, Lohoff FW, Loo SK, Lord C, Lowe JK, Lucae S, MacIntyre DJ, Madden PA, Maestrini E, Magnusson PK, Mahon PB, Maier W, Malhotra AK, Mane SM, Martin CL, Martin NG, Mattheisen M, Matthews K, Mattingsdal M, McCarroll SA, McGhee KA, McGough JJ, McGrath PJ, McGuffin P, McInnis MG, McIntosh A, McKinney R, McLean AW, McMahon FJ, McMahon WM, McQuillin A, Medeiros H, Medland SE, Meier S, Melle I, Meng F, Meyer J, Middeldorp CM, Middleton L, Milanova V, Miranda A, Monaco AP, Montgomery GW, Moran JL, Moreno-De-Luca D, Morken G, Morris DW, Morrow EM, Moskvina V, Muglia P, Muhleisen TW, Muir WJ, Muller-Myhsok B, Murtha M, Myers RM, Myin-Germeys I, Neale MC, Nelson SF, Nievergelt CM, Nikolov I, Nimgaonkar V, Nolen WA, Nothen MM, Nurnberger JI, Nwulia EA, Nyholt DR, O'Dushlaine C, Oades RD, Olincy A, Oliveira G, Olsen L, Ophoff RA, Osby U, Owen MJ, Palotie A, Parr JR, Paterson AD, Pato CN, Pato MT, Penninx BW, Pergadia ML, Pericak-Vance MA, Pickard BS, Pimm J, Piven J, Posthuma D, Potash JB, Poustka F, Propping P, Puri V, Quested DJ, Quinn EM, Ramos-Quiroga JA, Rasmussen HB, Raychaudhuri S, Rehnstrom K, Reif A, Ribases M, Rice JP, Rietschel M, Roeder K, Roeyers H, Rossin L, Rothenberger A, Rouleau G, Ruderfer D, Rujescu D, Sanders AR, Sanders SJ, Santangelo SL, Sergeant JA, Schachar R, Schalling M, Schatzberg AF, Scheftner WA, Schellenberg GD, Scherer SW, Schork NJ, Schulze TG, Schumacher J, Schwarz M, Scolnick E, Scott LJ, Shi J, Shilling PD, Shyn SI, Silverman JM, Slager SL, Smalley SL, Smit JH, Smith EN, Sonuga-Barke EJ, St Clair D, State M, Steffens M, Steinhausen HC, Strauss JS, Strohmaier J, Stroup TS, Sutcliffe JS, Szatmari P, Szelinger S, Thirumalai S, Thompson RC, Todorov AA, Tozzi F, Treutlein J, Uhr M, van den Oord EJ, Van Grootheest G, Van Os J, Vicente AM, Vieland VJ, Vincent JB, Visscher PM, Walsh CA, Wassink TH, Watson SJ, Weissman MM, Werge T, Wienker TF, Wijsman EM, Willemsen G, Williams N, Willsey AJ, Witt SH, Xu W, Young AH, Yu TW, Zammit S, Zandi PP, Zhang P, Zitman FG, Zollner S, Devlin B, kelsoe JR, Sklar P, Daly MJ, O'Donovan MC, Craddock N, Sullivan PF, Smoller JW, Kendler KS, Wray NR. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marona-Lewicka D, Nichols CD, Nichols DE. An animal model of schizophrenia based on chronic LSD administration: old idea, new results. Neuropharmacology. 2011;61:503–512. doi: 10.1016/j.neuropharm.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marona-Lewicka D, Thisted RA, Nichols DE. Distinct temporal phases in the behavioral pharmacology of LSD: dopamine D2 receptor-mediated effects in the rat and implications for psychosis. Psychopharmacology (Berl) 2005;180:427–435. doi: 10.1007/s00213-005-2183-9. [DOI] [PubMed] [Google Scholar]
- McGlothlin W, Arnold DO, Freedman DX. Organicity measures following repeated LSD ingestion. Arch Gen Psychiatry. 1969;21:704–709. doi: 10.1001/archpsyc.1969.01740240064008. [DOI] [PubMed] [Google Scholar]
- Muschamp JW, Regina MJ, Hull EM, Winter JC, Rabin RA. Lysergic acid diethylamide and [-]-2,5-dimethoxy-4-methylamphetamine increase extracellular glutamate in rat prefrontal cortex. Brain Res. 2004;1023:134–140. doi: 10.1016/j.brainres.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Nichols CD, Garcia EE, Sanders-Bush E. Dynamic changes in prefrontal cortex gene expression following lysergic acid diethylamide administration. Brain Res Mol Brain Res. 2003;111:182–188. doi: 10.1016/s0169-328x(03)00029-9. [DOI] [PubMed] [Google Scholar]
- Nichols CD, Sanders-Bush E. A single dose of lysergic acid diethylamide influences gene expression patterns within the mammalian brain. Neuropsychopharmacology. 2002;26:634–642. doi: 10.1016/S0893-133X(01)00405-5. [DOI] [PubMed] [Google Scholar]
- Nichols CD, Sanders-Bush E. Molecular genetic responses to lysergic acid diethylamide include transcriptional activation of MAP kinase phosphatase-1, C/EBP-beta and ILAD-1, a novel gene with homology to arrestins. J Neurochem. 2004;90:576–584. doi: 10.1111/j.1471-4159.2004.02515.x. [DOI] [PubMed] [Google Scholar]
- Nichols DE. Hallucinogens. Pharmacol Ther. 2004;101:131–181. doi: 10.1016/j.pharmthera.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlen M, Nielsen J. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:10084–10097. doi: 10.1093/nar/gks804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SD, Le-Niculescu H, Koller DL, Green SD, Lahiri DK, McMahon FJ, Nurnberger JI, Jr, Niculescu AB., 3rd Coming to grips with complex disorders: genetic risk prediction in bipolar disorder using panels of genes identified through convergent functional genomics. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:850–877. doi: 10.1002/ajmg.b.31087. [DOI] [PubMed] [Google Scholar]
- Pehek EA, Nocjar C, Roth BL, Byrd TA, Mabrouk OS. Evidence for the preferential involvement of 5-HT2A serotonin receptors in stress- and drug-induced dopamine release in the rat medial prefrontal cortex. Neuropsychopharmacology. 2006;31:265–277. doi: 10.1038/sj.npp.1300819. [DOI] [PubMed] [Google Scholar]
- Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, Wayland M, Freeman T, Dudbridge F, Lilley KS, Karp NA, Hester S, Tkachev D, Mimmack ML, Yolken RH, Webster MJ, Torrey EF, Bahn S. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. 643. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL, Gejman PV, O'Donovan MC, Andreassen OA, Djurovic S, Hultman CM, Kelsoe JR, Jamain S, Landen M, Leboyer M, Nimgaonkar V, Nurnberger J, Smoller JW, Craddock N, Corvin A, Sullivan PF, Holmans P, Sklar P, Kendler KS. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders-Bush E, Conn PJ. Effector systems coupled to serotonin receptors in brain: serotonin stimulated phosphoinositide hydrolysis. Psychopharmacol Bull. 1986;22:829–836. [PubMed] [Google Scholar]
- Seifuddin F, Pirooznia M, Judy JT, Goes FS, Potash JB, Zandi PP. Systematic review of genome-wide gene expression studies of bipolar disorder. BMC Psychiatry. 2013;13:213. doi: 10.1186/1471-244X-13-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya VA, Marek GJ, Aghajanian GK, Duman RS. 5-HT2A receptor-mediated regulation of brain-derived neurotrophic factor mRNA in the hippocampus and the neocortex. J Neurosci. 1997;17:2785–2795. doi: 10.1523/JNEUROSCI.17-08-02785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J, Kim Y, Moghaddam B. Disruption of prefrontal cortex large scale neuronal activity by different classes of psychotomimetic drugs. J Neurosci. 2012;32:3022–3031. doi: 10.1523/JNEUROSCI.6377-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing B, Guo J, Meng X, Wei SG, Li SB. The dopamine D1 but not D3 receptor plays a fundamental role in spatial working memory and BDNF expression in prefrontal cortex of mice. Behav Brain Res. 2012;235:36–41. doi: 10.1016/j.bbr.2012.06.035. [DOI] [PubMed] [Google Scholar]
- Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, Levy S, Gogos JA, Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro K, Philpot BD. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology. 2008;55:1081–1094. doi: 10.1016/j.neuropharm.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.