Abstract
Harnessing the RNA interference pathway offers a new therapeutic modality; however, solutions to overcome biological barriers to small interfering RNA (siRNA) delivery are required for clinical translation. This work demonstrates, by direct northern and quantitative PCR (qPCR) detection, stability, gastrointestinal (GI) deposition, and translocation into peripheral tissue of nonmodified siRNA after oral gavage of chitosan/siRNA nanoparticles in mice. In contrast to naked siRNA, retained structural integrity and deposition in the stomach, proximal and distal small intestine, and colon was observed at 1 and 5 hours for siRNA within nanoparticles. Furthermore, histological detection of fluorescent siRNA at the apical regions of the intestinal epithelium suggests mucoadhesion provided by chitosan. Detection of intact siRNA in the liver, spleen, and kidney was observed 1 hour after oral gavage, with an organ distribution pattern influenced by nanoparticle N:P ratio that could reflect differences in particle stability. This proof-of-concept work presents an oral delivery platform that could have the potential to treat local and systemic disorders by siRNA.
Keywords: chitosan, mucosal, nanoparticles, oral, siRNA, stability, systemic translocation
Introduction
RNA interference (RNAi) constitutes a fundamental pathway in eukaryotic organisms for the regulation of gene expression. The posttranscriptional silencing mechanism is mediated by small interfering RNA (siRNA) duplexes that, upon incorporation into the protein complex RNA-induced silencing complex, direct the cleavage, destabilization, or translational repression of the mRNA target.1,2 The capacity for exogenous applied synthetic siRNA to engage the RNAi pathway and facilitate specific gene silencing3,4 has driven research towards harnessing the RNAi pathway as a therapeutic modality. The susceptibility of siRNA to serum nuclease degradation, rapid renal clearance, and poor cellular uptake, however, are major challenges to clinical translation. Nanoparticle-based delivery solution is an exciting approach to overcome the extracellular and intracellular barriers required to allow target engagement of RNAi triggers at therapeutically relevant levels.5,6
Local application of RNAi based-therapeutics to the respiratory, genitourinary, and gastrointestinal (GI) tract mucosa7 is an attractive alternative to systemic administration by allowing direct access to mucosal pathologic sites. The oral route offers high patient compliance, direct access to GI tract diseases, and exploitation of intestinal adsorption for systemic translocation into peripheral tissue. The harsh chemical, enzymatic, and mechanical processes within the GI tract and poor adsorption of hydrophilic drugs, however, require nanoparticle-mediated delivery. Two studies have demonstrated local anti-inflammatory activity in ulcerative colitis mice using thioketal nanoparticles8 and a nanoparticles-in-microsphere oral system.9 Furthermore, a high-impact study by Aouadi et al.10 has shown systemic translocation after oral administration in the mice of siRNA within glucan-based particles; thought to occur by local macrophage capture and subsequent dissemination to peripheral tissue. In the aforementioned studies,8,9,10 indirect methods of siRNA detection either by silencing of the target gene or by microscopic/fluorometric detection of one of the particle components was used. Chitosan is an attractive biomaterial for mucosal drug delivery due to its mucopermeable and mucoadhesive properties11,12 that can facilitate paracellular drug transport and increase residence time at the epithelial surface, respectively.13,14 We have developed a chitosan/siRNA nanoparticle delivery system formed by electrostatic self-assembly.6 This system has shown effective delivery of siRNA into pulmonary mucosa and gene silencing of enhanced green fluorescent protein following nasal6 and intratracheal15 administration in transgenic enhanced green fluorescent protein mice. Furthermore, successful anti-tumor necrosis factor α–based treatment in murine disease models for rheumatoid arthritis16 and radiation induced fibrosis17 has been demonstrated with this chitosan system.
This work investigates the application of our chitosan/siRNA nanoparticle system for oral delivery of siRNA. Direct detection by northern and quantitative PCR (qPCR) analysis is used to evaluate the structural integrity of nonmodified siRNA, GI tract deposition, and systemic translocation to peripheral organs after oral gavage in mice.
Results
Stability and GI tract deposition after oral administration of chitosan/siRNA nanoparticles
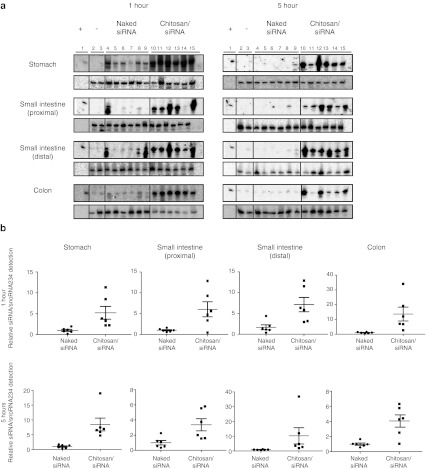
Nonchemically modified siRNA was used to identify the full protective and delivery properties of chitosan nanoparticles after oral administration. Twenty one-mer siRNA (78 µg), either naked or contained within chitosan nanoparticles ~150 nm in size at a NP ratio of 20 (ratio of chitosan bearing cationic amines to siRNA bearing anionic phosphates), were administrated by oral gavage to fasted mice (n = 6). GI tract tissue was harvested at 1 and 5 hours, and the presence and stability of siRNA was evaluated by northern blotting (Figure 1a) and qPCR analysis (Figure 1b). Northern blotting revealed low levels of siRNA within the stomach, proximal and distal small intestine, and colon, 1 hour after oral administration of naked siRNA (Figure 1a). Partial degradation products were clearly observed in the proximal (lane 4) and distal (lane 4 and 9) regions of the small intestine. Interestingly, nondegraded siRNA was almost exclusively present in the stomach. The low level of siRNA observed in the naked siRNA-treated mice was reduced further at 5 hours after administration, with traces only found in the colon.
Figure 1.
siRNA GI tract deposition and protection at 1 and 5 hours after oral gavage in C57BL/6J mice (N = 6). Animals received ~500 µl acetate buffer containing 78 µg of siRNA either naked or incorporated within chitosan/siRNA nanoparticles. (a) Northern blot analysis was performed with 10 µg total RNA from the stomach, small intestine, and colon of animals treated with acetate buffer (lanes 2–3), naked siRNA (lanes 4–9) or chitosan/siRNA nanoparticles (lanes 10–15). siRNA was detected by hybridization of the 32P 5′end-labeled sense strand (upper boxes). Levels of snoRNA U6 were used as a loading control (lower boxes). (b) Quantification of siRNA in intestinal tissue was performed by quantitative PCR analyses using a specific TaqMan gene expression assay. Expression of snoRNA234 was used for normalization purposes. For each tissue, normalized siRNA levels are expressed relative to the average siRNA level found in the animals treated with the naked siRNA.
In contrast, high levels of intact siRNA were detected in the stomach, small intestine, and colon in the animals dosed with chitosan/siRNA nanoparticles at both 1 and 5 hours (Figure 1a). Animal-to-animal variation could account for any differences in the amount of siRNA detected; however, a clear trend showing improved deposition and protection installed by the nanoparticles is evident. qPCR analysis (Figure 1b) supported the northern findings, with higher levels of siRNA detected in organs isolated from animals treated with the chitosan/siRNA nanoparticles compared with those with naked siRNA. Relative siRNA levels ranged from a ~3.4-fold increase in the proximal small intestine at 5 hours, to more than 11-fold for the distal small intestine and colon at 5 and 1 hour, respectively.
Intestinal gut loop studies were performed in fasted mice to visualize intestinal uptake of fluorescent chitosan/siRNA nanoparticles by confocal microscopy. Nanoparticles (NP 20) containing Cy5-labeled siRNA were injected into intestinal loops around the distal small intestine to ensure direct nanoparticle contact with the epithelial surface. A discontinuous fluorescence layer was observed at the apical surface of intestinal tissue harvested 45 minutes after injection (Figure 2). Interestingly, cells with morphological characteristics resembling goblet cells had no apparent surface staining that could reflect nanoparticle removal as a consequence of mucus secretions by this cell type. The punctate staining at the apical epithelial surface indicates nanoparticle presence and mucoadhesion provided by the chitosan. No distinct punctuate fluorescence was observed within the epithelial cells or submucosa that could indicate decomplexation of the chitosan/siRNA. The accumulation of siRNA in peripheral organs, however, is a clear evidence for intestinal translocation of the siRNA (refer to following section).
Figure 2.
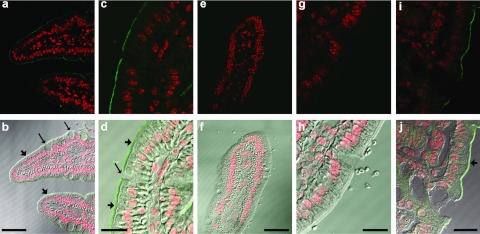
Fluorescent detection of chitosan/siRNA nanoparticles in the intestine, 45 minutes after injection within in situ gut loops performed in 8- to 10-week-old, female C57BL/6J mice (N = 2). Paraffin sections representative of in situ gut loops filled with Chitosan nanoparticles containing (a–d and i–j) 28 µg of Cy5-labeled siRNA (NP 20) or (e–h) acetate buffer. (a) Cy5-labeled siRNA (green) could be detected at the surface of epithelial cells in the small intestine. Nuclei are labeled red. (b) Differential interference contrast overlays highlight the siRNA at the brush border of epithelial cells (small arrows), with no siRNA in the sparse goblet cells (long arrows). (c,d) At a higher magnification, the siRNA labeling is more distinct. (e–h) No labeling was observed in tissue from mice injected with acetate buffer alone. (i,j) Similar chitosan/siRNA nanoparticle labeling was observed in colon epithelial cells. Scale bars: b, f = 50 µm, d, h, and j = 20 µm.
Systemic translocation and organ accumulation of siRNA following oral administration of chitosan/siRNA nanoparticles
The NP ratio of chitosan/siRNA nanoparticles has been shown previously to determine stability and biological interactions.18 A series of experiments were, therefore, performed in mice (N = 3) to investigate the effect of NP ratio (5, 20, 60, and 120) on intestinal deposition and systemic translocation of nonmodified siRNA (28 µg), 1 hour after oral gavage.
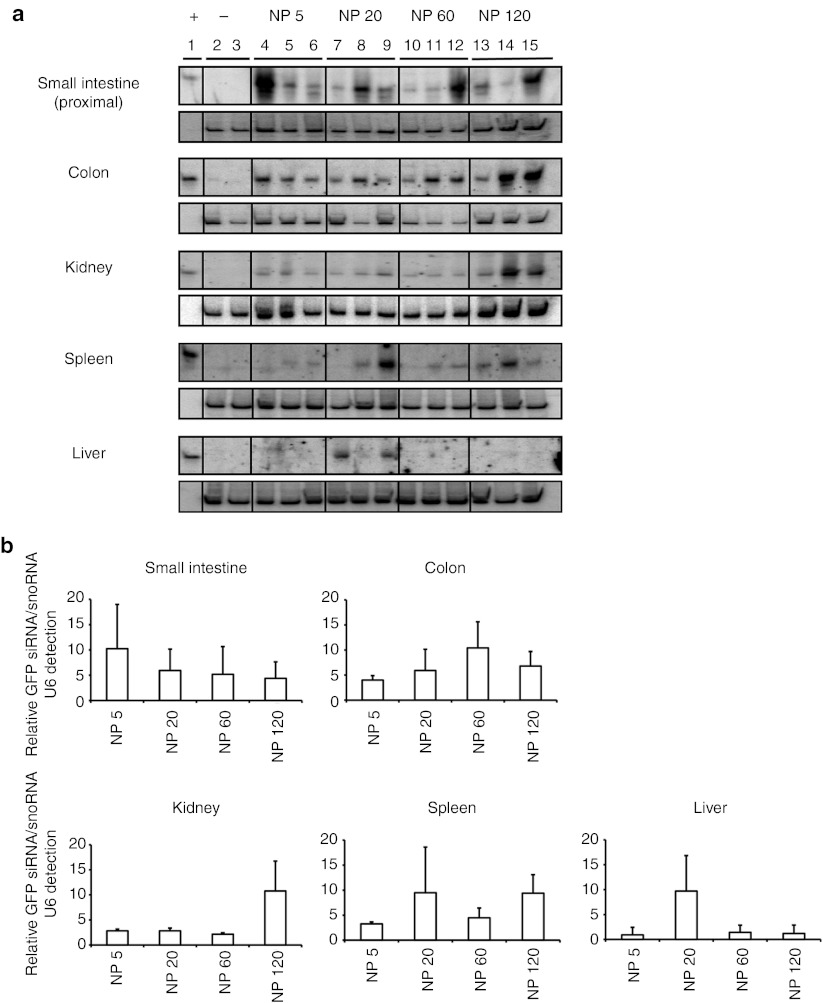
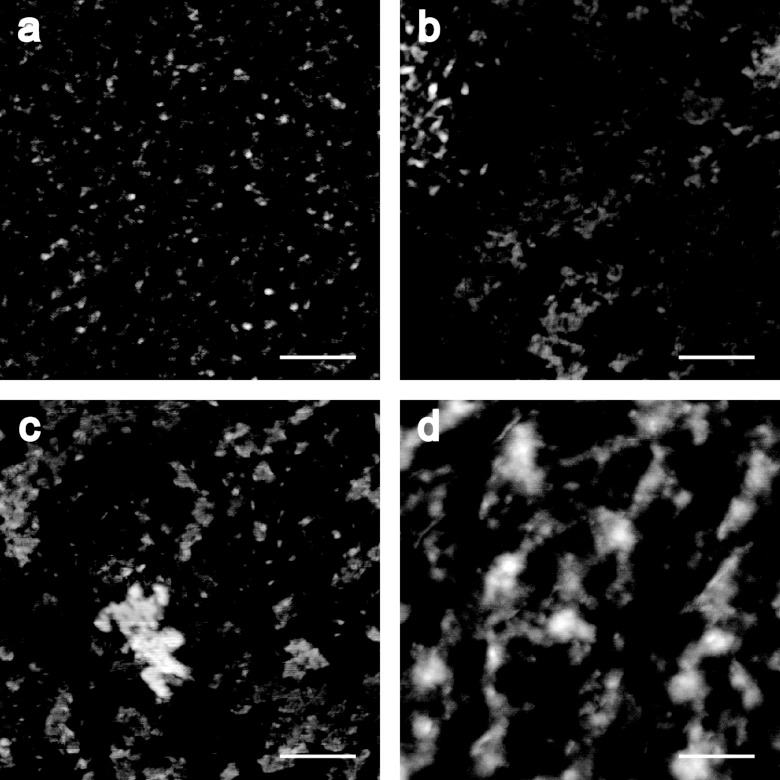
In agreement with the deposition data (Figure 1), northern analysis revealed intact siRNA in the small intestine and colon 1 hour after dosing for all NP ratios (Figure 3a). Although similar siRNA levels could be detected in the colon groups, considerable variability was observed in the intestinal tissue within the same groups (lanes 4,8,12, and 15). Interestingly, siRNA could be detected in liver, spleen, and kidney, at a level dependent on NP ratio verified by semiquantitative gel densitometry analysis (Figure 3b). Low levels of siRNA were only found in the liver with the NP 20 formulation; whereas, detection in the spleen and kidney was irrespective of NP, and in higher amounts. In general, higher levels of siRNA were detected in animals receiving the NP 120 chitosan/siRNA nanoparticles particularly in the kidney that could reflect greater stability and mucosal penetration for this formulation. Although the hydrodynamic size of the particles ~126 (NP 5), 124 (NP 20), 179 (NP 60), and 154 (NP 120), determined by nanoparticle tracking analysis, were similar over the NP ratio range (Table 1), differences in particle stability were observed using a gel mobility assay (Figure 4). The observed retarded siRNA migration suggests a gradual NP 120 > NP 60 > NP 20 > NP 5 stability of the chitosan/siRNA complexes in acetate buffer (Figure 4). Significantly, this tendency was also conserved when particles were incubated for 1 hour in solutions simulating the physicochemical characteristics such as pH and salt composition of intestinal (simulated intestinal fluid) and gastric (simulated gastric fluid) fluids. Morphological studies of the formulations by atomic force microscopy (Figure 5) revealed formation of nanoparticles over the NP ratio range with a tendency towards particle–particle interaction at a higher NP ratio (60 and 120) facilitated by the higher level of excess chitosan that may contribute to a greater stability.
Figure 3.
Systemic translocation of siRNA after oral gavage of chitosan/siRNA nanoparticles in C57BL/6J mice (N = 3). Animals were dosed with either acetate buffer (lanes 2–3) or nanoparticles at NP ratio 5 (lanes 4–6), NP ratio 20 (lanes 7–9), NP ratio 60 (lanes 10–12), or NP ratio 120 (lanes 13–15). Nanoparticle-dosed animals received 28 µg of siRNA for all formulations (NP ratios 5, 20, 60, and 120). (a) siRNA presence and integrity was evaluated by northern blot analysis in intestinal (proximal small intestine and colon) and nonintestinal tissue (spleen, kidney, and liver) (upper boxes) harvested 1 hour after dosing. Levels of snoRNA U6 were used as a loading control (lower boxes). (b) Semiquantitative evaluation by gel densitometry of siRNA deposition in small intestine, colon, kidney, spleen, and liver. For each tissue, relative GFP siRNA/ snoRNA U6 levels are indicated for the different nanoparticles (NP ratio 5, 20, 60, and 120). GFP, green fluorescent protein.
Table 1. Size determination of chitosan/siRNA nanoparticles by particle-tracking analysis.

Figure 4.
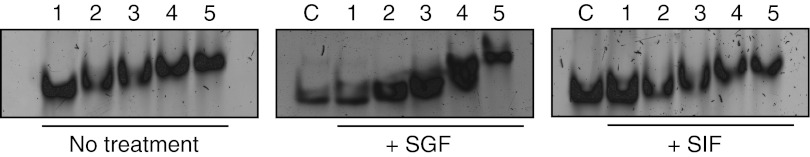
Evaluation of chitosan/siRNA nanoparticle stability using native polyacrylamide gel electrophoresis. Naked siRNA and nanoparticles were resolved by gel electrophoresis either directly or after incubation in simulated physiological fluids for 1 hour. Lane 1: Naked GFP siRNA; Lane 2: Nanoparticles NP ratio 5; Lane 3: Nanoparticles NP ratio 20; Lane 4: nanoparticles NP ratio 60; Lane 5: Nanoparticles NP ratio 120; C: naked GFP siRNA + corresponding simulated solution loaded without incubation period. GFP, green fluorescent protein; SIF, simulated intestinal fluid; SGF, simulated gastric fluid.
Figure 5.
Atomic Force Microscopy images of chitosan/siRNA nanoparticles prepared at NP ratio (a) 5, (b) 20, (c) 60, and (d) 120. Scale bar = 200 nm.
Discussion
This work demonstrates siRNA stability, GI tract deposition, and subsequent systemic translocation into liver, spleen, and kidney after oral administration in mice of a series of chitosan/siRNA nanoparticles. In contrast to the previous reports,8,9,10 the application of northern and qPCR analysis in our study allows the direct identification and stability assessment of orally administrated siRNA.
Retention of structural integrity within the harsh mechanical, chemical, and enzymatic environment of the GI tract is a prerequisite for oral-based RNAi therapeutics. Nonchemically modified siRNA, susceptible to breakdown in these conditions, was selected to fully evaluate the protective capability of the chitosan system. siRNA incorporation into nanoparticles resulted in much greater nucleic acid deposition throughout the stomach, small and large intestine at 1 and 5 hour after gavage compared with naked siRNA (Figure 1). The rapid degradation observed following naked siRNA administration may reflect the high nuclease activity and extreme physicochemical milieu present within the GI tract. Chemical modifications have been used to reduce nuclease degradation in serum conditions;19,20 however, limited studies have evaluated the stability of these modified siRNAs in GI tract conditions. Furthermore, the polyanionic and macromolecular nature of siRNA would seemingly restrict cellular entry even if stability was installed. The cationic charge exhibited by chitosan in acidic conditions could potentiate binding with polyanion siRNA and facilitate protection from gastric breakdown even to nonmodified siRNA. Nevertheless, chemical modifications of the siRNA will be required in subsequent gene silencing studies, even in the presence of chitosan, to avoid immune stimulation.21,22,23
In this work, we demonstrate the accumulation of siRNA in the liver, spleen and kidney at 1 hour after gavage (Figure 3). Two previous studies have suggested systemic distribution of siRNA after oral dosing.8,10 Aouadi et al.10 used confocal microscopy to identify fluorescently labeled glucan particles in macrophages of spleen, lung, and liver, in addition to the specific gene knockdown in isolated tissue macrophages at day 10. Whereas, in a study by Wilson et al.8 fluorometry detection of Cy3-labeled siRNA in the supernatant of spleen, liver, lung, and heart homogenates was used. The northern blot technique used in our work represents a direct method to determine both the presence and structural integrity of the siRNA. Interestingly, in contrast to the previous studies,8,10 we also show accumulation of siRNA in the kidney, suggesting a broad systemic distribution of the molecule. Previous work from our group has shown kidney accumulation after intravenous injection of chitosan/siRNA nanoparticles in mice.24 The level of accumulation in the liver, spleen, and kidney was dependent on the NP ratio of the nanoparticle that has been shown previously by ourselves to influence stability and cellular interactions.18 Accumulation in the spleen and kidney was observed over the range of formulations; however, only NP 20 resulted in detection in the liver. This observation could reflect a sampling bias due to the analysis of a liver section compared with the whole spleen and kidney analysis. The higher NP 120 nanoparticles seemed to facilitate greater accumulation of siRNA in tissue that could reflect higher stability and cellular interactions due to the excess chitosan.18 In a different experiment, siRNA detection in organs was absent at 24 hours after gavage (data not shown), that could indicate a necessity for chemical modifications using combinations of phosphorothioates, locked-nucleic acids or 2′OMe substitutions25,26,27,28,29 to reduce possible serum nuclease breakdown. We are presently conducting experiments to determine if the siRNA detected at distal sites remains associated with chitosan molecules.
Work is currently being carried out in our laboratory to quantify the amount of siRNA in organs and determine the mechanism of epithelial uptake of the chitosan/siRNA system. Aouadi et al.10 proposed the uptake across M cells within gut associated lymphoid tissue although no direct evidence was shown. The low number of M cells, however, makes the therapeutic exploitation of this route unrealistic, and uptake across adsorptive enterocytes or intraepithelial lymphocytes is likely needed to facilitate systemic translocation in sufficient amounts.
In conclusion, this work demonstrates, by direct northern blotting and qPCR analysis, siRNA stability, GI tract deposition, and systemic translocation after oral administration of chitosan/siRNA nanoparticles in mice; the pattern of distribution influenced by the NP ratio. In contrast to multicomponent carriers previously used for oral delivery of siRNA,8,9,10 the chitosan nanoparticle is a two-component system formed by electrostatic self-assembly upon mixing; the simplicity in design and production attractive features for clinical translation. This proof-of-concept work presents an oral delivery platform that could have the potential to treat local and systemic disorders with RNAi-based therapeutics.
Materials and Methods
Chemicals and siRNA. Chitosan was purchased from Novamatrix, FMC BioPolymer (Sandvika, Norway) (250 kDa, 86% deacetylation) and Kitozyme (Herstal, Belgium) (34 kDa, 82% deacetylation). siRNA duplex against enhanced green fluorescent protein with 5′-gacguaaacggccacaagugc; as the sense strand and 5′-acuuguggccguuuacgucgc; as the antisense was purchased from Dharmacon, Fisher Scientific (Slangerup, Denmark). Anti-tumor necrosis factor α siRNA containing a fluorescent Cy5-labeled sense strand 5′-gucucagccucuucucauuccugCT;Cy5 and 5′-agcaggaaugagaagaggcugagacau; as antisense strand were purchased from IDT (Iowa, IA), and used for microscopic studies. A DNA oligonucleotide of sequence 5′-GAATTTGCGTGTCATCCTTGCGCAGGGGCCATGCTAA; (DNA technology, Aarhus, Denmark) was used to probe for snoRNA U6.
Preparation and characterization of chitosan/siRNA nanoparticles. Chitosan particles were prepared as previously described.30 Briefly, chitosan was dissolved in sodium acetate buffer (300 mmol/l NaAc, pH 5.5) to obtain a 1–4.4 mg/ml working solution range. siRNA dissolved in nuclease-free water was slowly added to the filtrated chitosan solution and stirred for 1 hour. The hydrodynamic size of chitosan/siRNA complexes was determined by particle-tracking analysis using Nanosight equipment (NanoSight Limited, UK). Nanoparticles of 5, 20, 60, and 120 NP ratios were used for the systemic translocation studies. NP is defined as the molar ratio of chitosan amines and RNA phosphates.
Atomic force microscopy was used to investigate the morphology of the nanoparticles. Nanoparticles at different NP ratio were deposited onto freshly cleaved mica. Atomic force microscopy images of samples were obtained on a Nanoscope IV MultiMode AFM (Veeco Instruments, Santa Barbara, CA) in intermittent contact mode under ambient conditions with an OMCL-AC160TN cantilever (Olympus, Atomic Force F&E GmbH, Mannheim, Germany). All the images were analyzed by using the commercial Scanning Probe Image Processor software (Image Metrology ApS, Lyngby, Denmark).
Nanoparticle stability. Stability of the nanoparticles was evaluated using native polyacrylamide gel electrophoresis. Samples were incubated in a 1:1 volumetric ratio with simulated gastric fluid (without pepsin) or simulated intestinal fluid (without pancreatin) for 1 hour at room temperature. Electrophoresis analyses were performed in a 15% polyacrylamide gel (50 mmol/l Tris, 45 mmol/l boric acid, and 0.5 mmol/l EDTA) at 100–150 V for 5 hours. Gels were then stained with SBRY safe DNA gel stain (Invitrogen, Carlsbad, CA) and imaged using a Typhoon Trio+ scanner (GE Healthcare, Life Sciences, Copenhagen, Denmark). Simulated physiological fluids were prepared according the International Pharmacopoeia. Simulated gastric fluid, 34.2 mmol of sodium chloride were dissolved in water, adjusted to a final pH of 1.2 with approximately 7 ml of hydrochloric acid (420 g/l) and the solution was diluted with water to a final volume of 1 liter. For simulated intestine fluid, 50.3 mmol of potassium dihydrogen phosphate and 38 mmol of sodium hydroxide were dissolved in water, adjusted to final pH of 7.5 with sodium hydroxide (0.2 mol/l), and diluted to produce 1 l of aqueous solution.
Deposition and translocation studies. Acetate buffer, naked siRNA, or chitosan/siRNA nanoparticles was administrated by oral gavage to female, 8- to 10-week-old C57BL/6J mice (deposition experiment N = 6 and translocation experiment N = 3) (Taconic, Lille Skensved, Denmark). siRNA-dosed animals received 78 or 28 µg of the naked or formulated 21-mer in ~500 µl 300 mmol/l acetate buffer pH 5.5. Novomatrix chitosan was used for the GI deposition experiment and Kitozyme chitosan for the translocation experiment due to the availability of material. At the indicated time points, animals were euthanized by cervical dislocation and a laparotomy was performed to collect the desired tissue. Organs were then preserved in RNA later (Life Technologies Europe, Naerum, Denmark) for subsequent analysis. Before preservation, the GI tract lumen was intensively flushed with phosphate-buffered saline (PBS) to remove faeces and nonbound material. All animal experiments were performed with the approval from the Danish Experimental Animal Inspectorate and according to the Danish national and institutional regulations. During the experiment, mice were housed in type III Makrolon cages in rooms with controlled environment (21 ± 2 °C, 55 ± 5% relative humidity, 12–hour light/dark cycle, 12–16 changes of HEPA (Camfil, Skovlunde, Denmark) filtrated air per hour)
Northern blot analysis. Following tissue homogenization (TissueLyser II; Qiagen, Hilden, Germany), total RNA was isolated by Trizol reagent (Invitrogen) resolved in a 15% denaturing polyacrylamide and blotted to a positively charged nylon membrane (Hybon N+; Amersham, Uppsala, Sweden). Blots were then hybridized with the sense strand of the administrated siRNA (5′ labeled with 32P) and exposed in a storage phosphor-imaging cassette. For radioactive labeling, T4 polynucleotide kinase (New England Biolabs, Beverly, MA) was used. The probe was purified with a G50 spin column (Roche, Basel, Switzerland) to remove unincorporated [γ-32P] adenosine triphosphate. Homogeneous loading was investigated with an oligonucleotide probe against snoRNA U6. Semiquantitative evaluation of the relative siRNA deposition in systemic tissue was performed by gel densitometry using the GelEval software (FrogDance software, Dundee, UK).
RNA isolation and quantitative real-time PCR analysis. Total RNA was extracted from homogenized tissue with TRIzol (Life Technologies, Carlsbad, CA) according to the manufacturer's protocol. RNA was reverse transcribed with the TaqMan MicroRNA reverse transcription kit (Applied Biosystems, Foster City, CA) using the provided specific primers for green fluorescent protein and snoRNA 234. siRNA levels were measured by real-time PCR (Stratagen Mx3005P; Agilent Technologies, Santa Clara, CA) using specific TaqMan gene expression assays (Applied Biosystems, Foster City, CA). For the assay, 20 µl of reaction mix containing the cDNA template, the TaqMan Universal PCR Master Mix II, the provided specific primers and TaqMan MGB probe was subjected to an initial cycle of 95 °C for 10 minutes and then 40 cycles of 95 °C for 15 seconds and 60 °C for 60 seconds. Detected siRNA levels were normalized to the expression of the internal standard, snoRNA234. PCR efficiency for all reactions was within the acceptable margin of 90–110%.
In situ gut lops and fluorescent microscopy. Female, 8- to 10-week-old C57BL/6J mice (N = 2) were anaesthetized with a mixture of 2–4% isoflurane in air, and an incision was performed to access the abdominal cavity. Intestinal gut loops were then performed by tethering two strands of surgical cord upstream and downstream of the selected region and acetate buffer or nanoparticles (NP ratio 20) containing 28 µg Cy5-labeled siRNA injected on the lumen of the loop. After 45 minutes, animals were euthanized by cervical dislocation and the tissue removed and flushed thoroughly with PBS before immediate immersion fixation in 4% paraformaldehyde in PBS for 4–5 hours. Samples were then left overnight in a 0.4% paraformaldehyde/PBS solution at 4 °C. Tissue was prepared for embedding in paraffin as previously described.31 Two-micron sections were cut using a rotary microtome (Leica Microsystems, Ballerup, Denmark) followed by paraffin removal using xylene and rehydration in graded alcohols. Sections were rinsed in distilled water and incubated for 30 minutes in TOPRO3 (Invitrogen) in PBS/0.3% Triton-X 100. Sections were rinsed several times in PBS and mounted using antifade mounting media (Dako, Glostrup, Denmark). A Leica TCS SL (SP2) laser confocal microscope and Leica Confocal Software was used for imaging of the tissue sections. Images were taken using an HCX PL APO x63 oil objective lens (NA = 1.40). Microscope settings (photo multipier tube, offset and gain, sampling period, and averaging) were optimized for each section to provide optimal images.
Acknowledgments
The authors thank Bradley Whitehead, Anders Toftegaard Boysen, Inger Merete Paulsen and Bodil Kruse for excellent technical assistance. This project was partly sponsored by Nanoference ApS. K.A.H., J.K., and B.B.G. have association with Nanoference ApS. The work was also supported by the Danish Strategic Research Council and the Danish Medical Research Council. The authors declared no conflict of interest.
References
- Meister G., and, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA., and, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Davidson BL., and, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fougerolles A, Vornlocher HP, Maraganore J., and, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N.et al. (2008A combinatorial library of lipid-like materials for delivery of RNAi therapeutics Nat Biotechnol 26561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard KA, Rahbek UL, Liu X, Damgaard CK, Glud SZ, Andersen MØ.et al. (2006RNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle system Mol Ther 14476–484. [DOI] [PubMed] [Google Scholar]
- Ballarín-González B, Nielsen EB, Thomsen TB, Howard KA.2012Mucosal delivery of RNAi therapeutics Howard, KA.ed.). RNA Interference From Biology to Therapeutics Advances in Delivery Science and Technology, Springer: New York, 97–125 [Google Scholar]
- Wilson DS, Dalmasso G, Wang L, Sitaraman SV, Merlin D, Murthy N. Orally delivered thioketal nanoparticles loaded with TNF-α–siRNA target inflammation and inhibit gene expression in the intestines. Nat Mater. 2010;9:923–928. doi: 10.1038/nmat2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel C., and, Amiji M. Oral TNF-a gene silencing using a polymeric microsphere-based delivery system for the treatment of inflammatory bowel disease. J Control Release. 2011;150:77–86. doi: 10.1016/j.jconrel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouadi M, Tesz GJ, Nicoloro SM, Wang M, Chouinard M, Soto E.et al. (2009Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation Nature 4581180–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S, Sun W., and, Kissel T. Chitosan-based formulations for delivery of DNA and siRNA. Adv Drug Deliv Rev. 2010;62:12–27. doi: 10.1016/j.addr.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Illum L. Chitosan and its use as a pharmaceutical excipient. Pharm Res. 1998;15:1326–1331. doi: 10.1023/a:1011929016601. [DOI] [PubMed] [Google Scholar]
- Soane RJ, Frier M, Perkins AC, Jones NS, Davis SS., and, Illum L. Evaluation of the clearance characteristics of bioadhesive systems in humans. Int J Pharm. 1999;178:55–65. doi: 10.1016/s0378-5173(98)00367-6. [DOI] [PubMed] [Google Scholar]
- Aspden TJ, Mason JD, Jones NS, Lowe J, Skaugrud O., and, Illum L. Chitosan as a nasal delivery system: the effect of chitosan solutions on in vitro and in vivo mucociliary transport rates in human turbinates and volunteers. J Pharm Sci. 1997;86:509–513. doi: 10.1021/js960182o. [DOI] [PubMed] [Google Scholar]
- Nielsen EJ, Nielsen JM, Becker D, Karlas A, Prakash H, Glud SZ.et al. (2010Pulmonary gene silencing in transgenic EGFP mice using aerosolised chitosan/siRNA nanoparticles Pharm Res 272520–2527. [DOI] [PubMed] [Google Scholar]
- Howard KA, Paludan SR, Behlke MA, Besenbacher F, Deleuran B., and, Kjems J. Chitosan/siRNA nanoparticle-mediated TNF-alpha knockdown in peritoneal macrophages for anti-inflammatory treatment in a murine arthritis model. Mol Ther. 2009;17:162–168. doi: 10.1038/mt.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawroth I, Alsner J, Behlke MA, Besenbacher F, Overgaard J, Howard KA.et al. (2010Intraperitoneal administration of chitosan/DsiRNA nanoparticles targeting TNFa prevents radiation-induced fibrosis Radiother Oncol 97143–148. [DOI] [PubMed] [Google Scholar]
- Liu X, Howard KA, Dong M, Andersen MØ, Rahbek UL, Johnsen MG.et al. (2007The influence of polymeric properties on chitosan/siRNA nanoparticle formulation and gene silencing Biomaterials 281280–1288. [DOI] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W.et al. (2005Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs Nat Biotechnol 231002–1007. [DOI] [PubMed] [Google Scholar]
- Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- Cekaite L, Furset G, Hovig E., and, Sioud M. Gene expression analysis in blood cells in response to unmodified and 2'-modified siRNAs reveals TLR-dependent and independent effects. J Mol Biol. 2007;365:90–108. doi: 10.1016/j.jmb.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S.et al. (2005Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7 Nat Med 11263–270. [DOI] [PubMed] [Google Scholar]
- Karikó K, Buckstein M, Ni H., and, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Gao S, Dagnaes-Hansen F, Nielsen EJ, Wengel J, Besenbacher F, Howard KA.et al. (2009The effect of chemical modification and nanoparticle formulation on stability and biodistribution of siRNA in mice Mol Ther 171225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choung S, Kim YJ, Kim S, Park HO., and, Choi YC. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem Biophys Res Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Grünweller A, Wyszko E, Bieber B, Jahnel R, Erdmann VA., and, Kurreck J. Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2'-O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res. 2003;31:3185–3193. doi: 10.1093/nar/gkg409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N.et al. (2009A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity Nucleic Acids Res 372867–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y.et al. (2005Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality Nucleic Acids Res 33439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov AA, Kruglova NS, Meschaninova MI, Venyaminova AG, Zenkova MA, Vlassov VV.et al. (2009Selective protection of nuclease-sensitive sites in siRNA prolongs silencing effect Oligonucleotides 19191–202. [DOI] [PubMed] [Google Scholar]
- Andersen MØ, Howard KA., and, Kjems J. RNAi using a chitosan/siRNA nanoparticle system: in vitro and in vivo applications. Methods Mol Biol. 2009;555:77–86. doi: 10.1007/978-1-60327-295-7_6. [DOI] [PubMed] [Google Scholar]
- Moeller HB, Knepper MA., and, Fenton RA. Serine 269 phosphorylated aquaporin-2 is targeted to the apical membrane of collecting duct principal cells. Kidney Int. 2009;75:295–303. doi: 10.1038/ki.2008.505. [DOI] [PMC free article] [PubMed] [Google Scholar]