Abstract
Conventional experimental methods of studying the human genome are limited by the inability to independently study the combination of alleles, or haplotype, on each of the homologous copies of the chromosomes. We developed a microfluidic device capable of separating and amplifying homologous copies of each chromosome from a single human metaphase cell. Single-nucleotide polymorphism (SNP) array analysis of amplified DNA enabled us to achieve completely deterministic, whole-genome, personal haplotypes of four individuals, including a HapMap trio with European ancestry (CEU) and an unrelated European individual. The phases of alleles were determined at ~99.8% accuracy for up to ~96% of all assayed SNPs. We demonstrate several practical applications, including direct observation of recombination events in a family trio, deterministic phasing of deletions in individuals and direct measurement of the human leukocyte antigen haplotypes of an individual. Our approach has potential applications in personal genomics, single-cell genomics and statistical genetics.
The sequencing of the human reference genome and the development of high-throughput short-read sequencing technologies have enabled partial decoding of an increasing number of individual human genomes1–7. However, all of these ‘personal genomes’ are incomplete, and should essentially be regarded as rough draft genomes. Although they all suffer from imperfections, such as gaps, miscalled bases and difficulties in determining large-scale structural variation, they are missing fundamental information of the unique haploid structure of homologous chromosomes. Haplotypes, the combinations of alleles at multiple loci along a single chromosome, are difficult to measure with current technologies but are an essential feature of the genome. A simple example of how the lack of this information limits the interpretation of existing genomes is to consider an individual having two mutations in a certain gene. If both mutations are on the same allele, then this individual would have one normal (that is, putatively functional) version of the gene and one mutated version. If the mutations are on different alleles, this individual would have two mutated versions and no normal version of the protein. In the absence of haplotype information, it is impossible to distinguish between these two cases.
Knowledge of complete haplotypes of individuals (personal haplotypes) would therefore be useful in personalized medicine. Notably, several studies have linked specific haplotypes to drug response and to resistance or susceptibility to diseases. A well-known example is the association of human leukocyte antigen (HLA) haplotypes with autoimmune diseases and clinical outcomes in transplantations8–10. Haplotypes within the apolipoprotein gene cluster may influence plasma triglyceride concentrations and the risk toward atherosclerosis11. Research suggests that a specific β-globin locus haplotype is associated with better prognosis of sickle cell disease12, and other studies have linked haplotypes in the matrix metalloproteinase gene cluster with cancer development13. Haplotypes are also important in pharmacogenomics, an example being the association of β-2 adrenergic receptor to responses to drug treatment of asthma14. Deterministic haplotyping may greatly increase the power of genome-wide association studies in finding candidate genes associated with common but complex traits. It will also contribute to the understanding of population genetics and historical human migrations and the study of cis-acting regulation in gene expression.
Direct experimental determination of the haplotypes of an individual is challenging. The International HapMap Consortium has performed extensive SNP genotyping on different human populations, and by using family trios and statistical methods, has been able to catalog commonly occurring haplotype blocks in the human populations. However, in the best cases, when members of a family trio are analyzed, this approach leads to errors in resolving haplotype at approximately every ~3–8 megabases, and in the most general case, when an individual genome analyzed in the absence of family information, errors occur every 300 kilobases15,16. In the context of personalized genomics and medicine, the approaches used in the HapMap project have limited applicability, as materials from family members are not always available and computational approaches using statistical models have inherent statistical uncertainty and are limited to regions with strong linkage disequilibrium. Mate-pair shotgun genome sequencing has been demonstrated to achieve partial haplotype reconstruction of an individual but the haplotype blocks have limited sizes5,17. Other techniques have been demonstrated, including PCR in various forms18–22, atomic force microscopy with carbon nanotubes23, fosmid/cosmid cloning24 and hybridization of probes to single DNA molecules25. Weaknesses of these methods include the inability to phase SNPs (that is, determine their relative arrangement on homologous chromosomes) more than tens of kilobases apart and/or a limitation in the number of markers that could be phased in a single assay. Whole-genome haplotyping can in principle be achieved by chromosome microdissection26 or by the construction of somatic cell hybrids27. Yet the former is time-consuming and expensive, and the latter requires specialized and expensive equipment. So far, direct whole-genome haplotyping has not been accomplished for any individual. Here we address these issues using microfluidics.
RESULTS
Single-cell chromosome separation and amplification
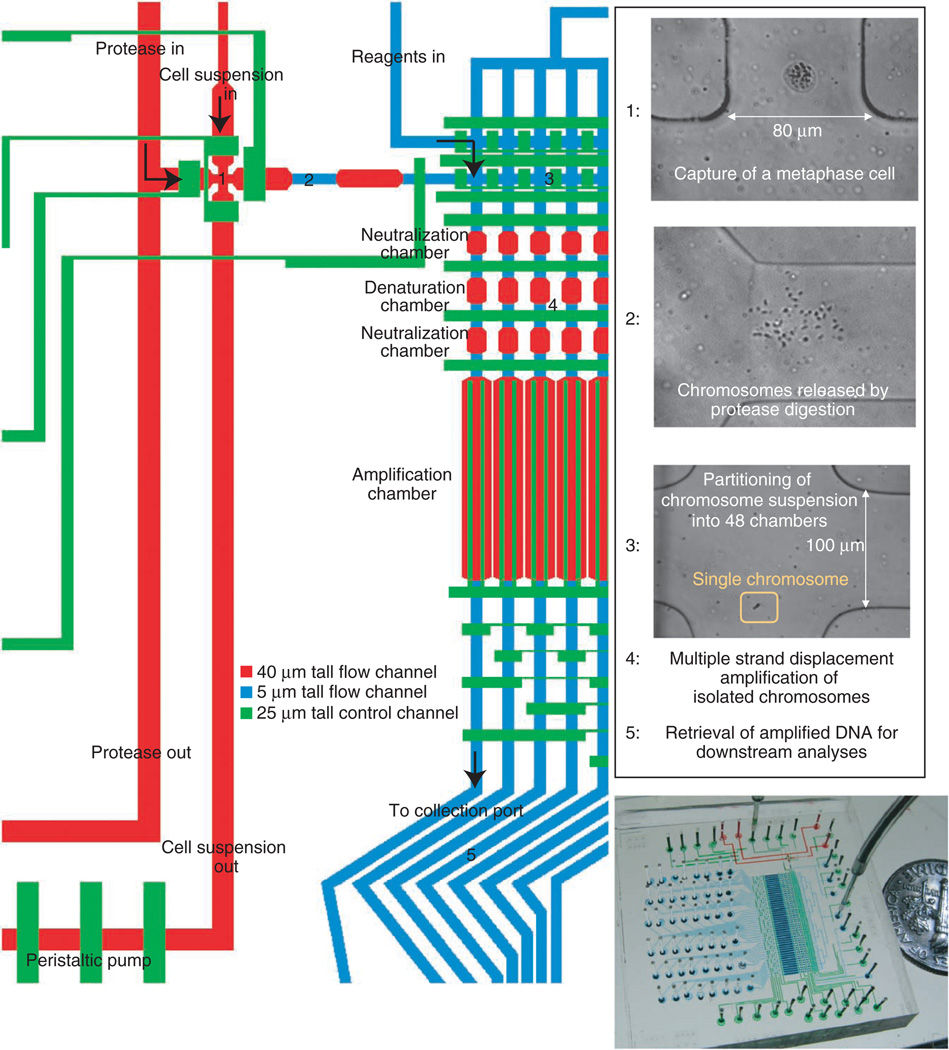
We developed an approach termed direct deterministic phasing (DDP) in which the intact chromosomes from a single cell are dispersed and amplified on a microfluidic device (Fig. 1). The device consists of a cell-sorting region, where a single metaphase cell is identified microscopically and captured from a cell suspension; a chromosome release region, where metaphase chromosomes are released by protease digestion of the cytoplasm; a chromosome partitioning region, where the chromosome suspension is randomly separated into 48 partitions of a long narrow channel; an amplification region, where isolated chromosomes are individually amplified by multiple strand displacement amplification; and a product retrieval region, where amplified products are collected. The products are recovered independently, thus allowing direct genetic interrogation and genome-wide determination of haplotypes without the need for family information or statistical inference.
Figure 1.
Microfluidic device designed for the amplification of metaphase chromosomes from a single cell. A single metaphase cell is recognized microscopically and captured in region 1. Protease (pepsin at low pH) is introduced to generate chromosome suspension in region 2. Chromosome suspension is partitioned into 48 units (region 3). Content in each partition is individually amplified (region 4). Specifically, chromosomes at low pH are first neutralized and treated with trypsin to digest chromosomal proteins. Chromosomes are denatured with alkali and subsequently neutralized for multiple strand displacement amplification to take place. As reagents are introduced sequentially into each air-filled chamber, enabled by the gas permeability of the device’s material, chromosomes are pushed into one chamber after the next and finally arrive in the amplification chamber. Amplified materials are retrieved at the collection ports (region 5). In the overview image of the device, control channels are filled with green dye. Flow channels in the cell-sorting region and amplification region are filled with red and blue dyes, respectively.
Whole-genome haplotyping of members in a HapMap CEU trio
We first verified DDP with three lymphoblastoid cell lines, GM12891, GM12892 and GM12878, representing a father-mother-daughter trio in the CEPH European (CEU) 1463 family. These cell lines have been extensively genotyped by the HapMap project.
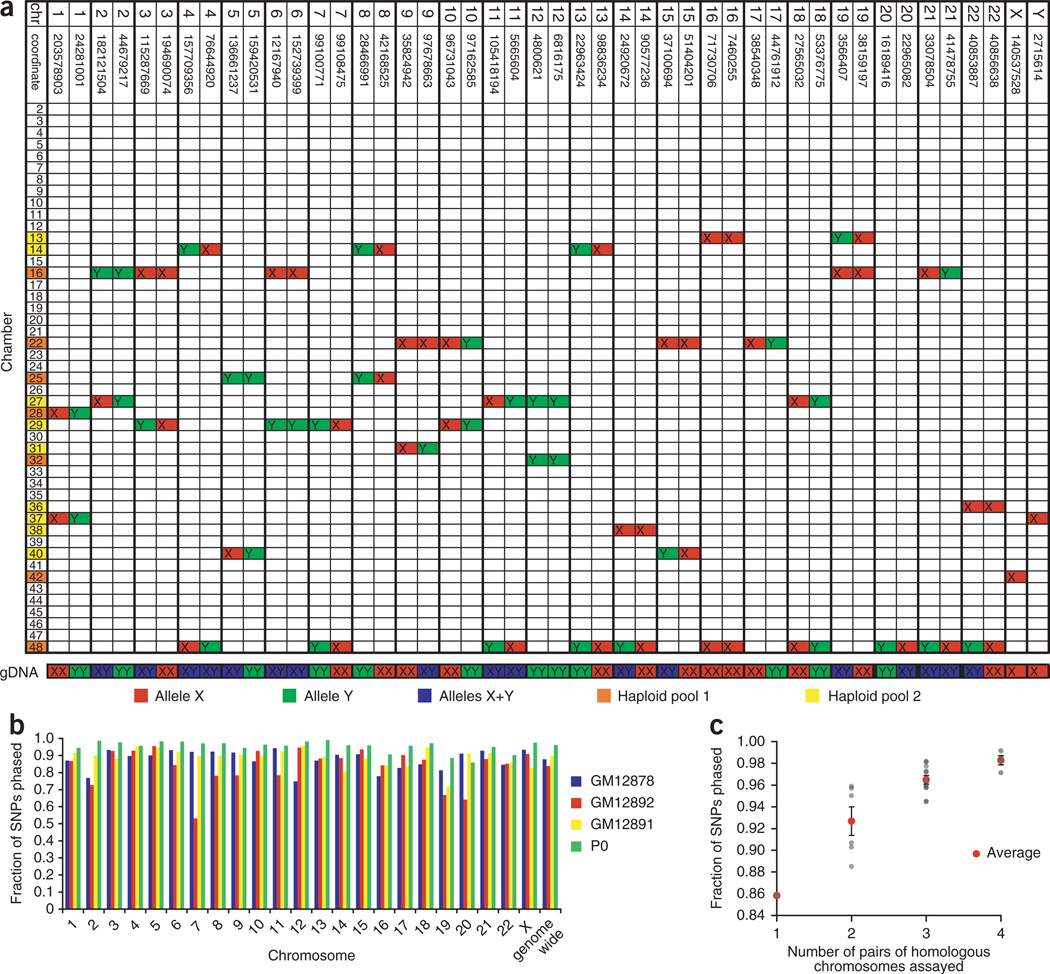
For each single-cell experiment, the chromosomal origins of the contents of each microfluidic chamber were established by a 46-loci Taqman genotyping PCR. In this stage of metaphase, the chromosomes have duplicated but sister chromatids are still bound together at the centromere; therefore each metaphase cell has 46 separable chromosomes and no more than two chambers should contain templates for a given PCR genotyping assay. As expected, for assays that yielded PCR signals in two chambers, the alleles for both chambers matched that of the genomic DNA if the individual was homozygous for the tested locus, and the alleles of the two chambers were different if the individual was heterozygous for the tested locus (Fig. 2). There was no obvious bias in the distribution and no chromosome pairs were particularly difficult to separate. Because the chromosomes are randomly dispersed into chambers, it is possible that both homologous copies of a chromosome will co-locate in the same chamber. This probability can be made arbitrarily small by increasing the number of chambers, and in practice when co-location occurs we simply repeat the experiment with another cell.
Figure 2.
Whole-genome haplotyping. (a) Determining the chromosomal origin of amplification products in a microfluidic device using 46-loci PCR. This table represents results from an experiment using a single metaphase cell of P0’s cultured whole blood. A row represents the content inside a chamber on the microfluidic device, and a column represents a locus, with specified chromosome and coordinate (NCBI Build 36.1). Each locus, except those on chromosomes 17 and 20, was found in two chambers. The two alleles of a SNP are highlighted in red and green. Heterozygous loci are labeled in blue. Chamber numbers labeled yellow were pooled together and genotyped on one HumanOmni1-Quad array, and chamber numbers labeled orange were pooled together and genotyped on another array. Genomic DNA extracted from cultured whole blood was also tested with the same 46-loci PCR. (b) Statistics of whole-genome haplotyping. The fraction of SNPs present on the array phased for each chromosome of each individual (GM12891, GM12892, GM12878 and a European individual ‘P0’) is shown as a colored bar. (c) Fraction of SNPs phased as a function of the number of pairs of homologous chromosomes assayed. This is based on the results from four single-cell experiments of P0. Each point represents the coverage of an autosome. The error bars represent s.e.m.
Products from multiple chambers were pooled together into two mixtures such that each mixture contained one of the two homologous copies of most chromosomes. The two ‘haploid’ mixtures were separately genotyped on whole-genome genotyping arrays (Illumina’s HumanOmni1-Quad BeadChip). For each individual, three to four single-cell experiments were performed, and each homologous chromosome had, on average, ~2 to 3 biological replicates. Phases were established for ~87.9%, ~89.9% and ~83.8% of ~970,000 refSNPs present on the array for GM12878, GM12891 and GM12892, respectively (Fig. 2 and Supplementary Data Sets 1–3). By counting the number of inconsistent allele calls among biological replicates of each chromosome homolog, we estimated the error originating from amplification and genotyping for a single phase measurement to be 0.2–0.4%. The actual phasing error per SNP was much smaller because the final phases of most SNPs were determined by the consensus among replicates (Supplementary Fig. 1) and can be made as small as desired by increasing the number of replicates.
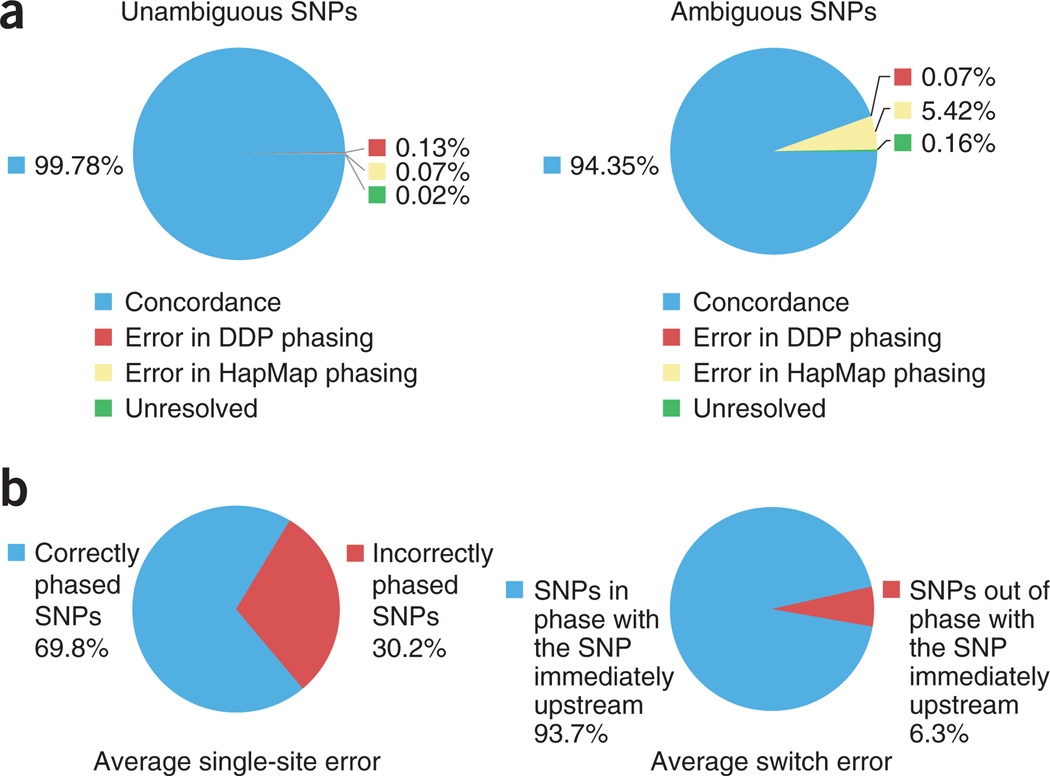
We compared our experimental phasing data of the child (GM12878) with haplotype data available from the HapMap project. In the HapMap project, haplotypes in the CEU population were obtained by studying the genotypes of family trios. About 80% of the heterozygous SNPs of the child can be unambiguously phased given that one parent is homozygous for the SNP. The remaining ~20% of heterozygous SNPs in the child are ambiguous and require statistical phasing because both parents are heterozygous. Comparison of DDP and HapMap data on unambiguous SNPs provides an estimate of the accuracy of DDP. The concordance rate between the two data sets was 99.8%. The small number of inconsistencies arose from either error in DDP genotyping or error in genotyping in HapMap data (Fig. 3a). When considering ambiguous SNPs alone, the incongruence rate between the two data sets was 5.7%. The majority of these inconsistencies (96.0%) came from incorrect statistical phasing in the HapMap project, as we could confirm the phases of these ambiguous SNPs in the child from the experimentally determined phases of the two parents (Fig. 3). These data agree with previous evaluations of the accuracies of statistical phasing in CEU trios15,28 and highlight the utility of direct experimental phasing even when family data are available.
Figure 3.
Comparison of statistically determined phases with experimentally determined phases. (a) Comparison of experimentally determined phases of ~160,000 heterozygous SNPs of GM12878 (child of the trio) and those determined by phase III of the HapMap project. Unambiguous SNPs refer to those that are homozygous for at least one parent and are deterministically phased using family data in HapMap. This comparison shows the accuracy of DDP. Ambiguous SNPs refer to those that are heterozygous for all members of the trio and statistical phasing is used in HapMap. This comparison provides an evaluation of statistical phasing. (b) Comparison of experimentally determined phases of P0 and those determined by PHASE. Seventy-six regions on the autosomal chromosomes were randomly selected and statistically phased three times. Each region carried 100 heterozygous SNPs and spanned an average of ~2 Mb. Switch error rate was calculated as the proportion of heterozygous SNPs with different phases relative to the SNP immediately upstream. Single-site error rate was calculated as the proportion of heterozygous SNPs with incorrect phase. A SNP was considered correctly phased if it had the dominant phase. For each region, the average values from the three runs were reported. Presented here are the average switch error and single-site error per region. The deterministic phases measured by DDP are taken as the ground truth.
Whole-genome haplotyping of a European individual
Having validated the DDP approach on well-characterized HapMap samples, we applied it to determine the haplotypes of an individual, labeled ‘P0’, whose genome has been sequenced6 and clinically annotated29. As only a few cells are required for DDP, we collected a blood sample from a finger prick. Whereas some of the early microfluidic devices used for experiments with the family trio contained defects leading to the failure to retrieve products from some chambers, refinement in device fabrication yielded fully functional devices and thus improved the number of SNPs phased per single-cell experiment for P0. The average number of pairs of autosomal chromosomes separated per single cell of P0 was 17.5. We obtained ~96.1% coverage of the ~1.2 million SNPs present on the HumanOmni1S array using four single cells (Fig. 2 and Supplementary Data Set 4). An additional ~861,000 SNPs were phased using materials from three single cells and the HumanOmni1-Quad array (Supplementary Data Set 5). For homologous chromosomes that were separated in all four single-cell replicates (that is, four biological replicates of each homologous copy), up to 99.2% of all SNPs assayed on a chromosome were phased (Fig. 2). We noticed that the SNPs that were not phased tended to cluster together and closer inspection revealed that they were usually located in regions with higher GC content (Supplementary Fig. 2). Stronger molecular associations between DNA strands at regions with higher GC content might have led to more difficult amplification, and such phenomena associated with phi29 have been previously reported30.
Phasing of SNPs was also achieved by direct sequencing. We lightly sequenced amplified material from three single copies of P0’s chromosome 6, at an average read depth of 3.5× to 7.7× per copy (Supplementary Table 1). About 46,000 heterozygous SNPs on chromosome 6 determined by previous genome sequencing were phased, including several of the medically relevant rare variants that were identified in the clinical annotation of the genome29. For alleles called by threefold or greater coverage, the concordance rate of phasing by sequencing and phasing by genotyping arrays was 99.8% (Supplementary Fig. 3a). This indicates that allele calling with haploid materials can be achieved accurately with relatively low coverage, an advantage over conventional genotyping by sequencing, which requires much higher fold coverage to guarantee accuracy of heterozygous SNPs. The amplification of minute amount of materials using the polymerase phi29 has been known to cause amplification bias and formation of nonspecific products that would undermine sequencing performance. Our group previously demonstrated improved performance of whole-genome amplification of single bacteria by reducing amplification volumes by ~1,000-fold using microfluidic devices similar to the one in this study31,32. The present sequencing experiments show that nonspecific products constituted a very small amount of the amplified materials and provide a characterization of the amplification bias for human chromosome–sized, single-molecule templates (Supplementary Table 1 and Supplementary Fig. 3b–d). The coverage across the chromosome was nonuniform, yet distribution of reads over most of the chromosome in all sequenced copies was within two orders of magnitude (Supplementary Fig. 3c).
Comparison of experimental and statistical phasing
Statistical inference has been commonly used to estimate haplotypes in unrelated individuals, yet the lack of true haplotypes means that few studies have been conducted to evaluate the accuracy of these computational approaches. We used the statistical inference software PHASE33–35 to infer haplotypes for P0 using CEU haplotypes, determined by family trios in the HapMap Project, as the background and compared the inferred haplotypes to the P0 haplotypes determined by DDP. Evaluation of a total of 76 ~2-Mb regions, each defined by 100 heterozygous SNPs, revealed an average of 6.3 block switches per region and an average block size of ~260 kb. An average of 30.2% of heterozygous SNPs were incorrectly phased using the statistical method (Fig. 3b and Supplementary Fig. 4). These results agree with two previous studies that compared statistical haplotype inference with real phases obtained from somatic cell hybrids and complete hydatidform moles36,37, and illustrate the importance of direct experimental phasing especially when family data are not available.
Direct measurement of recombination events within a family trio
The availability of parental haplotypes allowed us to directly measure the products of recombination events that led to an individual’s unique genome, which could previously only be inferred using three-generation families38 or two-generation families with large sibships39. We aligned each homologous chromosome of the child to the pair of chromosomes of the parent from whom the chromosome was inherited. Figure 4 illustrates the crossover events resulting from the paternal and maternal meioses. We detected 26 and 38 events in the male meiosis and female meiosis, respectively, with a median resolution of ~43 to 44 kb (Supplementary Table 2). The number of detected recombination events matched those in previous reports and supports the notion that the number of recombination events in females is generally higher than that in males38,40. At least 60% of these regions had recombination rates above the median sex average according to the deCODE genetic map39. In addition to the switchover of large blocks of homologous chromosomes as a result of recombination, we observed switchover at single sites, constituting ~0.4% of the total number of SNPs in each parent-child comparison; these are presumably products of gene conversion or cell culture–induced mutations, as well as DDP error.
Figure 4.
Direct observation of recombination events and deterministic phasing of heterozygous deletions in the family trio. Each allele with DDP data available for the child and the parent is represented by a colored line (blue, alleles transmitted to the child from the father; red, alleles transmitted to the child from the mother; black, untransmitted alleles). Centromeres and regions of heterochromatin are not assayed by genotyping arrays and are thus in white. Heterozygous deletions in the parents are represented as triangles along each homologous chromosome. A solid triangle represents one copy and a hollow triangle represents a null copy. The phases of deletions are determined for each parent independently. The triangles are color coded according to the state of transmittance as determined by the location of the deletion relative to spots of recombination. The phases of the deletions in the child are determined independent of the parents and are shown on top of the parental chromosomes. The integers on the left are the IDs of each region given by HapMap phase III. The numbers on the right are the copy number of a region in the child as determined by HapMap. Chromosomes are plotted with the same length.
Phasing of heterozygous deletions
Although copy number variants (CNVs) can be phased using statistical methods similar to those used to phase SNPs41–43, direct experimental phasing of structural variation such as copy number polymorphisms has largely been unexplored. We experimentally phased the heterozygous deletions accessible with genotyping arrays, as determined by the HapMap Project, of the three individuals in the family trio. We phased 12 and 6 heterozygous deletions present within the family trio using genotyping array data (Supplementary Table 3a) and real-time PCR (Supplementary Table 3b), respectively. All of the phased heterozygous deletions within the trio agreed with the inheritance pattern (Fig. 4). We also phased all eight heterozygous deletions that had been detected by genome sequencing of P0 (ref. 6) using data from genotyping arrays and real-time PCR. Results from all platforms among all single-cell replicates were consistent (Supplementary Table 4). Phasing of other types of CNVs with the current approach of genotyping array and PCR is challenging, but we envision that deep sequencing of amplified materials would eventually allow each chromosome to be assembled and thus enable phasing of all CNVs.
Direct determination of the HLA haplotypes of an individual
An important application of DDP is the determination of the HLA haplotypes within an individual. The HLA loci are highly polymorphic and are distributed over ~4 Mb on chromosome 6. The ability to haplotype the HLA genes within the region is clinically important because this region is associated with autoimmune and infectious diseases44, and the compatibility of HLA haplotypes between donor and recipient can influence the outcomes of transplantation8. Yet molecular techniques to measure HLA haplotypes in individuals are still limited45.
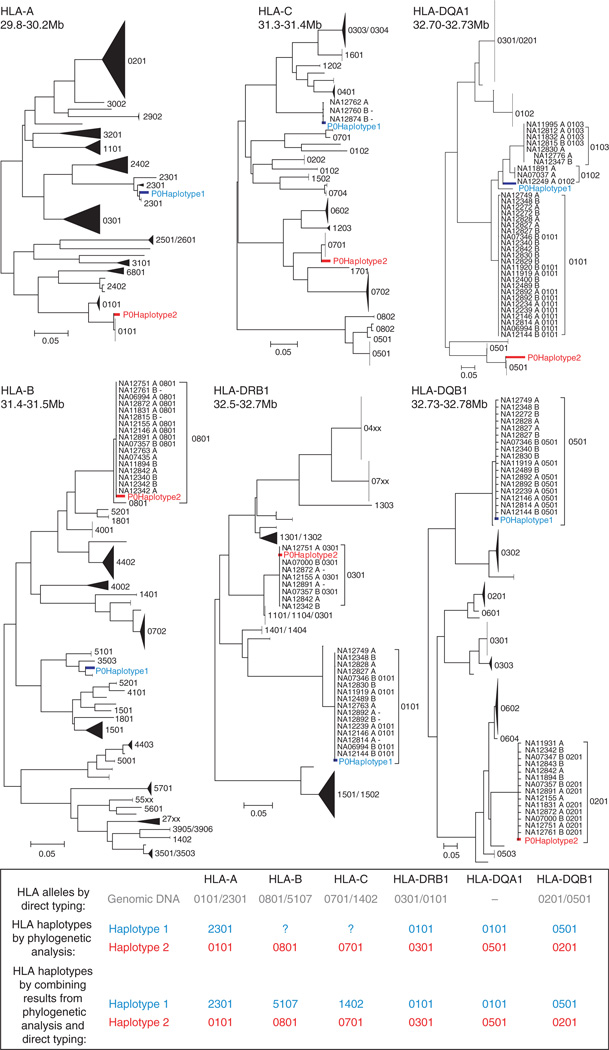
To determine the HLA haplotypes, we first had to determine the HLA allele at each locus, which is usually achieved by direct sequencing. Here, we sought a simpler approach to determine the allele at each HLA locus by taking advantage of the experimentally determined SNP haplotypes of P0. Briefly, we used phylogenetic analyses to compare the SNP haplotypes of P0 within each HLA gene to those of CEU individuals whose HLA genes were typed previously (Fig. 5). The combination of the alleles at each HLA locus determined by phylogenetic analyses agreed with direct HLA typing of genomic DNA. Combining the results from all loci yielded the two HLA haplotypes of P0. One of the HLA haplotypes is the 8.1 ancestral haplotype, which is one of the most frequently observed haplotypes in Caucasians46 and is associated with elevated risks of immunopathological diseases47.
Figure 5.
HLA haplotypes of P0 determined using DDP. At each of the six classical HLA loci, the experimentally phased SNP haplotypes of P0 and 176 phased SNP haplotypes of CEU trios available from HapMap phase III were placed on a neighbor-joining tree. The two haplotypes of P0 are labeled in red and blue. For haplotypes in the CEU panel with HLA typing data, the four-digit HLA allele is presented next to the sample label. Most of each tree is compressed. Each compressed subtree is labeled with the HLA allele associated with members inside the subtree, if HLA allele information is available. The allelic identities of HLA-B and HLA-C on haplotype 1 were not determined with DDP because CEU individuals with similar SNP haplotypes as P0’s SNP haplotypes did not have HLA typing data at these loci but could be inferred from the results of direct HLA typing of genomic DNA (first row of table). HLA-DQA1 was not directly typed.
DISCUSSION
The DDP approach is scalable. Multiple cells can be processed simultaneously by modifying device design. Currently, the most labor- intensive procedure is the manual identification of metaphase cells. We anticipate that automation of this with a relatively simple engineering solution, such as the combination of computer vision and fluorescent labeling of mitotic cells, will dramatically increase throughput. The majority of the cost of the project went to the genotyping arrays, and as sequencing costs continue to drop, it may become more cost effective to sequence rather than to genotype.
DDP requires the presence of metaphase chromosomes because during metaphase chromosomes are most condensed and can be physically separated. DDP therefore requires sources of cells that can undergo mitosis, such as blood samples and cell lines. Yet DDP requires as little as a single cell, and thus may also have important applications in single-cell genomics, in fields such as preimplantation genetic diagnosis, prenatal diagnosis, aging, and cancer diagnosis and research.
To our knowledge, the work described here represents the first demonstration of a molecular-based, whole-genome haplotyping technique amenable for personal genomics. Whereas the bulk of the experiments described here focus on direct deterministic phasing of ~1 million variants accessible by genotyping arrays, DDP can be used to phase all variants in the genome. DDP of tagSNPs present on the genotyping arrays inherently provides phasing information for common variants that are in strong linkage disequilibrium with the tagSNPs. In addition, we showed that amplified materials from separated chromosome homologs could be directly sequenced, yielding phasing information for variants, including the rare and private ones, which are absent on standard genotyping arrays. Combining DDP SNP analysis with shotgun genome sequencing could allow the determination of the complete personal haplotype of an individual, even in the absence of family information.
ONLINE METHODS
Microfluidic device design, fabrication and operation
The microfluidic device was made of polydimethylsiloxane (PDMS) and was fabricated using soft lithography by the Stanford Microfluidic Foundry. The two-layered device had rectangular 25-µm tall control channels at the bottom and rounded flow channels at the top. The device was bonded to a glass slide coated with a thin layer of PDMS. In the cell-sorting region of the device, flow channels were 40 µm high and 200 µm wide. In the amplification region of the device, flow channels were 5 µm and 100 µm wide and reaction chambers were 40 µm tall. A membrane valve was formed when a control channel crossed over with a flow channel and was actuated when the control channel was pressurized at 20–25 p.s.i. The area of each valve was 200 µm × 200 µm for the 40-µm flow channels, and 100 µm × 100 µm for the 5-µm flow channels. Membrane valves were controlled by external pneumatic solenoid valves that were driven by custom electronics connected to the USB port of a computer. A Matlab program was written to interface with the valves.
Cell culture
The Epstein-Barr virus–transformed lymphoblastoid cell lines GM12891, GM12892 and GM12878, belonging to the pedigree CEU 1463 (Coriell Cell Repositories), were cultured in RPMI 1640, supplemented with 15% FBS. Each culture was treated with 2 mM thymidine (Sigma) for 24 h at 37 °C to enrich the population of mitotic cells. Followed by multiple washings, cells were cultured in normal medium for 3 h and treated with 200 ng/ml nocodazole (Sigma) for 2 h at 37 °C to arrest cells at metaphase.
Whole blood (~250 µl) obtained from a finger-prick of Patient Zero (‘P0’) was treated with sodium heparin and cultured in PB-Max medium (Invitrogen) for 4 days. The culture was treated with 50 ng/ml colcemid (Invitrogen) for 6 h. The culture was layered on top of Accuspin System-Histopaque-1077 (Sigma) and centrifuged for 8 min at 590g. Nucleated cells at the interface was removed and washed once with HBSS.
Metaphase arrested cells incubated with 75 mM KCl at 25 °C for 10 to 15 min. Acetic acid was added to the cell suspension at a final concentration of 2% to fix the cells. After fixation on ice for 30 min, cells were washed multiple times and finally suspended in 75 mM KCl-1mM EDTA-1% Triton X-100. Cells were treated with 0.2 mg/ml RNaseA (Qiagen) before loading onto the microfluidic device.
Cell sorting, chromosome release and multiple strand displacement amplification
Before the loading of the cell suspension, the cell-sorting channel of the device was treated with Pluronic F127 (0.2% in PBS). Cell suspension was introduced into the device using an on-chip peristaltic pump and an off-chip pressure source. Metaphase cells could be distinguished from interphase cells microscopically by morphological differences. Once a single metaphase cell was recognized at the capture chamber, surrounding valves were actuated to isolate it from the remaining cell suspension. Pepsin solution (0.01% in 75 mM KCl, 1% Triton X-100, 2% acetic acid) was introduced to digest the cytoplasm and release the chromosomes. The chromosome suspension was pushed into a long narrow channel and partitioned into 48 180-pl compartments by actuating a series of valves along the channel. Trypsin (0.25%) in 150 mM Tris-HCl (pH 8.0) (1.2 nl) was introduced to neutralize the solution and to digest chromosomal proteins. Ten minutes later, denaturation buffer (Qiagen’s Repli-G Midi kit’s buffer DLB supplemented with 0.8% Tween-20; 1.4 nl) was introduced. The device was placed on a flat-topped thermal cycler set at 40 °C for 10 min. This was followed by the introduction of neutralization solution (Repli-G kit’s stop solution; 1.4 nl) and incubation at 25 °C for 10 min. A mixture of reaction buffer (Qiagen’s Repli-G Midi Kit), phi29 polymerase (Qiagen’s Repli-G Midi Kit), 1× protease inhibitor cocktail (Roche) and 0.5% Tween-20 (16 nl) was fed in. The total volume per reaction was 20 nl and the device was placed on the flat-topped thermal cycler set at 32 °C for about 16 h. Amplification products from each chamber was retrieved from its corresponding outlet by flushing the chamber with TE buffer (pH 8.0) supplemented with 0.2% Tween-20. About 3–5 µl of products were collected from each chamber. Products were incubated at 65 °C for 3 min to inactivate the phi29 enzyme.
Initial genotyping with 46 loci
To determine the identity of chromosomes in each chamber, we performed Taqman PCR using a set of 46 genotyping assays (two assays per autosome and one assay per sex chromosome) on the products of each chamber on the 48.48 Dynamic Array (Fluidigm). The assays used are listed in Supplementary Table 5.
Whole-genome phasing using genotyping arrays
To generate sufficient materials for genotyping array experiments, we amplified DNA products from the microfluidic device a second time in 10 µl volume using the Repli-G Midi Kit’s protocol for amplifying purified genomic DNA. Products from multiple chambers were pooled together into two mixtures such that each mixture contained one of the homologous copies of each chromosome. Each mixture, containing roughly one haploid genome of a cell, was genotyped on the HumanOmni1-Quad or HumanOmni1S BeadChips (Illumina). For GM12891, GM12878 and P0, four single cells were haplotyped. For GM12892, three single cells were haplotyped. Haplotyping data for cell lines were obtained from HumanOmni1-Quad array. Haplotyping data for P0 were obtained from both HumanOmni1-Quad and HumanOmni1S arrays. Genomic DNA extracted from each cell line was also genotyped on the HumanOmni1-Quad array. Genomic DNA of P0 was genotyped on the HumanOmni1S array.
For each chromosome homolog, the allelic identity of a SNP was determined from the consensus among the biological replicates. If equal numbers of both alleles were observed at the site, no consensus was drawn. We estimated the error of a single genotyping measurement by counting the number of inconsistent allele calls at sites typed more than once. For SNPs of which only one of the alleles was observed, the identity of the other allele was determined using the genotypes of genomic DNA. For the trio, the genotypes of genomic DNA were measured on the HumanOmni1-Quad BeadChip. The concordance rate of these genotypes with HapMap data was ~99.1% for each cell line. For P0, genotypes of genomic DNA were measured on the HumanOmni1S BeadChip. The combination of the consensus alleles from the two homologs at each SNP site should, in principle, agree with the genotype call of the genomic DNA control. SNPs that did not follow this rule (~0.3% for cell lines and ~0.4% for P0) were eliminated from downstream analyses.
Data files containing the phased haplotypes of the members of the trio and P0 are available as Supplementary Data Set 1 (GM12891), Supplementary Data Set 2 (GM12892), Supplementary Data Set 3 (GM12878), Supplementary Data Set 4 (P0 Omni1S) and Supplementary Data Set 5 (P0 Omni1Quad). Each file contains whole-genome haplotypes of each individual of the CEU trio (GM12891, GM12892, GM12878) and P0. For the trio, refSNPs present on the Omni1-Quad array (Illumina) that were phased by DDP are included in these files. For P0, refSNPs present on the Omni1-Quad and SNPs present on the Omni1S arrays phased by DDP are included. For P0’s data on Omni1-Quad arrays, only SNPs with both alleles directly observed were included. Alleles are presented relative to the forward strand, and SNPs with A/T and G/C alleles are not included. Column 1: SNP name; column 2: chromosome (chromosome X designated as ‘23’); column 3: position on chromosome (hg18); column 4: allele on homologous copy 1; column 5: allele on homologous copy 2.
Phasing of chromosome 6 using high-throughput sequencing
Three chambers containing amplified materials from a single copy of chromosome 6 were selected from the four single-cell experiments of P0 for paired-end sequencing on Illumina’s Genome Analyzer II. Two chambers contained materials from chromosome 6 only, whereas the third chamber contained materials from a homolog of chromosomes 6, 16 and 18. Second-round amplified materials from these chambers were fragmented through a 30-min 37 °C incubation with 4 µl dsDNA Fragmentase (New England Biolabs) in a 20-µl reaction. Fragmented DNA was end-repaired, tailed with a single ‘A’ base, and ligated with adaptors. A 12-cycle PCR was carried out and PCR products with sizes of 300–500 bp were selected using gel extraction. Sequencing libraries were quantified with digital PCR48. Thirty-six base pairs were sequenced on each end.
Image analysis, base calling and alignment were performed using Illumina’s GA Pipeline version 1.5.1. The first 32 bases on each read were aligned to the human genome (Build 36.1). SNP calling was carried out using Illumina’s CASAVA version 1.6.0. Positions covered at least three times according to the ‘sort.count’ intermediate files were used in downstream analyses. A list of heterozygous SNPs was obtained from the sequenced genome of P0, using quality score >2.8 and heterozygous score of 20 (ref. 6). The phases of heterozygous SNPs were determined either from the direct observation of both alleles in the different homologs, or by inferring the identity of the unobserved allele if only one allele was detected.
Data sources
Genotypes, CNVs and phasing data of the three lymphoblastoid cell lines were downloaded from the website of the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/). Genotypes of the merged phase I+II and III data were used. Phasing information from phase III was used. CNV data from phase III was used.
Comparison between experimentally determined phases of the family trio with HapMap data
We compared the experimentally determined phases of the heterozygous SNPs of the child (GM12878) to those determined by phase III of the HapMap Project. SNPs with A/T and G/C alleles were excluded from comparison. To determine the accuracy of experimental phasing and to locate spots of crossovers, the phases of heterozygous SNPs of each parent (GM12891, GM12892) were compared to those in the child (GM12878) inherited from that parent.
Phasing of heterozygous deletions
For the trio, a list of heterozygous deletions was obtained from phase III of the HapMap project. For P0, heterozygous deletions that were detected by previous genome sequencing and subsequently verified by digital PCR were studied6. For P0, the assays were the same as those used in a previous study6. For the trio, the sequences of primers and probes are listed in Supplementary Table 6. The assumption was that one of the chromosome homologs should give no calls for SNP markers or no PCR amplification within a region of heterozygous deletions. Digital PCR (Fluidigm’s 48.770 digital array) was also performed using genomic DNA of each member of the trio to verify copy numbers.
Statistical phasing of P0 using PHASE
We evaluated the accuracy of statistical phasing by comparing statistically phased haplotypes and experimentally determined haplotypes of P0. We inferred the haplotypes of P0 using PHASE 2.1 (a Bayesian method–based program for haplotype reconstruction)33–35. Due to computational capacity, we randomly chose four regions on each autosomal chromosome (except chromosomes 4, 20, 21), each having 100 biallelic SNPs that were heterozygous in P0. We only selected SNPs with both alleles directly haplotyped and with perfect concordance with genotype determined by whole-genome sequencing. Each region covered a range of ~0.7 to ~3.3 Mb, with an average SNP to SNP distance of ~20 kb. We used the 176 phased CEU haplotypes in phase III of the HapMap project as known haplotypes for the inference. For each region, we ran the reconstruction three times with the same default settings but different random seeds and compared the results with the experimentally determined haplotypes. Switch error rate was calculated as the proportion of heterozygous SNPs with different phases relative to the SNP immediately upstream. Single-site error rate was calculated as the proportion of heterozygous SNPs with incorrect phase. A SNP was considered correctly phased if it had the dominant phase. For each region, the average values from the three runs were reported.
Determination of HLA haplotypes of P0
A total of 176 phased CEU haplotypes obtained from phase III of the HapMap project, together with experimentally phased haplotypes of P0, were used to construct neighbor-joining trees at the six classical HLA loci on chromosome 6. The coordinate boundaries of which haplotyped SNPs were used for each locus are presented in Figure 5. Only SNPs with both alleles directly observed were used. The number of SNPs used for HLA-A, HLA-B, HLA-C, HLA-DRB, HLA-DQA, and HLA-DQB were 420, 139, 89, 59, 14 and 34, respectively. Allele sharing distances were computed for each pair of haplotypes as , where n is the number of loci and di equals 0 for matched alleles and 1 for unmatched alleles at the ith SNP locus. Trees were constructed using MEGA 4.1 (ref. 49). A list of HLA alleles of individuals in the CEU panel typed in a previous study9 was downloaded from http://www.inflammgen.org/. The allelic identity of each homologous chromosome of P0 at each HLA locus was determined by the allelic identities of its nearest neighbors in the tree.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Melin and the Stanford Microfluidic Foundry for fabrication of microfluidic devices. We thank N. Neff and G. Mantalas for performing sequencing experiments. We thank D. Pushkarev for providing data and primers from the genome sequencing project of P0. We thank M. Anderson and D. Tyan at Stanford Histocompatibility Laboratory for providing HLA typing results. We thank Y. Marcy, P. Blainey, J. Jiang and A. Wu for helpful discussions. The project was supported by the US National Institutes of Health (NIH) Pioneer Award and an NIH U54 award. H.C.F. was supported by a scholarship from the Siebel Foundation. J.W. was supported by a scholarship from the China Scholarship Council.
Footnotes
Accession code. Short-read sequence data have been deposited at the NCBI Sequence Read Archive (SRA) under accession no. SRA026722.
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
H.C.F. and S.R.Q. conceived the experiments. H.C.F. designed the microfluidic device. A.P. developed protocols for device fabrication. H.C.F. and J.W. performed the experiments. H.C.F., J.W. and S.R.Q. analyzed the data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/.
References
- 1.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 2.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn SM, et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JI, et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nat. Biotechnol. 2009;27:847–850. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuster SC, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersdorf EW, Malkki M, Gooley TA, Martin PJ, Guo Z. MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med. 2007;4:e8. doi: 10.1371/journal.pmed.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bakker PI, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart CA, et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–1187. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groenendijk M, Cantor RM, de Bruin TW, Dallinga-Thie GM. The apoAI-CIII-AIV gene cluster. Atherosclerosis. 2001;157:1–11. doi: 10.1016/s0021-9150(01)00539-1. [DOI] [PubMed] [Google Scholar]
- 12.Nagel RL, et al. The Senegal DNA haplotype is associated with the amelioration of anemia in African-American sickle cell anemia patients. Blood. 1991;77:1371–1375. [PubMed] [Google Scholar]
- 13.Sun T, et al. Haplotypes in matrix metalloproteinase gene cluster on chromosome 11q22 contribute to the risk of lung cancer development and progression. Clin. Cancer Res. 2006;12:7009–7017. doi: 10.1158/1078-0432.CCR-06-0464. [DOI] [PubMed] [Google Scholar]
- 14.Drysdale CM, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl. Acad. Sci. USA. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K, et al. Long-range polony haplotyping of individual human chromosome molecules. Nat. Genet. 2006;38:382–387. doi: 10.1038/ng1741. [DOI] [PubMed] [Google Scholar]
- 19.Mitra RD, et al. Digital genotyping and haplotyping with polymerase colonies. Proc. Natl. Acad. Sci. USA. 2003;100:5926–5931. doi: 10.1073/pnas.0936399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding C, Cantor CR. Direct molecular haplotyping of long-range genomic DNA with M1-PCR. Proc. Natl. Acad. Sci. USA. 2003;100:7449–7453. doi: 10.1073/pnas.1232475100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michalatos-Beloin S, Tishkoff SA, Bentley KL, Kidd KK, Ruano G. Molecular haplotyping of genetic markers 10 kb apart by allele-specific long-range PCR. Nucleic Acids Res. 1996;24:4841–4843. doi: 10.1093/nar/24.23.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruano G, Kidd KK, Stephens JC. Haplotype of multiple polymorphisms resolved by enzymatic amplification of single DNA molecules. Proc. Natl. Acad. Sci. USA. 1990;87:6296–6300. doi: 10.1073/pnas.87.16.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woolley AT, Guillemette C, Li Cheung C, Housman DE, Lieber CM. Direct haplotyping of kilobase-size DNA using carbon nanotube probes. Nat. Biotechnol. 2000;18:760–763. doi: 10.1038/77760. [DOI] [PubMed] [Google Scholar]
- 24.Burgtorf C, et al. Clone-based systematic haplotyping (CSH): a procedure for physical haplotyping of whole genomes. Genome Res. 2003;13:2717–2724. doi: 10.1101/gr.1442303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao M, et al. Direct determination of haplotypes from single DNA molecules. Nat. Methods. 2009;6:199–201. doi: 10.1038/nmeth.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma L, et al. Direct determination of molecular haplotypes by chromosome microdissection. Nat. Methods. 2010;7:299–301. doi: 10.1038/nmeth.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Douglas JA, Boehnke M, Gillanders E, Trent JM, Gruber SB. Experimentally-derived haplotypes substantially increase the efficiency of linkage disequilibrium studies. Nat. Genet. 2001;28:361–364. doi: 10.1038/ng582. [DOI] [PubMed] [Google Scholar]
- 28.Marchini J, et al. A comparison of phasing algorithms for trios and unrelated individuals. Am. J. Hum. Genet. 2006;78:437–450. doi: 10.1086/500808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashley EA, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bredel M, et al. Amplification of whole tumor genomes and gene-by-gene mapping of genomic aberrations from limited sources of fresh-frozen and paraffin-embedded DNA. J. Mol. Diagn. 2005;7:171–182. doi: 10.1016/S1525-1578(10)60543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcy Y, et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:e155. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcy Y, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. USA. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kukita Y, et al. Genome-wide definitive haplotypes determined using a collection of complete hydatidiform moles. Genome Res. 2005;15:1511–1518. doi: 10.1101/gr.4371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andres AM, et al. Understanding the accuracy of statistical haplotype inference with sequence data of known phase. Genet. Epidemiol. 2007;31:659–671. doi: 10.1002/gepi.20185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong A, et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 40.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su SY, et al. Inferring combined CNV/SNP haplotypes from genotype data. Bioinformatics. 2010;26:1437–1445. doi: 10.1093/bioinformatics/btq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 44.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J. Hum. Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 45.Guo Z, Hood L, Malkki M, Petersdorf EW. Long-range multilocus haplotype phasing of the MHC. Proc. Natl. Acad. Sci. USA. 2006;103:6964–6969. doi: 10.1073/pnas.0602286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum. Immunol. 2007;68:779–788. doi: 10.1016/j.humimm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 47.Price P, et al. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol. Rev. 1999;167:257–274. doi: 10.1111/j.1600-065x.1999.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 48.White RA, III, Blainey PC, Fan HC, Quake SR. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009;10:116. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.