Summary
There are few substantive methods to measure the health of the immune system, and the connection between immune strength and the viral component of the microbiome is poorly understood. Organ transplant recipients are treated with a post-transplant therapy that combines immunosuppressive and antiviral drugs, offering a window into the effects of immune modulation on the virome. We used sequencing of cell-free DNA in plasma to investigate drug-virome interactions in a cohort of organ transplant recipients (656 samples, 96 patients), and find that antivirals and immunosuppressants strongly affect the structure of the virome in plasma. We observe marked virome compositional dynamics at the onset of the therapy and find that the total viral load increases with immunosuppression, whereas the bacterial component of the microbiome remains largely unaffected. The data provide insight into the relationship between the human virome, the state of the immune system, and the effects of pharmacological treatment, and offer a potential application of the virome state to predict immunocompetence.
Introduction
The human microbiome is now recognized as an important component of human health (Turnbaugh et al., 2007). Community level analyses have shed light on factors that shape the structure of the bacterial component of the microbiome, such as age (Yatsunenko et al., 2012), diet (De Filippo et al., 2010; Muegge et al., 2011), geographical location (Yatsunenko et al., 2012), antibiotic treatment (Jakobsson et al., 2010) and disease (Clemente et al., 2012). The viral component of the microbiome, the human virome, remains relatively understudied (Wylie et al., 2012) and little is known about the effects of immune modulation and antiviral therapies on virome composition. It was previously shown that the healthy gut virome remains remarkably stable over time (Reyes et al., 2010), and that the predominant source of variation is due to differences between subjects, although an association between diet and the virome composition was found (Minot et al., 2011). Here, we study the dynamic response of the human virome in plasma to antiviral drugs and strong perturbations of the immune system as experienced by organ transplant recipients.
Immunosuppressive therapies significantly reduce the risk of graft rejection in organ transplantation but increase the susceptibility of recipients to infections (Fishman, 2007). Infections with viral pathogens, in particular the herpesvirus cytomegalovirus (CMV), occur frequently and increase the recipient's risk of graft failure (Fishman et al., 2006). Organ transplant recipients are therefore frequently subjected to antiviral prophylactic or preemptive therapies directed against CMV (Slifkin et al., 2004). The inverse relationship between the level of immunosuppression and the risks of infection and rejection leaves only a narrow therapeutic window available for patient treatment (see Fig. 1A). Post-transplant care is further complicated by numerous limitations of the currently available methods for the diagnosis of infection and rejection. Diagnosis of rejection mostly relies on invasive biopsies that suffer from interobserver variability, high cost and patient discomfort (Marboe et al., 2005; Saraiva et al., 2011; Snyder et al., 2011). Diagnosis of infections is challenging given the fact that the symptoms of infection are diminished following immunosuppression (Fishman, 2007), and commonly used diagnostic methods, such as antigen-detection and PCR-based molecular tests, rely on a specific target and therefore an a priori hypothesis for the source of the infection. As a final complication, patient-to-patient variability in the sensitivity to immunosuppressive drugs can give rise to over- and underimmunosuppression, increasing the risk of infection or rejection respectively (Budde et al., 2011; Wieland et al., 2010).
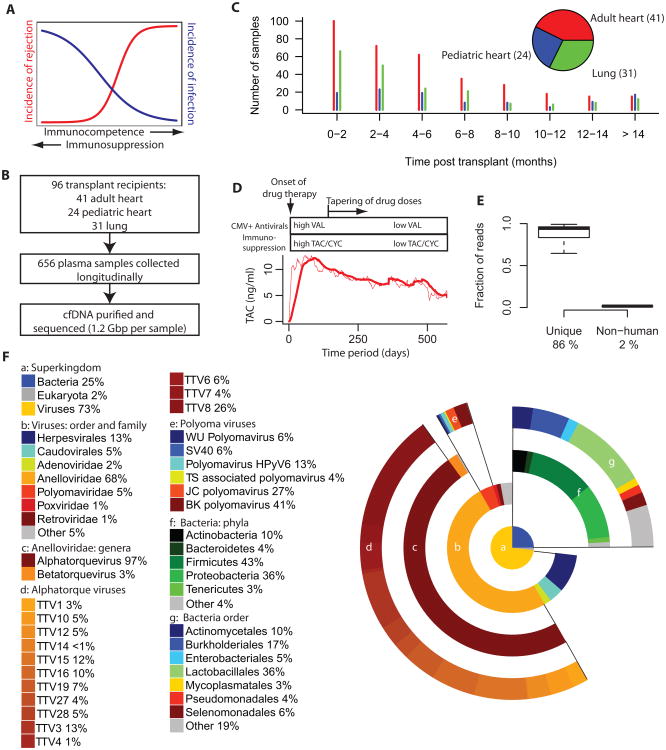
Fig. 1. Study design, read statistics and phylogenetic distribution.
A. Immunosuppression reduces the risk of rejection in transplantation but increases the risk of infection. B. Design of study. 656 plasma samples were collected, cell-free DNA was purified and sequenced to an average depth of 1.25 Gbp per sample. C. Number of samples collected as function of time for the different patient groups part of the study. D. Treatment protocol for patients in the study cohort, all patients are treated with maintenance immunosuppression (tacrolimus-based (TAC) for adult heart and lung transplant recipients and cyclosporine (CYC) for pediatric patients). CMV positive (donor or recipient, CMV+) transplant cases are treated with anti-CMV prophylaxis, valganciclovir (VAL). Mean level of tacrolimus measured in blood of transplant recipients treated with a TAC-based protocol (dashed line actual, solid line window average filter). E. Fraction of reads that remain after filtering of lower quality and duplicate reads (mean 86%, left) and after removal of human and low complexity reads (mean 2%, right). F. Relative genomic abundance at different levels of taxonomic classification after removal of human reads (average over all samples from all organ transplant recipients (n = 656)). The central piechart shows the composition at the superkingdom level of classification. Lower levels of classification are shown in donut charts with progressively larger radius. See also Figure S1.
In this work, we sequenced cell-free DNA circulating in plasma to investigate drug-microbiome interactions following organ transplantation. We studied the patterns of infection in heart and lung transplant recipients subjected to a combination of immunosuppressants and antiviral prophylaxis. We find that immunosuppressants and antivirals have a strong influence on the structure of the viral component of the microbiome but not the bacterial component. Strong compositional dynamics are observed at the onset of the drug therapy as the virome composition of different individuals converge to a similar, drug-determined state. The total viral load increases markedly in response to the therapy, as viruses, in particular the anelloviruses, take advantage of a reduction of immunocompetence. Finally, we show that measurement of the anellovirus burden enables stratification of rejecting and non-rejecting recipients.
Results
656 plasma samples were collected longitudinally from 96 solid organ transplant recipients (41 adult heart, 24 pediatric heart, 31 adult lung). Cell-free DNA was purified from plasma (Fan et al., 2008) and sequenced. In total, we obtained 820 gigabases (Gbp) of sequencing data, with an average of 1.25 Gbp per sample (Illumina HiSeq, 1×50 bp reads, Fig. 1B). Organ transplant recipients were continuously enrolled in the study over the course of more than 2 years and samples were collected from the recipients at regular time points post transplant, with the highest frequency of sample collection in the first months post transplant (see SI). Fig. 1C shows the number of samples analyzed as a function of time post transplant for the different patient classes.
The patients in the cohort were treated with antiviral prophylaxis and immunosuppression as part of a standardized post-transplant therapy (Fig. 1D, for more details of the protocol, see SI). Maintenance immunosuppression was tacrolimus-based for the adult heart and lung transplant recipients and was complemented with mycophenolate mofetil and prednisone. Pediatric patients were treated with a cyclosporine based anti-rejection therapy. CMV positive transplant recipients (prior CMV infection for recipient and/or donor), but not CMV negative recipients, were treated with antiviral prophylaxis. The protocol design entails high doses of immunosuppressants and antiviral drugs in the first few months post transplant, after which the doses are gradually reduced as the risks of rejection and infection diminish. Given the narrow therapeutic window available for immunosuppression and the large patient-to-patient variability in pharmacokinetics of tacrolimus (Jusko et al., 1995; Venkataramanan et al., 1995), the concentration of the tacrolimus is directly measured in the blood and the dose is adjusted to maintain a target drug level (see SI for details). Fig. 1D shows the mean level of tacrolimus measured in blood for the tacrolimus-treated patients and illustrates the design of the drug treatment protocol.
DNA sequence analysis
Microbiome-derived sequences were identified after computational subtraction of human-derived sequences (Weber et al., 2002; Xu et al., 2003). To this end, duplicate and low quality reads were removed and the remaining reads were mapped to the human reference genome, build hg19 (BWA (Li and Durbin, 2009), see methods). Unmapped reads were then collected and low complexity reads were removed (SEQCLEAN, http://www.tigr.org/tdb/tgi/software). Fig. 1E shows the distribution of the remaining read fraction after applying duplicate and quality filters (average of 86 %) and the distribution of the remaining fraction after subtraction of human reads (average of 2%).
To identify infectious agents, the remaining, high quality, unique, non-human reads were mapped using BLAST to a reference database of viral (n=1401), bacterial (n=1980) and fungal (n=32) genomes (downloaded from NCBI ftp://ftp.ncbi.nih.gov/genomes on June 14 2012, see Supplementary methods and Fig. S1A). 0.12% of the uniquely sequenced reads aligned to at least one of the target genomes (Fig. S1B, C). We used a quantitative PCR (qPCR) assay targeted to a subset of sequencing identified targets (herpesviruses 4,5,6 and parvovirus) to validate the positive hits identified by the sequencing-based approach. We found a quantitative agreement between viral counts as measured by sequencing and qPCR (Fig. S1D). We furthermore found that the sensitivity of the sequencing assay for the detection of herpesviruses is on par with qPCR measurements. The larger capture cross-section available to the sequencing assay – the complete target genome versus the PCR amplicon target region – is thus sufficient to overcome the signal loss in sequencing, caused by the finite efficiency of sequencing library preparation and library undersampling. The highest CMV loads measured using sequencing across all samples in the study corresponded to two adult heart transplant patients that suffered from a clinically diagnosed disseminated CMV infection (see Fig. S1E).
To test for the presence of potential contaminants in the reagents used for DNA extraction and sequencing library preparation, we performed two control experiments. In the first, we prepared 2 samples consisting of a defined template (Lambda gDNA, Pacbio Part no: 001-119-535), and purified DNA for sequencing using the above-described workflow (Illumina Miseq, 3.4 and 3.5 million reads). Lambda-derived sequences were removed and the remaining sequences (0.4%) were aligned to the BLAST reference database described above. No evidence was found for the various infectious agents discussed in this work, but we did detect sequences related to the Enterobacteriaceae bacterial family (phylum Proteobacteria), primarily E. coli (> 97%), and enterobacterial phages (<1%), which are likely a remnant of the lambda DNA culture. In a second control, we prepared a sample for sequencing from nuclease-free water. The sample was included in a sequencing run along with a sample unrelated to this study and recruited only a limited number of sequences, 15 in total, which mapped to genomes of two bacterial species (see SI). Again, no evidence was found for the infectious agents that are discussed below.
We studied the microbiome composition in plasma at different levels of taxonomic classification using GRAMMy (Xia et al., 2011), a tool that utilizes the sequence-similarity data obtained with BLAST to perform a maximum likelihood estimation of the relative abundance of species. GRAMMy accounts for differences in target genome size and the ambiguity of read assignments. This approach only allows estimating the abundance of species for which genomic data is available in the reference database. Figure 1F shows the relative abundance of species at different levels of taxonomic classification (average over all samples). We find that viruses (73%) are more abundantly represented than bacteria (25%) and fungi (2%) (Fig. 1F panel a). Among viruses, we find that ssDNA viruses occupy a larger fraction (72%) than dsDNA viruses (28%). Seven distinct viral families are found (abundance > 0.75%), with one dominant family, the Anelloviridae, which accounted for 68% of the total population (Fig. 1F, panel b). The anelloviridae fraction is mostly (97%) composed of viruses from the Alphatorquevirus genus (Fig. 1F, panel c). The Alphatorque genus is the genus of Torque Teno Viruses (TTVs), and sequences related to 14 different torque teno virotypes were identified (Fig1, panel d). Infections with polyomaviruses are widespread in the human population (Chesters et al., 1983), and polyomavirus DNAemia is not uncommon in the first year after solid organ transplantation (Razonable et al., 2005). Polyomavirus-derived sequences were found in 75 samples (11%) corresponding to 36 patients in the present cohort. Evidence for the presence of BK (41%), JC (27%), TS (4%), WU polyomavirus (6%), SV40 (6%) and the recently discovered HPyV6 (13 %) (Schowalter et al., 2010) was found (Fig. 1F, panel e). Among bacteria, Proteobacteria (36%), Firmicutes (50%), Actinobacteria (10%), Bacteroidetes (4%) are the phyla most abundantly represented in the sample (Fig. 1F, panel f).
To investigate potential incorrect assignments of the relatively short reads available to this study (50 bp), we examined the dependence of the abundance estimates on read length, based on longer, paired-end reads (2×100 bp) collected for a subset of samples (n=55). We found that the abundance estimates based on 50 bp subreads and 100 bp reads are similar for all levels of taxonomic classification reported here (fig. S1F).
Sensitivity of virome composition to drug dosage
The available clinical data on drug dosage was used to analyze drug-microbiome interactions. Here, we examined data for the adult heart and lung transplant patients that were treated with a tacrolimus-based anti-rejection protocol (47 patients and 380 observations), thereby excluding the pediatric patients that were treated with cyclosporine and patients that were switched from tacrolimus to cyclosporine immunosuppression due to drug-intolerance issues. Data on prescription antiviral drug doses (valganciclovir) and the measured levels of tacrolimus in blood were collected from individual patient records and the mean composition for samples corresponding to different drugs levels was extracted. To account for the delayed effect of microbiome composition on dose changes, the drug level and dose data were sliding window average filtered (see Fig. 1D and Fig.S2A-C; window size 45 days).
We find that the structure of the viral component of the microbiome is a sensitive function of drug dosage (47 patients, 380 samples, Figure 2A). However, the structure of the bacterial component of the microbiome was not significantly altered by the drug therapy, as discussed further below (Fig. S2D). Herpesvirales and caudovirales dominated the virome when patients received a low dose of valganciclovir and tacrolimus. In contrast, a high dose of immunosuppressants and antivirals gave rise to a virome structure that is dominated by anelloviridae (up to 94% occupation at high drug levels). The antiviral prophylaxis is intended to prevent CMV disease, but other herpesviruses are also susceptible to the drug (Razonable, 2011) so it is not surprising that a higher dose of valganciclovir gives rise to a lower fraction of viruses from the Herpesvirales order. The observation that anelloviridae take advantage of suppression of the host immune system is consistent with various observations from the literature: it was previously shown that the incidence of anelloviridae increases with progression towards AIDS in HIV patients (Madsen et al., 2002; Thom and Petrik, 2006), and that the total burden of the anellovirus TTV increases post liver transplantation (Thom and Petrik, 2006). Furthermore, an increased prevalence of anelloviridae in pediatric patients with fevers was reported recently (McElvania TeKippe et al., 2012).
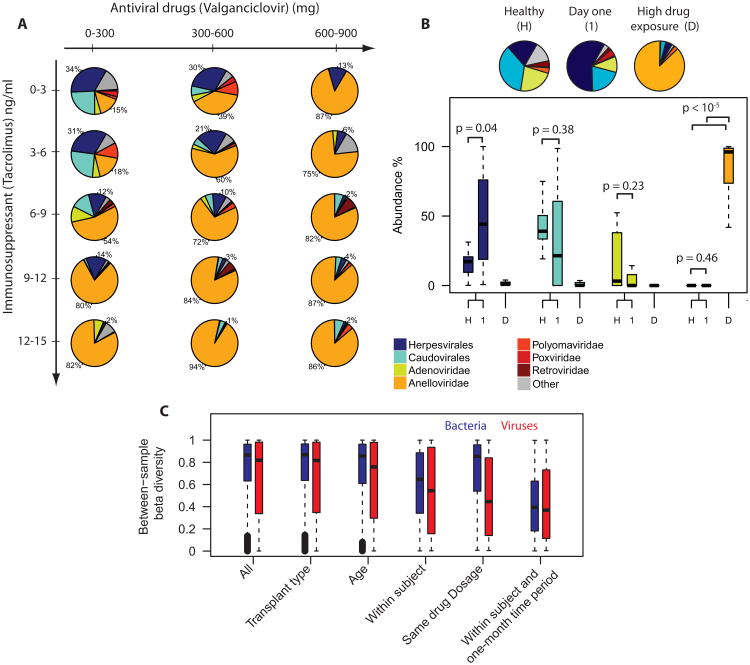
Fig. 2. Relative viral genomic abundance as a function of drug dose and comparison to healthy reference.
A. Mean virome composition for patients treated with the immunosuppressant tacrolimus (47 patients, 380 samples) as function of antiviral drug dose (valganciclovir) and concentration tacrolimus measured in blood. To account for the delayed effect of the virome composition on drug dose, the data on drug doses were window average filtered (window size 45 days, see fig. 1C). Herpesvirales and caudovirales dominate the virome when patients receive low doses of immunosuppressants and antiviral drugs. Conversely, anelloviridae dominate the virome when patients receive high doses of these drugs. B. Comparison of virome composition corresponding to healthy references (n = 9), post-transplant day one samples with low drug exposure, (n=13), and samples corresponding to high drug exposure (tacrolimus ≥ 9 ng/ml, valganciclovir ≥ 600 mg, n = 68). The virome structure for day one samples (1) and the virome structure measured for a set of healthy individuals (H) are distinct from the anellovirus-dominated distribution measured for samples corresponding to high drug doses (D). The piecharts show the mean fractions, p-values in boxplot based on the Mann-Whitney U test. C. Bray-Curtis beta diversity for all samples, among patients with the same transplant type (heart or lung), same age at time of transplant (10 year bins), within subjects, for patients treated with a similar drug dosage (tacrolimus level ± 0.5 ng/ml, valganciclovir ± 50 mg), and for samples collected from the same subjects within a one-month timespan. See also Figure S2.
We next compare the virome composition measured for organ transplant recipients to the composition observed in healthy individuals, not on immunosupressants or antivirals (n = 9, sequencing data available from a previous study). Here, we compare the healthy composition to the composition measured for organ transplant recipients at the start of the drug therapy (postoperative day one, n = 13), corresponding to a minimal drug exposure, and to the composition measured for transplant recipients exposed to high drug levels (well after the transplant procedure, tacrolimus ≥ 9 ng/ml. valgancicolvir ≥ 600 mg, n = 68). We find a similar composition of the virome for the healthy reference samples and samples corresponding to minimal drug exposure (Fig. 2B). However, the compositions of the healthy reference and minimal drug exposure samples are distinct from the anelloviridae-dominated composition measured for high drug exposure samples.
The tacrolimus-based immunosuppressive therapy is complemented with induction therapy in the first 3 days post transplant (with anti-thymocyte globulin, daclizumab, or basiliximab) and the patients furthermore receive the corticosteroid prednisone throughout the post transplant therapy (see supplemental materials). The time-dosage profile for prednisone and tacrolimus are similar: high doses at the onset of the therapy followed by a gradual dose reduction (Fig. S2A-C). The data in Fig. 2A thus reflect the combined effect of prednisone and tacrolimus. An analysis of the differential effect of prednisone and valganciclovir on the virome composition (Fig. S2E) shows the same trend observed in Fig. 2A: higher prednisone doses lead to a larger representation of anelloviruses. Lastly, we note that a subset of patients was not treated with antiviral drugs. The data corresponding to this subset of patients allowed us to further disentangle the differential effect of the antiviral drugs and the immunosuppressants on the composition of the virome, as described below.
Partitioning of microbiome diversity
We studied the diversity of the bacterial and viral components of the microbiome and found that the within-subject diversity was lower than the between-subject diversity, both for bacteria and viruses (Bray-Curtis beta diversity, bacterial composition at phylum level, viral composition at family and order level, Fig. 2C) (Oksanen et al., 2007). Partitioning the data for patients according to transplant type (heart or lung) did not reduce the diversity for bacteria and viruses (decrease in mean diversity < 0.001 for viruses, 95% confidence, Welch two sample t-test). Partitioning samples according to the age of the patient at time of transplant had a minor effect on sample-to-sample diversity (10 year bins, decrease in mean diversity < 0.035 for viruses, 95 % confidence, Welch two sample t-test). Within subjects, the diversity was lower for samples collected within a one month timespan, again both for bacteria and viruses. For viruses but not for bacteria, we find that the diversity is lower when comparing samples collected at a similar drug dosage (tacrolimus level ± 0.5 ng/ml, valganciclovir ± 50 mg). Taken together with the sensitivity of the population averages to drug dosage in Fig. 2A, we thus find that the composition of the virome for patients that are subject to the same drug therapy converges to a similar state.
Dynamic response of virome to drug dose changes
A strong temporal response of the virome to changes in drug dosage is observed, consistent with the sensitivity of the virome composition to drug dosage. Figure 3A shows the time dependence of the relative genomic abundance of ssDNA and dsDNA viruses (data from all patient groups and samples, n = 656). The fraction of ssDNA viruses expands rapidly during the first months post transplant followed by the opposite trend after 6 months. Figure 3B shows the time-dependent relative composition of the most abundant viruses grouped at the family and order level and provides more detail on the virome compositional dynamics (data from all patient groups and samples, n = 656). The dsDNA fraction consists of caudovirales, adenoviridae, polyomaviridae and herpesvirales, which together occupy 95% of the virome in the first week(s) post transplant. ssDNA viruses only occupy 5% of the initial virome and mainly consist of members of the anelloviridae family. The fraction occupied by adenoviridae, caudovirales and herpesvirales decreases strongly in the first few months as these virotypes are effectively targeted by the antiviral prophylaxis. In contrast, the relative abundance of anelloviridae increases rapidly as these virotypes largely escape targeting by the antiviral drugs and take advantage of the reduced immunocompetence of the patients (maximum of 84% during months 4.5-6). Six months after the organ transplant procedure, the opposite trends are observed, consistent with the reduction in antiviral and immunosuppressant drugs prescribed by the therapeutic protocol.
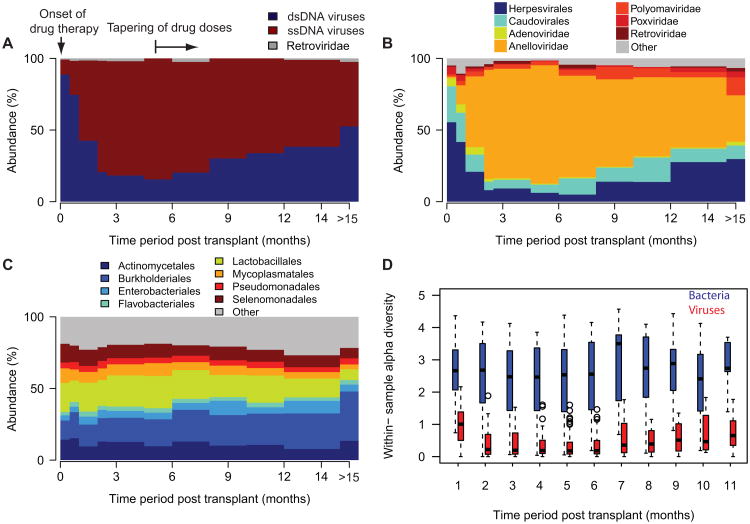
Figure 3. Temporal dynamics of the microbiome composition post transplant.
A. Relative abundance of dsDNA and ssDNA viruses for different time periods (average for all samples). The relative abundance of ssDNA viruses increases rapidly after the onset of the post-transplant drug therapy. After 6 months, the opposite trend is observed. B. Viral genome abundance at the family and order level of taxonomic classification for different time periods. Thefraction of anelloviridae expands rapidly in the first several months post-transplant. The fraction of herpesvirales,caudovirales and adenoviridae decreases in that same time period. After 6 months, the opposite trends are observed. C. Time-variation in the relative abundance of bacterial phyla. Compared to the viral abundance, the representationof different bacterial phyla remains relatively unchanged over the observed post transplant period. D. Shannonentropy as a measure of the within-sample alpha-diversity for bacterial and viral genera as function of time (datagrouped per one-month time periods). See also Figure S3.
Compared to the viral component, the bacterial component of the microbiome remains relatively stable over time, an observation that is made at the phylum, order and genus taxonomic levels (Fig. 3C, n = 656, and Fig. S3). Figure 3D shows the within-sample alpha diversity for the bacterial and viral genera as function of time (Shannon entropy, one month time periods, 590 bacterial genera, 168 viral genera examined). The diversity of observed viral genera decreases at the onset of the therapy (1.05 ± 0.5 in month 1 to 0.31 ± 0.33 in months 4-5, p ≪ 10-6, Mann-Whitney U test), whereas the alpha diversity of bacteria remains relatively unchanged during the course of the post transplant therapy (2.2 ± 1.14 in month 1 to 2.6 ± 0.85 in months 4-5, p = 0.1, Mann-Whitney U test).
Increase in total viral load at onset of post transplant therapy
To obtain insight into the effect of therapeutic drugs on total viral load, we extracted the absolute genomic abundance of all viruses relative to the number of human genome copies by normalizing the genome coverage of the viral targets to the coverage of the human genome (see SI). For all patient groups part of this study an increase in total viral load is observed at the onset of the therapy (Fig. 4A), regardless of transplant type (heart or lung) or age (adult or pediatric) (change in load, 7.4 ± 3, sigmoid fit, black line). Combined with relative abundance data, the total viral load data reveals a net reduction of the Herpesvirales load and a net increase in anelloviridae load in the first 3 months post-transplant for patients that are simultaneously treated with antivirals and immunosuppressants. The data thus show a differential effect of the combination of antivirals and immunosuppressants on different virotypes. The data also show a reduction in total adenoviridae load, indicating that adenoviridae replication is suppressed by valganciclovir, in agreement with previous studies (Avivi et al., 2004; Bruno et al., 2003). Figure 4B summarizes data for all transplant types, but the same trends are observed when stratifying according to different transplant types: adult heart transplant recipients (n = 268, Fig. S4A), adult lung transplant recipients (n = 166, Fig. S4B), and the pediatric patients that are treated with cyclosporine as opposed to tacrolimus (n = 99, Fig. S4C).
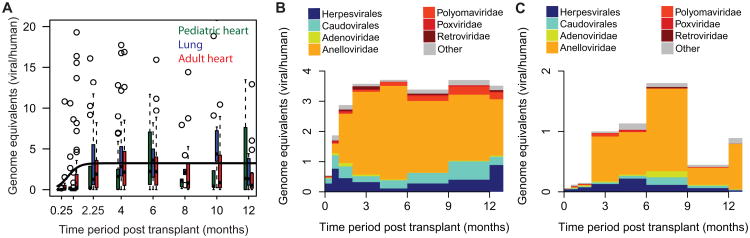
Figure 4. Virome composition and total viral burden in the absence and presence of antiviral prophylaxis.
A. Absolute viral load as a function of time, measured as viral genome copies per human genome copies detected by sequencing. Box plots are shown for different time periods with centers of the time periods marked on the x-axis. For all patient classes, the total viral load increases in the first weeks post transplant (black line is sigmoid fit, change in load 7.4 ± 3). B. Viral load and composition for CMV positive patients (prior CMV infection recipient and/or donor) that are treated with both immunosuppressants and antiviral drugs (78 patients, 543 samples). C. Viral load and composition for CMV negative patients, only treated with immunosuppressants (12 patients, 75 samples). See also Figure S4.
Not all patients in the study cohort received both antiviral and immunosuppressant drugs: for transplant cases where both the donor and recipient do not show evidence of a prior CMV infection in a CMV antibody assay, it is judged that the risks of complications due to antiviral prophylaxis outweigh the potential risk of a newly acquired CMV infection, and the patients are accordingly not treated with antiviral prophylaxis. These patients are thus solely treated with immunosuppressants. Figure 4C shows the time dependent viral load and composition of the CMV negative cases (n = 75). The net effect of immunosuppressant-only therapy is an expansion of all virotypes, including Herpesvirales and adenoviridae. Tapering of immunosuppression leads to a reduction of the total viral load.
Lower anellovirus burden in patients suffering from a graft rejection episode
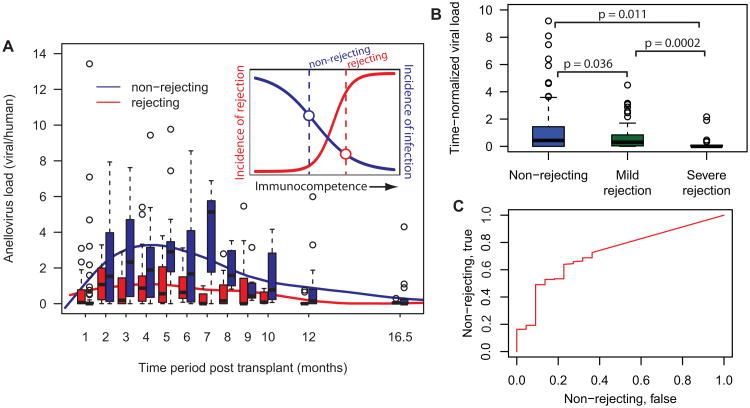
Given the correlation of the anellovirus burden with the extent of immunosuppression (see Fig. 2A and Fig. 4), and given the association between immunocompetence and the risk of rejection, we asked whether the anellovirus burden can be used for the classification of rejecting and non-rejecting graft recipients. Figure 5A shows the anellovirus load measured for rejecting and non-rejecting patients as function of time post transplant. Here, patients are classified as rejecting in case they suffer from at least one biopsy-determined moderate or severe rejection episode, biopsy grade ≥ 2R/3A (Stewart et al., 2005) (in red; 20 patients, 177 data points). The rejection-free patients correspond to patients that are not diagnosed to suffer from a moderate or severe graft damage throughout their post transplant course (in blue; biopsy grades < 2R/3A, 40 patients, 285 data points). Figure 5A shows that the anellovirus burden is significantly lower for the rejecting individuals at almost every time point. We next directly compared the anellovirus burden for patients at rejection with the burden measured for patients in the absence of rejection. To account for the time dependence of the anellovirus load described above (Fig. 5A), we extract the anellovirus load relative to the mean load measured for all samples at the same time point. Figure 5B shows the time-normalized load for non-rejecting patients (N = 208) compared to the load measured for patients suffering from a mild rejection event (biopsy grade 1R, N = 102) and patients suffering from a severe rejection episode (biopsy grade ≥ 2R/3A, N = 22). The figure shows that the time-normalized loads are significantly lower for the patients at greater risk of rejection. P-values were calculated by random sampling of the population with a greater amount of measurement points, p = sum(median(Arej) > median(Anon-rej))/N, where N = 104 and Arej and Anon-rej are the relative viral loads for the populations at greater and lesser risk of rejection and non-rejecting respectively (p = 0.011, p = 0.0002 and p = 0.036).
Figure 5. Lower anellovirus burden in patients that suffer from graft rejection.
A. Time dependence of the anellovirus load in the subgroup of patients that suffer from a severe rejection episode (biopsy grade ≥ 2R/3A, red data, 20 patients, 177 time points) and in the subgroup of patients with a rejection-free post transplant course (biopsy grades < 2R/3A, blue data, 40 patients, 285 time points). Box plots are shown for different time periods with centers of the time periods marked on the x-axis. Solid lines are cubic splines (smoothing parameter 0.75). The inset shows a cartoon of the expected inverse relationship between the level of immunosuppression and the incidence of rejection and infection. B. Anellovirus load relative to the average load measured for all samples at the same time point. The time-normalized load for non-rejecting patients (N = 208) is compared to the load measured for patients suffering from a mild rejection event (biopsy grade 1R, N = 102) and patients suffering from a severe rejection episode (biopsy grade ≥ 2R/3A, N = 22). The p-values reflect the probability that the median viral load is higher for the subgroups at greater risk of rejection. The p-values are calculated by random sampling of the population with a greater amount of measurement points. N-fold random sampling, p = sum(median(Arej) > median(Anon-rej))/N), where N=104 and Arej and Anon-rej are the relative viral loads for the populations at greater and lesser risk of rejection and non-rejecting respectively. C Test of the performance of the relative anellovirus load in classifying patients as non-rejecting vs. severely rejecting, receiver-operating characteristic curve, area under the curve = 0.72.
These observations are in line with a view that the risk of rejection and the incidence of infection have an opposite association with the patients' immunocompetence (see inset Figure 5A). The lower viral load observed for rejecting patients is thus indicative of a higher level of immunocompetence in this subgroup of patients, even though these patients are treated with the same immunosuppressive protocol. Patient-to-patient variability in the sensitivity to suppression of immune function is known to occur (Wieland et al., 2010) and the lack of predictability in immunosuppression is an important risk factor in transplantation. A commercial assay for the measurement of immunocompetence is available but was found not to be predictive of acute rejection or significant infections (Rossano et al., 2009). The development of methods for the direct measurement of immunocompetence that can replace or complement existing assays will therefore be important (Wieland et al., 2010). The total anellovirus load recorded in organ transplant recipients could serve as an alternative marker (Touinssi et al., 2001). Figure 5C shows a receiver-operating characteristic (Sing et al., 2005) and tests the performance of the relative anellovirus load in classifying non-rejecting and rejecting patients (area under the curve = 0.72).
Discussion
We have studied drug-microbiome interactions following solid organ transplantation by sequencing cell-free DNA in the recipients' plasma. The data reveal much about the fundamental structure of the human virome in plasma and how it responds to pharmacological perturbation; they also show the relative insensitivity to immunosuppression of the composition of the bacterial component of the microbiome. These data may help in the future design and optimization of post-transplant therapeutic protocols. For example, they show that tapering of antiviral prophylaxis from initial high doses leads to a resurgence of the herpesvirales fraction. CMV DNA load has previously been shown to predict CMV disease relapse and rejection (Potena et al., 2007; Sia et al., 2000), raising the question of whether patients would benefit from longer-term prophylactic therapy.
The marked expansion in abundance of anelloviridae upon immunosuppression is also worth further consideration. Anelloviruses are ubiquitous in the human population and, although no pathogenicity has been established (Hino and Miyata, 2007), anelloviruses are currently under investigation as potential cofactors in carcinogenesis (zur Hausen and de Villiers, 2009). The sensitivity of anelloviridae to immunosuppression makes organ transplantation an ideal setting for the study of the properties of anelloviridae, particularly in the light of the increased incidence of cancer seen in transplant recipients. The observation of a lower-than-average burden of anelloviruses in patients that suffer from a rejection episode is indicative of insufficient immunosuppression in this subgroup of patients, even though these patients were subject to the immunosuppressant levels prescribed per protocol. This suggests that there would be value in designing assays that allow directly gauging the level of a patient's immunocompetence, in addition to measurements of circulating drug levels. The total burden of anelloviruses identified in a transplant recipient's blood may serve as one such marker of the overall state of immunosuppression of the individual patient.
We expect that high throughput DNA sequencing will find future applicability in the hypothesis-free diagnosis of infections. This approach may be of particular relevance in the context of transplantation given the fact that infections occur frequently in transplantation and are difficult to diagnose in immunocompromised individuals, and given that sequence analysis can additionally provide information on the graft health through the quantification of donor-derived human DNA circulating in plasma (Snyder et al., 2011). In other areas of infectious disease, it may be of value to develop subtractive methods to eliminate the human DNA and enrich for DNA of viral and microbial origin.
Experimental procedures
Clinical sample collection
Patients were enrolled at Stanford University Hospital (SUH) or Lucile Packard Children's Hospital (LPCH), and were excluded if they were recipients of multi-organ transplants. This study was approved by the Stanford University Institutional Review Board (protocol # 17666) and enrollment commenced in March 2010. For details on patient recruitment and post transplant treatment of the patients see the extended experimental procedures section.
Plasma processing and DNA extraction
Plasma was extracted from whole blood samples within three hours of sample collection, as previously described (Fan et al., 2008), and stored at - 80°C. When required for analysis, plasma samples were thawed and circulating DNA was immediately extracted from 0.5-1 ml plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen).
Sequencing library preparation and sequencing
Sequencing libraries were prepared from the purified patient plasma DNA using the NEBNext DNA Library Prep Master Mix Set for Illumina with standard Illumina indexed adapters (purchased from IDT), or using a microfluidics-based automated library preparation platform (Mondrian ST, Ovation SP Ultralow library system). Libraries were characterized using the Agilent 2100 Bioanalyzer (High sensitivity DNA kit) and quantified by qPCR. Samples were part of 26 different sequencing runs and were sequenced over the course of 22 months. On average 6 samples were sequenced per lane.
Data deposition
The sequence data generated in this study has been deposited in the Sequence Read Archive
Supplementary Material
Highlights.
Virome-drug interactions were measured in a cohort of organ transplant recipients
The structure of the virome is strongly affected by immune modulation and antiviral
The total viral load increases markedly at the onset of the therapy
A potential application of the virome state in predicting immune strength is reported
Acknowledgments
This work was supported by NIH grant RC4AI092673 and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avivi I, Chakrabarti S, Milligan DW, Waldmann H, Hale G, Osman H, Ward KN, Fegan CD, Yong K, Goldstone AH. Incidence and outcome of adenovirus disease in transplant recipients after reduced-intensity conditioning with alemtuzumab. Biology of Blood and Marrow Transplantation. 2004;10:186–194. doi: 10.1016/j.bbmt.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Bruno B, Gooley T, Hackman RC, Davis C, Corey L, Boeckh M. Adenovirus infection in hematopoietic stem cell transplantation: effect of ganciclovir and impact on survival. Biology of blood and marrow transplantation: journal of the American Society for Blood and Marrow Transplantation. 2003;9:341. doi: 10.1016/s1083-8791(03)00102-2. [DOI] [PubMed] [Google Scholar]
- Budde K, Matz M, Dürr M, Glander P. Biomarkers of over-immunosuppression. Clinical Pharmacology & Therapeutics. 2011;90:316–322. doi: 10.1038/clpt.2011.111. [DOI] [PubMed] [Google Scholar]
- Chesters PM, Heritage J, McCance DJ. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. Journal of Infectious Diseases. 1983;147:676–684. doi: 10.1093/infdis/147.4.676. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proceedings of the National Academy of Sciences. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman JA. Infection in solid-organ transplant recipients. New England Journal of Medicine. 2007;357:2601–2614. doi: 10.1056/NEJMra064928. [DOI] [PubMed] [Google Scholar]
- Fishman JA, Emery V, Freeman R, Pascual M, Rostaing L, Schlitt HJ, Sgarabotto D, Torre-Cisneros J, Uknis ME. Cytomegalovirus in transplantation–challenging the status quo. Clinical transplantation. 2006;21:149–158. doi: 10.1111/j.1399-0012.2006.00618.x. [DOI] [PubMed] [Google Scholar]
- Hino S, Miyata H. Torque teno virus (TTV): current status. Rev Med Virol. 2007;17:45–57. doi: 10.1002/rmv.524. [DOI] [PubMed] [Google Scholar]
- Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5:0009836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusko WJ, Piekoszewski W, Klintmalm GB, Shaefer MS, Hebert MF, Piergies AA, Lee CC, Schechter P, Mekki QA. Pharmacokinetics of tacrolimus in liver transplant patients. Clin Pharmacol Ther. 1995;57:281–290. doi: 10.1016/0009-9236(95)90153-1. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen CD, Eugen-Olsen J, Kirk O, Parner J, Kaae Christensen J, Brasholt MS, Ole Nielsen J, Krogsgaard K. TTV viral load as a marker for immune reconstitution after initiation of HAART in HIV-infected patients. HIV Clin Trials. 2002;3:287–295. doi: 10.1310/8c94-vypq-ng1h-4cnw. [DOI] [PubMed] [Google Scholar]
- Marboe CC, Billingham M, Eisen H, Deng MC, Baron H, Mehra M, Hunt S, Wohlgemuth J, Mahmood I, Prentice J, et al. Nodular endocardial infiltrates (Quilty lesions) cause significant variability in diagnosis of ISHLT Grade 2 and 3A rejection in cardiac allograft recipients. J Heart Lung Transplant. 2005;24:S219–226. doi: 10.1016/j.healun.2005.04.001. [DOI] [PubMed] [Google Scholar]
- McElvania TeKippe E, Wylie KM, Deych E, Sodergren E, Weinstock G, Storch GA. Increased prevalence of anellovirus in pediatric patients with fever. PLoS One. 2012;7:30. doi: 10.1371/journal.pone.0050937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Research. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O'Hara B, Stevens MHH, Oksanen MJ, Suggests M. The vegan package. Community ecology package. Version 2.0-7 2007 [Google Scholar]
- Potena L, Holweg CT, Vana ML, Bashyam L, Rajamani J, McCormick AL, Cooke JP, Valantine HA, Mocarski ES. Frequent occult infection with Cytomegalovirus in cardiac transplant recipients despite antiviral prophylaxis. J Clin Microbiol. 2007;45:1804–1810. doi: 10.1128/JCM.01362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razonable RR. Antiviral drugs for viruses other than human immunodeficiency virus. Mayo Clin Proc. 2011;86:1009–1026. doi: 10.4065/mcp.2011.0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razonable RR, Brown RA, Humar A, Covington E, Alecock E, Paya CV. A longitudinal molecular surveillance study of human polyomavirus viremia in heart, kidney, liver, and pancreas transplant patients. Journal of Infectious Diseases. 2005;192:1349–1354. doi: 10.1086/466532. [DOI] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossano JW, Denfield SW, Kim JJ, Price JF, Jefferies JL, Decker JA, Smith EOB, Clunie SK, Towbin JA, Dreyer WJ. Assessment of the Cylex ImmuKnow Cell Function Assay in Pediatric Heart Transplant Patients. The Journal of Heart and Lung Transplantation. 2009;28:26–31. doi: 10.1016/j.healun.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Saraiva F, Matos V, Goncalves L, Antunes M, Providencia LA. Complications of endomyocardial biopsy in heart transplant patients: a retrospective study of 2117 consecutive procedures. Transplant Proc. 2011;43:1908–1912. doi: 10.1016/j.transproceed.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Sia IG, Wilson JA, Groettum CM, Espy MJ, Smith TF, Paya CV. Cytomegalovirus (CMV) DNA load predicts relapsing CMV infection after solid organ transplantation. J Infect Dis. 2000;181:717–720. doi: 10.1086/315242. [DOI] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Slifkin M, Doron S, Snydman DR. Viral prophylaxis in organ transplant patients. Drugs. 2004;64:2763–2792. doi: 10.2165/00003495-200464240-00004. [DOI] [PubMed] [Google Scholar]
- Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proceedings of the National Academy of Sciences. 2011;108:6229–6234. doi: 10.1073/pnas.1013924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Winters GL, Fishbein MC, Tazelaar HD, Kobashigawa J, Abrams J, Andersen CB, Angelini A, Berry GJ, Burke MM, et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant. 2005;24:1710–1720. doi: 10.1016/j.healun.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Thom K, Petrik J. Progression towards AIDS leads to increased Torque teno virus and Torque teno minivirus titers in tissues of HIV infected individuals. Journal of medical virology. 2006;79:1–7. doi: 10.1002/jmv.20756. [DOI] [PubMed] [Google Scholar]
- Touinssi M, Gallian P, Biagini P, Attoui H, Vialettes B, Berland Y, Tamalet C, Dhiver C, Ravaux I, De Micco P, et al. TT virus infection: prevalence of elevated viraemia and arguments for the immune control of viral load. J Clin Virol. 2001;21:135–141. doi: 10.1016/s1386-6532(01)00157-3. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataramanan R, Swaminathan A, Prasad T, Jain A, Zuckerman S, Warty V, McMichael J, Lever J, Burckart G, Starzl T. Clinical pharmacokinetics of tacrolimus. Clin Pharmacokinet. 1995;29:404–430. doi: 10.2165/00003088-199529060-00003. [DOI] [PubMed] [Google Scholar]
- Weber G, Shendure J, Tanenbaum DM, Church GM, Meyerson M. Identification of foreign gene sequences by transcript filtering against the human genome. Nature genetics. 2002;30:141. doi: 10.1038/ng818. [DOI] [PubMed] [Google Scholar]
- Wieland E, Olbricht CJ, Süsal C, Gurragchaa P, Böhler T, Israeli M, Sommerer C, Budde K, Hartmann B, Shipkova M. Biomarkers as a tool for management of immunosuppression in transplant patients. Therapeutic drug monitoring. 2010;32:560–572. doi: 10.1097/FTD.0b013e3181efb3d2. [DOI] [PubMed] [Google Scholar]
- Wylie KM, Weinstock GM, Storch GA. Emerging view of the human virome. Transl Res. 2012;160:283–290. doi: 10.1016/j.trsl.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia LC, Cram JA, Chen T, Fuhrman JA, Sun F. Accurate genome relative abundance estimation based on shotgun metagenomic reads. PloS one. 2011;6:e27992. doi: 10.1371/journal.pone.0027992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Stange-Thomann N, Weber G, Bo R, Dodge S, David RG, Foley K, Beheshti J, Harris NL, Birren B, et al. Pathogen discovery from human tissue by sequence-based computational subtraction. Genomics. 2003;81:329–335. doi: 10.1016/s0888-7543(02)00043-5. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H, de Villiers EM. TT viruses: oncogenic or tumor-suppressive properties? Curr Top Microbiol Immunol. 2009;331:109–116. doi: 10.1007/978-3-540-70972-5_7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.