The human genome project will be completed in the early part of the next decade. For example, most of the approximately 105 human mRNAs have already been at least partially cloned and sequenced as cDNAs. These cDNAs, when completed, will provide valuable tools for studying the protein products of their respective mRNAs. The challenge in the postgenomic era will be to apply advances in molecular oncology to select from among this diverse set of proteins the most suitable candidates for anti-cancer drug discovery programs. Here, I outline some considerations that may be helpful in choosing rational drug targets for cancer therapy.
Pathways controlling cellular proliferation
Cancer is caused by the accumulation of genetic damage in a susceptible cell. Some of these mutations play a direct causal role in the development and maintenance of the malignant phenotype. Mathematical models suggest that the number of such causal, rate-limiting mutations in a given tumor is finite and probably on the order of 5–8 for the common tumors observed in adults (1). Among these mutations are dominantly acting mutations that convert proto-oncogenes to oncogenes, as well as recessive mutations that inactivate so-called gatekeeper tumor suppressor genes (2). Such gatekeeper genes play relatively direct roles in preventing abnormal cellular proliferation. During malignant progression, tumor cells also accumulate additional genetic alterations that play no direct role with respect to tumor behavior and might therefore be considered epiphenomenal. For example, a large deletion affecting a tumor suppressor gene might also lead to the loss of a contiguous gene (3). The accumulation of genetic damage, both causal and epiphenomenal, is fostered because, in many cases, cancer cell genomes are unstable as a result of inactivation of genes that monitor the fidelity of DNA replication and mitosis. Among these genes are the so-called caretaker tumor suppressor genes (2). The majority of hereditary cancer syndromes are due to germline mutations affecting either gatekeeper or caretaker tumor suppressor genes.
The need for multiple causal mutations within a given tumor almost certainly reflects the fact that cancer cells must overcome multiple obstacles to behave in a malignant fashion and still survive. Thus, tumor cells need to enter and traverse the cell cycle in the absence of normal mitogenic signals; to silence programmed cell death (apoptotic) signals arising from such illegitimate cell-cycle passages or due to genomic damage; to reactivate telomerase, which allows them to avoid progressive telomere erosion that would otherwise result from repeated DNA duplication; and to invade surrounding tissues and induce angiogenesis. In keeping with this notion, Weinberg and coworkers recently showed that normal human cells could be rendered tumorigenic by the introduction of telomerase in conjunction with 2 oncogenes that together provide a mitogenic stimulus, inhibit apoptosis, and induce angiogenesis (4).
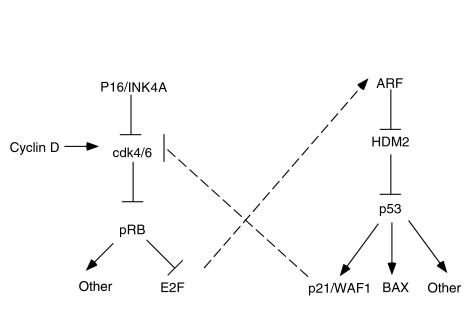
It is becoming clear that many of the genes that are altered in human cancer define molecular pathways that are central to the control of the processes just described. For example, most human cancers harbor mutations that directly or indirectly inactivate the retinoblastoma tumor suppressor protein (pRB) (5). Examples of the latter include mutations affecting genes such as p16/INK4A, cdk4, and cyclin D1, which lead to the untimely phosphorylation, and hence functional inactivation, of pRB (Figure 1). Altered growth factor receptors, as well as oncogenic Ras, likely also impinge on this pRB pathway (6). pRB serves as an important negative regulator of cell-cycle progression and serves to integrate positively and negatively acting mitogenic signals. Loss of pRB therefore contributes to the mitogen independence of cancer cells.
Figure 1.
Simplified views of the pRB and p53 pathways. The cdk4 and cdk6 kinases are positively regulated by G1 cyclins such as the D-type cyclins and negatively regulated by cdk inhibitors such as p16/INK4A. Cdk4 and Cdk6 phosphorylate, and thus inhibit, the retinoblastoma protein (pRB). pRB forms complexes with members of the E2F transcription factor family. These complexes repress transcription from E2F-responsive promoters. p53 is a sequence-specific DNA-binding protein that transcriptionally activates target genes such as p21/WAF1 and BAX. HDM2 silences the p53 transcriptional activation domain and targets p53 for degradation. ARF antagonizes HDM2. As indicated by the dashed lines, these apparently linear pathways are subject to cross-talk, which links them into a more complex regulatory network.
Likewise, the p53 tumor suppressor protein sits at a nodal point in an apoptotic pathway (Figure 1) (7, 8). p53 is mutated in approximately 50% of all human tumors, and many human tumors that retain a wild-type p53 allele harbor mutations in other genes that regulate p53. For example, as shown in Figure 1, ARF inhibits HDM2, which, in turn, negatively regulates p53. ARF loss has been described in a variety of human tumors, and there is a strong selection pressure to mutate either ARF or p53 during murine carcinogenesis (7, 9). Similarly, amplification and overexpression of HDM2 have been described in a subset of tumors that retain wild-type p53 (10, 11). Among the downstream targets of p53 is the apoptosis inducer BAX (12). BAX function is antagonized by Bcl-2, which is overexpressed in a variety of tumors including nodular lymphomas. For more on BAX and Bcl-2, see Sellers and Fisher in this Perspective series.
The importance of pRB and p53 in carcinogenesis is underscored by the fact that a variety of unrelated DNA tumor viruses have independently evolved the means to inactivate both pRB and p53 (13). To replicate, these viruses must induce quiescent cells to re-enter the cell cycle and, in the process, must avoid the rapid induction of apoptosis. One such virus, the human papillomavirus (HPV), has been implicated in human cervical carcinogenesis. The oncogenic HPVs produce E7 and E6 proteins that inactivate pRB and p53, respectively. Thus, inactivation of the pRB pathway and p53 pathway appears to be a common theme in carcinogenesis.
The identification of molecular pathways such as these is important for a variety of reasons. First, it aids in the identification of potential drug targets that are genetically validated in the sense that they are recurrently altered in tumors and play a causal role in maintenance of the malignant phenotype. Indeed, for some of these potential targets, both human hereditary cancer syndromes and the phenotypes of genetically engineered mice support their role in cancer causation. Second, the understanding of these pathways allows one to appreciate that certain genetic alterations can be viewed as phenocopies of other mutations. Thus, loss of ARF might be equated with loss of p53, and overproduction of cyclin D1 might be equated with loss of pRB (Figure 1). In this respect, some of the heterogeneity between different tumors that one gleans from genotypic analysis is more apparent than real. For this reason, it may be possible to choose the most suitable drug target from a variety of structurally unrelated molecules that all function in the same signaling pathway. It needs to be borne in mind, however, that in reality, these pathways are not strictly linear but, rather, constitute molecular networks. This is again illustrated by examination of the pRB and p53 pathways. Loss of pRB leads to derepression of the E2F transcription factor family, which, in turn, leads to the induction of ARF and subsequent activation of p53. Conversely, one p53-target, the cdk inhibitor p21/WAF1, can block pRB phosphorylation and thus lock pRB in its active, growth suppressing mode. As discussed later here, the potential for molecular cross-talk between pathways has implications for the development of drugs that specifically target tumor cells while sparing normal cells.
Challenges for rational drug design
We already possess many drugs that will kill cancer cells. The problem is that currently available anti-cancer drugs have among the narrowest therapeutic indices (toxic dose per therapeutic dose) of drugs in current use. The remainder of this article will focus on the impact of genomics and advances in molecular oncology on 2 issues: (a) the search for targets that are, at least in principle, amenable to pharmacological manipulation, and (b) the search for targets that, when manipulated, will kill cancer cells with relative sparing of normal cells. To help frame the discussion, I consider 3 concerns that are commonly advanced in arguing that anti-cancer drugs (even when rationally designed) will fail to control the disease or to spare healthy tissue. As discussed here, I believe that each of these pessimistic views is flawed.
First, given that cancer cells harbor multiple mutations, will it be possible to correct all of the consequent defects by drug therapies?
Although it is true that cancer cells harbor many mutations, some of these changes are epiphenomenal, as already argued here; the number of mutations that actually sustain the malignant phenotype is limited. However, even accepting that multiple mutations conspire to induce malignancy, it appears that not all of the abnormalities within a cancer cell would have to be corrected to have a therapeutic effect. For example, restoring p53 function in p53-defective tumor cells is sufficient to induce cell death, to induce a cell-cycle block, or, in some cases, to restore sensitivity to conventional chemotherapy (14). Likewise, restoration of pRB function in pRB-defective tumor cells or restoration of pVHL (the product of the von Hippel-Lindau tumor suppressor gene) function in pVHL-defective tumor cells suppresses tumorigenesis (15, 16). One interpretation of these observations is that the requirement for multiple causal mutations reflects inherent redundancies in the molecular pathways that prevent tumorigenesis. In this view, the different causal genetic alterations might be viewed somewhat akin to tumblers on a lock. Reversing any 1 of the tumblers of a lock prevents it from opening. A somewhat stronger interpretation would be that some genetic abnormalities (whether causal or epiphenomenal) are advantageous to the cancer cell — or, indeed, even tolerated by it — only because the cell already harbors other specific mutations. In this case, correcting genetic alterations that occurred early during the development of a malignant tumor might cause profound antitumor effects. I return to this concept at the end of the article.
Second, given that most cancer-causing mutations lead to the loss or inactivation of a protein and that drugs typically inhibit the function of a protein rather than promoting it, can drug therapies restore the critical missing functions?
It is true that the majority of cancer-causing mutations — like most mutations of all kinds — induce a loss, rather than a gain, of function. Fortunately, as already indicated here, the loss of function of 1 molecule is often similar in consequence to a gain of function of another. For example, loss of the cdk inhibitor p16/INK4A might be viewed as a gain of Cdk4 and Cdk6 (17). Similarly, the vascular tumors in VHL disease can be understood in light of the recent finding that pVHL degrades HIF-1α and HIF-2α (18), transcription factors that induce the expression of VEGF. Thus, loss of pVHL results in a gain of HIF and subsequent gain of VEGF. On the basis of these considerations, one might treat loss of INK4A with a small molecule inhibitor of Cdk4/6 or VHL-associated neoplasms with small molecule inhibitors of HIF or VEGF.
It is also clear from the explosive growth of gene databases that most genes are actually members of gene families. For example, 2 homologues of p53, called p63 and p73, were recently discovered (19). Unlike p53, however, these 2 genes are rarely mutated in human cancers, but, under experimental conditions, the p63 and p73 proteins can substitute for p53 to induce apoptosis in p53-defective tumor cells. Therefore, one approach to restoring p53 function in tumor cells would be to identify small molecules that can activate p63 or p73. Developing such a strategy would require a detailed understanding of the signals or pathways that impinge on these genes.
Finally, the concept of synthetic lethality, which has been applied in yeast genetics for many years, may provide a strategy to exploit defects in critical regulatory genes in tumor cells. Two genes are said to be synthetically lethal if disruption of either gene singly is compatible with viability but if loss of both causes death. As Hartwell and Friend and their coworkers (20) have proposed, tumor cells that have lost the function of some gene A might be especially sensitive to mutations that cripple another gene, gene B, and this sensitivity suggests another avenue for anticancer drug design. If loss of function in gene B is harmless by itself but fatal in cells with mutations in gene A, then a small molecule inhibitor of gene B’s protein product should act as a cytotoxic drug specific for tumor cells. For example, cells that have lost pRB function (and thus have derepressed E2F) appear to be exquisitely sensitive to inhibitors of cyclin A/cdk2 activity (ref. 21; for more on this interaction, see Shapiro and Harper in this Perspectives series). Peptidic cyclin A/cdk2 antagonists kill cells with high levels of E2F activity but not their normal counterparts.
Third, it may be objected that the majority of cancer-causing mutations affect proteins that regulate essential cellular functions such as cell division, differentiation, and apoptosis. Will drugs that target such proteins be associated with unacceptable levels of toxicity?
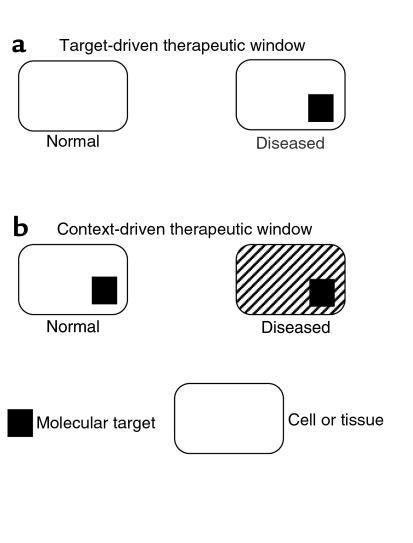
To be sure, virtually all drugs produce unwanted effects if given at high enough concentration. The critical issue, again, is one of therapeutic index, which can be improved in at least 2 ways, as indicated in Figure 2. First, one could target a molecule that is unique to the diseased tissue. This approach (Figure 2a) has been very successful in the treatment of infectious diseases, for example, but so far has been of limited use in oncology. For instance, it has not been possible to develop drugs that distinguish between normal and oncogenic Ras proteins. Likewise, because the oncogenic fusion proteins described in some cancers typically include functional domains from normal proteins, drugs that target such fusion proteins are unlikely to be completely specific. An alternative way to obtain drugs with high therapeutic indices, however, is to identify situations in which the requirement for a given target is quantitatively or qualitatively altered by other genetic abnormalities within the diseased tissue (Figure 2). In the simplest case, the relative contextual change might represent a single genetic abnormality that behaves in a synthetic lethal fashion with the target. More broadly, the contextual changes could be the sum of all of the genetic abnormalities, causal and epiphenomenal, that mark the malignant clone.
Figure 2.
Basis for therapeutic window. A therapeutic window can either be target-driven or context-driven. In target-driven drug design (a), the target molecule is unique to the diseased tissue. The context-driven strategy (b), on the other hand, is directed at a target that is present in both normal and diseased tissue. This alternative approach takes advantage of mutations or other physiological changes (hatched marks) that occur during tumorigenesis and that alter the cell’s or tissue’s requirement for the target protein. Such changes may include quantitative effects, as when a tumor cell becomes dependent on high expression of the target protein, or they may be qualitative, if the target protein takes on novel functions that sustain the growth of the diseased tissue.
This emphasis on the altered physiological context of the tumor cell provides some grounds for optimism that more specific drugs can be devised to control or eliminate cancers. The aggressive clinical behavior of most cancers probably accounts for the usual view that successive genetic changes in cancer cells serve to make them ever more invincible. However, it is important to remember that cancer cells are the product of genetic selection in vivo, and the alterations they undergo reflect the different selective pressures they encounter. Mutations that were selected because they were essential for their survival therefore flag potential “Achilles heels,” weaknesses of these cells that could be exploited by a suitable drug.
References
- 1.Renan MJ. How many mutations are required for tumorigenesis? Implications from human cancer data. Mol Carcinog. 1993;7:139–146. doi: 10.1002/mc.2940070303. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler K, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 3.Frei E., III Gene deletion: a new target for cancer chemotherapy. Lancet. 1993;342:662–664. doi: 10.1016/0140-6736(93)91764-d. [DOI] [PubMed] [Google Scholar]
- 4.Hahn W, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 5.Sellers WR, Kaelin WG. Role of the retinoblastoma protein in the pathogenesis of human cancer. J Clin Oncol. 1997;15:3301–3312. doi: 10.1200/JCO.1997.15.11.3301. [DOI] [PubMed] [Google Scholar]
- 6.Peeper D, et al. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature. 1997;386:177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- 7.Sherr C. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 8.Adams P, Kaelin WG. Negative control elements of the cell cycle in human tumors. Curr Opin Cell Biol. 1998;10:791–797. doi: 10.1016/s0955-0674(98)80123-3. [DOI] [PubMed] [Google Scholar]
- 9.Kamijo T, et al. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliner J, et al. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 11.Leach F, et al. p53 Mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res. 1993;53:2231–2234. [PubMed] [Google Scholar]
- 12.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–300. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 13.Moran E. DNA tumor virus transforming proteins and the cell cycle. Curr Opin Genet Dev. 1993;3:63–70. doi: 10.1016/s0959-437x(05)80342-9. [DOI] [PubMed] [Google Scholar]
- 14.Harris CC. p53 Tumor suppressor gene: from the basis research laboratory to the clinic-an abridged historical perspective. Carcinogenesis. 1996;17:1187–1198. doi: 10.1093/carcin/17.6.1187. [DOI] [PubMed] [Google Scholar]
- 15.Huang H-JS, et al. Suppression of the neoplastic phenotype by replacement of the RB gene in human cancer cells. Science. 1988;242:1563–1566. doi: 10.1126/science.3201247. [DOI] [PubMed] [Google Scholar]
- 16.Iliopoulos O, Kibel A, Gray S, Kaelin WG. Tumor suppression by the human von Hippel-Lindau gene product. Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 17.Palmero I, Peters G. Perturbation of cell cycle regulators in human cancer. Cancer Surv. 1996;27:351–367. [PubMed] [Google Scholar]
- 18.Maxwell P, et al. The von Hippel-Lindau gene product is necessary for oxygen-dependent proteolysis of hypoxia-inducible factor α subunits. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 19.Kaelin WG. The emerging p53 gene family. J Natl Cancer Inst. 1999;91:594–598. doi: 10.1093/jnci/91.7.594. [DOI] [PubMed] [Google Scholar]
- 20.Hartwell L, Szankasi P, Roberts C, Murray A, Friend S. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, et al. Selective killing of transformed cells by cyclin/cyclin-dependent kinase 2 antagonists. Proc Natl Acad Sci USA. 1999;96:4325–4329. doi: 10.1073/pnas.96.8.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]