The emergence of new microbial infections is ever more likely with the globalization of trade and travel, changes to agricultural practices and climate change. However, as Lipkin describes in this Essay, this threat is being met by dramatic technological advances in pathogen discovery, surveillance and modelling.
Subject terms: Epidemiology, Infectious diseases, Public health, Pathogens, Clinical microbiology, Pathogenesis
Abstract
The pace of pathogen discovery is rapidly accelerating. This reflects not only factors that enable the appearance and globalization of new microbial infections, but also improvements in methods for ascertaining the cause of a new disease. Innovative molecular diagnostic platforms, investments in pathogen surveillance (in wildlife, domestic animals and humans) and the advent of social media tools that mine the World Wide Web for clues indicating the occurrence of infectious-disease outbreaks are all proving to be invaluable for the early recognition of threats to public health. In addition, models of microbial pathogenesis are becoming more complex, providing insights into the mechanisms by which microorganisms can contribute to chronic illnesses like cancer, peptic ulcer disease and mental illness. Here, I review the factors that contribute to infectious-disease emergence, as well as strategies for addressing the challenges of pathogen surveillance and discovery.
Main
When the H1N1 influenza virus struck in 1918, little was known about how infectious diseases emerge or their routes of transmission. Indeed, even the identification of the causative agent as a virus (that is, capable of passing through a filter) rather than the bacterium Haemophilus influenzae (championed by some leading microbiologists at the time) was in dispute until late in the course of the pandemic1. The 1918 virus had an estimated case fatality rate of 10–20%, spread to six continents, infected ∼500 million people and killed approximately 3% of the world's population2,3. The severe acute respiratory syndrome (SARS) coronavirus pandemic of 2002–2003, the first pandemic of the twenty-first century, had a case fatality rate of near 10%, but infected far fewer people (8,096) than the H1N1 influenza virus of the 1918 pandemic4. Transmissibility, as estimated by R0 (the basic reproduction number; that is, the number of cases generated through contact with one infected individual) was similar for the influenza pandemic of 1918 (R0 = 2–3)5 and the SARS pandemic of 2003 (R0 = 2–5)6,7. However, the global response to SARS was facilitated by advances in epidemiology and microbiology that enabled the rapid containment and identification of the causative agent as a novel coronavirus. The discovery process is faster still today. Whereas two large teams invested weeks using classical dideoxy sequencing techniques to characterize SARS coronavirus genetic material that had been amplified in tissue culture8,9, the high-throughput sequencing platforms that have been used in more recent outbreaks, such as the Lujo virus outbreak in South Africa in 2008, have allowed the identification of novel agents in clinical materials in only 48–72 hours10.
As a consequence of the globalization of travel and trade, infectious agents are expanding their geographical ranges and appearing in new contexts. Thus, clinicians and public health officials must be prepared to detect and respond to the unexpected. The ongoing development of new antimicrobial drugs, therapeutic antibodies, vaccines and probiotics means that early and accurate disease diagnosis can have profound implications for medical management and public health. This is particularly true for viral infections, for which, until recently, opportunities for effective intervention were limited to HIV, hepatitis C virus and herpesvirus infections. Surveillance and discovery efforts are bearing fruit in chronic disorders and in studies of normal physiology, as well as in the investigation of acute diseases such as pneumonia, diarrhoea, meningitis, encephalitis and haemorrhagic fevers. The role of Helicobacter pylori in causing peptic ulcers11, the role of human papillomavirus in cervical cancer12 and the role of polyomaviruses in Merkel cell carcinoma13 are prominent examples of the microbial contributions to the pathogenesis of disorders that were idiopathic only a few years ago. Insights into the role of the human microbiome in nutrition, allergies and autoimmunity have led to the implementation of the same surveillance and discovery platforms that are used to investigate classic infectious diseases14,15. Finally, although there have been no recent examples of bioterrorism, the risk has only increased with political instability and with the accessibility of synthetic genomics techniques that enable the creation or re-creation of virulent pathogens.
In this Essay, I discuss the factors that contribute to the emergence (and re-emergence) of infectious diseases, the evolution of strategies and tools for pathogen surveillance and discovery, and future prospects for the field. To guide the reader, I provide a timeline of the events and innovations described in the text (Timeline).
Factors in microbial emergence
Globalization of travel and trade. Travel and trade are increasingly global. For instance, the number of international airline flights has nearly doubled over the past 15 years from just fewer than 500,000 in 1996 to close to 850,000 in 2011 (see US Bureau of Transportation Statistics flight information; commercial flights by US domestic airlines). John F. Kennedy International Airport, for example, one of two international airports in the greater New York metropolitan area, USA, hosts non-stop flights to 100 international destinations and serves nearly 12 million international customers annually (see US airlines and foreign airlines US passengers continue to increase from 2009). Similar data apply worldwide for airports serving large urban centres. This means that an infected individual or mosquito can travel around the world in less than 24 hours, so it is not surprising that air travel has been implicated in the global dissemination of HIV, SARS coronavirus, West Nile virus, chikungunya virus, influenza viruses and Mycobacterium tuberculosis16,17,18.
Indeed, it is perhaps more remarkable that so few outbreaks of infectious disease have been attributed to air travel, especially given that the transportation of plants and animals has continued to increase dramatically with the development of global agribusinesses and urbanization. Whereas the global population and food production have increased at comparable rates since 1975 (74% and 100%), the international food trade has burgeoned by more than 200% (see World Population Prospects, the 2010 Revision; Table 'Total Population, Both Sexes' and FAOSTAT trade data). One hundred years ago, most fresh food was produced and consumed within a radius of a few kilometres, whereas it is now not unusual for individuals to consume plants and animals that were harvested thousands of kilometres away19.
Agricultural practices. Contamination of meat has affected the international trade of livestock on a number of occasions20, with examples including contamination by agents that can threaten humans, such as prions, influenza viruses and Rift Valley fever virus, or agents that threaten the livestock themselves, such as foot-and-mouth disease virus and Schmallenberg virus21. Furthermore, bacteria, viruses and parasites, particularly those present in faeces, can contaminate fruits and vegetables to cause disease in humans and other animals, resulting in costly food recalls and affecting consumer demand22,23. High-density farming of livestock, poultry and fish is frequently associated with the use of antibiotics as growth promoters, and this can result in the emergence of antibiotic-resistant bacteria24,25.
The centralization of food production and processing — particularly for ground meats, raw fruits and raw vegetables — has resulted in outbreaks of infectious diseases that have been distributed over large geographical areas. Furthermore, it is difficult to monitor the illegal trafficking of wildlife to the US, which is estimated to exceed US$10 billion and $15 billion per annum in pet and food sales, respectively (see US Department of State Wildlife Trafficking). Nonetheless, there is evidence that these activities are associated with the introduction of microorganisms into new environs, and that this might pose a threat to public health. An analysis of bat, rodent and non-human primate bushmeat that was confiscated in major ports has revealed evidence of foamy viruses, herpesviruses and pathogenic bacteria in these samples26. Imported pets have been linked to outbreaks of human infection with poxviruses and Salmonella, as well other pathogens27,28.
Although plant pathogens do not infect humans or other animals, the infection of food crops can have dire economic consequences. Recently, with a greater appreciation of the importance of pollinators in food production and with the recognition of colony collapse disorder, increased attention has been directed towards the potential for emerging infections of honeybees (Apis mellifera) and other pollinators, particularly with regard to viruses, fungi and external parasites29. Mariculture (ocean aquaculture) is also at risk of emerging infectious diseases, as demonstrated by recent reports of novel viruses in farmed salmon30,31,32. In addition, attention has become increasingly focused on the role of land use dynamics in infectious-disease emergence33. Deforestation and the expansion of both agriculture and the extractive industries, particularly in tropical regions with high wildlife biodiversity, have led directly or indirectly to the emergence of HIV/AIDS, Nipah virus and filoviruses34,35.
Climate change and mass migration. Global warming is already extending the geographical range of mosquitoes and ticks that harbour and transmit Plasmodium spp. and arboviruses, resulting in outbreaks of malaria, dengue fever and yellow fever in new locations36. Recent examples in the United States include the appearance of dengue fever in Florida from 2009 to 2010 (Refs 37, 38) and a surge in cases of West Nile encephalitis in Texas in 2012 (Ref. 39). Mass migration (owing to war, natural disaster, poverty and desertification) can lead to increases in the population density not only of humans, but also of disease vectors, such as rodents and ectoparasites, which carry pathogenic viruses and bacteria. These factors, along with poor sanitation, malnutrition, a lack of access to vaccines, and exposure to contaminated food and water create a perfect storm for the emergence and transmission of infectious diseases40,41.
Laboratory analyses
Culture — once the mainstay for the detection of organisms in the laboratory — is still emphasized in some public health organizations and remains vital to clinical microbiology, chiefly as a tool for testing the utility of drugs. However, genetic methods have moved to the forefront in microbial surveillance and diagnostics42,43,44,45,46. The foundation for most of these methods is PCR, which was developed in 1983 by Kary Mullis (Timeline). PCR requires minimal equipment and operator training, can be completed in minutes rather than days, is less expensive than culture and has been adapted to portable instruments that can be used in the field in developing countries or near a patient's bedside. Furthermore, like other genetic methods, PCR might succeed in detecting an organism that has fastidious requirements which confound cultivation. Most PCR assays that are approved for clinical applications test for the presence of a single type of bacterium or virus. Such assays, described as singleplex, are used to screen for any evidence of infection (for example, to find hepatitis B virus in blood products to be used for transfusion) or to quantify microorganism levels when assessing a response to therapy (for example, to determine the HIV burden in the serum or plasma of subjects receiving antiretroviral medication).
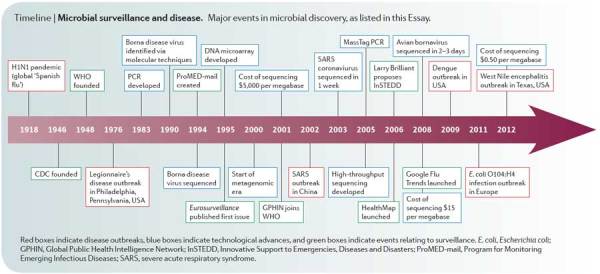
Timeline | Microbial surveillance and disease. Major events in microbial discovery, as listed in this Essay.
Multiplex assays. Multiplex PCR, which was initially implemented for screening human genetic polymorphisms, has now been extended to the field of microbiology, wherein assays have been developed that allow simultaneous screening for the presence of up to 30 different microorganisms42. Such assays are particularly important for differential diagnosis in medicine, in cases for which many distinct infectious agents can be implicated (for example, diseases like pneumonia, diarrhoea, meningitis or encephalitis). Thus, although multiplex PCR is rarely used other than in research and public health laboratories, there is reason to believe that this technique will ultimately gain wider acceptance.
An even broader platform is the DNA microarray, in which millions of genetic probes are bound to glass or silicon wafers and tested for their capacity to bind complementary sequences in clinical and environmental samples. Binding is typically detected through the measurement of fluorescent molecules attached to the nucleic acid amplified from sample extracts. Such microarrays have the potential to survey the entire known microbial world; however, their implementation in this capacity has been hampered by their low sensitivity and the cumbersome processing required. Recently developed prototypes indicate that it might be possible to circumvent these obstacles through the use of portable devices that use nanofluidics and electronic nanocircuitry47,48.
Genomics and metagenomics. The most dramatic advance in microbial surveillance has been achieved in DNA sequencing. The emergence of high-throughput sequencing over the past decade has enabled the discovery of new microorganisms, the rapid resolution of the causes of infectious-disease outbreaks and the development of metagenomics, a field in which investigators inventory the complex microbial communities found in humans, domesticated animals, wildlife, plants and various environments. Although high-throughput sequencing was initially confined to specialized laboratories owing to the high costs of instruments and supplies, along with the requirements for specially trained personnel, recent technical improvements have increased access to this technology to a broader segment of the research community. One index for the evolution of sequencing technology is the per-base cost, which decreased from $5,000 per megabase in 2001 (using classical dideoxy methods), to $15 per megabase in 2008 (using pyrosequencing; see DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP)), to $0.50 per megabase in 2012 (using the Illumina platform49). Another index is the time required to obtain sequence data. Whereas the SARS coronavirus genome was sequenced over the course of 1 week by a large team in 2003 (Ref. 8), a single investigator could sequence that same genome in a few hours in 2012. Recent examples of the power of advancements in genomic-sequencing methods include reports on the evolution of influenza viruses50, hepatitis C virus51 and HIV52, on the human origin of livestock-associated methicillin-resistant Staphylococcus aureus53 and on the spread of antibiotic-resistant Klebsiella pneumoniae in and between health care institutions54.
Metagenomic analyses42,55,56 have revealed dynamic microorganism–host relationships that influence normal physiological processes — for example, digestion57,58 and the immune response59, which might be factors in the pathogenesis of autoimmune diseases59 and cancer56 — and that probably also contribute to global climate regulation through effects on marine plankton60. The challenge now is not in obtaining sequence data but in analysing it. Millions of sequence reads must be assembled into continuous strings of genetic information and identified as originating from a particular microorganism or host by using algorithms that search for similarity between the sequences obtained from metagenomics and those already catalogued in existing databases. Few investigators currently have the in-house processing power and expertise required for these types of analyses; however, access to large computer clusters can be achieved through cloud computing and high-throughput sequencing software that is rapidly becoming more user friendly.
Microbial surveillance and forecasting
Passive and active surveillance. Surveillance is broadly divided into two categories: passive and active. Whereas passive surveillance uses data that already exist or are collected routinely, active surveillance involves a new investment in and/or new processes for microorganism collection and analysis. A classic example of passive surveillance is the concept of reportable diseases. Most regional and national public health authorities maintain lists of infectious diseases for which laboratory tests indicative of infection must be reported (see CDC National Notifiable Diseases Surveillance System). These diseases include those characterized by human-to-human transmission, such as sexually transmissible diseases (for example, syphilis or gonorrhoea) and vaccine-preventable diseases (for example, measles), as well as those for which detection indicates the presence, in the environment, of an infectious agent that poses a substantial threat to public health, such as haemorrhagic fever viruses or highly pathogenic bacteria (for example, Yersinia pestis, the causative agent of bubonic plague). On the non-human side, agricultural authorities monitor infections such as foot-and-mouth disease, which have important economic implications. Passive surveillance is informative and inexpensive, but might underestimate the true frequency of an agent or a disease. Furthermore, by definition, it cannot detect the risk of infection with a particular pathogen before the onset of symptoms.
Surveillance using social media. Today, Internet-based infectious-disease surveillance is well established, largely owing to the influence of Joshua Lederberg61, a pioneer in microbial genetics and the use of computers for communication as well as data analysis. ProMED-mail (Program for Monitoring Emerging Infectious Diseases), created in 1994, provides continuous free e-mail updates about new or evolving outbreaks and epidemics62. Submissions from a grass-roots network of readers are curated by a panel of experts, who post submissions with commentary in five languages to a listserv comprising more than 60,000 subscribers in 185 different countries. GPHIN (Global Public Health Intelligence Network)63 scans news services in nine languages across the globe for information concerning outbreaks. Unlike ProMED-mail, GPHIN is a fee-based, private subscription and does not systematically validate its posts, although the WHO began providing verification services when GPHIN was added to the Global Outbreak Alert Response Network (GOARN) in 2001.
HealthMap64 is a hybrid of the passive and active surveillance strategies that are used by ProMED-mail and GPHIN, respectively. HealthMap integrates reports from news media, ProMED-mail and official documents into a user-friendly map that displays real-time updates of disease emergence. HealthMap also allows public submission of georeferenced observations, or 'crowd sourcing' of apparent disease occurrences, via its website or a number of smart phone-based apps, such as Outbreaks Near Me. Google Flu Trends is similar, but aggregates search data to estimate global influenza virus activity.
InSTEDD (Innovative Support to Emergencies, Diseases and Disasters), which was proposed during Larry Brilliant's talk at the 2006 TED Conference, is developing open-source tools to improve global information collection and exchange. What is anticipated, although not yet achieved, is the development of systems that aggregate data about the use of medical services, data about prescription and over-the-counter drug purchases, and other chatter that could promote situational awareness and focus epidemiological investigations. Zoonoses (that is, infections that originate in wildlife or domestic animals) account for more than 70% of emerging infectious diseases65; thus, to be proactive, a substantive surveillance system66 for humans must also include surveillance of animals.
Modelling infectious-disease emergence. Quantitative analyses of emerging and re-emerging infectious diseases have enabled the identification of both geographical hot spots of infectious-disease emergence and the underlying drivers (primarily human activities) that facilitate the process33,67,68,69,70. The recent development of high-quality, global-scale data sets for human demographics, agricultural production, land-use change, travel patterns, trade patterns, climate and wildlife distribution has greatly improved the resolution and specificity of predictive modelling40. This has resulted in substantial advances in risk analyses from the original hotspot maps40 (Fig. 1). These risk algorithms are being used to focus passive and active surveillance programmes on the sites, populations, professions and domestic or wild animals for which there is an increased probability of known or novel high-threat pathogen emergence.
Figure 1. Hot spots of outbreaks for recently emerging and re-emerging infectious diseases.

Zoonotic infections are highlighted in red text. Data from Ref. 103 and P. Daszak (EcoHealth Alliance; personal communication). E. coli, Escherichia coli; S. aureus, Staphylococcus aureus; vCJD, variant Creutzfeldt–Jakob disease.
An example is the Emerging Pandemic Threats programme of the US Agency for International Development, which uses hotspot models to prioritize regions and countries for investments in surveillance, laboratory diagnostics, outbreak responses, and collaboration in 20 African, South American and Asian countries. The algorithms used to build hotspot models are continuously tested and modified in light of experimental data derived from human, domestic-animal and wildlife sample analysis.
Causation and mechanisms of pathogenesis
Finding footprints of a microorganism is only the first step in establishing a causative role for that microorganism in a disease. In some instances, the connection is immediately apparent because precedent supports plausibility — for example, finding a new type of Ebola virus in an individual with a haemorrhagic fever, or a new strain of Vibrio cholerae in a diarrhoea outbreak. However, in other instances, the link is more tenuous. Host factors can have a profound impact on susceptibility to infection and the consequences thereof. Agents that are normally innocuous can have high morbidity and mortality rates in individuals with immunological deficits, whether those deficits are due to genetic mutations, age, malnutrition, a co-occurring infection (for example, HIV/AIDS), or complications of cancer treatment or transplantation.
Mechanisms of disease can vary (Box 1). Microorganisms might cause damage at the site of infection, as a direct result of replication or an indirect effect of host innate or adaptive immune responses to microbial gene products. Microorganisms can also induce neoplasia through interference with cell cycle controls. Although not yet confirmed in human disease, work in animal models indicates that viruses can reduce the production of hormones or neurotransmitters that are vital to normal host physiology, and that they can do so without causing any apparent cell or organ damage71. Linkage of a disease to infection with a specific pathogen is facilitated in each of the forementioned examples because the microorganism, its nucleic acid or its protein is found at the site of pathology. More difficult to recognize are instances in which the expression of microbial toxins has remote effects, or infection induces immune responses to the microorganism that break tolerance to self. Clostridium spp., for example, can infect the skin or gastrointestinal tract and produce toxins that act on the nervous system to cause spasms (Clostridium tetani)72 or flaccid paralysis (Clostridium botulinum)73. Streptococcal infection of the skin or the oropharynx can result in autoimmunity, culminating in cardiac damage (known as rheumatic heart disease) and brain dysfunction (known as Sydenham chorea)74.
The best established criteria for proof of causation were formulated by Loeffler and Koch in the 1880s. Popularly known as Koch's postulates75, these criteria require that an agent be present in every case of the disease, be specific for the disease and be sufficient to reproduce the disease after culture and inoculation into a naive host. Rivers76 modified these postulates by acknowledging that the presence of neutralizing antibodies to an agent is evidence of infection. Fredericks and Relman77 noted that pathogens can often be recognized by molecular methods before they can be cultured, and therefore allowed as evidence the presence of microbial sequences as well as of infectious microorganisms. Thus, although Koch's postulates remain the gold standard, they need not be fulfilled to implicate an agent in a disease. Indeed, a focus on Koch's postulates might impede the successful discovery of, and response to, emerging pathogens and the development of models for infectious disease.
Through the discovery and characterization of nearly 500 viruses, my colleagues and I have developed a three-level scoring system for establishing the level of confidence in a particular association, from possible to definitive (Box 2). Poor design or execution of pathogen discovery projects can lead to spurious links being made between infectious agents and diseases, and this can result in the use of inappropriate and potentially dangerous treatments or the rejection of health-promoting interventions such as vaccines42. The effort required to break these links can be greater than that invested in building them, particularly for disorders with a grim prognosis and/or limited treatment options. From our experience with amyotrophic lateral sclerosis (linked to enteroviruses), mental illness (linked to bornaviruses), autism (linked to the measles, mumps and rubella (MMR) vaccine) and myalgic encephalomyelitis–chronic fatigue syndrome (linked to both xenotropic murine leukaemia virus-related virus and polytropic murine leukaemia virus), we have developed a strategy to try and acquit microorganisms by addressing social as well as scientific considerations (Box 3).
Progress in microbial detection: field cases
Over the past three decades, innovations in genetic and information technologies have enhanced and expedited the rate of detection and solution of infectious-disease outbreaks. The following examples illustrate the progress that has been made in methods for acquiring public health intelligence.
In 1976, the CDC was alerted that 11 war veterans had died from pneumonia after returning from the US Bicentennial Convention of the American Legion in Philadelphia, Pennsylvania78. A case definition was established78,79, and Pennsylvania health officials were notified of a potential state-wide epidemic. Public health personnel searched hospitals, news reports and obituaries, and a telephone hotline was established to accept tips from the general public. Ultimately, 221 cases of Legionnaire's disease, as it was called, were identified. All affected individuals had visited the lobby of the hotel hosting the convention or had walked along the adjacent street. Initial efforts to culture an infectious agent failed78,80. Histological staining of lungs from individuals with Legionnaire's disease revealed inflammation but no microorganisms.
The breakthrough came when Joseph McDade, a rickettsia expert at the CDC, recognized liver disease in some victims and in guinea pigs inoculated with extracts from patients. He inoculated embryonated chicken eggs with liver extracts from guinea pigs and then reproduced the disease by inoculating additional guinea pigs with the extracts from the embryonic chicken eggs. The pathogen enrichment through passage in these model systems resulted in the discovery of a novel fastidious Gram-negative bacterium, Legionella pneumophila.
The 4 months that elapsed between the onset of the outbreak in Philadelphia and the identification of the causative agent would not be required today. Whereas almost 2 weeks passed before the CDC was notified in 1976, alerts are now distributed in near real time through services like ProMED-mail and HealthMap. In addition, access to an electronic registry of the convention guests would have obviated the need for a state-wide, grass-roots search for cases. Modern culture-independent methods of pathogen discovery would also enhance response time.
Recent improvements in surveillance and microbial forensic science were illustrated by the 2011 European outbreak of the Shiga toxin-producing bacterium Escherichia coli O104:H4 (Ref. 81, 82). In May 2011, the German national public health agency — the Robert Koch Institute — sent representatives to investigate a cluster of haemolytic–uraemic syndrome cases associated with bloody diarrhoea in Hamburg. By the time the outbreak resolved, more than 3,000 people had been infected in Europe, and 40 had died. Economic losses were substantial, particularly in Spain, as an early inaccurate link to Spanish cucumbers led to an almost Europe-wide import ban on produce from Spain83,84.
The initial clues to the identity of the causal agent were obtained using PCR85,86. Genomic characterization was rapidly achieved using high-throughput sequencers87. Within 3 days of receipt of a clinical sample, sequence data were released into the public domain for global, crowd-sourced bioinformatic analysis88. Genome assembly was completed in the next 24 hours, which enabled the development of specific diagnostic tests and provided insights into the pathogenesis and phylogenetic origin of the bacterium. The integration of laboratory findings with patient surveys ultimately led to implication of bean sprouts from a single farm in Lower Saxony81.
Another comparison involves the discovery of Borna disease virus (BDV)89 and the related but distinct avian bornavirus (ABV). Borna disease is named after a town in Saxony where a characteristic fatal meningoencephalitis was described in horses, and the disease has been known in the veterinary literature since the 1700s90,91,92. The transmissibility of the disease was first demonstrated in the 1920s; however, it took nearly 60 years to establish methods for BDV culture and another 10 years to classify the virus as a novel non-segmented, negative-strand RNA virus93. The capacity to culture the virus led to the development of serological assays, and in 1983, these assays provided evidence of a connection to human neuropsychiatric diseases94.
Intrigued by the potential importance of BDV in human disease and challenged by the failure of efforts to characterize the virus by electron microscopy or to isolate BDV nucleic acids, I initiated a subtractive-cloning project that culminated in the determination of the first BDV sequences in 1990 (Ref. 95). It took an additional 4 years to determine the genomic organization of the virus96,97. With specific cloned reagents in hand, I reasoned that it would be straightforward to determine whether BDV was indeed a human pathogen; however, the issue lingered until 2012, when blinded multicentre analyses, both molecular and serological, ultimately revealed no evidence of human infection98.
By contrast, ABV, the causative agent of proventricular dilatation disease (a wasting syndrome in parrots), was identified in only a few days99,100. The breakthrough was enabled by access to genome databases and the availability of culture-independent methods for pathogen discovery, including viral microarrays and high-throughput sequencing. Subsequently, ABV PCR assays and serology have allowed the investigation and containment of outbreaks in aviaries101.
Future prospects
Although we will continue to see instances in which classical approaches to microorganism hunting, like culture and the pursuit of Koch's postulates, will succeed, pathogen discovery has evolved from a 'whodunit' exercise carried out by solitary investigators to a team effort involving microbiologists, cellular and systems biologists, geographers, mathematicians and other specialists. Models of diseases have expanded from simple one-to-one relationships between organ damage and the presence of a single agent therein to consider more complex mechanisms that might enable the recognition of links between microorganisms and mental illness, obesity, vascular disease, cancer and autoimmunity.
The increase in international travel and trade has led to the globalization of infectious diseases. It has also fostered a new appreciation of the relationship between land use, particularly in the developing world, and the appearance of zoonoses. This globalization of risk across national and species boundaries has promoted the development of international health regulations102 that emphasize technology transfer and data sharing, as well as programmes that proactively survey not only humans, but also the entire animal kingdom for insights into potential threats to public health and economic welfare.
The integration of human and animal medicine, the advent of tools for the rapid and efficient molecular characterization of microorganisms and hosts, and the emphasis on the use of social media to promote early detection of risk together have great potential for the development of a truly global immune system.
Box 1 | Mechanisms of microbial pathogenesis.
Direct damage at the site of microbial replication, owing to host cell lysis, apoptosis or autophagy.
Indirect damage at the site of microbial replication, owing to the expression of proteins that serve as targets for host humoral or cell-mediated immune responses.
The elaboration of toxins or other products that have deleterious local or systemic effects.
The induction of host cytokines and chemokines that have deleterious local or systemic effects.
The abrogation of host tolerance for self, resulting in host autoimmunity.
Immunosuppression of the host, resulting in opportunistic infection.
Induction of host cell neoplasia.
Disturbing the functions of differentiated cells.
Disruption of embryogenesis.
Box 2 | Levels of certainty in pathogen discovery.
Level 1: a possible causative relationship
The initial clue in pathogen discovery is evidence of exposure to a microorganism in one or more individuals with a disease. This evidence might be the isolation and growth (on media or in cultured cells or animals) of a microorganism that is present in the blood, other body fluids, faeces or tissues of such individuals. It might alternatively be detection of a nucleic acid (by PCR, DNA microarray or sequencing) or protein (by immunological methods or mass spectroscopy) component of a microorganism, a specific adaptive immune response to a microorganism (that is, detection of antibodies through immunological methods), or visualization of the microorganism (by light microscopy, immunomicroscopy or electron microscopy).
Level 2: a probable causal relationship
More confidence in the clinical significance of the association between a pathogen and a disease is achieved when a causal relationship is biologically plausible. Evidence of biological plausibility can include the presence of microbial nucleic acid, microbial protein or microorganism-specific antibody in or adjacent to host cells showing signs of disease, or precedent for a similar disease caused by a similar agent in either the same or a similar host. The strength of the association is increased when the concentration of the microorganism (or nucleic acid, protein or antibody) is high, when the antibody response indicates recent exposure (that is, when immunoglobulin M (IgM) is present and/or there has been a recent increase in the IgG titre) and when there is evidence of infection in other individuals, all of whom have the disease. However, a microorganism can also be implicated in a disease without a robust immune response, particularly in chronic infections.
Different microorganisms can cause similar diseases. Although clusters of a disease are the ideal proving ground, many opportunities for pathogen discovery involve only one or a few cases of a disease; indeed, clusters might not be appreciated as such until details of common exposure (for example, through travel, food, water or intermediary hosts) become apparent.
Level 3: a confirmed causal relationship
Proof of causation can be achieved through fulfilment of Koch's postulates or by the mitigation or the prevention of the disease (that is, a reduction in the levels of the microorganism, its nucleic acids or proteins, or the immune response to the microorganism) through the use of microorganism-specific drugs, antibodies or vaccines. Although not formally required, my colleagues and I insist on the replication of results by independent investigators.
Box 3 | The road to pathogen de-discovery.
Step 1: the initial finding
A report links a disease or syndrome to an infectious agent, toxin or other factor.
Step 2: a failure to reproduce the finding
Independent efforts by other investigators fail to confirm the statistically significant association described in the initial report. Members of the scientific community begin to question the validity of the association, but cannot exclude the possibility that the apparent difference in results reflects variability in the samples or assays used.
Step 3: plausible doubt
A biologically plausible explanation is developed to account for the findings in the initial report that might have led to a misinterpretation. The mainstream scientific community rejects the validity of the association. However, some investigators, patients and advocates might continue to believe the merits of the initial report.
Step 4: comprehensive de-discovery
A well-powered, blinded analysis — involving the investigators responsible for the initial report and using a strategy that is approved in advance by the investigators, representatives of patients and patient advocacy groups — tests samples from well-characterized subjects and fails to replicate the findings of the initial report. The association between the pathogen, toxin or other factor and the disease is rejected by both the scientific and non-scientific communities.
Acknowledgements
The author thanks N. Akpan for assistance with the manuscript; P. Daszak, W. Karesh, C. Firth, M. Hornig and E. Holmes for thoughtful comments; and the US National Institutes of Health (grant AI57158), the US Agency for International Development (the PREDICT programme) and the US Defense Threat Reduction Agency for financial support.
Biography
W. Ian Lipkin is the John Snow Professor of Epidemiology and Professor of Neurology and Pathology at Columbia University, New York, USA. He is internationally recognized for the development and implementation of molecular methods for microbial surveillance and discovery. He directs the Center for Infection and Immunity at Columbia University (a World Health Organization Collaborating Centre on Diagnostics, Surveillance and Immunotherapeutics for Emerging Infectious and Zoonotic Diseases) and the Northeast Biodefense Center, USA, and is co-chair of the National Biosurveillance Advisory Subcommittee. A graduate of Sarah Lawrence College, Yonkers, New York, he obtained his M.D. at Rush Medical College, Chicago, Illinois, USA, followed by a Residency in Medicine at the University of Washington, Seattle, USA, a Residency in Neurology at the University of California, San Francisco, USA, and a fellowship with Michael Oldstone at The Scripps Research Institute, La Jolla, California. His contributions include the first identification of an infectious agent using purely molecular methods; implicating West Nile virus as the cause of the encephalitis outbreak in New York in 1999; the development of MassTag PCR and panmicrobial microarrays; the use of deep sequencing in pathogen discovery; and the discovery or molecular characterization of more than 500 viruses. He served in Beijing, China, as an intermediary between the WHO and the Chinese government during the severe acute respiratory syndrome (SARS) outbreak of 2002–2003 and co-directed SARS research efforts in China with the current Minister of Health, Chen Zhu. His honours include the following awards: Pew Scholar in the Biomedical Sciences, Japanese Human Science Foundation Visiting Professor, Columbia College of Physicians and Surgeons Visiting Bruenn Professor, Ellison Medical Foundation Senior Scholar in Global Infectious Disease, Fellow of the New York Academy of Sciences, Fellow of the American Society for Microbiology, John Courage Professor National University of Singapore, Fellow of the Wildlife Conservation Society, Fellow of the American Association for the Advancement of Science, Member of the Association of American Physicians and Fellow of the UK Royal Geographical Society.
Related links
FURTHER INFORMATION
The Center for Infection and Immunity, Columbia University
CDC National Notifiable Diseases Surveillance System
DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP)
US airlines and foreign airlines US passengers continue to Increase from 2009
US Bureau of Transportation Statistics flight information
US Department of State Wildlife Trafficking
World Population Prospects, the 2010 Revision; Table 'Total Population, Both Sexes'
Competing interests
The author declares no competing financial interests.
References
- 1.Taubenberger JK, Hultin JV, Morens DM. Discovery and characterization of the 1918 pandemic influenza virus in historical context. Antiviral Ther. 2007;12:581–591. [PMC free article] [PubMed] [Google Scholar]
- 2.Durand JD. Historical Estimates of World Population: an Evaluation. 1974. [Google Scholar]
- 3.Taubenberger JK, Morens DM. 1918 influenza: the mother of all pandemics. Emerg. Infect. Dis. 2006;12:15–22. doi: 10.3201/eid1209.05-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003 (WHO, 2003).
- 5.Mills CE, Robins JM, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riley S, et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 7.Lipsitch M, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marra MA, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 9.Rota PA, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 10.Briese T, et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurata JH, Nogawa AN. Meta-analysis of risk factors for peptic ulcer. Nonsteroidal antiinflammatory drugs, Helicobacter pylori, and smoking. J. Clin. Gastroenterol. 1997;24:2–17. doi: 10.1097/00004836-199701000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 13.Schrama D, Ugurel S, Becker JC. Merkel cell carcinoma: recent insights and new treatment options. Curr. Opin. Oncol. 2012;24:141–149. doi: 10.1097/CCO.0b013e32834fc9fe. [DOI] [PubMed] [Google Scholar]
- 14.Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci. Transl. Med. 2012;4:137rv7. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosseini P, Sokolow SH, Vandegrift KJ, Kilpatrick AM, Daszak P. Predictive power of air travel and socio-economic data for early pandemic spread. PLoS ONE. 2010;5:e12763. doi: 10.1371/journal.pone.0012763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowdall NP, Evans AD, Thibeault C. Air travel and TB: an airline perspective. Travel Med. Infect. Dis. 2010;8:96–103. doi: 10.1016/j.tmaid.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Mangili A, Gendreau MA. Transmission of infectious diseases during commercial air travel. Lancet. 2005;365:989–996. doi: 10.1016/S0140-6736(05)71089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maki DG. Don't eat the spinach — controlling foodborne infectious disease. N. Engl. J. Med. 2006;355:1952–1955. doi: 10.1056/NEJMp068225. [DOI] [PubMed] [Google Scholar]
- 20.Newell DG, et al. Food-borne diseases — the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food. Microbiol. 2010;139(Suppl. 1):S3–S15. doi: 10.1016/j.ijfoodmicro.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eurosurveillance Editorial Team. European Food Safety Authority publishes its second report on the Schmallenberg virus. Euro Surveill.17, 20140 (2012). [PubMed]
- 22.Beuchat LR. Surface decontamination of fruits and vegetables eaten raw: a review. 1998. [Google Scholar]
- 23.Berger CN, et al. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 2010;12:2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Alvarez L, Dawson S, Cookson B, Hawkey P. Working across the veterinary and human health sectors. J. Antimicrob. Chemother. 2012;67(Suppl. 1):i37–i49. doi: 10.1093/jac/dks206. [DOI] [PubMed] [Google Scholar]
- 25.McEwen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 2002;34(Suppl. 3):S93–S106. doi: 10.1086/340246. [DOI] [PubMed] [Google Scholar]
- 26.Smith KM, et al. Zoonotic viruses associated with illegally imported wildlife products. PLoS ONE. 2012;7:e29505. doi: 10.1371/journal.pone.0029505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris JR, Neil KP, Behravesh CB, Sotir MJ, Angulo FJ. Recent multistate outbreaks of human salmonella infections acquired from turtles: a continuing public health challenge. Clin. Infect. Dis. 2010;50:554–559. doi: 10.1086/649932. [DOI] [PubMed] [Google Scholar]
- 28.Hutson CL, et al. Monkeypox zoonotic associations: insights from laboratory evaluation of animals associated with the multi-state US outbreak. Am. J. Trop. Med. Hyg. 2007;76:757–768. doi: 10.4269/ajtmh.2007.76.757. [DOI] [PubMed] [Google Scholar]
- 29.Cox-Foster DL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 30.Finstad OW, Falk K, Lovoll M, Evensen O, Rimstad E. Immunohistochemical detection of piscine reovirus (PRV) in hearts of Atlantic salmon coincide with the course of heart and skeletal muscle inflammation (HSMI) Vet. Res. 2012;43:27. doi: 10.1186/1297-9716-43-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovoll M, et al. A novel totivirus and piscine reovirus (PRV) in Atlantic salmon (Salmo salar) with cardiomyopathy syndrome (CMS) Virol. J. 2010;7:309. doi: 10.1186/1743-422X-7-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palacios G, et al. Heart and skeletal muscle inflammation of farmed salmon is associated with infection with a novel reovirus. PLoS ONE. 2010;5:e11487. doi: 10.1371/journal.pone.0011487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karesh WB, et al. The ecology of zoonoses: their natural and unnatural histories. Lancet. 2012;380:1936–1945. doi: 10.1016/S0140-6736(12)61678-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chua KB, et al. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet. 1999;354:1257–1259. doi: 10.1016/S0140-6736(99)04299-3. [DOI] [PubMed] [Google Scholar]
- 35.Pulliam JR, et al. Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis. J. R. Soc. Interface. 2012;9:89–101. doi: 10.1098/rsif.2011.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuman EK. Global climate change and infectious diseases. N. Engl. J. Med. 2010;362:1061–1063. doi: 10.1056/NEJMp0912931. [DOI] [PubMed] [Google Scholar]
- 37.CDC. Locally acquired Dengue — Key West, Florida, 2009–2010. Morbid. Mortal. Wkly Rep.59, 577–581 (2010). [PubMed]
- 38.Adalja AA, Sell TK, Bouri N, Franco C. Lessons learned during dengue outbreaks in the United States, 2001–2011. Emerg. Infect. Dis. 2012;18:608–614. doi: 10.3201/eid1804.110968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roehr B. Texas records worst outbreak of West Nile virus on record. BMJ. 2012;345:e6019. doi: 10.1136/bmj.e6019. [DOI] [PubMed] [Google Scholar]
- 40.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–994. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morse SS. Factors and determinants of disease emergence. Rev. Sci. Tech. 2004;23:443–451. doi: 10.20506/rst.23.2.1494. [DOI] [PubMed] [Google Scholar]
- 42.Lipkin WI. Microbe hunting. Microbiol. Mol. Biol. Rev. 2010;74:363–377. doi: 10.1128/MMBR.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casas I, Tenorio A, Echevarria JM, Klapper PE, Cleator GM. Detection of enteroviral RNA and specific DNA of herpesviruses by multiplex genome amplification. J. Virol. Methods. 1997;66:39–50. doi: 10.1016/S0166-0934(97)00035-9. [DOI] [PubMed] [Google Scholar]
- 44.Nichol ST, et al. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 45.Shirato K, et al. Diagnosis of human respiratory syncytial virus infection using reverse transcription loop-mediated isothermal amplification. J. Virol. Methods. 2007;139:78–84. doi: 10.1016/j.jviromet.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tumpey TM, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 47.Rosenstein JK, Wanunu M, Merchant CA, Drndic M, Shepard KL. Integrated nanopore sensing platform with sub-microsecond temporal resolution. Nature Methods. 2012;9:487–492. doi: 10.1038/nmeth.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaittanis C, Santra S, Perez JM. Emerging nanotechnology-based strategies for the identification of microbial pathogenesis. Adv. Drug Delivery Rev. 2010;62:408–423. doi: 10.1016/j.addr.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loman NJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nature Biotech. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 50.Bhatt S, Holmes EC, Pybus OG. The genomic rate of molecular adaptation of the human influenza A virus. Mol. Biol. Evol. 2011;28:2443–2451. doi: 10.1093/molbev/msr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bull RA, et al. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog. 2011;7:e1002243. doi: 10.1371/journal.ppat.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poon AF, et al. Reconstructing the dynamics of HIV evolution within hosts from serial deep sequence data. PLoS Comput. Biol. 2012;8:e1002753. doi: 10.1371/journal.pcbi.1002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fitzgerald JR. Human origin for livestock-associated methicillin-resistant. Staphylococcus aureus. mBio. 2012;3:e00082–12. doi: 10.1128/mBio.00082-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Snitkin ES, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 2012;4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Relman DA. Microbial genomics and infectious diseases. N. Engl. J. Med. 2011;365:347–357. doi: 10.1056/NEJMra1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Streit, W. R. & Daniel, R. Metagenomics: Methods and Protocols. (Humana, 2010).
- 57.Faust K, Raes J. Microbial interactions: from networks to models. Nature Rev. Microbiol. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 58.Claesson MJ, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 59.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Danovaro R, et al. Marine viruses and global climate change. FEMS Microbiol. Rev. 2011;35:993–1034. doi: 10.1111/j.1574-6976.2010.00258.x. [DOI] [PubMed] [Google Scholar]
- 61.Institute of Medicine (US) Forum on Microbial Threats. Microbial evolution and co-adaptation: a tribute to the life and scientific legacies of Joshua Lederberg, workshop summary. (The National Academies Press, 2009). [PubMed]
- 62.Madoff LC. ProMED-mail: an early warning system for emerging diseases. Clin. Infect. Dis. 2004;39:227–232. doi: 10.1086/422003. [DOI] [PubMed] [Google Scholar]
- 63.Mykhalovskiy E, Weir L. The Global Public Health Intelligence Network and early warning outbreak detection: a Canadian contribution to global public health. Can. J. Public Health. 2006;97:42–44. doi: 10.1007/BF03405213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freifeld CC, Mandl KD, Reis BY, Brownstein JS. HealthMap: global infectious disease monitoring through automated classification and visualization of Internet media reports. J. Am. Med. Inform. Assoc. 2008;15:150–157. doi: 10.1197/jamia.M2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woolhouse ME, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karesh WB. One world – one health. Clin. Med. 2009;9:259–260. doi: 10.7861/clinmedicine.9-3-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lederberg J, Shope RE, Oaks SC. Emerging Infections: Microbial Threats to Health in the United States. 1992. [PubMed] [Google Scholar]
- 68.Morse SS. Factors in the emergence of infectious disease. Emerg. Infect. Dis. 1995;1:7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- 70.Smolinski MS, Hamburg MA, Lederberg J. Microbial Threats to Health: Emergence, Detection, and Response. 2003. [PubMed] [Google Scholar]
- 71.Lyte M. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol. 2004;12:14–20. doi: 10.1016/j.tim.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 72.Bruggemann H, et al. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl Acad. Sci. USA. 2003;100:1316–1321. doi: 10.1073/pnas.0335853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Segelke B, Knapp M, Kadkhodayan S, Balhorn R, Rupp B. Crystal structure of Clostridium botulinum neurotoxin protease in a product-bound state: evidence for noncanonical zinc protease activity. Proc. Natl Acad. Sci. USA. 2004;101:6888–6893. doi: 10.1073/pnas.0400584101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Allos BM. Association between Campylobacter infection and Guillain-Barre syndrome. J. Infect. Dis. 1997;176(Suppl. 2):S125–S128. doi: 10.1086/513783. [DOI] [PubMed] [Google Scholar]
- 75.Koch R. Verhandl. des X. Interna. Med. Congr., Berlin 1890. 1891. [Google Scholar]
- 76.Rivers TM. Viruses and Koch's postulates. J. Bacteriol. 1937;33:1–12. doi: 10.1128/jb.33.1.1-12.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fredericks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin. Microbiol. Rev. 1996;9:18–33. doi: 10.1128/CMR.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fraser DW. The challenges were legion. Lancet Infect. Dis. 2005;5:237–241. doi: 10.1016/S1473-3099(05)70054-2. [DOI] [PubMed] [Google Scholar]
- 79.Fraser DW, et al. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 1977;297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 80.McDade JE, et al. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 1977;297:1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- 81.Frank C, et al. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 2011;365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 82.Karch H, et al. The enemy within us: lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO Mol. Med. 2012;4:841–848. doi: 10.1002/emmm.201201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.BBC. E. coli cucumber scare: Spain angry at German claims. BBC News[online] (2011)
- 84.Busemann, H.-E. Germany says beansprouts may be behind E.coli. Reuters[online] (2011).
- 85.Bielaszewska M, et al. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 2011;11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 86.Scheutz F, et al. Characteristics of the enteroaggregative Shiga toxin/verotoxin-producing Escherichia coli O104:H4 strain causing the outbreak of haemolytic uraemic syndrome in Germany, May to June 2011. Euro Surveill. 2011;16:19889. doi: 10.2807/ese.16.24.19889-en. [DOI] [PubMed] [Google Scholar]
- 87.Mellmann A, et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS ONE. 2011;6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rohde H, et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N. Engl. J. Med. 2011;365:718–724. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 89.Hornig M, Briese T, Lipkin WI. Borna disease virus. J. Neurovirol. 2003;9:259–273. doi: 10.1080/13550280390194064. [DOI] [PubMed] [Google Scholar]
- 90.Abildgaard P. Pferde- und Vieharzt in einem kleinen Auszüge. 1795. [Google Scholar]
- 91.Trichtern V. Pferd-Anatomie, oder Neu-auserlesen-vollkommen- verbessert-und ergänztes Roß-Artzeney-Buch. 1716. [Google Scholar]
- 92.von Sind J. Der im Feld und auf der Reise geschwind heilende Pferdearzt, welcher einen gründlichen Unterricht von den gewöhnlichsten Krankheiten der Pferde im Feld und auf der Reise wie auch einen auserlesenen Vorrath der nützlichsten und durch die Erfahrung bewährtesten Heilungsmitteln eröffnet. 1767. [Google Scholar]
- 93.Lipkin W, Briese T. Fields Virology. 2007. pp. 829–1851. [Google Scholar]
- 94.Rott R, et al. Detection of serum antibodies to Borna disease virus in patients with psychiatric disorders. Science. 1985;228:755–756. doi: 10.1126/science.3922055. [DOI] [PubMed] [Google Scholar]
- 95.Lipkin WI, Travis GH, Carbone KM, Wilson MC. Isolation and characterization of Borna disease agent cDNA clones. Proc. Natl Acad. Sci. USA. 1990;87:4184–4188. doi: 10.1073/pnas.87.11.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Briese T, de la Torre JC, Lewis A, Ludwig H, Lipkin WI. Borna disease virus, a negative-strand RNA virus, transcribes in the nucleus of infected cells. Proc. Natl Acad. Sci. USA. 1992;89:11486–11489. doi: 10.1073/pnas.89.23.11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Briese T, et al. Genomic organization of Borna disease virus. Proc. Natl Acad. Sci. USA. 1994;91:4362–4366. doi: 10.1073/pnas.91.10.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hornig M, et al. Absence of evidence for bornavirus infection in schizophrenia, bipolar disorder and major depressive disorder. Mol. Psychiatry. 2012;17:486–493. doi: 10.1038/mp.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Honkavuori KS, et al. Novel borna virus in psittacine birds with proventricular dilatation disease. Emerg. Infect. Dis. 2008;14:1883–1886. doi: 10.3201/eid1412.080984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kistler AL, et al. Recovery of divergent avian bornaviruses from cases of proventricular dilatation disease: identification of a candidate etiologic agent. Virol. J. 2008;5:88. doi: 10.1186/1743-422X-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kistler AL, Smith JM, Greninger AL, Derisi JL, Ganem D. Analysis of naturally occurring avian bornavirus infection and transmission during an outbreak of proventricular dilatation disease among captive psittacine birds. J. Virol. 2010;84:2176–2179. doi: 10.1128/JVI.02191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Committee on Anticipating Biosecurity Challenges of the Global Expansion of High-Containment Biological Laboratories, US National Academy of Sciences, US National Research Council. Biosecurity Challenges of the Global Expansion of High-Containment Biological Laboratories (The National Academies Press, 2012). [PubMed]
- 103.Fauci AS. Infectious diseases: considerations for the 21st century. Clin. Infect. Dis. 2001;32:675–685. doi: 10.1086/319235. [DOI] [PubMed] [Google Scholar]


