Abstract
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous in the environment as components of fossil fuels and by-products of combustion. These multi-ring chemicals differentially activate the Aryl Hydrocarbon Receptor (AHR) in a structurally dependent manner, and induce toxicity via both AHR-dependent and-independent mechanisms. PAH exposure is known to induce developmental malformations in zebrafish embryos, and recent studies have shown cardiac toxicity induced by compounds with low AHR affinity. Unraveling the potentially diverse molecular mechanisms of PAH toxicity is essential for understanding the hazard posed by complex PAH mixtures present in the environment. We analyzed transcriptional responses to PAH exposure in zebrafish embryos exposed to benz(a)anthracene (BAA), dibenzothiophene (DBT) and pyrene (PYR) at concentrations that induced developmental malformations by 120 hours post-fertilization (hpf). Whole genome microarray analysis of mRNA expression at 24 and 48 hpf identified genes that were differentially regulated over time and in response to the three PAH structures. PAH body burdens were analyzed at both time points using GC-MS, and demonstrated differences in PAH uptake into the embryos. This was important for discerning dose-related differences from those that represented unique molecular mechanisms. While BAA misregulated the least number of transcripts, it caused strong induction of cyp1a and other genes known to be downstream of the AHR, which were not induced by the other two PAHs. Analysis of functional roles of misregulated genes and their predicted regulatory transcription factors also distinguished the BAA response from regulatory networks disrupted by DBT and PYR exposure. These results indicate that systems approaches can be used to classify the toxicity of PAHs based on the networks perturbed following exposure, and may provide a path for unraveling the toxicity of complex PAH mixtures.
Keywords: toxicity, AHR, microarray, dibenzothiophene, pyrene, benz(a)anthracene, network analysis
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are a diverse class of chemicals, comprised of at least two fused benzene rings, which are of concern to human health due to their ubiquity in the environment and toxicological effects. Both petrogenic and pyrogenic sources contribute to PAHs in the environment, but increasing concentrations, particularly in urban areas, have been attributed to anthropogenic activity such as fossil fuel burning, automobile exhaust, oil refining and coal tar seal coating (Van Metre and Mahler, 2005; Polidori et al., 2010; Van Metre and Mahler, 2010). PAHs are present in the ultrafine particulate fraction as well as the gas phase of ambient air and are considered carcinogenic components of cigarette smoke, vehicle exhaust, wood smoke and other emissions (Bostrom et al., 2002; Ramirez et al., 2011). The primary routes of human exposure are inhalation and ingestion. PAHs associated with ultrafine particulate matter can accumulate in the bronchial epithelium, while more volatile PAHs are readily absorbed through the alveolar epithelium (Ramirez et al., 2011). For non-smoking individuals, ingestion via foods and unintentional consumption of household dust (of particular concern for young children) is a primary contributor to PAH exposure (Menzie et al., 1992; Ramesh et al., 2004; Williams et al., 2012). PAH-containing coal tar mixtures are carcinogenic in humans (International Agency for Research on Cancer) (Collins et al., 1998). Seven non-substituted PAHs are considered possible carcinogens (group 2B) by the US EPA, and 16 PAHs are listed as priority pollutants because of their prevalence in urban and suburban air (EPA, 2012). Human exposure to PAHs almost always occurs within complex mixtures, which may contain multiple PAHs and often include other chemicals such as halogenated hydrocarbons and metals. Because of complex exposure patterns, it is difficult to associate health effects in human populations with individual PAHs.
While the bulk of research on PAHs has focused on mutagenic and carcinogenic properties, exposure to PAH mixtures and ultrafine particulate matter is associated with a diverse array of other health effects in humans, including immune system deficiency, cardiovascular disease and impaired development (Burstyn et al., 2005; Choi et al., 2006; Hertz-Picciotto et al., 2008; Lee et al., 2011; Ren et al.). Activation of the aryl hydrocarbon receptor (AHR) and generation of reactive oxygen species (ROS) are key modes of action initiated by some PAHs, but the full extent of the molecular responses that result from exposure to this diverse set of compounds has not been characterized. A number of PAHs, including benzo(a)pyrene (BaP) and dimethylbenz(a)anthracene (DMBA) , bind the AHR and induce expression of phase I and II metabolizing genes, such CYP1A1, GSTA1, NQO1 and UGT1A6, along with many other downstream transcripts (Guengerich, 2000; Nebert et al., 2000). Activation of the AHR pathway and metabolism of PAHs can result in a protective effect against PAH toxicity. In many cases, however, AHR activation and metabolism by CYP enzymes increases PAH reactivity and toxicity, which is consequential to the PAH, route of exposure, and exposure concentration (Nebert et al., 2004; Shi et al., 2010; Kerley-Hamilton et al., 2012). The low molecular weight PAHs (2–3 rings) are generally poor AHR ligands and less potent carcinogens, but are often detected at higher levels in environmental samples and human urine than their higher molecular weight counterparts (Durant et al., 1996; Naumova et al., 2002; Ciganek et al., 2004; Hecht et al., 2010).
Several studies have associated PAH exposure during pregnancy with adverse birth outcomes such as reduced fetal growth and neural tube defects (Choi et al., 2006; Ren et al., 2011). In rodents, exposure to BaP and DMBA induces abnormal vasculature in the placenta and interferes with fetal growth (Detmar et al., 2008; Rennie et al., 2011). Developmental exposure to BaP also impairs cardiac function later in life (Jules et al., 2012).
In zebrafish embryos, BaP-induced cardiac toxicity is mediated by the aryl hydrocarbon receptor (AHR2) (Incardona et al., 2011). However, other PAH structures induce cardiac toxicity and developmental effects via distinct mechanisms that are not AHR-dependent (Incardona et al., 2005). Analyses of global mRNA transcriptional responses to individual PAH exposures demonstrate that structurally-distinct PAHs induce unique gene expression patterns in both human macrophage leukemia (THP-1) cells and circulating leukocytes of rats (Wan et al., 2008; Jung et al., 2011). Little is known, however, about the toxicity pathways and molecular signatures of these diverse exposures during embryonic development.
We used whole genome mRNA microarrays to investigate transcriptional responses that lead to developmental toxicity of three distinct PAHs in developing zebrafish. Dibenzothiophene (DBT), pyrene (PYR) and benz(a)anthracene (BAA) all induce developmental abnormalities by 5 days post fertilization, but have different proposed toxicity mechanisms. DBT (3 rings) induces cardiac toxicity that is independent of the Ahr (Incardona et al., 2004). BAA (4 rings) induces Cyp1a expression and developmental toxicity via activation of AHR2, while PYR (4 rings) toxicity was shown to be metabolism-dependent (Incardona et al., 2006). We determined PAH body burden and corresponding transcriptional profiles in PAH-exposed zebrafish embryos at 24 and 48 hours post-fertilization, before toxicity could be visibly identified. We found that DBT, PYR and BAA induce mRNA expression profiles that differentially implicate AHR activity, and highlight multiple pathways that can be disrupted by exposure to PAHs over the course of development.
Methods
Zebrafish lines and embryos
Adult wild type 5D zebrafish were housed at the Sinnhuber Aquatic Research Laboratory on a recirculating system maintained at 28±1°C with a 14 h light/10 h dark schedule. Embryos were collected from group spawns of adult zebrafish as described previously (Reimers et al., 2006) and all experiments were conducted with fertilized embryos according to Oregon State University Institutional Animal Care and Use Protocols.
Chemical Exposures and Developmental Toxicity Assessment
Dibenzothiophene (>99%), pyrene (99%) and 1,2-benzanthracene (99%), were purchased from Sigma-Aldrich and dissolved in DMSO (J.T. Baker) at 50 mM, 50 mM and 25 mM concentrations, respectively. Embryos were cleaned, developmentally staged, and batch-exposed in glass vials at 6 hours post fertilization (hpf) (chorions intact) to PAHs or vehicle control with 1% final DMSO concentration in E2 embryo medium (Kimmel et al., 1995). For all experiments, exposures were conducted on a rocker and embryos were protected from light until experimental time points. For developmental toxicity experiments, PAH solutions were removed at 48 hpf and embryos were rinsed 4× and incubated in fresh embryo medium until 120 hpf, when they were assessed visually for malformations as previously described (Truong et al., 2011). Preliminary range-finding studies were conducted with each PAH and all further developmental toxicity assessments were conducted at 25 µM with 20 embryos per vial in 2 ml exposure solution. Microarray and uptake exposures were conducted with 40 embryos per vial in 4 ml solution.
Analysis of Developmental Toxicity Endpoints
Embryos were anesthetized with tricaine methanesulfonate and visually assessed at 120 hpf for yolk sac, axis, trunk, somite, fin, cardiac, eye, snout, jaw, otic vesicle, brain and pigment malformations. Mortality and the percentage of embryos with each malformation were calculated for each treatment group with the vial (20 embryos) as the experimental unit. The experiment was repeated 3 times. A generalized linear model (binomial distribution, logit link) one-way ANOVA was performed for the 8 endpoints which were observed in at least 3 embryos across all treatment groups. If the overall p-value indicated differences among the treatment percentages, individual comparisons were conducted using Tukey’s all pairwise post hoc test in R version 2.12.
Detection of PAH uptake in zebrafish embryos
Embryos were exposed to 0, 1, 5, 10 and 25 µM PAH (1% DMSO) solutions in glass vials as described previously, with 40 embryos per vial in 4 mL exposure solution. As with all exposures in this study, embryos were exposed at 6 hpf with chorions intact and incubated at 28°C on a rocker. Control embryos hatched just before 48 hpf; exposure to 10 and 25 µM PAH delayed hatching by 3–4 hours, but all treatment groups hatched on their own by 72 hpf. Because several exposure concentrations are above solubility for PAHs in embryo medium, PAH precipitate accumulated on the outside of the chorion. In order to measure the amount of PAH internalized by the embryos, chorions were removed immediately following exposure and before analysis as described below. For each biological replicate, 2 vials were combined after exposure.
For analysis at 24 hpf, embryos were rinsed with fish water and transferred to a clean glass petri dish. They were incubated in 82 µg/mL pronase (Sigma-Aldrich) at room temperature, gently agitated for 3 min, then rinsed thoroughly using an automated dechorionating system as previously described (Mandrell et al., 2012). Following rinsing, embryos were placed in a 28°C incubator for 20 minutes, after which >95% of chorions were removed by gentle agitation of the dish.
At 48 hpf, the majority of embryos had hatched and did not require batch dechorionation. They were chilled on ice to reduce activity, PAH solution was removed, and embryos were transferred to a clean glass petri dish with cold fish water. Chorions were removed from any remaining embryos with forceps, and embryos were gently agitated and rinsed 4X with fish water.
Immediately following dechorionation, 50 embryos from each treatment group were loaded into microcentrifuge tubes with approximately 80 mg 1 mm glass beads and placed on ice for at least 10 min. Embryos were homogenized in 500 µl ethyl acetate with a bullet blender (Next Advance, Averill Park, NY). Samples were then vortexed and incubated 15 min before centrifuging for 5 min at 16,000 RCF. 400 µl of supernatant was stored in amber vials at 4°C until analysis.
Percent PAH recovery for this method was calculated from 4 replicates of unexposed 24 and 48 hpf embryo samples loaded into microcentrifuge tubes as above and spiked with 12.5 µl PAH stock in DMSO. Samples were processed identically to experimental samples.
Zebrafish extracts were analyzed using an Agilent 5975B Gas Chromatograph-Mass Spectrometer (GC-MS) with a DB-5MS column (30 m × 0.25 mm × 0.25 µm) in electron impact mode (70 eV) using selective ion monitoring (SIM). The GC parameters were as follows: injection port maintained at 300 °C, 1.0 mL min−1 helium flow, 70 °C initial temperature, 1 min hold, 10 °C min−1 ramp to 300 °C, 4 min hold, 10 °C min−1 ramp to 310 °C, 4 min hold. The MS temperatures were operated at 150, 230 and 280 °C for the quadrupole, source and transfer line respectively. Standards for BAA, DBT and PYR (>97% purity) were purchased from Accustandard (New Haven, CT). Isotopically labeled chrysene-D12 and acenaphthylene-D8 were purchased from C/D/N incorporated (Quebec, Canada). A nine point calibration curve (10 pg/ µl to 10 ng/µl) was conducted to determine relative response ratios of PAHs to deuterated surrogate standards; chrysene-D12 was used as the deuterated surrogate for PYR (r2 = 0.9992) and BAA (r2 = 0.9982), acenaphthylene-D8 was used for DBT (r2 = 0.9991).
Calibration verification standards for target analytes, surrogates were analyzed at least every 22 samples and reported values within ±20% of the true value were considered to meet our data quality objectives (DQO). Only results from samples run between two calibration verifications that met the DQO were accepted; the majority were within ±10% of the true value. PAHs in all laboratory blanks (solvent-exposed embryos) were below detection except for 5 samples in which DBT and BAA were detected. This possible contamination was < 10% of the levels detected in our lowest exposure sample groups, and deemed negligible. Body burden (µmol/g embryo) was calculated using average embryo weights of 0.4 mg at 24 hpf and 0.3 mg at 48 hpf.
Pairwise comparisons were conducted between PAH-exposed samples and time-matched controls, as well between 24 and 48 hpf at each exposure concentration with Mann-Whitney Rank Sum tests using Sigmaplot software.
Microarray analysis of mRNA expression
Embryos batch-exposed in groups of 40 to 25 µM DBT, PYR, BAA or 1% DMSO control were homogenized in TriReagent (Molecular Research Center, Cincinnati, OH) at 24 and 48 hpf for RNA isolation. Four independent biological replicates were prepared for each treatment. Total RNA was isolated with phenol-chloroform extraction, and RNA was quantified and quality confirmed with a Nanodrop ND-1000 UV-Vis spectrophotometer and Agilent Bioanalyzer 2100. Microarray analysis was performed by the University of Wisconsin McArdle Laboratory of Cancer Research Microarray Facility. Briefly, cDNA was synthesized from 1.2 µg of total RNA from each sample and labeled with cy3 (experimental samples) or cy5 (pooled control sample) according to the Agilent protocol with minor modifications. Equal amounts of cy3 and cy5 labeled samples were mixed, fragmented, and hybridized to Agilent Zebrafish V2 array chips. Slides were scanned immediately with an Agilent microarray scanner (Agilent Technologies, Santa Clara, CA). Microarray files were submitted to the NCBI Gene Expression Omnibus, accession number GSE44130 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE44130.
Microarray analysis
Raw intensity data were processed by Agilent Feature Extraction software using Lowess normalization. Quality control analysis was performed on preprocessed data in GeneSpring v.11 (Silicon Genetics, Redwood City, CA) software using feature intensity distributions from Box-whisker plots to determine interquartile range span and median intensity value across the experiment. The intra-group versus between-group comparisons were made using correlation matrix plots, followed with principle components analysis to determine potential outliers. One biological replicate from the DBT 48 hpf treatment group was removed as an outlier, resulting in an N=3 for that treatment group. Normalized data were transformed to time-specific controls and analyzed by one-way ANOVA for unequal variances (Welch’s ANOVA) with Tukey’s posthoc test and 5% false discovery rate calculation (Benjamini and Hochberg, 1995). Values are reported as fold change (log2) in each treatment group compared to time-matched control. Correlation analysis between treatment groups was performed by linear regression of log2 fold change values, using the union of significant genes from both groups. Based on the significant correlation between DBT and PYR treatments at both time points, these datasets were further filtered to identify the subset of genes that were significantly different between them (p<0.05, 1.5-fold change). Genes that did not meet these criteria were considered similar between the DBT and PYR treatments for functional and transcription factor analysis.
Bioinformatic analysis
Unsupervised hierarchical clustering of microarray data was performed using Euclidean distance metric and centroid linkage clustering to group gene expression patterns by similarity. The clustering algorithms, heat map visualizations and centroid calculations were performed with Multi-Experiment Viewer (Saeed et al., 2003) software based on log2 expression ratio values. For downstream bioinformatic analysis, zebrafish identifiers on the Agilent platform were converted to human orthologs using Bioinformatics Resource Manager v. 2.3 (Tilton et al., 2012). Genes that did not have human orthologs were still included in the bioinformatic analysis using their zebrafish identifier. Both Metacore (GeneGO) and DAVID software recognize mixed identifiers (Entrez Gene ID) from human and zebrafish. Significant targets from the microarray and genes of interest are referred to by zebrafish gene identifiers, where zebrafish-derived information was available in the literature. Functional annotation and network information, however, was primarily derived from other species, and data for many genes of interest were only available in the mammalian literature; we present this information with human gene identifiers throughout the results and discussion. Functional enrichment statistics were determined using the DAVID functional annotation tool (Huang da et al., 2009), which utilizes the Fisher Exact test to measure gene enrichment in biological process Gene Ontology (GO) category terms for significant genes compared to background, which consisted of all genes on the Agilent platform. GO biological process categories from levels 3, 4, and 5 were included for enrichment calculation with a statistical cut-off of p<0.05. Since the DAVID functional annotation tool clusters GO terms by similarity to reduce redundancy, the biological processes are presented in the results with a representative process (p<0.05) from each significant cluster. To identify major transcriptional regulators of gene expression by PAHs, the Statistical Interactome tool was used in MetaCore to measure the interconnectedness of genes in the experimental dataset relative to all known interactions in the background dataset. Statistical significance of over-connected interactions was calculated using a hypergeometric distribution, where the p value represents the probability of a particular mapping arising by chance for experimental data compared to the background (Nikolsky et al., 2009). Networks were constructed in MetaCore for experimental data using an algorithm that identifies the shortest path to directly connect nodes in the dataset to transcription factors. Network visualizations were generated in Cytoscape (Shannon et al., 2003).
Quantitative RT-PCR
Validation of gene expression changes identified in the microarray analysis was conducted for a group of transcripts selected to represent differential regulation patterns by the three PAHs at 24 and 48 hpf. Gene-specific primers (MWG Operon) for qRT-PCR amplification are listed in Table S1. Sub aliquots of 10 µg total RNA from the microarray analysis were reverse transcribed using Superscript III (Invitrogen) according to manufacturer instructions. All qRT-PCR assays were performed in 20 µl reactions consisting of 10 µl Power SYBR Green PCR master mix (Applied Biosystems), 0.4 µl each primer, 9.2 µl H2O and 50 ng equivalents of cDNA. Amplification (Step One Plus, Applied Biosystems) was performed with cycling parameters as follows: 95°C for 10 min; 40 cycles of 95°C for 15 s, 60°C for 1 min; 95°C for 15 sec and 60°C for 1 min. A melt curve was performed at 3° increments to assess for multiple products. Relative fold change values in PAH-treated samples compared to vehicle controls were calculated for genes of interest, normalized to β-actin, by the method described by Pfaffl (Pfaffl, 2001). Three independent biological replicates were assessed and statistically analyzed by One-way ANOVA with Tukey’s post-hoc test using Sigmaplot software.
Results and Discussion
Dibenzothiophene, pyrene, and benz(a)anthracene induce developmental toxicity in zebrafish embryos
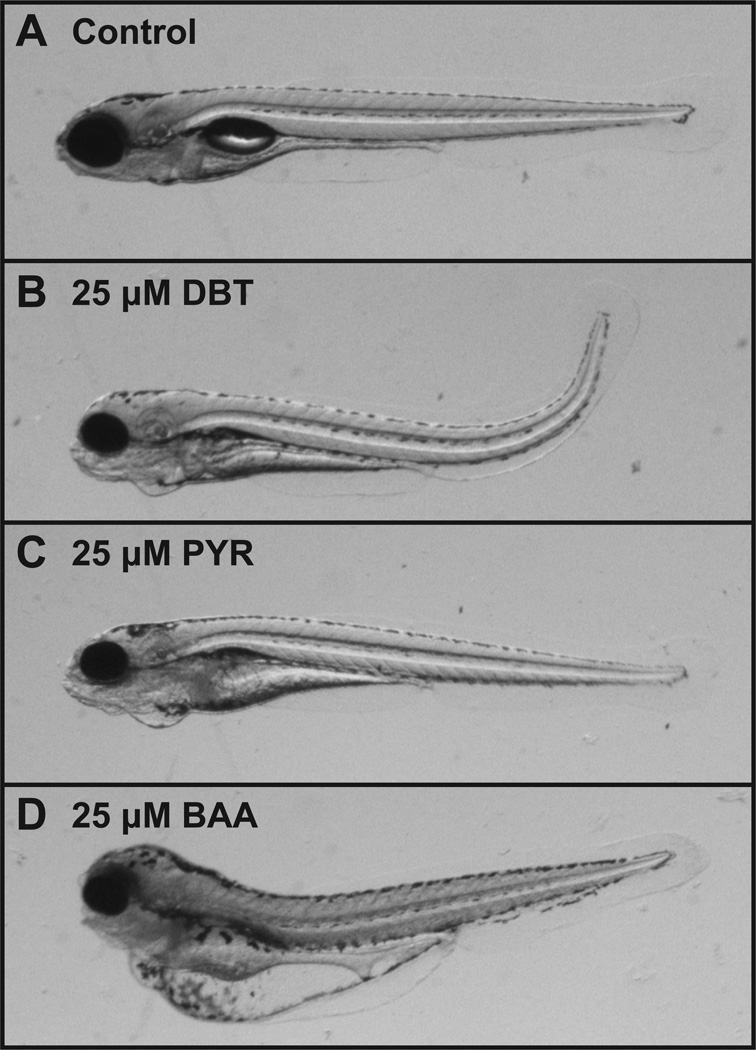
Exposure to DBT, PYR or BAA caused a significant increase in the incidence of abnormal embryos compared to the vehicle control exposure at 120 hpf. All three compounds induced pericardial edema, snout and jaw malformations (Figure 1). PYR and BAA exposures caused significant increases in yolk sac edema, while DBT did not. DBT, however, induced distinct axis malformations (Figure 1B) which were not present in BAA- or PYR-exposed embryos (Tables 1, 2). Interestingly, the 25 µM concentration of all three PAHs induced malformations in >80% of embryos by 120 hpf, while mortality was < 10%, not significantly different from control (Table 1). The phenotypes induced by these PAHs suggested different underlying pathologies. BAA induced more severe edema, while necrotic tissue was observed in PYR-exposed embryos, particularly in the anterior yolk sac, liver and digestive tract (Figure 1C); these embryos did not survive more than a couple hours past 120 hpf. This was observed previously by Incardona et al, who demonstrated that DBT, PYR and BAA induced malformations in zebrafish that were differentially dependent on activation of the AHR and metabolism by Cyp1a (Incardona et al., 2004; Incardona et al., 2005). The differential proposed mechanisms of these PAHs presented an ideal opportunity to investigate the diversity of molecular pathways that lead to developmental effects of PAH exposure. The objective of this study was not to mimic environmental exposures, but rather to identify molecular pathways that precede morphological changes induced by different PAH structures. Based on our developmental toxicity data, 25 µM was identified as an appropriate concentration for microarray analysis of early gene expression changes elicited by DBT, PYR and BAA exposure.
Figure 1.
Representative images of 120 hpf larvae after exposure to (A) 1% DMSO control, (B) 25 µM DBT, (C) 25 µM PYR, and (D) 25 µM BAA from 6–48 hpf.
Table 1.
Mean percentage of embryos (95 percentile) with malformations observed at 120 hpf following exposure to 25 µM BAA, DBT, PYR or DMSO control from 6–48 hpf.
| Effect |
||||||||
|---|---|---|---|---|---|---|---|---|
| Treatment | Mortality | Axis | Eye | Jaw | Pericardial Edema |
Snout | Yolk Sac | Any Malformation |
| Control | 4 (1, 13) | 3.1 (1, 10) | 4.2 (1, 16) | 7.3 (2, 21) | 4.2 (1, 13) | 4.2 (1, 15) | 4.2 (1, 15) | 11 (5, 22) |
| DBT | 3.3 (0, 58) | 44.8 (9, 87)* | 15.5 (1, 78) | 75.9 (21, 97)* | 55.2 (12, 92)* | 19 (2, 76)* | 15.5 (1, 75) | 83.3 (39, 97)* |
| PYR | 5 (0, 57) | 12.3 (1, 61) | 5.3 (0, 72) | 98.3 (33, 100)* | 68.4 (19, 95)* | 22.8 (2, 79)* | 36.8 (4, 88)* | 98.3 (33, 100)* |
| BAA | 1.7 (0, 67) | 8.5 (1, 55) | 10.2 (0, 74) | 81.4 (25, 98)* | 74.6 (23, 97)* | 32.2 (4, 85)* | 54.2 (9, 94)* | 85 (41, 98)* |
Significantly different than DMSO Control, p < 0.05, one-way ANOVA with Dunnett's post-hoc test.
Table 2.
Comparison of malformations induced by individual PAH treatments.
| Test | Effect | |||||||
|---|---|---|---|---|---|---|---|---|
| Mortality | Axis | Eye | Jaw | Pericardial Edema |
Snout | Yolk Sac | Any Malformation |
|
| DBT v Control | 8.53E-01 | 4.30E-06 | 1.41E-01 | 2.70E-06 | 2.50E-06 | 4.66E-02 | 1.01E-01 | 7.20E-09 |
| PYR v Control | 7.09E-01 | 6.02E-02 | 6.53E-01 | 1.50E-04 | 3.10E-08 | 2.22E-02 | 1.87E-03 | 2.10E-05 |
| BAA v Control | 9.47E-01 | 1.83E-01 | 3.31E-01 | 8.20E-07 | 4.60E-09 | 3.44E-03 | 3.10E-05 | 6.10E-09 |
| DBT v PYR | 9.31E-01 | 1.91E-03 | 4.84E-01 | 1.75E-01 | 4.63E-01 | 9.27E-01 | 1.67E-01 | 1.85E-01 |
| BAA v DBT | 8.93E-01 | 5.10E-04 | 8.34E-01 | 8.83E-01 | 1.80E-01 | 4.59E-01 | 8.26E-03 | 9.80E-01 |
| BAA v PYR | 7.23E-01 | 8.07E-01 | 7.93E-01 | 2.58E-01 | 8.22E-01 | 6.87E-01 | 3.78E-01 | 2.21E-01 |
p values of Tukey’s all pairwise post hoc test are displayed for each comparison, malformations that were significantly different between treatment groups are shaded (p < 0.05).
Zebrafish exposed to dibenzothiophene, pyrene and benz(a)anthracene in embryo medium accumulate differential PAH body burdens
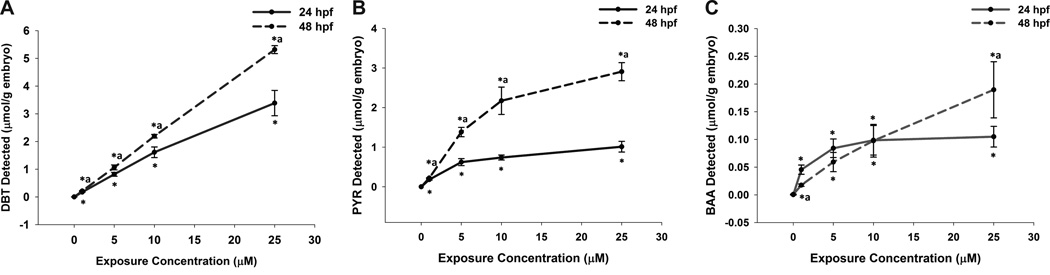
The internal body burden of PAH in embryos was measured after exposures as described for the microarray analysis. To allow for structure-toxicity comparisons between the three compounds, as well as to relate gene expression data to other model systems, DBT, PYR and BAA were detected by GC-MS in zebrafish embryos exposed to a range of concentrations (1–25 µM). PAH recovery averaged between 80–125% (Table S2). Measured PAH values were therefore reported as detected in experimental samples, unadjusted for recovery. The amount of PAH detected in embryos revealed stark differences between the three PAH structures. At all concentrations and time points, DBT body burden in embryos was the highest, averaging 3.4 and 5.3 µmol/g at 24 and 48 hpf, respectively, following exposure to 25 µM DBT (Figure 2A). DBT had the highest solubility in water, and uptake did not plateau in the range of concentrations tested here. PAH body burden of embryos exposed to 25 µM PYR averaged 1.0 and 2.9 µmol/g at 24 hpf and 48 hpf, respectively, and uptake appeared to reach a plateau, likely because of low compound solubility in embryo medium (Figure 2B). BAA body burden was markedly lower than the other two PAHs at all exposure concentrations. An apparent saturation was reached at 0.10 µmol/g embryo at 24 hpf, an order of magnitude lower than PYR (Figure 2C). Water solubility of BAA was the lowest of these PAHs, and the high concentrations employed in this study were above solubility with 1% DMSO in embryo medium. At 48 hpf, BAA concentrations averaged 0.19 µmol/g in embryos from the 25 µM exposure group.
Figure 2.
PAH detected in embryos exposed to (A) DBT, (B) PYR, and (C) BAA from 6 until 24 (solid lines) or 48 (dashed lines) hpf. *Significantly different than time-matched DMSO control (Mann-Whitney rank sum test, p < 0.05). aSignificant difference between 48 and 24 hpf samples at the same exposure concentration (Mann-Whitney rank sum test, p < 0.05).
Log Kow values of these compounds are 4.38 (DBT), 4.88 (PYR) and 5.79 (BAA) (Hansch, 1995). Studies of early-life exposure to PAHs showed that the bioconcentration factor (BCF) in fish embryos correlated with Kow (Mathew et al., 2008). BAA would therefore be predicted to have the highest BCF of the PAHs in our study. BCFs have primarily been calculated for larvae (post-hatch), however, and the short duration of acute exposures employed in our study did not allow steady-states to be achieved. Steady-state PAH concentrations were similarly not attained in zebrafish eggs in a study reported by Petersen and colleagues (Petersen and Kristensen, 1998). Metabolism could also explain differences in parent PAH concentration, but is expected to be low during the developmental stages chosen for gene expression analysis in this study (Petersen and Kristensen, 1998). Metabolism increases upon hatching in Atlantic killifish embryos, and zebrafish exhibit greater inducibility of cyp1a starting at 48 hpf, the approximate time of hatching in our laboratory (Binder and Stegeman, 1984; Andreasen et al., 2002). While some metabolism could potentially explain the small decrease in BAA concentration between 24 and 48 hpf at the lower concentrations, it is unlikely to explain the large difference in body burdens observed between PAH structures. Differences between PAHs in this study appear to be driven by their solubility in embryo medium, rather than their BCFs or metabolism. The 25 µM exposures for the microarray study represent an acute, relatively high-dose exposure intended to identify mRNA expression changes that precede appearance of morphological abnormalities. While total dose and maximum exposure calculations were beyond the scope of this study, the measurement of parent PAH in the embryos at the time of gene expression analysis provided important information for mechanistic comparison between the PAHs.
mRNA expression profiles induced by PAH exposure are different at 24 and 48 hours post fertilization
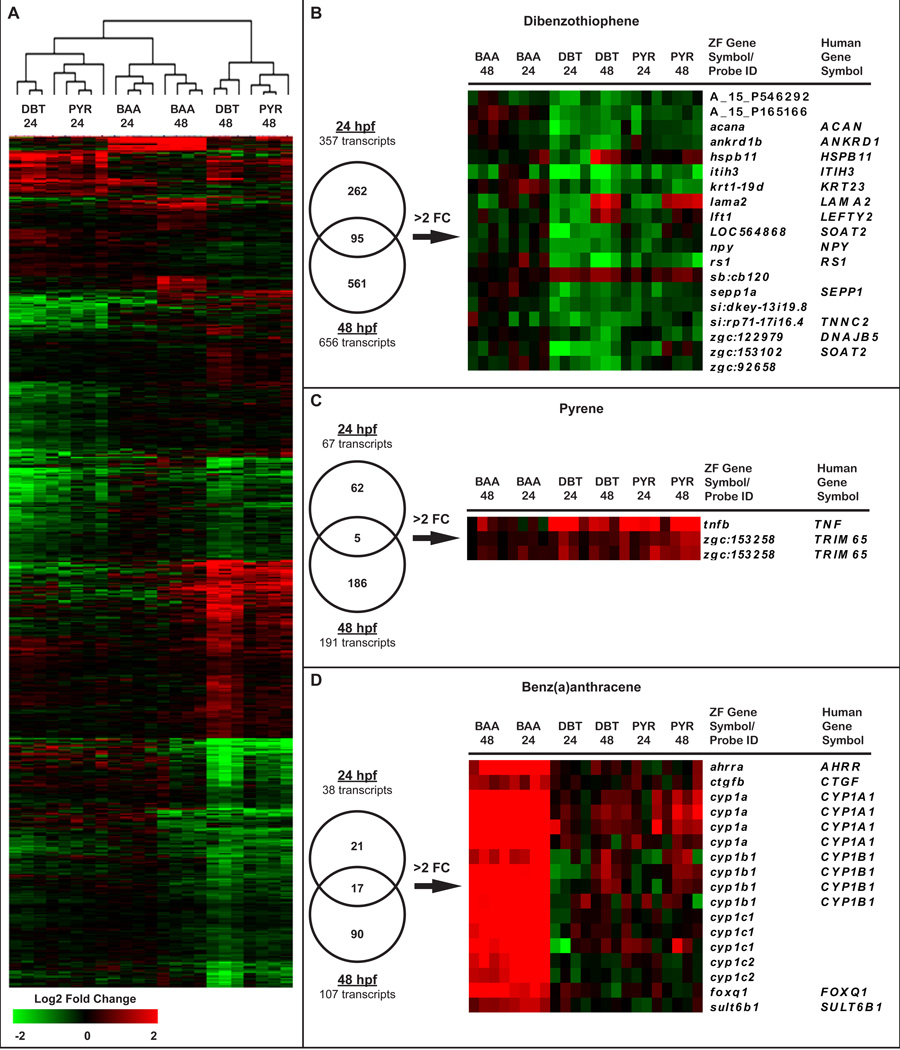
Pairwise analysis of variance across all exposure groups identified significant expression changes in 1079 transcripts compared to time-matched controls (Table S3). Entrez or Ensdart IDs were identified for 935 of these in the Ensembl zebrafish genome assembly (Zv9). Unsupervised bidirectional clustering of all experimental groups indicated a strong developmental time point effect, and revealed unique gene expression patterns in response to the three PAHs. At 24 hpf, DBT and PYR exposure groups clustered closely, while BAA induced a strikingly different expression pattern (Figure 3A, D). DBT and PYR exposure groups also clustered at 48 hpf, but with distinct separation from the 24 hpf samples and with a notably larger group of down-regulated transcripts. The expression profile induced by BAA at 48 hpf clustered more closely with 24 hpf BAA than with the other 48 hpf PAH samples (Figure 3A). At 24 hpf, DBT, PYR and BAA exposures induced significant changes in 357, 67 and 38 transcripts, respectively. As reflected in the heatmap, more transcripts were differentially expressed at 48 hpf, but relative quantities of differentially expressed transcripts were maintained; DBT induced changes in 656, PYR in 191 and BAA in 107 transcripts (Figure 3B–D). Fifteen genes that were significantly differentially regulated by at least one of the PAHs were selected for QPCR validation of the differential regulatory patterns observed in the array. PAH- and time-dependent expression changes were confirmed for the majority of genes examined (Table 3). Fold change values were similar between the microarray and QPCR for most genes, demonstrating good reliability of the microarray for identifying meaningful changes in gene expression induced by PAH exposure. mstnb, which was identified as a significantly decreased transcript at 24 hpf, was not decreased in BAA-exposed samples analyzed by QPCR. Upon further investigation, however, we identified 5 probes on the Agilent array that target mstnb, only one of which identified a significant expression difference (p value 0.04). This suggested nonspecificity of that probe for mstnb, or potentially differential splicing. As others have reported previously, we observed lower correlation between the microarray and QPCR for down-regulated transcripts; QPCR identified fewer changes that met statistical significance (p < 0.05), but trends in regulation were consistent (Morey et al., 2006).
Figure 3.
A) Bidirectional hierarchical clustering heatmap of log2 fold change (FC) values of all 1079 genes significantly differentially expressed compared to control (ANOVA, p < 0.05). Comparison of significant genes (p<0.05) between 24 and 48 hpf for (B) DBT, (C) PYR and (D) BAA exposure groups are shown by Venn diagram. Heatmap enlargements show transcripts differentially expressed (p<0.05, >2-FC) at both time points for each PAH.
Table 3.
Mean log2 fold change values of differentially regulated transcripts from the microarray, compared with log2 fold change (mean ± SD) detected with QPCR.
| 24 hpf | 48 hpf | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe ID | Ensembl Transcript ID | Zebrafish Gene Symbol |
Human Gene Symbol |
DBT Array log2 FC |
DBT QPCR log2 FC |
PYR Array log2 FC |
PYR QPCR log2 FC |
BAA Array log2 FC |
BAA QPCR log2 FC |
DBT Array log2 FC |
DBT QPCR log2 FC |
PYR Array log2 FC |
PYR QPCR log2 FC |
BAA Array log2 FC |
BAA QPCR log2 FC |
| A_15_P212366 | ENSDART00000010918 | agt | AGT | 2.6a | 2.71 ± 0.71b | 1.5 | 2.07 ± 1.25 | −0.02 | 0.05 ± 0.87 | --- | --- | --- | --- | --- | --- |
| A_15_P104804 | ENSDART00000105896 | atp2a1l | ATP2A1 | −1.13a | −1.82 ± 0.67 | −0.58 | −0.85 ± 0.76 | −0.12 | 0.06 ± 0.85 | --- | --- | --- | --- | --- | --- |
| A_15_P120653 | ENSDART00000077511 | ccr9a | CCR9 | 0.04 | 0.41 ± 0.95 | 0.17 | 0.78 ± 0.53 | 0.73 | 1.65 ± 0.49 | −0.23 | 0.2 ± 0.49 | −0.05 | −0.24 ± 0.52 | 1.75a | 1.3 ± 0.27a |
| A_15_P170406 | ENSDART00000066439 | ch25h | CH25H | 2.21 | 2.71 ± 1.28b | 2.6a | 2.95 ± 1.09b | 0.07 | 0.96 ± 0.53 | --- | --- | --- | --- | --- | --- |
| A_15_P140876 | ENSDART00000017756 | ctsl.1 | CTSL1 | --- | --- | --- | --- | --- | --- | 0.47 | −0.17 ± 0.35 | −0.08 | −0.39 ± 0.09 | 2.08a | 1.68 ± 0.16b |
| A_15_P131751 | ENSDART00000038200 | cyp1a | CYP1A1 | 0.61 | −0.03 ± 0.47 | 0.73 | 0.47 ± 0.54 | 4.11a | 4.77 ± 0.43b | 0.25 | 0.81 ± 0.23 | 1.1 | 1.43 ± 0.47b | 5.11a | 6.63 ± 0.32b |
| A_15_P554522 | ENSDART00000099870 | cyp1b1 | CYP1B1 | 0.24 | −0.79 ± 0.24 | 0.33 | −0.52 ± 0.51 | 2.87a | 2.37 ± 0.34b | −0.18 | 0.55 ± 0.33 | 0.02 | 0.05 ± 0.61 | 2.46a | 2.55 ± 0.25b |
| A_15_P130722 | ENSDART00000019953 | cyp1c1 | −0.78 | −0.34 ± 0.37 | 0.53 | −0.05 ± 0.53 | 2.49a | 2.22 ± 0.41b | 0.18 | −0.22 ± 0.18 | 0.65 | −0.23 ± 0.34 | 4.67a | 4.14 ± 0.44b | |
| A_15_P287511 | ENSDART00000016487 | cyp1c2 | −0.21 | −0.29 ± 0.21 | 0.31 | 0 ± 0.19 | 1.74a | 1.59 ± 0.25b | −0.37 | −0.17 ± 0.27 | −0.23 | −0.3 ± 0.3 | 2.51a | 3.12 ± 0.34b | |
| A_15_P402330 | ENSDART00000109464 | g0s2 | G0S2 | −1.32a | −1.62 ± 0.38b | −0.77 | −0.47 ± 0.29 | −0.22 | 0.09 ± 0.23 | −0.21 | −0.38 ± 0.52 | −1.02a | −0.75 ± 0.4 | −1.06a | −0.61 ± 0.22 |
| A_15_P175986 | ENSDART00000100386 | mstnb | MSTN | −1.84a | −0.66 ± 0.1 | −0.64 | −0.16 ± 0.27 | −1.5a | −0.3 ± 0.24 | --- | --- | --- | --- | --- | --- |
| A_15_P168906 | ENSDART00000103784 | si:ch211-202b2.2 | EDN2 | --- | --- | --- | --- | --- | --- | −0.02 | −0.01 ± 0.14 | −0.34 | 0.03 ± 0.31 | −0.94a | −0.75 ± 0.18b |
| A_15_P143451 | ENSDART00000025669 | slc16a9b | SLC16A9 | 2.96a | 3.31 ± 0.3b | 2.59a | 2.9 ± 0.42b | 0.16 | 0.28 ± 0.84 | --- | --- | --- | --- | --- | --- |
| A_15_P153096 | ENSDART00000130131 | sult6b1 | SULT6B1 | 0.06 | 0.11 ± 0.09 | −0.2 | 0.18 ± 0.29 | 1.02a | 0.96 ± 0.22 | 0.03 | −0.27 ± 0.26 | 0.04 | −0.28 ± 0.18 | 1.42a | 1.03 ± 0.24b |
| A_15_P206556 | ENSDART00000017569 | tnfb | TNF | 2.74a | 2.88 ± 1.02b | 3.2a | 3.2 ± 0.63b | 0.59 | 0 ± 0.15 | 1.04 | 0.82 ± 0.24 | 2.47a | 2.3 ± 0.66b | 0.05 | −0.17 ± 0.17 |
Significantly different from vehicle control (One-way ANOVA with Tukey’s post hoc test and 5% FDR, p<0.05)
Significantly different from vehicle control (One-way ANOVA with Tukey’s post hoc test, p< 0.05).
For each PAH, we compared transcripts significantly differentially expressed at 24 hpf with transcripts that were significant at 48 hpf. These experimental time points encompass a period of rapid development in zebrafish, during which fin morphogenesis begins, the circulatory system forms, tactile sensitivity and swimming behavior are initiated, and pigment develops (Kimmel et al., 1995). Developmental progression is reflected in broad changes in endogenous gene expression between the time points, unrelated to chemical exposure, and by the stronger influence of time points than the different PAH structures in the bidirectional clustering (Figure 3A).
While 95 transcripts were misregulated by DBT at both 24 and 48 hpf, they represented only 27% of the 24 hpf significant gene set, and did not include the most highly misexpressed transcripts from either time point. The most differentially expressed transcripts across both time points were acana, ankrd1b and hspb11 (Figure 3B). ankrd1b and hspb11 are both involved in myogenesis; hspb11 is specifically expressed in muscle pioneers, up-regulated by intracellular calcium, and involved in muscle fiber organization in developing zebrafish (Kluver et al., 2011; Kojic et al., 2011). Expression of three genes, hspb11, lama2, and lft1, was reduced at 24 hpf, but significantly elevated at 48 hpf.
PYR induced a modest transcriptional response which, similar to DBT, had a low percentage of transcripts conserved between 24 and 48 hpf. The two significant genes that were differentially expressed >2 fold were zgc:153258 (uncharacterized transcript) and tnfb, a member of the tumor necrosis factor family of proinflammatory cytokines (Figure 3C). These robust responses, conserved over time, represent potential biomarkers of exposure to the individual PAHs. The transcripts with the largest fold changes were different at 24 and 48 hpf, however, which suggested that separate analysis of the time points could provide better insight into mechanisms driving the DBT-PYR response.
The BAA transcriptional response is consistent from 24 to 48 hpf, and distinct from responses induced by DBT and PYR
In contrast to DBT and PYR, 45% of transcripts differentially expressed by BAA at 24 hpf were also significant at 48 hpf, and those with the largest fold changes were conserved between time points (Figure 3D). The most highly misexpressed genes at 24 hpf were cyp1a, cyp1b1, cyp1c1, cyp1c2, ahrra and foxq1. All of these genes were elevated and, along with sult6b1 and ctgfb, remained elevated at 48 hpf. The cyp1 genes and aryl-hydrocarbon receptor repressor (ahrra) are well-known targets of AHR. sult6b1 is a recently identified sulfotransferase with unknown specific function, and ctgfb is a member of the CCN family of intercellular signaling proteins, hypothesized to antagonize WNT signaling when overexpressed (Fernando et al., 2010). Together, these genes represent a consistent signature of the transcriptional response to BAA exposure in zebrafish embryos from 24 to 48 hpf.
We conducted between-PAH comparisons separately at 24 and 48 hpf to identify significant transcripts unique to each PAH exposure. Though BAA exposures induced the smallest number of significant transcriptional changes, they were highly induced and formed a distinct cluster (Figure 3D) that overlapped minimally with the DBT and PYR transcriptional profiles. Only 7 of the significant genes in the 24 hpf BAA exposure group were similarly differentially expressed in response to DBT or PYR. At 48 hpf, the BAA expression pattern remained distinct, where only 27 of the 107 differentially expressed transcripts were similarly regulated by another PAH. The entire set of significant BAA transcripts was therefore used for analysis of pathways and biological functions disrupted by BAA exposure at 24 and 48 hpf (discussed below).
DBT and PYR induce a similar dose-dependent transcriptional profile
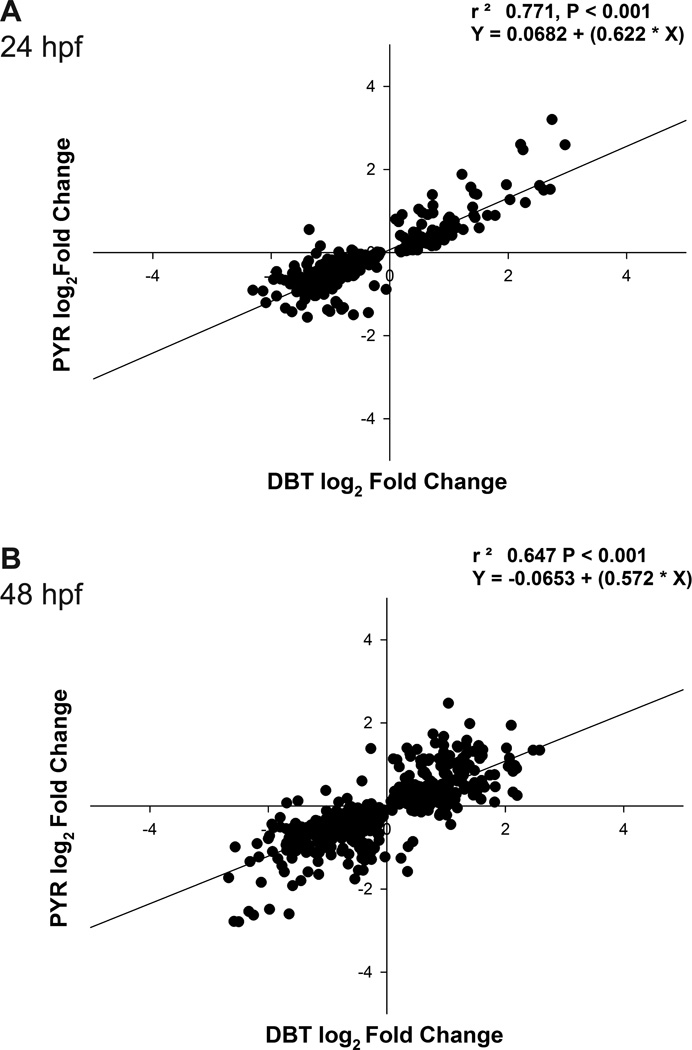
Common patterns in gene expression between DBT and PYR exposure groups are apparent in the heatmap in Figure 3A at both time points; however, the magnitude of the PYR-induced transcriptional response is visibly lower. This trend was reflected in linear regression analysis of PYR vs. DBT log2 expression values for all genes significantly misregulated by either compound at 24 hpf (Figure 4A). A strong positive correlation was observed between PYR and DBT log2 expression values (r2 = 0.77, p < 0.001). The regression slope, however, demonstrated that DBT-induced expression changes were on average 1.6 fold greater than PYR-induced changes in these transcripts. This apparent dose-effect suggested that solubility and uptake may have been the primary drivers of differential transcriptional responses between these compounds rather than unique molecular targets. Exaggerated differences in response were inferred from the ANOVA analysis because many PYR-induced transcriptional changes did not reach statistical significance (p< 0.05 compared to control). The dose-effect is supported by the uptake data, which showed that following the 25 µM exposure, DBT body burdens were 3.4 times higher than PYR body burdens at 24 hpf. This trend persisted with a similar correlation at 48 hpf (r2 = 0.647, p< 0.001), wherein DBT on average induced 1.75-fold greater expression changes than PYR (Figure 4B). DBT body burdens were 1.8 times greater than PYR at that time point. Because of this correlation, we employed a direct statistical comparison of DBT and PYR to better define the conserved transcriptional response, as well as identify transcripts with meaningful expression differences between the two groups.
Figure 4.
Comparison of gene expression between PYR and DBT treatment groups at 24 and 48 hpf. Linear regressions of log2 FC values for the union of transcripts significantly (p<0.05) misregulated by DBT or PYR compared to control (n = 712). Linear associations at (A) 24 hpf and (B) 48 hpf were both significant (p < 0.001).
Direct pairwise comparison of DBT and PYR log2 FC values at 24 hpf identified 343 similarly expressed transcripts, and only 42 that were significantly different (p< 0.05). At 48 hpf, 139 were significantly different, while 572 transcripts were similar between the two PAHs. Because of the overwhelming conservation of response, functional analysis was performed using the set of similarly expressed genes at each time point to identify biological processes disrupted by DBT and PYR exposure. We focused further mechanistic analysis on the DBT-PYR vs. the BAA responses.
Disruption of ion transport, muscle function, and metabolism by DBT and PYR at 24 hpf
Of the 343 transcripts representing the conserved DBT and PYR responses at 24 hpf, approximately 70% were under-expressed compared to control. 308 had sufficient annotation, which translated to 256 unique DAVID IDs. Fatty acid biosynthesis, ion transport, skeletal muscle contraction, steroid biosynthesis and oxoacid metabolism were the most enriched of the 12 significant biological processes identified by DAVID functional analysis, which together depict wide disruption of molecular signaling by 24 hpf (Table 4). We used the MetaCore Statistical Interactome tool to identify major transcription factors predicted to regulate significant genes in this dataset. JUN, RELA, SP1, PPARA, RXRA, ESR1, ESR2, and NR3C1 (glucocorticoid receptor) were predicted to regulate the largest sets of differentially expressed transcripts (Table S4). There was considerable overlap of the predicted targets of these transcription factors, which included approximately equal numbers of up and down-regulated transcripts. The extensive molecular responses to these PAHs highlighted a complex network of regulatory processes involved in normal embryo development that were unlikely to be mediated through one primary transcription factor but rather were responsive to chemical-induced perturbations such as oxidative stress, inflammation, altered metabolism and disruption of ion balance and cardiac function.
Table 4.
Significantly enriched biological functions identified by DAVID analysis of all transcripts differentially regulated (p<0.05) by BAA exposure or by DBT and PYR at 24 hpf.
| Biological Process | GO Term | Downregulated genes | Upregulated genes | E Score | % | P Value | |
|---|---|---|---|---|---|---|---|
| BAA | hormone metabolic process | GO:0042445 | cyp1a, cyp1b1, cyp1c1, cyp1c2, si:dkey-94e7.2 | 2.06 | 15.79 | 5.12E-03 | |
| tissue development | GO:0009888 | mstnb | foxq1l, ptn, ctgfb | 1.21 | 21.05 | 2.77E-02 | |
| DBT and PYR | fatty acid biosynthetic process | GO:0006633 | elovl6, fads2, ptgds, si:ch73-131e21.5, tpi1b | ch25h, elovl7a | 2.67 | 3.02 | 6.10E-04 |
| ion transport | GO:0006811 | atp2a1l, cpt1b, gabra1, grin1b, KCNAB1, kcnip1b, kcnip3, LOC100004247, rhbg, sfxn4, si:ch211-195b13.1, si:ch211-221p4.4, slc24a5, zgc:101827, zgc:113361, zgc:158296 | LOC571584, si:ch211-244h7.4, slc22a18, slc31a1, tmem38b, zgc:162356, zgc:162495 | 2.32 | 8.30 | 7.86E-03 | |
| skeletal muscle contraction | GO:0003009 | homer1, mb, si:rp71-17i16.4, tnni2b.2 | 2.18 | 1.51 | 1.10E-03 | ||
| steroid biosynthetic process | GO:0006694 | cyp17a1, hmgcs1, hsd17b7, lss, nsdhl, rdh8l | ch25h, dhcr7 | 2.14 | 3.02 | 9.43E-04 | |
| oxoacid metabolic process | GO:0043436 | acsf3, cpt1b, elovl6, fabp11b, fads2, ghra, hibadhb, mdh1b, ptgds, rbp1a, rnpep, si:ch73-131e21.5, tpi1b, tyrp1b, zgc:113076, zgc:154046 | ch25h, elovl7a, mthfd1 | 1.93 | 7.17 | 1.27E-02 | |
| intermediate filament organization | GO:0045109 | dnajb6b, krt1-19d, krt23, nefm | 1.80 | 1.13 | 6.71E-03 | ||
| negative regulation of cell proliferation | GO:0008285 | bdnf, cd9a, cx43, smad3b, tnfrsf9a, wfdc1, zgc:114127, zgc:158296 | agt, msxe, notch2, tbx16, tnfb | 1.77 | 4.91 | 1.67E-02 | |
| muscle cell development | GO:0055001 | homer1, LOC796577, myoz1a, zgc:158296 | myog | 1.71 | 1.89 | 1.89E-02 | |
| sterol biosynthetic process | GO:0016126 | hmgcs1, lss, nsdhl | ch25h, dhcr7 | 1.56 | 1.89 | 5.49E-03 | |
| cellular amide metabolic process | GO:0043603 | ghra, hibadhb, mdh1b, tpi1b, zgc:113076 | 1.41 | 1.89 | 2.64E-02 | ||
| monosaccharide catabolic process | GO:0046365 | hibadhb, mdh1b, pfkma, tpi1b, zgc:162337 | 1.28 | 1.89 | 3.16E-02 | ||
| regulation of erythrocyte differentiation | GO:0045646 | inhbaa, mafba, spi1l | 1.09 | 1.13 | 4.65E-02 |
E score: overall cluster enrichment score, %: percentage of total gene list involved in functional cluster, p-values determined by modified Fischer’s Exact test (EASE score).
The ion transport biological process contained the largest number of misregulated transcripts at 24 hpf, and was composed of a diversity of both under- and over-expressed transporters (Table 4). These are discussed further below, as disrupted ion transport was common to all PAH exposures in this study. Ion balance is important for muscle development and function, which was also significantly affected by DBT and PYR at 24 hpf. Deficiency in the Ca2+ transporter atp2a1, for example, causes zebrafish to develop abnormal musculature (Gleason et al., 2004). Transcripts in the skeletal muscle contraction and muscle cell development biological processes were primarily under-expressed, and included myoglobin, which is also required for angiogenesis in zebrafish (Table 4)(Vlecken et al., 2009). Members of the ion transport cluster may interact with these transcripts or themselves be important for muscle and cardiac function in the context of zebrafish development.
Down-regulated genes similarly predominated in the significant fatty acid biosynthesis, steroid biosynthesis and oxoacid metabolism biological processes (Table 4). A notable exception was cholesterol 25-hydroxylase (ch25h), which encodes a cholesterol metabolizing enzyme involved in the inflammatory response, and was elevated > 4 fold by both DBT and PYR (Park and Scott, 2010). The functions of these genes and their roles during development have yet to be characterized in zebrafish, but together they highlight disruption of metabolic processes.
Transcription factors RELA and JUN are predicted to regulate renin-angiotensin system-related genes misexpressed in DBT and PYR exposed embryos
In contrast to the previously-discussed biological processes, the majority of genes in the negative regulation of cell proliferation process (Table 4) have known roles in zebrafish development, and many are predicted downstream targets of the NF-κB family member RELA (Table S4; bdnf, tnfrsf9a, zgc:114127, agt, msxe, tnfb) as well as JUN (Table S4; cx43, smad3b, tnfrsf9a, agt, tnfb) (Rutenberg et al., 2006; Jia et al., 2008; Jiang et al., 2010; Mueller et al., 2010; Zuniga et al., 2011). These processes are likely mediated through multiple interacting transcription factors. Of the 15 transcription factors that were significant at 24 hpf, RELA and JUN were predicted to regulate the most highly induced genes in the DBT-PYR dataset agt and tnfb, as well as many other genes involved in the significant biological processes. The inflammatory cytokine tnfb (also known as tnf2) is one of two TNF homologs in zebrafish, both of which are highly induced in larvae in response to LPS stimulation (Wiens and Glenney, 2011). Angiotensinogen (AGT) is the precursor of angiotensin (ANG II), a potent regulator of blood pressure and water homeostasis in the Renin-Angiotensin (RAS) signaling network (Wu et al., 2011a). Transcription of AGT is induced by glucocorticoids through the glucocorticoid receptor, as well as by TNF and other inflammatory cytokines via activation of NF-κB (Brasier and Li, 1996). ANG II induces AGT transcription in a positive feedback loop involving NF-κB, and also induces JUN activation via JNK signaling in cardiac myocytes and vascular smooth muscle cells (Brasier et al., 2000) (Brasier and Li, 1996). As an initiator of tissue inflammation, TNF activates NF-κB, induces inflammatory and anti-apoptotic gene expression in a cell-type dependent manner, and also activates JUN via JNK (Tian et al., 2005). In this study, DBT and PYR exposure led to increased expression of tnfb and complement component 7, along with macrophage-related genes mpeg1 and mst1, all of which are involved in innate inflammatory response. Both Tnf and Agt are expressed in the myocardium of rats following ischemia, remodel ATP-dependent calcium channels, and have been implicated in atherosclerosis and hypertension (Isidoro Tavares et al., 2009). In developing rat embryos, activation of angiotensin receptors with exogenous ANG II disrupts cardiac looping (Price et al., 1997). These two genes, in combination with the significant enrichment of other genes downstream of RELA, JUN, and the glucocorticoid receptor, suggest that inflammatory response and RAS signaling may play a role in the cascade of effects observed in response to DBT and PYR exposure. Cardiovascular functions of the RAS system are conserved in teleosts, and agt was similarly increased along with acute phase response genes in 24 hpf zebrafish exposed to mercury (Le Mevel et al., 2008; Ung et al., 2010). Though not yet explored in zebrafish embryos, the RAS system has been identified as important for fetal cardiovascular response, body fluid balance, and neuroendocrine regulation, and may be involved in fetal programming of hypertension later in life (Mao et al., 2009).
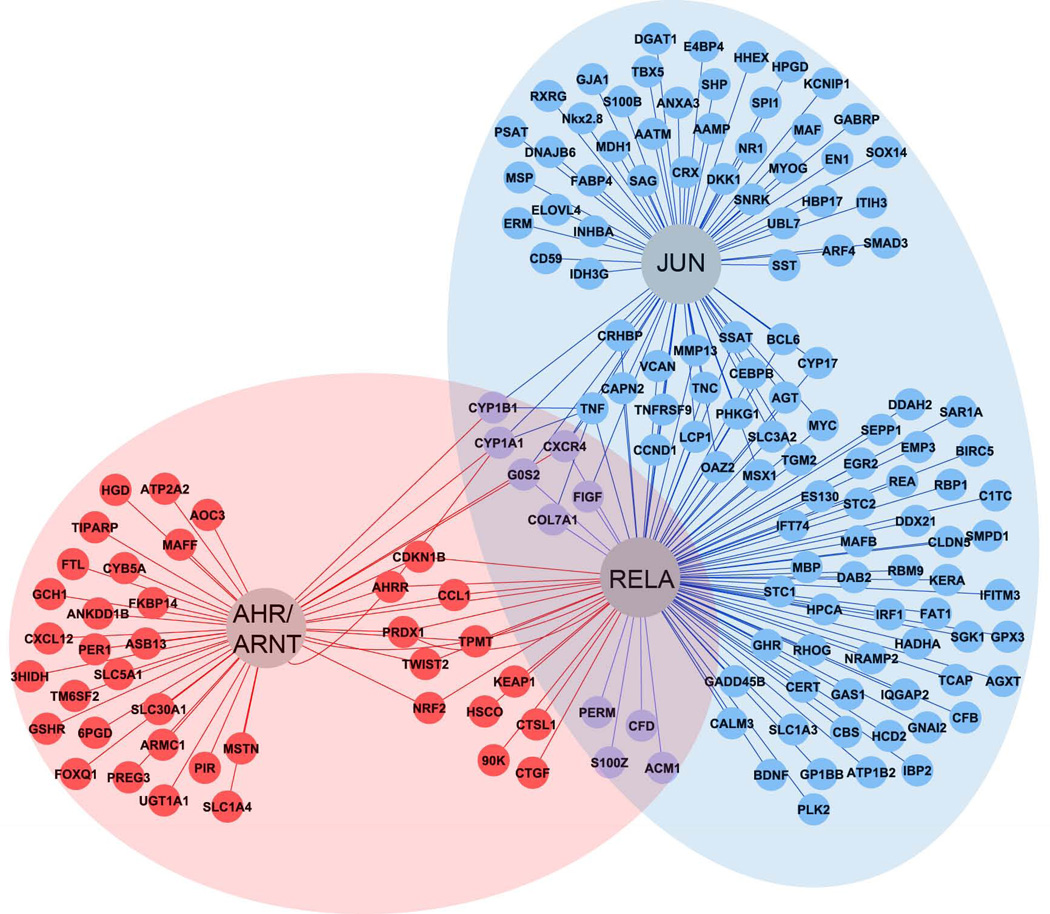
We created a map of key predicted transcription factors, including RELA and JUN, and their downstream targets that were significantly misregulated in the three PAH exposure groups at 24 or 48 hpf (Figure 5). A substantial number of transcripts, including TNF and AGT, are predicted to be regulated by both transcription factors, but RELA is predicted to regulate the largest number of genes that were induced by DBT/PYR exposure at 24 or 48 hpf. While RELA was also identified as a significant transcriptional regulator of BAA genes (discussed below), only 10 of its targets overlapped between the DBT-PYR and BAA exposure networks (Figure 5, purple). Figure 5 therefore highlights the distinct nature of RELA regulatory roles in the toxicity pathways of different PAH structures.
Figure 5.
Networks of transcripts under regulatory control of AHR, RELA and JUN, that were misregulated in response to BAA (red), or DBT/PYR (blue) exposure. Transcripts that overlapped between the two PAH networks are represented in purple. Significant transcripts (p<0.05) from both the 24 and 48 hpf time points were combined to create the regulatory network.
Developmental processes in DBT and PYR-exposed embryos are widely misregulated at 48 hpf
By 48 hpf, 572 transcripts were differentially expressed in DBT and PYR embryos compared to controls, 478 of which were annotated. DAVID functional analysis identified 25 biological processes that were significantly affected by DBT and PYR exposure; oxoacid metabolic process was again significant and the most enriched functional cluster, but was composed of different genes than at 24 hpf (Table 5). Many of the most significant processes misregulated at 48 hpf were directly related to embryonic development. Embryonic development ending in birth or egg hatching consisted primarily of elevated transcripts, and was the second most enriched process. The regionalization, neurogenesis and central nervous system development functions together highlight widespread disruption of central nervous system development (Table 5). Thirty-five transcription factors were predicted to regulate significantly enriched groups of genes within this dataset; JUN, PPARA, RELA, RXRAa and SP1 were significant at both 24 and 48 hpf (Table S4). NR3C1 and ER, which were significant at 24 hpf, were no longer enriched at 48 hpf, whereas CREB1, P53, YY1 and TBP became significant with the largest numbers of misregulated downstream targets. The breadth of changes that occurred by 48 hpf lack specificity to a singular toxicity pathway but rather represent a profile of development gone awry. These and the many other misexpressed transcripts may result from a cascade of processes downstream of the genes disrupted at 24 hpf, but also reflect the vast molecular changes that occur in a normally developing zebrafish between these two time points.
Table 5.
Significantly enriched biological functions identified by DAVID analysis of all transcripts differentially regulated (p<0.05) in BAA exposed embryos, and common transcripts disrupted by DBT and PYR at 48 hpf.
| Biological Process | GO Term | Downregulated Genes | Upregulated Genes | E Score | % | P Value | |
|---|---|---|---|---|---|---|---|
| BAA | cellular homeostasis | GO:0019725 | edn2, cxcl12b, atp2a2a | slc30a1, cxcr4a, zgc:92066, gsr, prdx1, ccl1, ccr9a | 2.75 | 14.29 | 4.50E-04 |
| chemotaxis | GO:0006935 | cxcl12b, edn2 | ccr9a, cxcr4a, ccl1 | 2.50 | 7.14 | 2.18E-03 | |
| hormone metabolic process | GO:0042445 | lrata | ugt1b5, ugt1b7, cyp1b1, cyp1c2, cyp1a, cyp1c1 | 2.16 | 5.71 | 1.26E-02 | |
| tetrapyrrole metabolic process | GO:0033013 | - | zgc:77234, ugt1b5, ugt1b7, cyp1a | 1.92 | 4.29 | 1.25E-02 | |
| vasculature development | GO:0001944 | cx39.4, cxcl12b | ctgfb, cxcr4a, figf, tiparp | 1.89 | 8.57 | 1.01E-02 | |
| hydrogen peroxide metabolic process | GO:0042743 | - | cyp1a, prdx1 | 1.59 | 4.29 | 5.65E-03 | |
| cation transport | GO:0006812 | armc1l, atp2a2a, cx39.4, slc5a1 | cdkn1bl, slc30a1, zgc:92066 | 1.32 | 10.00 | 3.83E-02 | |
| organ development | GO:0048513 | atp2a2a, col7a1, cx39.4, cxcl12b, edn2, prl, slc5a1 | cdkn1bl, ctgfb, cxcr4a, cyp1a, figf, foxq1, foxf2a, tiparp, ved | 1.29 | 21.43 | 4.12E-02 | |
| DBT and PYR | oxoacid metabolic process | GO:0043436 | acaa1, acadl, adipor1a, adipor1b, agxt2l1, agxta, agxtb, amt, cpt1b, elovl4b, glsa, got2a, hadha, hadhb, idh3b, LOC565975, lta4h, mdh1b, padi2, rbp2b, rnpep, sc5dl, sgpl1, zgc:113076, zgc:136850, zgc:154046 | aspg, cbsb, cyp1a, cyp26c1, gldc, npc1, phgdh, ppat, psat1, slc1a3a | 4.72 | 8.35 | 2.66E-05 |
| embryonic development ending in birth or egg hatching | GO:0009792 | ift52, ric8a, zfpm2b | capn2b, cebpb, col4a3bp, cyp1a, eng1b, evx1, foxa, gas1b, gata2a, hoxa2b, hoxa4a, hoxb2a, hoxc1a, hoxc6b, msxe, nkx2.7, pax1a, phgdh, si:ch211-204c21.1, tcap, tgfbr1a | 3.61 | 5.90 | 1.01E-04 | |
| regionalization | GO:0003002 | egr2b, ift52, neurod | cyp26c1, egr2a, eng1b, evx1, foxa, gas1b, hhex, hoxa2b, hoxa4a, hoxb2a, hoxc4a, hoxc6b, pax1a, tcap, tgfbr1a | 3.38 | 4.18 | 2.75E-04 | |
| neurogenesis | GO:0022008 | bdnf, clic5, crx, egr2b, gnao1b, LOC799290, mbp, mbpa, neurod, otx5, rab3aa, rnd1, spon2b, vcanb | ascl1a, btg4, cebpb, cxcr4a, egr2a, eng1b, epha2, evx1, foxa, gas1b, gata2a, gdf7, her15.1, hoxa2b, mag, nr2f6b, phgdh, slc1a3a, tgfbr1a, unc5b, uts1 | 2.77 | 7.62 | 3.27E-03 | |
| embryonic organ development | GO:0048568 | clic5, neurod, zfpm2b | cebpb, gas1b, gata2a, hoxa2b, hoxa4a, hoxb2a, hoxc4a, myca, otop1, tcap, tgfbr1a | 2.43 | 3.44 | 2.40E-03 | |
| positive regulation of macromolecule metabolic process | GO:0010604 | crx, egr2b, fkbp1ab, ift52, klf2a, LOC570917, maf, neurod, npas4, otx5, psmd4b, psmd7, psmd8, rxrga, tnni2b.2, zfpm2b, zgc:110116 | ascl1a, cask, cebpb, cebpg, egr2a, evx1, foxa, gata2a, gdf7, her15.1, hhex, hoxa2b, im:7162084, irf11, myca, pth1a, sox19a, tgfbr1a, tnfb, uts1, vgll2b, zgc:158781 | 2.35 | 9.34 | 2.19E-03 | |
| negative regulation of cell communication | GO:0010648 | gnai2, hcrt, rgs11, zgc:136569, bcl6ab | cyp26c1, dkk1b, gas1b, hhex, im:7162084, npc1, onecut1, rgs4 | 1.99 | 3.44 | 1.01E-02 | |
| cellular component morphogenesis | GO:0032989 | bbs7, bdnf, clic5, cryaa, egr2b, rab3aa, spon2b, vcanb | bcl6ab, col4a3bp, cxcr4a, egr2a, gas1b, gdf7, hoxa2b, LOC796577, onecut1, sfrp5, si:ch211-204c21.1, slc1a3a, tcap, unc5b | 1.93 | 5.16 | 9.16E-03 | |
| rhombomere 3 development | GO:0021569 | egr2b | egr2a, hoxa2b, hoxb2a | 1.89 | 0.74 | 7.08E-03 | |
| central nervous system development | GO:0007417 | egr2b, faim2, gnao1b, LOC799290, mbpa, mbpa, neurod, sepp1a, sh3gl2 | ascl1a, cxcr4a, cyp26c1, dkk1b, egr2a, eng1b, evx1, foxa, gas1b, gata2a, hhex, hoxa2b, hoxb2a, msxe, nkx2.7, phgdh | 1.87 | 5.41 | 1.27E-02 | |
| hormone metabolic process | GO:0042445 | lrata, rbp2b | crhbp, cyp1a, cyp1b1, cyp26c1, scarb1, | 1.82 | 1.97 | 1.51E-02 | |
| organ morphogenesis | GO:0009887 | agc1, bbs7, clic5, crx, cryaa, cryaa, fkbp1ab, ift52, neurod, otx5, sgpl1, six6a | cmlc1, gas1b, hhex, hoxa2b, hoxa4a, hoxb2a, hoxc4a, im:7162084, msxe, myca, otop1, tcap, tgfbr1a, tnfb, zgc:158781 | 1.79 | 6.39 | 1.62E-02 | |
| fatty acid oxidation | GO:0019395 | acaa1, adipor1a, adipor1b, cpt1b, hadha, hadhb, zgc:154046 | 1.66 | 1.47 | 7.21E-03 | ||
| muscle tissue development | GO:0060537 | fkbp1ab, LOC100536295, rxrga, zfpm2b | cmlc1, LOC796577, nkx2.7, tcap, vgll2b | 1.65 | 2.21 | 2.45E-02 | |
| regulation of transmission of nerve impulse | GO:0051969 | bdnf, cspg5b, egr2b, gnai2, hcrt, rab3aa, zgc:136569 | egr2a, slc1a3a, tnfb, uts1 | 1.58 | 2.46 | 2.56E-02 | |
| positive regulation of cellular process | GO:0048522 | bdnf, crx, egr2b, fkbp1ab, gnai2, hcrt, ift52, klf2a, LOC556700, LOC570917, maf, mfge8a, neurod, npas4, otx5, pnp4b, psmd4b, psmd7, psmd8, rxrga, si:ch211-135f11.1, tnni2b.2, trim35, zfpm2b, zgc:100906, zgc:110116, zgc:110680 | ascl1a, bcl6ab, cask, cebpb, cebpg, cyp1a, egf, egr2a, evx1, flt4, foxa, GAS1 (3 of 3), gas1b, gata2a, gdf7, her15.1, hhex, hoxa2b, im:7162084, irf11, LOC794658, myca, ncs1a, nkx2.7, onecut1, plk2b, pth1a, scarb1, si:dkey-24p1.4, slc1a3a, sox19a, sst1.1, tgfbr1a, tnfb, uts1, vgll2b, zgc:154093, zgc:158781, zgc:85939 | 1.57 | 15.5 | 2.29E-02 | |
| oxidoreduction coenzyme metabolic process | GO:0006733 | coq3, idh3b, idh3g, itgb1bp3, mdh1b, pgls, pnp4b, zgc:113076 | 1.55 | 1.72 | 3.73E-03 | ||
| positive regulation of catalytic activity | GO:0043085 | gadd45bb, gnai2, gnao1b, gng13b, hcrt, psmd4b, psmd7, psmd8, ptplad1, rgn, si:ch211-135f11.1, zgc:110116 | cmlc1, cxcr4a, egf, LOC794658, myca, npr3, pth1a, scarb1, slc11a2, tgfbr1a, tnfb | 1.52 | 5.9 | 2.22E-02 | |
| negative regulation of ion transport | GO:0043271 | gnai2, gnao1b, hcrt | tnfb | 1.50 | 0.98 | 3.17E-02 | |
| endocrine pancreas development | GO:0031018 | neurod | foxa, insm1a, onecut1 | 1.49 | 0.98 | 1.19E-02 | |
| pyridine nucleotide metabolic process | GO:0019362 | idh3b, itgb1bp3, mdh1b, pgls, pnp4b, zgc:113076 | 1.44 | 1.47 | 9.14E-03 | ||
| cellular amino acid metabolic process | GO:0006520 | agxt2l1, agxta, agxtb, amt, glsa, got2a, padi2 | aspg, cbsb, gldc, phgdh, ppat, psat1, slc1a3a | 1.43 | 3.19 | 2.33E-02 | |
| cell morphogenesis involved in differentiation | GO:0000904 | bdnf, clic5, cryaa, egr2b, rab3aa, spon2b, vcanb | cxcr4a, egr2a, gas1b, gdf7, hoxa2b, si:ch211-204c21.1, slc1a3a, unc5b | 1.38 | 3.44 | 3.54E-02 | |
| diterpenoid metabolic process | GO:0016101 | LOC556575, lrata, rbp2b | cyp1a, cyp26c1 | 1.38 | 0.98 | 3.17E-02 | |
| ear development | GO:0043583 | bdnf, clic5 | gas1b, her15.1, hoxa2b, myca, otop1, tcap | 1.35 | 1.97 | 2.53E-02 |
E score: overall cluster enrichment score, %: percentage of total gene list involved in functional cluster, p-values determined by modified Fischer’s Exact test (EASE score).
Biological functions of BAA-misregulated genes are consistent with AHR-dependent toxicity
Human or mouse homologues were available for 29 of the 38 transcripts significantly misregulated by BAA at 24 hpf, which translated to 19 unique DAVID IDs. Several genes, including cyp1a, were represented by multiple probes within this significant transcript set. Functional analysis of genes misregulated by BAA at 24 hpf identified two biological processes, hormone metabolism and tissue development, that were significantly enriched within this dataset (Table 4). Metabolic process genes were up-regulated, and included well-known biomarkers of AHR activation such as cyp1a, as well as si:dkey-94e7.2, a predicted homolog of retinol dehydrogenase 11 (RDH11). Expression of genes involved in tissue development was also primarily increased (Table 4), likely via AHR signaling. Two homologues of foxq1 have been identified in zebrafish, and Entrez and Ensembl annotation were not in agreement as to the identity of the transcript in our dataset. We determined the Agilent probe, however, to be the same as was reported by Planchart et al. to target foxq1b, an AHR-dependent TCDD-inducible gene expressed in zebrafish jaw primordium (Planchart and Mattingly, 2010). Tissue development genes ptn and ctgfb (Table 4) are not known to be directly regulated by the AHR, but may be important mediators of AHR-dependent developmental toxicity; ctgfb is expressed in notochord, cartilage and retina of zebrafish larvae, and was also induced in developing jaws of zebrafish exposed to TCDD (Xiong et al., 2008; Fernando et al., 2010).
Transcription factor prediction identified AHR as significant at 24 hpf, along with its dimerization partner, ARNT, and C/EBPδ (Table S4). The large fold changes in a relatively small number of significant transcripts suggest BAA interacts with one primary transcription factor at 24 hpf, and the transcriptional profile supports previous demonstration of AHR-dependent toxicity induced by BAA (Incardona et al., 2006).
BAA transcriptional response indicates oxidative and metabolic stress at 48 hpf
The BAA transcriptional response expanded to 107 misexpressed transcripts at 48 hpf, 99 of which were sufficiently annotated. Though the cyp1 genes remained the most strongly elevated, they were joined by ahrra, wfikkn1, and cathepsin L.1 (ctsl.1), which was elevated 4-fold. Ctsl.1 encodes a widely expressed protease important for blood pressure regulation, and was recently identified as dioxin-responsive (Szlama et al., 2010; Mbewe-Campbell et al., 2012). DAVID functional annotation clustering of the 70 unique targets identified eight significantly enriched biological functions (Table 5). Hormone metabolic process was again significant, and included two phase 2 metabolizing enzymes, ugt1b5 and ugt1b7, along with the cyp1 transcripts. Transcripts in the cation transport process, in contrast, were not known AHR targets, and are discussed further within the ion transport response common to all three PAHs. Known AHR targets were similarly scarce in the enriched biological process, cellular homeostasis, which was composed of antioxidant-related genes (gsr, prdx1, and zgc:92066, a homolog of FTMT), and transcripts involved in blood pressure regulation and chemokine signaling (Table 5). Homologs of slc30a1 and edn2 (elevated and reduced, respectively) are involved in blood pressure regulation in other species, and atp2a2a (reduced expression) is required for heart looping in zebrafish (Saida et al., 2000; Ebert et al., 2005; Aguilar-Alonso et al., 2008).
Vascular development genes are misexpressed in BAA-exposed embryos
Genes involved in vasculature development were over-represented amongst transcripts affected by BAA at 48 hpf. They included chemokine receptor cxcr4a, which had increased expression, and its ligand, cxcl12b, which was under-expressed. Interestingly, this expression pattern was also observed in a microarray analysis of TCDD-induced transcriptional changes in zebrafish jaw, suggesting misregulated chemokine signaling may indeed be involved in AHR-mediated toxicity in the developing embryo (Xiong et al., 2008). cxcr4a is required for arterial-venous network formation, development of the trunk lymph system, and is expressed in response to low blood flow and in unperfused blood vessels in developing zebrafish (Bussmann et al., 2011; Cha et al., 2012). Other vasculature development genes included connexin 39.4 (cx39.4), connective tissue growth factor b (ctgfb), c-fos induced growth factor (figf, previously vegf-d) and TCDD-inducible poly(ADP-ribose) polymerase (tiparp) (Table 5). Together these transcriptional changes convey disruption of vascular development and circulatory system function. This is in agreement with blood pressure misregulation and endothelial dysfunction in rats developmentally exposed to benzo(a)pyrene, another PAH known to induce AHR signaling (Jules et al., 2012).
The previously-demonstrated dependence of BAA toxicity on the AHR suggests vascular development genes in our study are downstream of the receptor. The molecular signaling that precipitates reduced expression of transcripts, however, is not yet defined. An intriguing study of AHR binding sites using chip-seq in hepatoma cells found that AHR targets in unexposed cells were significantly enriched for angiogenesis, blood vessel patterning, and other developmental functions (Sartor et al., 2009). Upon exposure to AHR ligands, the targets of the AHR changed dramatically to favor genes involved in metabolism (Sartor et al., 2009). Reduced expression of some transcripts within our dataset could reflect the absence of AHR at endogenous transcription sites in BAA-treated embryos.
RELA is a significant transcription factor in the BAA regulatory network
Transcription factor analysis at 48 hpf predicted involvement of multiple transcription factors (Table S4). AHR was interestingly no longer significant, though its dimerization partner ARNT was predicted to regulate a significantly enriched cluster of genes. SP1, TP53, CREB1 and RELA were upstream of the largest number of genes misregulated by BAA at 48 hpf (Table S4). RELA interacts directly with AHR and is an important regulator of inflammatory immune and oxidative stress responses (Tian et al., 1999; Tian, 2009). The RELA and AHR regulatory networks are displayed in Figure 5, which shows that the AHR regulates a set of genes that were distinct to the BAA exposures and were not predicted to be under direct regulation by RELA. The large number of genes downstream of RELA, however, which includes some of the most highly misexpressed genes such as CTGF and CTSL.1, suggests RELA may play an important role in the BAA toxicity pathway (Figure 5).
Differential affinities for the AHR result in few transcripts common to PAH exposure
The few genes similarly misregulated by all three PAHs represent potential general biomarkers of PAH exposure. At 24 hpf, only 5 transcripts were similarly affected by all 3 PAHs. The most highly elevated probes (approximately 3-fold for all PAHs), A_15_P477220 and A_15_P247256, both target ESTs that are not yet annotated in the zebrafish V9 genome. Decreased transcripts at 24 hpf included slco5a1, a solute carrier organic transporter family member, and a non-specific probe. By 48 hpf, 23 transcripts were similarly expressed in response to all three PAHs. The most elevated genes across all three PAHs were cyp1a, cyp1b1, wfikkn1, LOC794658 (similar to chrm3), s100z and cxcr4a. The largest decreases were observed in g0s2, kif20a, cfdl, and two uncharacterized genes, zgc:171318 and zgc:153311. Though the molecular toxicity pathways of BAA and DBT/PYR are, on the whole, very different, these genes highlight some commonalities.
The most commonly used biomarker of AHR activation, cyp1a, was elevated by all three PAHs at 48 hpf. However, DBT and PYR only induced 1.2 and 2.1 fold changes, respectively, whereas BAA induced cyp1a 34.5-fold. The minimal and delayed cyp1a induction suggests that it occurs via metabolites or very weak AHR activation by DBT and PYR. Barron and colleagues reported the potency of BAA as an AHR agonist as 519 times greater than PYR, and though DBT was not analyzed, 3-ring PAHs included in the study were less potent than PYR or were inactive in assay systems (Barron et al., 2004). Though Cyp1a protein expression is induced by PYR exposure in zebrafish embryos, Incardona et al. reported a markedly different expression pattern than was observed with BAA, and suggested Cyp1a metabolism and hepatic toxicity were drivers of the developmental effects (Incardona et al., 2006). DBT, in contrast, has been reported to induce developmental toxicity via disruption of early cardiac function, as well as act as a Cyp1a inhibitor (Incardona et al., 2004; Wassenberg et al., 2005). In light of these proposed different mechanisms of action, the overlap of transcripts misregulated by DBT and PYR at both 24 and 48 hpf in this study is striking. Indeed, the different malformations observed in DBT and PYR-treated embryos, despite similar molecular response profiles, may be a result of metabolic processes that are more active after the 48 hpf time point employed in this study. The marked effects of DBT exposure on axis formation, which were not observed in response to PYR, could also be explained by the dramatic differences in PAH body burden at these equivalent exposure concentrations. Signaling that directs axis formation occurs early in development; the uptake of DBT was relatively rapid, whereas the lower solubility and uptake of PYR potentially did not achieve a threshold concentration to induce such effects.
Disruption of ion transport and calcium signaling is common across all PAHs
Ion transport and homeostatic processes were misregulated by all three PAHs in this study, though the significant transcripts in the DBT-PYR response are largely different from those affected by BAA. All three exposures induced differential expression of genes involved in calcium homeostasis, suggesting that calcium signaling plays a role in PAH-induced developmental toxicity, as has been shown previously in dioxin-exposed zebrafish at 48 hpf (Alexeyenko et al., 2010). In a separate study of TCDD effects on heart development, transcriptional changes related to calcium homeostasis preceded the development of cardiac malformations in zebrafish, suggesting that they may be causal for malformations rather than simply a result of reduced blood flow (Carney et al., 2006). Though early calcium influx is a well-known response to several AHR ligands, the dependence of this response on AHR binding and the consequence within the developmental context is unknown.
PAHs have previously been shown to increase intracellular calcium through protein tyrosine kinases, inhibiting SERCA activity, and activating RYR receptors, though the intensity and duration of the response is dependent on PAH and cell type (Archuleta et al., 1993; Krieger et al., 1995; Gao et al., 2005). BaP induces a Ca2+ increases in endothelial cells, which is independent of the AHR but required for CYP1B1 induction (Mayati et al., 2012). Common mechanisms that do not require AHR signaling have also been observed with CYP1B1 and TGFB-related genes in AHR-null vascular smooth muscle cells exposed to BaP and TCDD (Karyala et al., 2004). All three PAHs in our study increased transcription of wfikkn1, which binds TGFB (Szlama et al., 2010), as well as calcium binding protein s100z. The S100 family of EF-hand calcium binding proteins regulates a diverse range of cellular functions in a calcium-dependent manner, and is associated with many pathological conditions including inflammation, atherosclerosis, diabetes, and neurodegeneration (Hermann et al., 2012). Future investigation of the dependence of these transcriptional changes on AHR signaling will provide insight into whether they represent a common mechanism, or are induced via different molecular responses to the PAHs in our study
Differential AHR activation and uptake result in distinct RELA regulatory responses to PAH exposures
RELA was a predicted transcriptional regulator of both the BAA and DBT-PYR toxicological responses. Despite this, there was little overlap in the transcriptional networks (Figure 5). Differential AHR activation can explain the AHR gene battery that was uniquely induced by BAA at 24 hpf. However, a large portion of the RELA network expressed in response to DBT and PYR was not affected by BAA exposure (Figure 5). This difference could potentially be explained by uptake. BAA is the least soluble in water, and uptake was an order of magnitude lower that the other PAHs. We therefore cannot exclude the possibility that BAA would activate the DBT-PYR transcriptional network at an equivalent internal concentration. Body burdens of DBT and PYR-exposed embryos are within the range reported to induce toxicity through mechanisms classified under “nonpolar narcosis”, such as interference with lipid fluidity and membrane function (Vanwezel and Opperhuizen, 1995). The general pattern of narcotic response, including lost sense of balance, response to stimuli, and reduced ventilation frequency, is not applicable to the early developmental stages of embryos analyzed in this study. However, DBT concentrations in embryos averaged 3.4 µmol/g at 24 hpf, and PYR reached 2.9 µmol/g by 48 hpf following exposure to 25 µM waterborne concentrations. Toxicity from nonpolar narcosis has been reported to occur at 2–8 µmol/g body weight, depending on the compound and organism (Vanwezel and Opperhuizen, 1995). The data presented here characterize the extensive molecular response to these relatively high internal concentrations, and our analysis identified RELA as a significant mediator in the DBT-PYR transcriptional network. Interestingly, though BAA toxicity was induced by a lower body burden concentration of 0.12 µmol/g at 48 hpf, network analysis also identified RELA as a significant regulator of BAA-induced transcriptional changes. This suggests that RELA involvement in PAH toxicity is modulated by both AHR activation and PAH concentration. Future studies with multiple PAHs would be useful for identifying whether transcriptional networks identified here are differentially induced by diverse PAH structures.
Transcriptional responses to PAH exposures are conserved across species
We compared the profiles of genes differentially regulated by three PAHs in the developing zebrafish embryo and found disrupted biological processes with surprising overlap with studies in other model systems. The genes differentially regulated by BAA were consistent with previous reports of AHR activation by this PAH, and many of them were identified in array studies with other known AHR ligands in zebrafish. All three PAHs misregulated genes important in vasculature development and cardiac function. This has been observed in BaP-exposed rats, as well as in previous studies of fish exposed to a number of PAHs (Incardona et al., 2009; Incardona et al., 2011; Huang et al., 2012; Jules et al., 2012). Oxidative stress was a component of the toxic response, as has also been reported previously, and we observed differential regulation of immune-related genes, particularly by DBT and PYR. Though fewer studies have examined PAHs that are not strong AHR agonists, PAHs that do not induce CYP1A have similarly been observed to induce inflammatory cytokines in cells in culture (Suresh et al., 2009; Ovrevik et al., 2010). PAHs are known immunotoxicants in fish, with well-established effects on lymphocytes (Krieger et al., 1994; Reynaud and Deschaux, 2006). The gene expression changes observed in this study, however, primarily represent innate immune responses, as the adaptive immune system is not mature until weeks 4–6 of development (Meeker and Trede, 2008). We therefore would not expect to see substantial overlap between the genes observed in this study and others conducted with tissues from adult organisms. Nevertheless, calcium binding and immune response were identified as important differentially expressed gene clusters in human macrophage leukemia cells exposed to diverse PAHs in vitro, and metal ion binding and transport was the most significant biological process associated with occupational PAH exposure in peripheral blood of coke-oven workers (Wan et al., 2008; Wu et al., 2011b). Chronic PAH exposure in coke-oven workers has also been associated with altered immunological parameters, including increased TNFα in serum, as well increased markers of lipid peroxidation and oxidative stress (Jeng et al., 2011). Increased malondialdehyde and decreased reduced glutathione were similarly observed in bronchial asthma patients, and correlated with blood phenanthrene levels, providing further evidence of PAH-induced oxidative stress in human populations (Suresh et al., 2009).
We identified multiple potential biomarkers of individual PAHs over time, as well as genes commonly misregulated by PAHs with differential AHR affinity. Many of the significant biological processes disrupted in this study, such as ion homeostasis, have been observed previously in other models, and provide insight into fundamental molecular pathways that are sensitive to PAH exposure and conserved between organ systems and species. Further investigation of these pathways in response to more structurally diverse PAHs in the environment will be invaluable to understanding the hazard potential of PAH exposure during development.
Supplementary Material
Highlights.
We compared global mRNA expression changes induced by developmental exposure to PAHs
PAH body burden was determined to discern molecular vs. uptake-driven toxicity differences
Genes uniquely induced by benz(a)anthracene included known targets of the AHR and RELA
Dibenzothiophene and pyrene perturbed a RELA network distinct from benz(a)anthracene
Transcriptional networks reveal differential mechanisms of PAH toxicity in zebrafish
Acknowledgements
This work was supported by the NIEHS Superfund Research Program P42 ES016465, Core Center Grant P30 ES00210 and the NIEHS Training Grant T32ES7060. Bradley Stewart processed the microarrays at University of Wisconsin McArdle Laboratory of Cancer Research Microarray Facility. We are grateful to Cari Buchner, Carrie Barton and the staff at the Sinnhuber Aquatic Research Laboratory for fish husbandry expertise. We would also like to Tanguay laboratory members Jane La Du and Michael Simonich for their helpful feedback and critical review of the manuscript. Pacific Northwest National Laboratory is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under contract number DE-AC05-76RLO1830.
References
- Aguilar-Alonso P, Martinez-Fong D, Pazos-Salazar NG, Brambila E, Gonzalez-Barrios JA, Mejorada A, Flores G, Millan-Perezpena L, Rubio H, Leon-Chavez BA. The increase in zinc levels and upregulation of zinc transporters are mediated by nitric oxide in the cerebral cortex after transient ischemia in the rat. Brain research. 2008;1200:89–98. doi: 10.1016/j.brainres.2007.11.077. [DOI] [PubMed] [Google Scholar]
- Alexeyenko A, Wassenberg DM, Lobenhofer EK, Yen J, Linney E, Sonnhammer EL, Meyer JN. Dynamic zebrafish interactome reveals transcriptional mechanisms of dioxin toxicity. PLoS One. 2010;5:e10465. doi: 10.1371/journal.pone.0010465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen EA, Spitsbergen JM, Tanguay RL, Stegeman JJ, Heideman W, Peterson RE. Tissue-specific expression of AHR2, ARNT2, and CYP1A in zebrafish embryos and larvae: effects of developmental stage and 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure. Toxicol Sci. 2002;68:403–419. doi: 10.1093/toxsci/68.2.403. [DOI] [PubMed] [Google Scholar]
- Archuleta MM, Schieven GL, Ledbetter JA, Deanin GG, Burchiel SW. 7,12-Dimethylbenz[a]anthracene activates protein-tyrosine kinases Fyn and Lck in the HPB-ALL human T-cell line and increases tyrosine phosphorylation of phospholipase C-gamma 1, formation of inositol 1,4,5-trisphosphate, and mobilization of intracellular calcium. Proc Natl Acad Sci U S A. 1993;90:6105–6109. doi: 10.1073/pnas.90.13.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron MG, Heintz R, Rice SD. Relative potency of PAHs and heterocycles as aryl hydrocarbon receptor agonists in fish. Mar Environ Res. 2004;58:95–100. doi: 10.1016/j.marenvres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Binder RL, Stegeman JJ. Microsomal electron transport and xenobiotic monooxygenase activities during the embryonic period of development in the killifish, Fundulus heteroclitus. Toxicol Appl Pharmacol. 1984;73:432–443. doi: 10.1016/0041-008x(84)90096-6. [DOI] [PubMed] [Google Scholar]
- Bostrom CE, Gerde P, Hanberg A, Jernstrom B, Johansson C, Kyrklund T, Rannug A, Tornqvist M, Victorin K, Westerholm R. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ Health Perspect. 2002;110(Suppl 3):451–488. doi: 10.1289/ehp.110-1241197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasier AR, Jamaluddin M, Han Y, Patterson C, Runge MS. Angiotensin II induces gene transcription through cell-type-dependent effects on the nuclear factor-kappaB (NF-kappaB) transcription factor. Mol Cell Biochem. 2000;212:155–169. [PubMed] [Google Scholar]
- Brasier AR, Li J. Mechanisms for inducible control of angiotensinogen gene transcription. Hypertension. 1996;27:465–475. doi: 10.1161/01.hyp.27.3.465. [DOI] [PubMed] [Google Scholar]
- Burstyn I, Kromhout H, Partanen T, Svane O, Langard S, Ahrens W, Kauppinen T, Stucker I, Shaham J, Heederik D, Ferro G, Heikkila P, Hooiveld M, Johansen C, Randem BG, Boffetta P. Polycyclic aromatic hydrocarbons and fatal ischemic heart disease. Epidemiology. 2005;16:744–750. doi: 10.1097/01.ede.0000181310.65043.2f. [DOI] [PubMed] [Google Scholar]
- Bussmann J, Wolfe SA, Siekmann AF. Arterial-venous network formation during brain vascularization involves hemodynamic regulation of chemokine signaling. Development. 2011;138:1717–1726. doi: 10.1242/dev.059881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney SA, Chen J, Burns CG, Xiong KM, Peterson RE, Heideman W. Aryl hydrocarbon receptor activation produces heart-specific transcriptional and toxic responses in developing zebrafish. Mol Pharmacol. 2006;70:549–561. doi: 10.1124/mol.106.025304. [DOI] [PubMed] [Google Scholar]
- Cha YR, Fujita M, Butler M, Isogai S, Kochhan E, Siekmann AF, Weinstein BM. Chemokine signaling directs trunk lymphatic network formation along the preexisting blood vasculature. Dev Cell. 2012;22:824–836. doi: 10.1016/j.devcel.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Jedrychowski W, Spengler J, Camann DE, Whyatt RM, Rauh V, Tsai WY, Perera FP. International studies of prenatal exposure to polycyclic aromatic hydrocarbons and fetal growth. Environ Health Perspect. 2006;114:1744–1750. doi: 10.1289/ehp.8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciganek M, Neca J, Adamec V, Janosek J, Machala M. A combined chemical and bioassay analysis of traffic-emitted polycyclic aromatic hydrocarbons. Sci Total Environ. 2004;334–335:141–148. doi: 10.1016/j.scitotenv.2004.04.034. [DOI] [PubMed] [Google Scholar]
- Collins JF, Brown JP, Alexeeff GV, Salmon AG. Potency equivalency factors for some polycyclic aromatic hydrocarbons and polycyclic aromatic hydrocarbon derivatives. Regul Toxicol Pharmacol. 1998;28:45–54. doi: 10.1006/rtph.1998.1235. [DOI] [PubMed] [Google Scholar]
- Detmar J, Rennie MY, Whiteley KJ, Qu D, Taniuchi Y, Shang X, Casper RF, Adamson SL, Sled JG, Jurisicova A. Fetal growth restriction triggered by polycyclic aromatic hydrocarbons is associated with altered placental vasculature and AhR-dependent changes in cell death. Am J Physiol Endocrinol Metab. 2008;295:E519–E530. doi: 10.1152/ajpendo.90436.2008. [DOI] [PubMed] [Google Scholar]
- Durant JL, Busby WF, Jr., Lafleur AL, Penman BW, Crespi CL. Human cell mutagenicity of oxygenated, nitrated and unsubstituted polycyclic aromatic hydrocarbons associated with urban aerosols. Mutat Res. 1996;371:123–157. doi: 10.1016/s0165-1218(96)90103-2. [DOI] [PubMed] [Google Scholar]
- Ebert AM, Hume GL, Warren KS, Cook NP, Burns CG, Mohideen MA, Siegal G, Yelon D, Fishman MC, Garrity DM. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc Natl Acad Sci U S A. 2005;102:17705–17710. doi: 10.1073/pnas.0502683102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA, U. Integrated Risk Information System. 2012. [Google Scholar]
- Fernando CA, Conrad PA, Bartels CF, Marques T, To M, Balow SA, Nakamura Y, Warman ML. Temporal and spatial expression of CCN genes in zebrafish. Dev Dyn. 2010;239:1755–1767. doi: 10.1002/dvdy.22279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Voss AA, Pessah IN, Lauer FT, Penning TM, Burchiel SW. Ryanodine receptor-mediated rapid increase in intracellular calcium induced by 7,8-benzo(a)pyrene quinone in human and murine leukocytes. Toxicol Sci. 2005;87:419–426. doi: 10.1093/toxsci/kfi265. [DOI] [PubMed] [Google Scholar]
- Gleason MR, Armisen R, Verdecia MA, Sirotkin H, Brehm P, Mandel G. A mutation in serca underlies motility dysfunction in accordion zebrafish. Dev Biol. 2004;276:441–451. doi: 10.1016/j.ydbio.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Metabolism of chemical carcinogens. Carcinogenesis. 2000;21:345–351. doi: 10.1093/carcin/21.3.345. [DOI] [PubMed] [Google Scholar]
- Hansch C, Albert L, Hoekman D. Exploring QSAR: Volume 2: Hydrophobic, Electronic, and Steric Constants. American Chemical Society; 1995. [Google Scholar]
- Hecht SS, Carmella SG, Villalta PW, Hochalter JB. Analysis of phenanthrene and benzo[a]pyrene tetraol enantiomers in human urine: relevance to the bay region diol epoxide hypothesis of benzo[a]pyrene carcinogenesis and to biomarker studies. Chem Res Toxicol. 2010;23:900–908. doi: 10.1021/tx9004538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann A, Donato R, Weiger TM, Chazin WJ. S100 calcium binding proteins and ion channels. Frontiers in pharmacology. 2012;3:67. doi: 10.3389/fphar.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz-Picciotto I, Park HY, Dostal M, Kocan A, Trnovec T, Sram R. Prenatal exposures to persistent and non-persistent organic compounds and effects on immune system development. Basic Clin Pharmacol Toxicol. 2008;102:146–154. doi: 10.1111/j.1742-7843.2007.00190.x. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang L, Wang C, Zhang Y, Li J, Zhong Y, Zhou Y, Chen Y, Zuo Z. Benzo[a]pyrene exposure influences the cardiac development and the expression of cardiovascular relative genes in zebrafish (Daniorerio) embryos. Chemosphere. 2012;87:369–375. doi: 10.1016/j.chemosphere.2011.12.026. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Carls MG, Day HL, Sloan CA, Bolton JL, Collier TK, Scholz NL. Cardiac arrhythmia is the primary response of embryonic Pacific herring (Clupea pallasi) exposed to crude oil during weathering. Environ Sci Technol. 2009;43:201–207. doi: 10.1021/es802270t. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Carls MG, Teraoka H, Sloan CA, Collier TK, Scholz NL. Aryl hydrocarbon receptor-independent toxicity of weathered crude oil during fish development. Environ Health Perspect. 2005;113:1755–1762. doi: 10.1289/ehp.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona JP, Collier TK, Scholz NL. Defects in cardiac function precede morphological abnormalities in fish embryos exposed to polycyclic aromatic hydrocarbons. Toxicol Appl Pharmacol. 2004;196:191–205. doi: 10.1016/j.taap.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Day HL, Collier TK, Scholz NL. Developmental toxicity of 4-ring polycyclic aromatic hydrocarbons in zebrafish is differentially dependent on AH receptor isoforms and hepatic cytochrome P4501A metabolism. Toxicol Appl Pharmacol. 2006;217:308–321. doi: 10.1016/j.taap.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Linbo TL, Scholz NL. Cardiac toxicity of 5-ring polycyclic aromatic hydrocarbons is differentially dependent on the aryl hydrocarbon receptor 2 isoform during zebrafish development. Toxicol Appl Pharmacol. 2011;257:242–249. doi: 10.1016/j.taap.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Isidoro Tavares N, Philip-Couderc P, Baertschi AJ, Lerch R, Montessuit C. Angiotensin II and tumour necrosis factor alpha as mediators of ATP-dependent potassium channel remodelling in post-infarction heart failure. Cardiovasc Res. 2009;83:726–736. doi: 10.1093/cvr/cvp162. [DOI] [PubMed] [Google Scholar]
- Jeng HA, Pan CH, Diawara N, Chang-Chien GP, Lin WY, Huang CT, Ho CK, Wu MT. Polycyclic aromatic hydrocarbon-induced oxidative stress and lipid peroxidation in relation to immunological alteration. Occup Environ Med. 2011;68:653–658. doi: 10.1136/oem.2010.055020. [DOI] [PubMed] [Google Scholar]
- Jia S, Ren Z, Li X, Zheng Y, Meng A. smad2 and smad3 are required for mesendoderm induction by transforming growth factor-beta/nodal signals in zebrafish. J Biol Chem. 2008;283:2418–2426. doi: 10.1074/jbc.M707578200. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Liu D, Sun S, Hu J, Tan L, Wang Y, Gui Y, Yu M, Song H. Critical role of connexin43 in zebrafish late primitive and definitive hematopoiesis. Fish physiology and biochemistry. 2010;36:945–951. doi: 10.1007/s10695-009-9371-1. [DOI] [PubMed] [Google Scholar]
- Jules GE, Pratap S, Ramesh A, Hood DB. In utero exposure to benzo(a)pyrene predisposes offspring to cardiovascular dysfunction in later-life. Toxicology. 2012 doi: 10.1016/j.tox.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Noh JH, Eun JW, Kim JK, Bae HJ, Xie H, Jang JJ, Ryu JC, Park WS, Lee JY, Nam SW. Molecular signature for early detection and prediction of polycyclic aromatic hydrocarbons in peripheral blood. Environ Sci Technol. 2011;45:300–306. doi: 10.1021/es101840s. [DOI] [PubMed] [Google Scholar]
- Karyala S, Guo J, Sartor M, Medvedovic M, Kann S, Puga A, Ryan P, Tomlinson CR. Different global gene expression profiles in benzo[a]pyrene- and dioxin-treated vascular smooth muscle cells of AHR-knockout and wild-type mice. Cardiovascular toxicology. 2004;4:47–73. doi: 10.1385/ct:4:1:47. [DOI] [PubMed] [Google Scholar]
- Kerley-Hamilton JS, Trask HW, Ridley CJ, Dufour E, Lesseur C, Ringelberg CS, Moodie KL, Shipman SL, Korc M, Gui J, Shworak NW, Tomlinson CR. Inherent and Benzo[a]pyrene-Induced Differential Aryl Hydrocarbon Receptor Signaling Greatly Affects Life Span, Atherosclerosis, Cardiac Gene Expression, and Body and Heart Growth in Mice. Toxicol Sci. 2012;126:391–404. doi: 10.1093/toxsci/kfs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kluver N, Yang L, Busch W, Scheffler K, Renner P, Strahle U, Scholz S. Transcriptional response of zebrafish embryos exposed to neurotoxic compounds reveals a muscle activity dependent hspb11 expression. PLoS One. 2011;6:e29063. doi: 10.1371/journal.pone.0029063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic S, Radojkovic D, Faulkner G. Muscle ankyrin repeat proteins: their role in striated muscle function in health and disease. Critical reviews in clinical laboratory sciences. 2011;48:269–294. doi: 10.3109/10408363.2011.643857. [DOI] [PubMed] [Google Scholar]
- Krieger JA, Born JL, Burchiel SW. Persistence of calcium elevation in the HPB-ALL human T cell line correlates with immunosuppressive properties of polycyclic aromatic hydrocarbons. Toxicol Appl Pharmacol. 1994;127:268–274. doi: 10.1006/taap.1994.1161. [DOI] [PubMed] [Google Scholar]
- Krieger JA, Davila DR, Lytton J, Born JL, Burchiel SW. Inhibition of sarcoplasmic/endoplasmic reticulum calcium ATPases (SERCA) by polycyclic aromatic hydrocarbons in HPB-ALL human T cells and other tissues. Toxicol Appl Pharmacol. 1995;133:102–108. doi: 10.1006/taap.1995.1131. [DOI] [PubMed] [Google Scholar]
- Le Mevel JC, Lancien F, Mimassi N. Central cardiovascular actions of angiotensin II in trout. General and comparative endocrinology. 2008;157:27–34. doi: 10.1016/j.ygcen.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Lee MS, Magari S, Christiani DC. Cardiac autonomic dysfunction from occupational exposure to polycyclic aromatic hydrocarbons. Occup Environ Med. 2011;68:474–478. doi: 10.1136/oem.2010.055681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrell D, Truong L, Jephson C, Sarker MR, Moore A, Lang C, Simonich MT, Tanguay RL. Automated zebrafish chorion removal and single embryo placement: optimizing throughput of zebrafish developmental toxicity screens. Journal of laboratory automation. 2012;17:66–74. doi: 10.1177/2211068211432197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Shi L, Xu F, Zhang L, Xu Z. Development of fetal brain renin-angiotensin system and hypertension programmed in fetal origins. Progress in neurobiology. 2009;87:252–263. doi: 10.1016/j.pneurobio.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, McGrath JA, Di Toro DM. Modeling polycyclic aromatic hydrocarbon bioaccumulation and metabolism in time-variable early life-stage exposures. Environmental Toxicology and Chemistry. 2008;27:1515–1525. doi: 10.1897/07-355.1. [DOI] [PubMed] [Google Scholar]
- Mayati A, Levoin N, Paris H, N'Diaye M, Courtois A, Uriac P, Lagadic-Gossmann D, Fardel O, Le Ferrec E. Induction of intracellular calcium concentration by environmental benzo(a)pyrene involves a beta2-adrenergic receptor/adenylyl cyclase/Epac-1/inositol 1,4,5-trisphosphate pathway in endothelial cells. J Biol Chem. 2012;287:4041–4052. doi: 10.1074/jbc.M111.319970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbewe-Campbell N, Wei Z, Zhang K, Friese RS, Mahata M, Schork AJ, Rao F, Chiron S, Biswas N, Kim HS, Mahata SK, Waalen J, Nievergelt CM, Hook VY, O'Connor DT. Genes and environment: novel, functional polymorphism in the human cathepsin L (CTSL1) promoter disrupts a xenobiotic response element (XRE) to alter transcription and blood pressure. Journal of hypertension. 2012 doi: 10.1097/HJH.0b013e328356b86a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker ND, Trede NS. Immunology and zebrafish: spawning new models of human disease. Developmental and comparative immunology. 2008;32:745–757. doi: 10.1016/j.dci.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Menzie CA, Potocki BB, Santodonato J. Exposure to Carcinogenic Pahs in the Environment. Environmental Science & Technology. 1992;26:1278–1284. [Google Scholar]
- Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biological procedures online. 2006;8:175–193. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RL, Huang C, Ho RK. Spatio-temporal regulation of Wnt and retinoic acid signaling by tbx16/spadetail during zebrafish mesoderm differentiation. BMC Genomics. 2010;11:492. doi: 10.1186/1471-2164-11-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova YY, Eisenreich SJ, Turpin BJ, Weisel CP, Morandi MT, Colome SD, Totten LA, Stock TH, Winer AM, Alimokhtari S, Kwon J, Shendell D, Jones J, Maberti S, Wall SJ. Polycyclic aromatic hydrocarbons in the indoor and outdoor air of three cities in the U.S. Environ Sci Technol. 2002;36:2552–2559. doi: 10.1021/es015727h. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, Dalton TP. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- Nikolsky Y, Kirillov E, Zuev R, Rakhmatulin E, Nikolskaya T. Functional analysis of OMICs data and small molecule compounds in an integrated "knowledge-based" platform. Methods Mol Biol. 2009;563:177–196. doi: 10.1007/978-1-60761-175-2_10. [DOI] [PubMed] [Google Scholar]
- Ovrevik J, Arlt VM, Oya E, Nagy E, Mollerup S, Phillips DH, Lag M, Holme JA. Differential effects of nitro-PAHs and amino-PAHs on cytokine and chemokine responses in human bronchial epithelial BEAS-2B cells. Toxicol Appl Pharmacol. 2010;242:270–280. doi: 10.1016/j.taap.2009.10.017. [DOI] [PubMed] [Google Scholar]
- Park K, Scott AL. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. Journal of leukocyte biology. 2010;88:1081–1087. doi: 10.1189/jlb.0610318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen GI, Kristensen P. Bioaccumulation of lipophilic substances in fish early life stages. Environmental Toxicology and Chemistry. 1998;17:1385–1395. [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchart A, Mattingly CJ. 2,3,7,8-Tetrachlorodibenzo-p-dioxin upregulates FoxQ1b in zebrafish jaw primordium. Chem Res Toxicol. 2010;23:480–487. doi: 10.1021/tx9003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polidori A, Kwon J, Turpin BJ, Weisel C. Source proximity and residential outdoor concentrations of PM(2.5), OC, EC, and PAHs. Journal of exposure science & environmental epidemiology. 2010;20:457–468. doi: 10.1038/jes.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RL, Carver W, Simpson DG, Fu L, Zhao J, Borg TK, Terracio L. The effects of angiotensin II and specific angiotensin receptor blockers on embryonic cardiac development and looping patterns. Dev Biol. 1997;192:572–584. doi: 10.1006/dbio.1997.8764. [DOI] [PubMed] [Google Scholar]
- Ramesh A, Walker SA, Hood DB, Guillen MD, Schneider K, Weyand EH. Bioavailability and risk assessment of orally ingested polycyclic aromatic hydrocarbons. Int J Toxicol. 2004;23:301–333. doi: 10.1080/10915810490517063. [DOI] [PubMed] [Google Scholar]
- Ramirez N, Cuadras A, Rovira E, Marce RM, Borrull F. Risk assessment related to atmospheric polycyclic aromatic hydrocarbons in gas and particle phases near industrial sites. Environ Health Perspect. 2011;119:1110–1116. doi: 10.1289/ehp.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers MJ, La Du JK, Periera CB, Giovanini J, Tanguay RL. Ethanol-dependent toxicity in zebrafish is partially attenuated by antioxidants. Neurotoxicol Teratol. 2006;28:497–508. doi: 10.1016/j.ntt.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Ren A, Qiu X, Jin L, Ma J, Li Z, Zhang L, Zhu H, Finnell RH, Zhu T. Association of selected persistent organic pollutants in the placenta with the risk of neural tube defects. Proc Natl Acad Sci U S A. 2011;108:12770–12775. doi: 10.1073/pnas.1105209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie MY, Detmar J, Whiteley KJ, Yang J, Jurisicova A, Adamson SL, Sled JG. Vessel tortuousity and reduced vascularization in the fetoplacental arterial tree after maternal exposure to polycyclic aromatic hydrocarbons. American journal of physiology. Heart and circulatory physiology. 2011;300:H675–H684. doi: 10.1152/ajpheart.00510.2010. [DOI] [PubMed] [Google Scholar]
- Reynaud S, Deschaux P. The effects of polycyclic aromatic hydrocarbons on the immune system of fish: a review. Aquat Toxicol. 2006;77:229–238. doi: 10.1016/j.aquatox.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Rutenberg JB, Fischer A, Jia H, Gessler M, Zhong TP, Mercola M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 2006;133:4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saida K, Hashimoto M, Mitsui Y, Ishida N, Uchide T. The prepro vasoactive intestinal contractor (VIC)/endothelin-2 gene (EDN2): structure, evolution, production, and embryonic expression. Genomics. 2000;64:51–61. doi: 10.1006/geno.1999.6083. [DOI] [PubMed] [Google Scholar]
- Sartor MA, Schnekenburger M, Marlowe JL, Reichard JF, Wang Y, Fan Y, Ma C, Karyala S, Halbleib D, Liu X, Medvedovic M, Puga A. Genomewide analysis of aryl hydrocarbon receptor binding targets reveals an extensive array of gene clusters that control morphogenetic and developmental programs. Environ Health Perspect. 2009;117:1139–1146. doi: 10.1289/ehp.0800485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Dragin N, Galvez-Peralta M, Jorge-Nebert LF, Miller ML, Wang B, Nebert DW. Organ-specific roles of CYP1A1 during detoxication of dietary benzo[a]pyrene. Mol Pharmacol. 2010;78:46–57. doi: 10.1124/mol.110.063438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh R, Shally A, Mahdi AA, Patel DK, Singh VK, Rita M. Assessment of association of exposure to polycyclic aromatic hydrocarbons with bronchial asthma and oxidative stress in children: A case control study. Indian journal of occupational and environmental medicine. 2009;13:33–37. doi: 10.4103/0019-5278.50722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szlama G, Kondas K, Trexler M, Patthy L. WFIKKN1 and WFIKKN2 bind growth factors TGFbeta1, BMP2 and BMP4 but do not inhibit their signalling activity. The FEBS journal. 2010;277:5040–5050. doi: 10.1111/j.1742-4658.2010.07909.x. [DOI] [PubMed] [Google Scholar]
- Tian B, Nowak DE, Brasier AR. A TNF-induced gene expression program under oscillatory NF-kappaB control. BMC Genomics. 2005;6:137. doi: 10.1186/1471-2164-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y. Ah receptor and NF-kappaB interplay on the stage of epigenome. Biochem Pharmacol. 2009;77:670–680. doi: 10.1016/j.bcp.2008.10.023. [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA. Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J Biol Chem. 1999;274:510–515. doi: 10.1074/jbc.274.1.510. [DOI] [PubMed] [Google Scholar]
- Tilton SC, Tal TL, Scroggins SM, Franzosa JA, Peterson ES, Tanguay RL, Waters KM. Bioinformatics resource manager v2.3: an integrated software environment for systems biology with microRNA and cross-species analysis tools. BMC Bioinformatics. 2012;13:311. doi: 10.1186/1471-2105-13-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong L, Harper SL, Tanguay RL. Evaluation of embryotoxicity using the zebrafish model. Methods Mol Biol. 2011;691:271–279. doi: 10.1007/978-1-60761-849-2_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ung CY, Lam SH, Hlaing MM, Winata CL, Korzh S, Mathavan S, Gong Z. Mercury-induced hepatotoxicity in zebrafish: in vivo mechanistic insights from transcriptome analysis, phenotype anchoring and targeted gene expression validation. BMC Genomics. 2010;11:212. doi: 10.1186/1471-2164-11-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Metre PC, Mahler BJ. Trends in hydrophobic organic contaminants in urban and reference lake sediments across the United States, 1970–2001. Environ Sci Technol. 2005;39:5567–5574. doi: 10.1021/es0503175. [DOI] [PubMed] [Google Scholar]
- Van Metre PC, Mahler BJ. Contribution of PAHs from coal-tar pavement sealcoat and other sources to 40 U.S. lakes. Sci Total Environ. 2010;409:334–344. doi: 10.1016/j.scitotenv.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Vanwezel AP, Opperhuizen A. Narcosis Due to Environmental-Pollutants in Aquatic Organisms - Residue-Based Toxicity, Mechanisms, and Membrane Burdens. Critical Reviews in Toxicology. 1995;25:255–279. doi: 10.3109/10408449509089890. [DOI] [PubMed] [Google Scholar]
- Vlecken DH, Testerink J, Ott EB, Sakalis PA, Jaspers RT, Bagowski CP. A critical role for myoglobin in zebrafish development. Int J Dev Biol. 2009;53:517–524. doi: 10.1387/ijdb.082651dv. [DOI] [PubMed] [Google Scholar]
- Wan B, Yarbrough JW, Schultz TW. Structure-related clustering of gene expression fingerprints of thp-1 cells exposed to smaller polycyclic aromatic hydrocarbons. SAR QSAR Environ Res. 2008;19:351–373. doi: 10.1080/10629360802083798. [DOI] [PubMed] [Google Scholar]
- Wassenberg DM, Nerlinger AL, Battle LP, Di Giulio RT. Effects of the polycyclic aromatic hydrocarbon heterocycles, carbazole and dibenzothiophene, on in vivo and in vitro CYP1A activity and polycyclic aromatic hydrocarbon-derived embryonic deformities. Environ Toxicol Chem. 2005;24:2526–2532. doi: 10.1897/04-440r1.1. [DOI] [PubMed] [Google Scholar]
- Wiens GD, Glenney GW. Origin and evolution of TNF and TNF receptor superfamilies. Developmental and comparative immunology. 2011;35:1324–1335. doi: 10.1016/j.dci.2011.03.031. [DOI] [PubMed] [Google Scholar]
- Williams ES, Mahler BJ, Van Metre PC. Coal-tar pavement sealants might substantially increase children's PAH exposures. Environ Pollut. 2012;164:40–41. doi: 10.1016/j.envpol.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Wu C, Lu H, Cassis LA, Daugherty A. Molecular and Pathophysiological Features of Angiotensinogen: A Mini Review. North American journal of medicine & science. 2011a;4:183–190. doi: 10.7156/v4i4p183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MT, Lee TC, Wu IC, Su HJ, Huang JL, Peng CY, Wang W, Chou TY, Lin MY, Lin WY, Huang CT, Pan CH, Ho CK. Whole genome expression in peripheral-blood samples of workers professionally exposed to polycyclic aromatic hydrocarbons. Chem Res Toxicol. 2011b;24:1636–1643. doi: 10.1021/tx200181q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong KM, Peterson RE, Heideman W. Aryl hydrocarbon receptor-mediated down-regulation of sox9b causes jaw malformation in zebrafish embryos. Mol Pharmacol. 2008;74:1544–1553. doi: 10.1124/mol.108.050435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E, Rippen M, Alexander C, Schilling TF, Crump JG. Gremlin 2 regulates distinct roles of BMP and Endothelin 1 signaling in dorsoventral patterning of the facial skeleton. Development. 2011;138:5147–5156. doi: 10.1242/dev.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.