Abstract
Epigenetic histone modifications play critical roles in the control of gene transcription. Recently, an increasing number of histone H2A deubiquitinases have been identified and characterized. However, the physiological functions for this entire group of histone H2A deubiquitinases remain unknown. In this study, we revealed that the histone H2A deubiquitinase MYSM1 plays an essential and intrinsic role in early B-cell development. MYSM1 deficiency results in a block in early B-cell commitment and a defect of B-cell progenitors in expression of EBF1 and other B-lymphoid genes. We further demonstrated that MYSM1 de-represses EBF1 transcription in B-cell progenitors by orchestrating histone modifications and transcription factor recruitment to the EBF1 locus. Thus, this study not only uncovers the essential role for MYSM1 in gene transcription during early B cell development, but also underscores the biological significance of reversible epigenetic histone H2A ubiquitination.
Introduction
Protein ubiquitination plays a critical role in a variety of cellular processes, including protein degradation and trafficking, cell cycle regulation, DNA repair, and transcriptional regulation (Komander et al., 2009). Nucleosomes, the basic units of chromatin, are composed of genomic DNA wrapped around the core histone (H2A, H2B, H3, and H4) octamer. Histone ubiquitination was discovered in 1975, and has since been found to be a relatively abundant modification in eukaryotic organisms (Goldknopf et al., 1975; Komander et al., 2009). Histone H2A at the conserved residue lysine (K) 119 was found to be mono-ubiquitinated (Komander et al., 2009). Polyubiquitination of a protein is often marked for its degradation, whereas monoubiquitination of histones usually represents a non-degradative signal (Komander et al., 2009). Ring1B and Bmi1, the core components of the Polycomb Repressive Complex 1 (PRC1), can ubiquitinate histone H2A (Cao et al., 2005; de Napoles et al., 2004; Wang et al., 2004a; Wang et al., 2004b). Another H2A ubiquitinase, hRUL138, in the N-CoR and HDAC1 corepressor complex was recently found to suppress chemokine genes (Zhou et al., 2008). H2A ubiquitination is generally associated with gene silencing and X chromosome inactivation (Cao et al., 2005; de Napoles et al., 2004; Wang et al., 2004a).
Recently, an increasing number of histone H2A deubiquitinases, including ubiquitin-specific proteases (USP) 16 (Ubp-M), USP21, USP22, MYSM1, and PR-DUB, have been identified and characterized (Joo et al., 2007; Nakagawa et al., 2008; Scheuermann et al., 2010; Zhao et al., 2008; Zhu et al., 2007). USP16 plays a role in regulating the mitotic phase of the cell cycle and homeobox gene expression (Cai et al., 1999; Joo et al., 2007). USP22 is able to deubiquitinate histone H2A and H2B in vitro and is required for androgen receptor transcription activation (Zhao et al., 2008). Zhu et al. identified Myb-like, SWIRM, and MPN domain-containing protein 1 (MYSM1) as a histone H2A deubiquitinase (2A-DUB) (Zhu et al., 2007). The JAMM and MPN metalloenzyme domain possesses an intrinsic metalloprotease activity that hydrolyzes the isopeptide bonds of ubiquitin chains (Sato et al., 2008). The SANT (switching-defective protein 3, adaptor 2, nuclear receptor co-repressor, and transcription factor IIIB) domain is similar to the DNA-binding domain of Myb-related proteins and is a motif that exists in many transcription regulators and is capable of binding to DNA and histones (Boyer et al., 2004). The SWIRM domain is named for its presence in the proteins Swi3, Rsc8, and Moira, which are members of the SWI/SNF-family of ATP-dependent chromatin remodeling complexes, and favors interactions with linker DNA and histone H3 (Yoneyama et al., 2007). MYSM1 is required for the activation of several target genes in prostate cancer cells and is a component of a complex that included the histone acetyltransferase PCAF (Zhu et al., 2007). Although the mechanisms by which histone ubiquitinases regulate gene transcription are unclear, it was proposed that MYSM1 forms a regulatory complex to regulate transcription by a stepwise coordination of histone acetylation, H2A deubiquitination, and linker histone H1 disassociation from the nucleosome (Zhu et al., 2007). Among various histone modifications, histone ubiquitination remains the least understood despite the early discovery. In particular, the physiological functions for this entire group of histone H2A deubiquitinases remain unknown. The present study revealed that the histone H2A deubiquitinase MYSM1 is essential for early B-cell development by de-repressing EBF1 transcription.
Results
MYSM1 is essential for B-cell development
To investigate the physiological role of MYSM1, we generated Mysm1 mRNA truncation-first floxed mice from the Mysm1-targeted sperms in the C57BL/6J background provided by the Knockout Mouse Project (KOMP) Repository (Supplementary Fig. S1a). The Mysm1 mRNA truncation-first floxed mice were characterized using genomic polymerase chain reaction (PCR) and Southern blot analyses. To avoid potential transcriptional leakage of the splice acceptor-capture and RNA polyA termination strategy, Mysm1-/- mice were generated by crossing the Mysm1 mRNA truncation-first floxed mice with MMTV-cre mice in the B6129F1 background to delete the floxed Mysm1- exon (Fig. S1a). MMTV-cre mice have a widespread pattern of Cre expression in various cells including B and T cells and their progenitors (Kasper et al., 2006; Kim et al., 2008). Homozygous Mysm1-/- mice were fertile and viable, although they had truncated tails and growth retardation (Fig. S1b). Heterozygous mice did not differ in morphology, growth and viability from their wild-type (WT) littermates. MYSM1 protein expression was drastically down-regulated in the spleen and thymus as well as other tissues of the Mysm1-/- mice, as determined by Western blot analysis (Fig. S1c). MYSM1 mRNA in sorted B-cells, T-cells and hematopoietic stem cells (HSC) was reduced by more than 80% in the Mysm1-/- mice, as detected by qRT-PCR (Fig. S1d).
We analyzed the effect of MYSM1 deficiency on the hematopoietic system by comparing the Mysm1-/- mice with WT littermates. Several tissues and organs of ~6 week-old mice were analyzed by flow cytometry to enumerate B-cells and other hematopoietic cells. In the bone marrow (BM), spleen (SP), peripheral blood (PB), and lymph nodes (LN), Mysm1-/- mice showed a drastic decrease in the percentage of B220+CD19+ B-cells, compared to WT MMTV-cre littermates (Fig. 1a). Moreover, in Mysm1-/-mice, peripheral B-cells that expressed surface IgM and IgD were markedly reduced in various tissues (Figs. 1b&c). In contrast to the drastic reduction in B-cell frequency, the frequencies of CD3+ T and CD4+ T lymphocytes in various tissues from Mysm1-/-mice were largely normal or increased compared to WT littermates (Fig. 1d&e, Fig. S2a). Moreover, the frequencies of other cell lineages, including Gr1+CD11b+ myeloid cells, Ter119+ erythrocytes, and CD41+Ter119 -megakaryocytes were largely normal or elevated in various tissues of Mysm1-/-mice (Fig. S2b-e). Absolute numbers of all hematopoietic lineage cells were reduced by various degrees (Fig. S2f). However, the reduction in B220+CD19+ B-cell number was most dramatic in Mysm1-/-mice, with only 0.25% of B cell numbers in WT mice (Fig. 1f). Collectively, these data demonstrate that MYSM1 is essential for B-cell development.
Fig. 1. MYSM1 is essential for B-cell development.

a-d. Representative flow cytometry profiles of bone marrow (BM), spleen (SP), lymph node (LN) and peripheral blood (PB) from homozygous Mysm1-/-mice and WT littermates (n = 8 per group, ~6 weeks old) to identify total B-cells (B220+CD19+) (a), percent cells expressing IgM and IgD (b) or immature B-cells (B220+IgM+) (c), and T cells (CD3+) (d). Numbers in quadrants indicate percent cells in each.
e. Percents of B220+CD19+ B-cells, CD3+ T-cells, CD3+CD4+ and CD3+CD8+ T-cells in indicated tissues of WT and Mysm1-/-mice (n=4 per group).
f. Percentages of indicated cell numbers from BM of Mysm1-/- mice compared to those from WT mice (n=4). Error bars indicate +SD.
MYSM1 is required for early B-cell lineage commitment
Next we examined the B-cell compartment of the bone marrow. Figs. 2a&b show that frequencies of pre-B cells (CD19+CD25+IgM -CD43 -) and pro-B cells (B220+CD43+CD19+c-Kit+) were dramatically reduced in the bone marrow of Mysm1-/-mice. To examine the pre-pro-B cell population, we used Ly6C and DX5 markers to gate out natural killer (NK) cells (DX5+) and plasmacytoid dendritic cells (pDCs) (Ly6C+) (Rumfelt et al., 2006) in order to better identify B220+CD19-CD93+ Sca1loc-KitloIL7Rα+ pre-pro-B cells. Fig. 2c shows a reduction in frequencies of the pre-pro-B cell population in Mysm1-/-BM. In agreement, absolute cell numbers of pro-B and pre-B cells were drastically reduced (Figs. 2d&e). Cell numbers of pre-pro-B cells were reduced at a lesser degree. We also examined the HSC compartment of Mysm1-/-and WT mice and observed that frequencies of Lin-Sca1+c-Kit+ (LSK) HSCs and common lymphoid progenitors (CLPs) (Lin-IL-7Rα+cKit+Sca1+) were not apparently compromised in Mysm1-/-mice (Fig. 2f). We noticed that most of CLP cells in the BM of Mysm1-/-mice were Ly6D-all-lymphoid progenitors (ALPs) with only few Ly6D+ B-cell-biased lymphoid progenitors (BLPs) (Inlay et al., 2009) (Fig. 2f). We also examined T cell development and found that frequencies of both CD4+CD8+ double positive (DP) and double negative (DN) T-cell precursor populations were roughly comparable in WT and Mysm1-/-thymus, although the DN1 population frequency was increased and DN2 frequency was reduced (data not shown). In addition, MYSM1 was broadly expressed in B-cells and their precursors and other hematopoietic cells, as detected by quantitative reverse transcriptase (qRT)-PCR (Fig. S3). Collectively, these data demonstrate a block in the early B-cell development in Mysm1-/-mice.
Fig. 2. Block in early B-cell development in the absence of MYSM1.
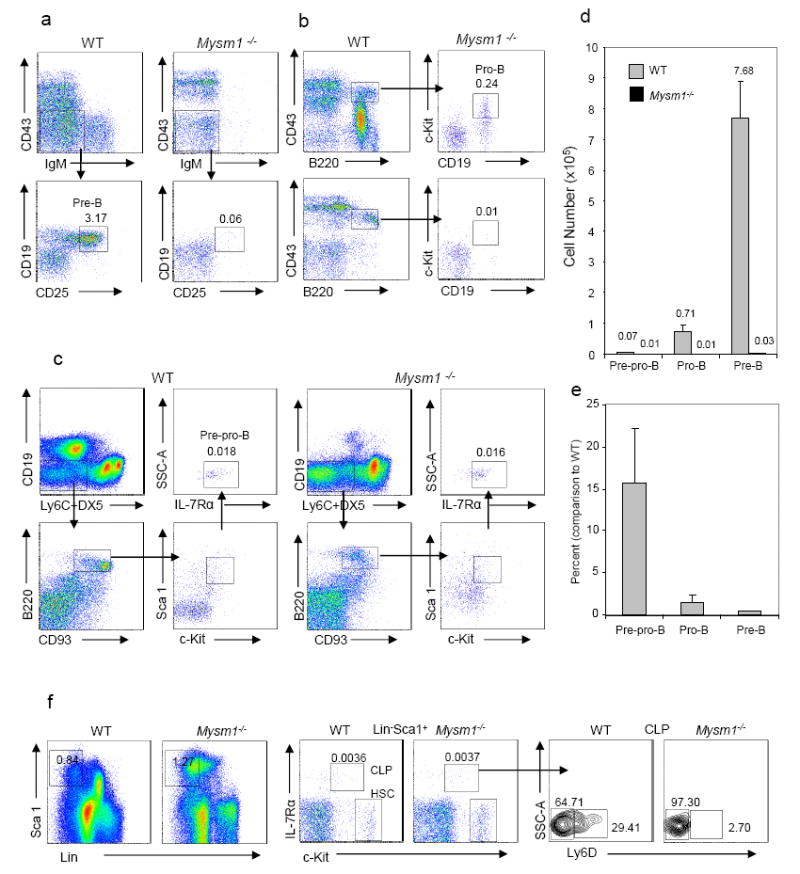
a-c. Representative flow cytometry of the bone marrow of ~6-week-old homozygous Mysm1-/-mice and WT littermates (n = 3-6 per group). Numbers adjacent indicate percentage of pre-B cells (CD19+CD25+IgM-CD43-) (a), pro-B cells (CD43+B220+CD19+c-Kit+) (b), and pre-pro-B cells (B220+CD19 -Ly6C-DX5-Sca1loc-KitloIL7Rα+CD93+) (c) in total BM nucleated cells.
d & e. Absolute numbers (d) of indicated cells in BM of Mysm1-/-mice and WT littermates per femur and percentages (e) of Mysm1-/-cell numbers compared to WT cell numbers (n = 3-6 per group) from one of three independent experiments.
f. Representative flow cytometry plots of bone marrow HSC (Lin-Sca1+c-Kit+), CLP (Lin-IL-7Rα+Sca1+c-Kit+) and Ly6D-or Ly6D+ CLP populations of Mysm1-/-mice and WT littermates (n = 4). Numbers adjacent indicate percents of indicated subpopulations.
MYSM1 has an intrinsic role in B-cell development
To investigate whether MYSM1 has an intrinsic role, in vitro pre-B cell colony forming assays using MethoCult M3630 (StemCell) supplemented with interleukin-7 (IL-7) were performed. Fig. 3a shows that no B-cell colonies were formed from Mysm1-/-bone marrow cells in repeated attempts. To rule out the possibility that the inability of forming pre-B cell colonies is due to the lack of hematopoietic stem cells, we first sorted LSK cells and then performed B-cell colony formation assays on the OP9 stromal cell culture in the presence of stem cell factor (SCF), Fms-like tyrosine kinase-3 (Flt3), and IL-7 (Greig et al., 2010). Fig. 3b also shows the inability of isolated Mysm1-/-LSK cells to form B-cell colonies in repeated experiments. To confirm these results, we further examined the propagation of sorted pro-B cells on OP9 stromal cells in the presence of IL-7. Fig. 3c also shows that the Mysm1-/-pro-B cells failed to expand in IL-7-conditioned cultures. However, we noticed that expression of IL-7Rα chain on CLP and pre-pro-B cells (Figs. 2c&f), as well as on thymic DN and DP T-cells (data not shown) was not apparently perturbed in Mysm1-/-mice.
Fig. 3. MYSM1 has an intrinsic role in B-cell development.
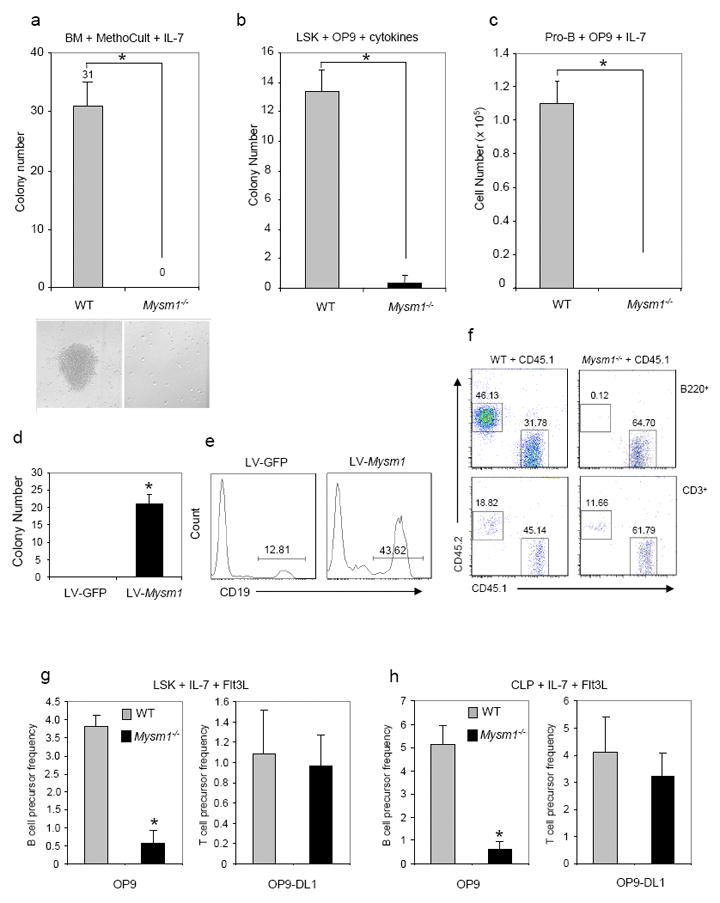
a. Pre-B colony assays of bone marrow cells from Mysm1-/-mice and WT littermates. B-cell colony numbers (n=3) and representative colony morphology (bottom) on day 7 of MethoCult M3630 culture (StemCell Technologies) containing IL-7 are shown from one of three repeated experiments. *P, <0.01, Mysm1-/-vs. WT.
b. B-cell colony formation assays of sorted Lin-c-Kit+Sca1+ (LSK) hematopoietic stem cells on the OP9 stromal cell culture in the presence of SCF, Flt3, and IL-7 for 10 days. B-cell colony numbers per well (n=3) are shown from one of two repeated experiments. *P, <0.01, Mysm1-/-vs. WT.
c. In vitro proliferation of sorted pro-B cells (B220+CD43+IgM-) from the bone marrow on OP9 stromal cells supplemented with IL-7 for 5 days. B-cell numbers per well (n=3) are shown from one of two repeated experiments.
d & e. MYSM1 rescue assays. Bone marrow cells were transduced with a recombinant lentiviral vector LV-Mysm1 or a control LV-GFP. The transduced BM cells were then subject to pre-B colony assays and B-cell colony numbers were scored on day 7 (d). Flow cytometric analysis was performed on day 10 of the culture (e). Data are showed from one of three repeated experiments. *P, < 0.01, LV-Mysm1 vs. LV-GFP.
f. Bone marrow transplantation. A 1:1 mixture of whole BM cells from CD45.2 Mysm1-/-mice or WT littermates and CD45.1 congenic mice were injected retro-orbitally into irradiated (9.5 Gy) CD45.1 congenic mice (2 × 106 cells/mouse, n=6 per group). After 3 wks, the relative percentages of B220+ B-cells or CD3+ T cells expressing donor (CD45.2+) or recipient (CD45.1+) derived population were determined by flow cytometry analysis. Results are representative of two independent experiments.
g & h. B-cell and T-cell colony formation assays of HSC (LSK) (g) and CLP (h) cells sorted from BM of WT and Mysm1-/-mice on the OP9 or OP-DL1 stromal cell culture in the presence of Flt3 and IL-7 for 10 days. B-cell and T-cell colony numbers per 100 seeded WT or Mysm1-/-HSC or CLP cells are shown from one of two repeated experiments. *P, < 0.01, Mysm1-/-vs. WT OP9 culture.
To further investigate the role for MYSM1 in B-cell development, we produced a recombinant lentiviral vector (LV) that coexpressed a full-length MYSM1 and GFP marker for rescue assays (Fig. S4). Figs. 3d&e shows that Mysm1-/-BM cells transduced with LV-MYSM1 vector, but not with LV-GFP, were able to form B-cell colonies in the IL-7-conditioned culture and that differentiated cells were CD19+, indicating that the defect in B-cell differentiation of Mysm1-/-BM can be rescued by the forced expression of MYSM1. Moreover, we performed bone marrow (BM) transplantation assays to assess the intrinsic effects of MYSM1 in vivo. Lethally irradiated congenic CD45.1 WT mice received a 1:1 mixture of CD45.2 bone marrow cells from Tek-cre:Mysm1 mRNA truncation-first floxed mice or WT mice and CD45.1 WT mice. Upon analyzing the mice at 3 wk post transplantation, we observed the inability of the donor Mysm1-/-BM cells to generate B220+ B-cells in the recipient mice (Fig. 3f). To determine that MYSM1 has a different role in B-cell and T-cell commitment further, we then cultured LSKs from BM of WT and Mysm1-/-mice with OP9 stromal cells and OP9-DL1 that express Delta-like 1 (DL1) in the presence of cytokines Flt3L (FMS-like tyrosine kinase 3 ligand) and IL-7 to induce T cell, as well as B-cell differentiation in vitro (Li et al., 2010). Fig. 3g shows that T cell development of Mysm1-/-LSK was largely normal in vitro while B-cell development was abrogated. To verify the results of LSK culture, we also cultured CLPs sorted from WT and Mysm1-/-mice on either OP9 or OP9-DL1 stromal cells. The relatively normal T cell development and inability of B-cell development of Mysm1-/-CLP was also observed (Fig. 3h). Collectively, these data demonstrate that MYSM1 is critical and intrinsic for instructing B-cell, but not T-cell lineage commitment.
MYSM1 de-represses Ebf1 transcription
B-cell lineage commitment and early B-cell development is governed by a small set of transcription factors (Nutt and Kee, 2007). Given that histone H2A ubiquitination is generally involved in the repression of target gene transcription (Zhou et al., 2008; Zhu et al., 2007), we set out to examine whether MYSM1 regulates the transcription of transcription factors that are important for B-cell lineage commitment and development. We examined the expression of a panel of transcription factors in CLPs and pre-pro-B cells by qRT-PCR analysis. CLPs (Lin-IL-7Rα+Sca1+c-Kit+) and pre-pro-B cells (B220+CD43+CD19-CD24-Ly6C-DX5-c-Kit+IL7Rα+CD93+) were isolated from BM cells by flow cytometric sorting. Figs. 4a&b show a drastic reduction in the mRNA of EBF1, Pax5, and other transcription factors that are critical for B-cell lineage commitment and development in sorted Mysm1-/-CLPs and pre-pro-B cells. In contrast, the expression of the genes important for T-cell lineage commitment and development were normal or elevated in Mysm1-/-CLPs and thymic CD4+ CD8+ DN T-cells (Figs. 4a&c). Consistent with the observations by flow cytometric analyses, these qRT-PCR data indicate that MYSM1 is required for the transcription of B-cell lineage-promoting genes, such as EBF1, Pax5, and downstream genes.
Fig. 4. MYSM1 is required for the transcription of B-lymphoid genes.
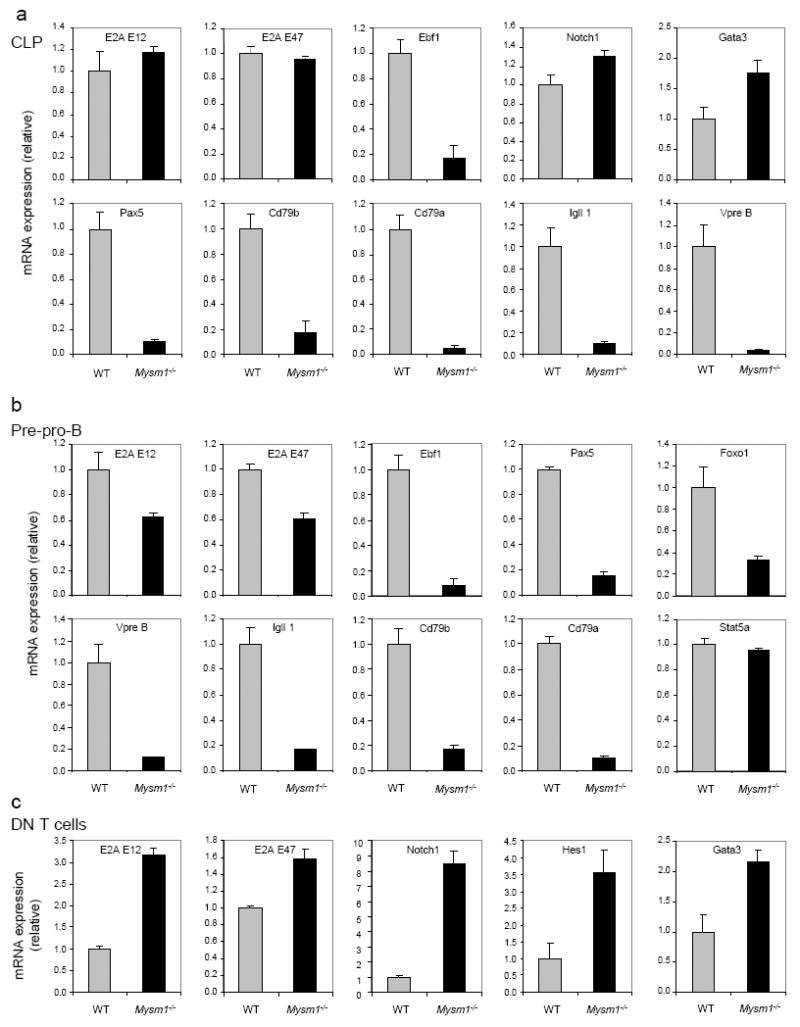
Expression of representative transcription factors in sorted CLP (Lin-IL-7Rα+Sca1+c-Kit+) (a), pre-pro-B cells (B220+CD43+CD19-CD24-Ly6C-DX5-c-Kit+IL7Rα+CD93+) (b) and thymic CD4+ and CD8+ DN T-cells (c) of WT and Mysm1-/-mice detected by qRT-PCR assays. mRNA expression was normalized to hypoxanthine-guanine phosphoribosyl transferase (Hprt) mRNA amount and presented related to their expression in WT cells, set as 1. Data are representative of three independent experiments. Each sample was analyzed in triplicate, and error bars indicate ±SD.
Since EBF1 is a master transcription factor for B-cell lineage commitment and early development (Lin and Grosschedl, 1995), we investigated the association of MYSM1 with the EBF1 locus using chromatin immunoprecipitation (ChIP) assays. A panel of PCR primer pairs to encompass the enhancer, promoter and 5’ coding region of the Ebf1 locus (Lin and Grosschedl, 1995; Pongubala et al., 2008) were used (Fig. 5a). Immunoprecipitation with the MYSM1-specific antibody, but not control IgG, enriched the sequences located at the Ebf1α and Ebf1β promoter and enhancer regions (Fig. 5b). In addition, we observed that anti-MYSM1 ChIP did not enrich for the Ebf1 promoter sequences in thymic T cells (Fig. 5c). These data further demonstrate the direct association of MYSM1 with the Ebf1 promoter region in B-cells and their precursors. To test whether MYSM1 is directly involved in activating Ebf1 transcription, Ebf1α and Ebf1β promoter and enhancer region DNA fragments, as well as the promoter region fragments of Pax5 and Cd79a were cloned into the pGL3-Basic Luc reporter vector (Promega), respectively (Fig. S5a). Subsequently, we performed transcription assays by co-transfecting one of these Luc reporter vectors and the MYSM1 expression vector into Ba/F3 cells (Sharabi et al., 2008; Zhu et al., 2007). MYSM1 expression activated transcription of the Ebf1α promoter and modestly activated the Ebf1β promoter, but only marginally activated the Pax5 promoter and Cd79a promoter. Moreover the activation of the Ebf1α and Ebf1β promoters occurred in a dose-dependent manner (Fig. S5b-d). To further demonstrate the role of MYSM1-controlled EBF1 expression in B-cell development, we performed a rescue assay with a retroviral vector expressing EBF1 (MIG-Ebf1) (Greig et al., 2010). Figs. 5d&e show that Mysm1-/-BM cells transduced with MIG-Ebf1, but not with control retroviruses, were able to form B-cell colonies and to develop into CD19+ B-cells in vitro. However, Mysm1-/-BM cells transduced with MIG-Pax5 and MIG-Il7ra were still unable to form B-cell colonies. Moreover, higher endogenous EBF1 expression was detected in WT Lin- BM cells transduced with the lentiviral vector expressing MYSM1 (LV-Mysm1), compared to WT cells transduced with a control LV-GFP vector (Fig. S5e). Collectively, these data demonstrate that MYSM1 is essential for activating Ebf1 transcription, thus controlling B-cell lineage commitment and development.
Fig. 5. MYSM1 targets the Ebf1 locus.
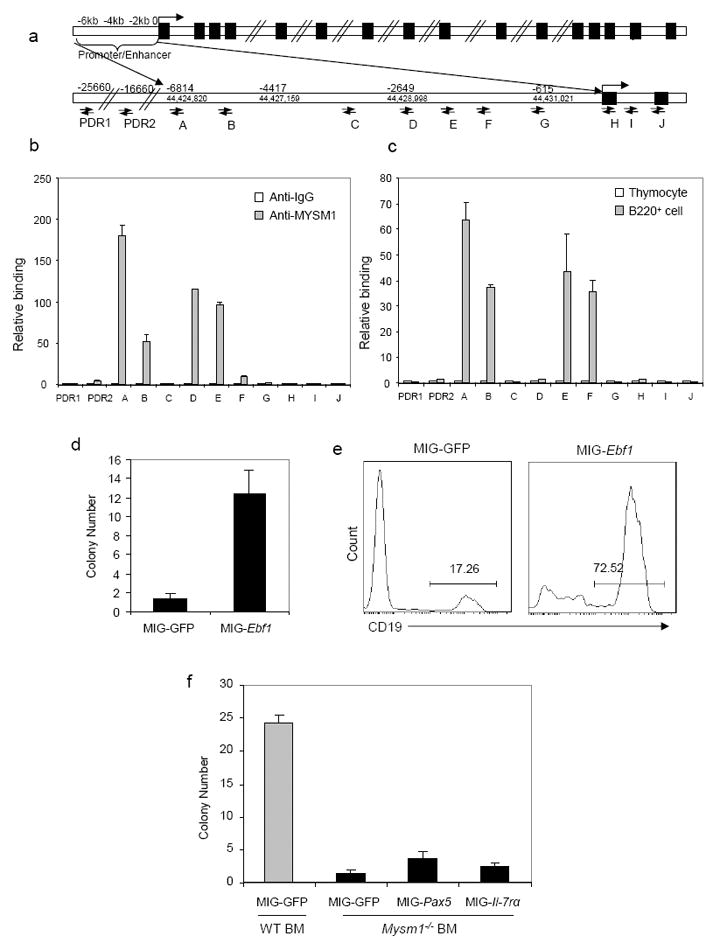
a. Schematic diagram of the Ebf1 gene and its promoter and enhancer region illustrating the positions of the primers used for ChIP assays.
b & c. ChIP assays of WT B220+ B cells using a MYSM1 antibody probing for the Ebf1 gene and regulatory sequence pull-down or IgG non-specific pull-down (control) (b). ChIP assays of WT B220+ B-cells and WT thymocytes using the MYSM1 antibody (c). Quantitative PCR was used to analyze the enrichment. The fold changes of enriched DNA are presented from one of three independent experiments. PDR: primers for a “promoter desert” region of genomic DNA upstream of the Ebf1 promoter.
d-f. EBF1, Pax5 and IL-7Rα rescue assay. WT or Mysm1-/-lin-BM cells were transduced with a recombinant retroviral vector that expresses mouse EBF1 (MIG-Ebf1) (d & e), Pax5, IL-7Rα (f) or a control vector (MIG). The transduced BM cells were then subject to B-cell colony formation assays. Colony numbers were examined and flow cytometric analysis performed on day 10 (e) of the culture. Data are showed from one of two independent experiments.
MYSM1 orchestrates histone modifications and transcription factor recruitment at the Ebf1 locus
In an attempt to investigate how MYSM1 activates Ebf1 transcription, we first determined whether MYSM1 deubiquitinates ubH2A at the Ebf1 promoter locus of B-cell precursors. To get sufficient numbers of B-cell precursors for ChIP assays, we isolated BM lineage marker-negative (Lin−) progenitors, which still contain CLPs in Mysm1-/-mice (Fig. 2f). Fig. 6a shows that ubH2A was increased in the Ebf1 α and β promoter locus of Mysm1-/-Lin− progenitors, compared with that in WT Lin− progenitors. In contrast, there was no apparent difference in ubH2A in EBF1 promoter desert region (PDR) and exon 2 regions, as well as in the osteocalcin (OC) promoter locus of WT and Mysm1-/-Lin− progenitors, suggesting the selective deubiquitination of uH2A at the Ebf1 locus by MYSM1. Histone ubiquitination and deubiquitination has been shown to coordinate histone modifications and regulate gene transcription by interacting with other histone modifiers (Campos and Reinberg, 2009). We then tested whether MYSM1 may orchestrate histone modifications at the Ebf1 locus. Figs. 6b-c show that repressive trimethylated histone H3 at lysine 27 (H3K27me3) at the Ebf1α and β promoter locus significantly elevated in Mysm1-/-BM Lin− progenitors, whereas the permissive trimethylated histone H3 at lysine 4 (H3K4me3) levels at the Ebf1 promoter locus were decreased. We also tested whether the recruitment transcriptional factors to the Ebf1 locus was compromised due to the altered histone modifications in Mysm1-/-BM progenitors. Fig. 6d shows that the recruitment of several key transcription factors for Ebf1 transcription, such as E2A, to the Ebf1 locus was reduced in Mysm1-/-BM Lin− progenitors. We also examined the binding of E2A to other target loci (Lin et al., 2010) in Mysm1-/-BM Lin− progenitors. It was observed that the binding of E2A to its target locus Dntt was also significantly reduced in Mysm1-/-cells (Fig. S6). However, to our surprise, E2A binding to other target loci, including Pou2af1, Tcf7 and Notch1, was not reduced or even slighted increased in Mysm1-/-cells. This unexpected observation suggests a different degree of dependence on MYSM1 for the binding of E2A to different target loci. Moreover, we observed that recruitment of Ring1b and Bmi1, which are components of the repressive polycomb group (PcG) complex 1 and act as an ubiquitin E3 ligase towards histone H2A at lysine 119 (Cao et al., 2005; de Napoles et al., 2004; Wang et al., 2004a; Wang et al., 2004b), to the EBF1 locus was significantly increased in Mysm1-/-Lin− progenitors (Fig. 6d). Collectively, these data indicate that MYSM1 not only deubiquitinates ubH2A, but also orchestrates histone modifications at the Ebf1 locus and that its deficiency results in the repressive histone modifications and impaired recruitment of the key transcription factors to the Ebf1 locus.
Fig. 6. Altered histone modifications and transcription factor recruitment at the Ebf1 promoter locus in Mysm1-/-B-cell progenitors.
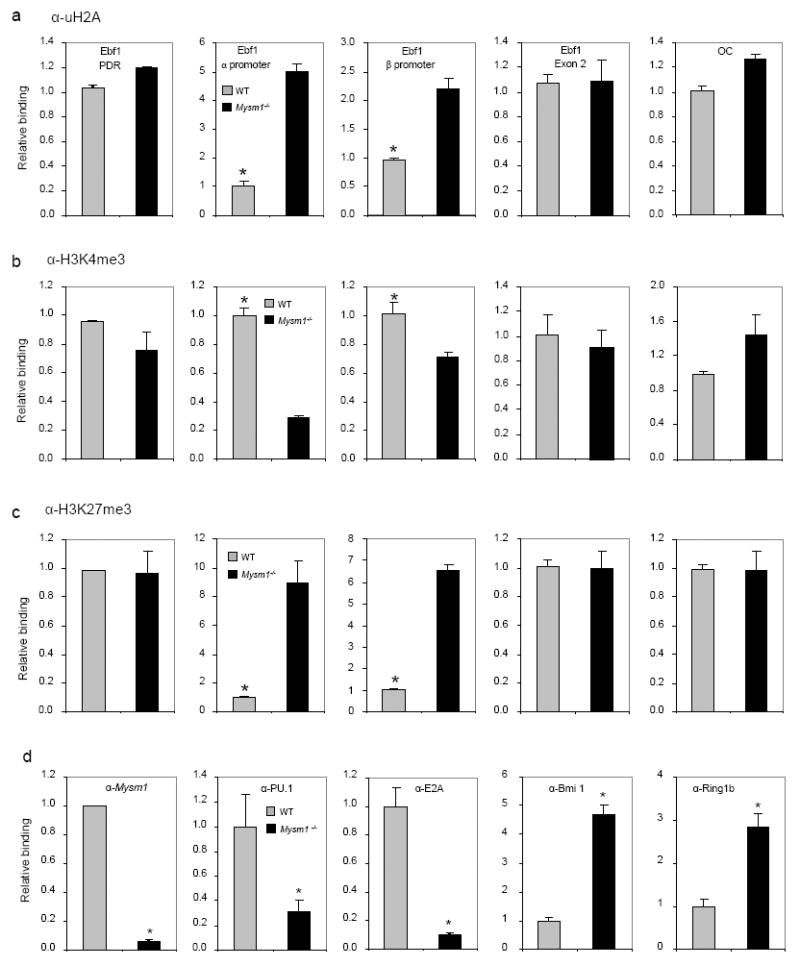
a-c. ChIP analyses of WT or Mysm1-/-BM Lin− cells using uH2A (a), H3K4me3 (b), H3K27me3 antibody (c), or IgG control. The precipitated DNA was analyzed by quantitative PCR using primers amplifying the EBF1α or EBF1β promoter regions, the EBF1 PDR region, the EBF1 exon 2 region, or the control OC promoter region, and normalized with the input DNA before compared to WT (set as 1). *p < 0.05, WT vs. Mysm1-/-.
d. ChIP analyses of the occupancy of E2A, PU.1, Bmi1 and Ring1B in WT or Mysm1-/-BM Lin− cells using a panel of antibodies indicated. The precipitated DNA was analyzed by quantitative PCR using primers (primer pair A) amplifying the EBF1α promoter region for ChIP with antibodies against E2A, Bmi1, Ring1B and MYSM1 and using primers (primer pair E) amplifying the EBF1β promoter region for ChIP with anti-PU.1, and normalized with the input DNA before compared to WT (set as 1). *p < 0.05.
Association with the transcription factor E2A and the SWI/SNF chromatin remodeling complex for EBF1 transcription
To probe the possible interaction of endogenous MYSM1 and the transcription factors of Ebf1, we performed co-immunoprecipitation assays. A weak association of endogenous MYSM1 protein and endogenous E2A (E47) protein was detected in WT BM Lin-progenitor cells, although the association of MYSM1 and PU.1, Ikaros, Stat5, or Runx1 was not positively identified (data not shown). To further investigate the interaction of MYSM1 and E2A, sequential two-step ChIP assays were performed (Zhou et al., 2008). Fig. 7a shows an association of both MYSM1 and E2A at the Ebf1 locus of WT BM Lin− progenitors. These data demonstrate an association of MYSM1 and E2A, which may be involved in the selective targeting of MYSM1 to the EBF1 locus.
Fig. 7. MYSM1 interacts with the transcription factor E2A and BRM.
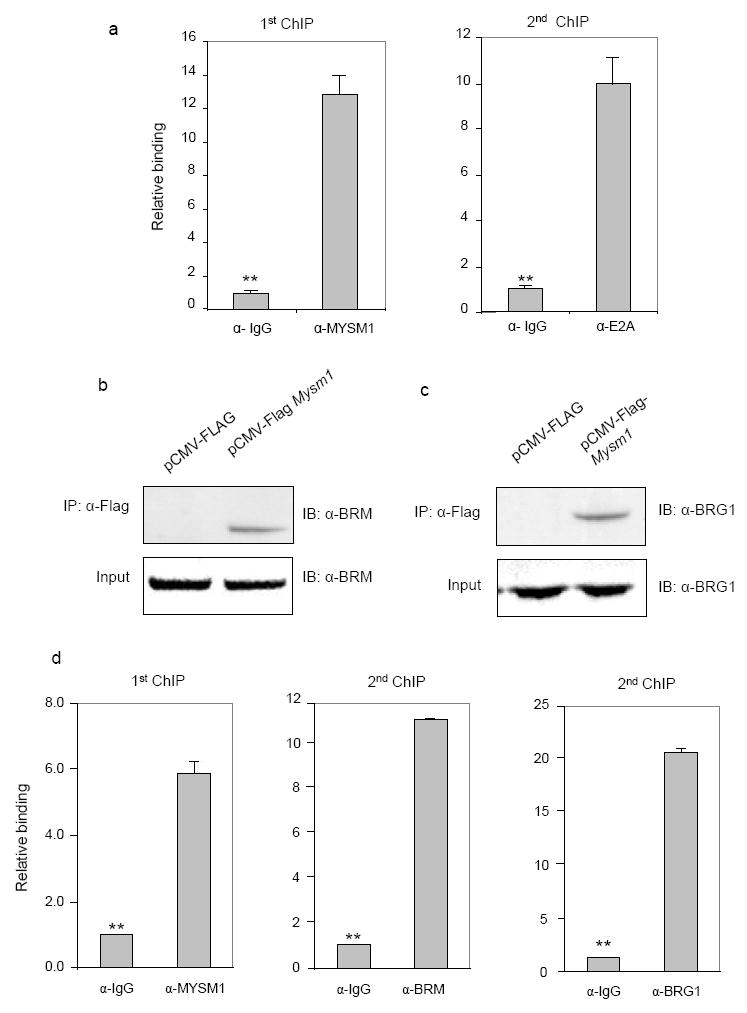
a. Sequential two-step ChIP assays of WT BM Lin-progenitors were performed, showing the recruitment of the endogenous E2A and MYSM1 to the Ebf1 promoter from one of two independent experiments. The relative binding was defined by determining the immunoprecipitation level (ratio of the amount of immunoprecipitated DNA to that of the input sample) and then comparing to corresponding 1st ChIP or 2nd ChIP control IgG immunoprecipitation level, which was set as 1.0. **p < 0.01.
b & c. Endogenous BRM and BRG1 coimmunoprecipitated with MYSM1. Cell lysates from HEK293T cells transfected with a pCMV-Flag-Mysm1, or pCMV-FLAG expression plasmid were incubated with an anti-Flag antibody, and immunoprecipitated proteins were analyzed by a BRM (b) or BRG1 (c) antibody. Ten percent of the input was loaded.
d. Sequential two-step ChIP assays of WT BM Lin-progenitors were performed from one of two repeated experiments. **p < 0.01.
To investigate how MYSM1 orchestrates histone modifications to regulate EBF1 transcription, we set out to identify MYSM1-binding partners by yeast two-hybrid screens. We used the full length mouse MYSM1, the N-terminal region containing the SANT and SWIRM domain, or the C-terminal region containing the JAMM domain, and a generic LXXLL motif as bait (Fig. S7a). This approach of using a full-length molecule, as well as its domains as bait provides an increased specificity and a broad spectrum of binding partner selection, based on our previous experience (Lee et al., 2009). Several MYSM1-binding partners that were not reported previously (Zhu et al., 2007) were identified using a human leukocyte matchmaker cDNA library for screening, including Brahma (BRM), also known as SMARCA2. The SWI/SNF (Switch/sucrose non-Fermenter) chromatin-remodeling complex contains either BRM or Brahma-related gene 1 (BRG-1) as its central ATPase subunit and is important for controlling gene transcription (Osipovich et al., 2007; Reisman et al., 2009). To confirm the interaction of MYSM1 with BRM or BRG-1, we performed co-immunoprecipitation assays. Figs. 7b&c show that MYSM1 was co-precipitated with endogenous BRM and BRG-1 in 293 cells that were transfected with the Flag-MYSM1 expression vector (Fig. S7b). To further determine the association of MYSM1 and BRM or BRG-1 at the EBF1 promoter locus, sequential two-step ChIP assays were performed. Fig. 7d shows the association of both MYSM1 and BRM and/or BRG-1 at the Ebf1 promoter locus of WT BM Lin− progenitors. We also examined whether BRM and BRG-1 were involved in MYSM1-mediated Ebf1 transcription. Early hematopoietic progenitor cells Ba/F3 were cotransfected with the Ebf1α promoter-Luc reporter, siRNA and MYSM1 expression plasmids and the transfection of BRM siRNA or BRG-1 siRNA resulted in the reduction of BRM or BRG-1 mRNA (Fig. S7c). BRM or BRG-1 silencing reduced MYSM1-mediated Ebf1 promoter transcription (Fig. S7d). In addition, we examined the effects of BRM and BRG1 down-regulation by siRNA on the expression of the endogenous Ebf1 locus. It was observed that BRM and BRG1 down-regulation of WT Lin-BM cells reduced endogenous EBF1 expression (Fig. S7e). Collectively, these data demonstrate that MYSM1 interacts with E2A and BRM and/or BRG-1 and its association is likely involved in MYSM1-mediated Ebf1 transcription in B-cell precursors.
Discussion
Understanding the physiological roles of histone H2A deubiquitination is of fundamental importance in illustrating the epigenetic regulation of gene transcription and cell development. So far, the physiological roles for the entire group of histone H2A deubiquitinases are largely unknown (Joo et al., 2007; Nakagawa et al., 2008; Scheuermann et al., 2010; Zhao et al., 2008; Zhu et al., 2007). Here, we have revealed that the histone H2A deubiquitinase MYSM1 plays an essential and intrinsic role in B-cell development by activating the pivotal target EBF1 and possibly other transcription factors in B cell progenitors. Further studies revealed that MYSM1 derepresses Ebf1 transcription as a key transcription switch by orchestrating histone modifications and transcription factor recruitment to the locus. Thus, this study uncovers the essential biological role of the histone H2A deubiquitinase MYSM1 in B cell development, as well as underscores the biological importance of histone H2A ubiquitinases in general.
The differentiation of lymphocytes from HSCs is controlled by orchestrated expression of a set of shared and lineage-specific transcription factors (Zon, 2008). Lymphopoiesis initiates with the generation of CLPs, a heterogeneous cell population with multiple differentiation potential into T, B, DC and NK cells (Inlay et al., 2009), by graded expression of the transcription factors PU.1, Ikaros and E2A, which induce the expression of components of signaling pathways, including IL-7 and Flt3, for lymphoid development (Nutt and Kee, 2007). T and B cell developmental pathways diverge, due to their expression of distinct lineage-specific transcription factors (Rothenberg et al., 2008). B-cell lineage commitment from CLPs is determined by E2A and two lineage-specific transcription factors, EBF1 and Pax5 (Cobaleda et al., 2007; Lin and Grosschedl, 1995), for their differentiation into pre-pro-B and pro-B cells (Hu et al., 2006; Johnson, 2008; Ma et al., 2006; Nutt and Kee, 2007). In contrast, T-cell lineage differentiation is driven by the expression of Notch1 and Notch-Delta signaling in lymphoid progenitors (Rothenberg et al., 2008). We found that MYSM1 is essential for B-cell lineage, but not T lineage, commitment. We identified the transcription factor EBF1 as a pivotal target gene of MYSM1 in lymphoid precursors and demonstrated that MYSM1 epigenetically de-represses EBF1 transcription by orchestrating histone modifications and regulating recruitment of the transcription factors to the EBF1 locus. EBF1 is a master transcription factor for B-cell lineage commitment and development, since its deficiency leads to a block in early pro-B cell development (Lin and Grosschedl, 1995). EBF1 activates many genes, such as Pax5, Foxo1, Cd79a, Cd79b, Igll1 (λ5), and others, important for B-cell development (Igarashi et al., 2002; Zandi et al., 2008). Although EBF1 is expressed first in CLPs, it is not essential for early lymphoid development, given that there are no defects in EBF1-deficient early progenitors including the HSC, CLPs and pre-pro-B cell populations (Zandi et al., 2008). In this study, we demonstrated that early B cell development was blocked at the ALP stage in MYSM1-deficient mice, as Ly6D+ BLPs and pro-B cells were drastically reduced. However, a considerable number of Ly6D -ALPs and pre-pro-B cells, although reduced, were still detected in MYSM1-deficient mice. The scarcity of Ly6D+ BLP cells suggests a developmental block at the ALP stage and/or a speedy transition from ALP to pre-pro-B cells in Mysm1-/-mice. It was recently reported that ALPs and BLPs were formed in normal numbers in EBF1-deficient mice, while the deficiency in IL-7 signaling blocked early B cell development at the ALP stage (Tsapogas et al., 2011). Collectively, the data convincingly show that MYSM1 is essential for early B cell development and that EBF1 is one important MYSM1 target gene. However, MYSM1 likely regulates other critical target genes, upstream of EBF1, which are responsible for the defect in the earliest stage of B cell development in MYSM1-/-mice. During the course of this study, Oguro et al. recently reported that Bmi1, which is associated with Ring1B and other components of Polycomb group (PcG) complexes and is involved in H2A ubiquitination (Cao et al., 2005; de Napoles et al., 2004; Wang et al., 2004a; Wang et al., 2004b), was not only critical for HSC maintenance, but also for lymphocyte development by repressing B-cell lineage developmental genes, Ebf1 and Pax5 (Oguro et al., 2010). Ebf1, Pax5, and downstream target genes were derepressed, while the expression of upstream genes of Ebf1, such as Sfpi1, Tcf3, and Il7ra, was not altered in Bmi1-deficient hematopoietic progenitors. Bmi1 deficiency enhanced B-cell lineage differentiation at the expense of T cell lineage via de-repressing Ebf1 and Pax5 transcription. Taken together, these data presented in this study demonstrate that MYSM1 plays an essential role in B-cell lineage commitment and development by epigenetically derepressing the transcription of Ebf1 and possibly other early lymphoid lineage developmental factors.
Transcription factors are key determinants in the complex orchestration of hematopoiesis; however, little is known about the underlying mechanisms by which transcription of these transcription factors is regulated. Due to the highly compact nature of genomic DNA in the chromatin of eukaryotic cells, a transcription complex that consists of chromatin remodelers, DNA and histone modifiers and transcription factors need to work in concert to reconfigure nucleosomes at specific locations, as well as to alter the promoter structure to expose regulatory sites for gene activation or repression (Cairns, 2009). Despite that at least five known histone H2A deubiquitinases can deubiquitinate mono-ubiquitinated K119 H2A (Joo et al., 2007; Nakagawa et al., 2008; Scheuermann et al., 2010; Zhao et al., 2008; Zhu et al., 2007), MYSM1 plays a non-redunant, essential biological role in B-cell development. We also found that MYSM1 not only deubiquitinated uH2A, but also orchestrated histone modifications at the Ebf1 locus to activate its transcription. These data suggest that MYSM1 interacts with other histone modifiers to regulate chromatin status at the Ebf1 locus.
At present, a major question of how MYSM1 is targeted to the Ebf1 gene locus remains. However, the results of this study provide some clues. MYSM1 was found to interact with the transcription activator E2A, as well as BRM and/or BRG-1, the key ATPase component of the SWI/SNF chromatin-remodeling complex (Reisman et al., 2009). Both BRM and BRG-1 are ATPases with 80% homology in their amino acid sequence and have shared activities, although they also have distinct biological roles (Reisman et al., 2009). These results, together with the data of others (Campos and Reinberg, 2009; Zhu et al., 2007), suggest that the selective gene locus targeting of MYSM1 may be guided by its partners such as transcription factors and chromatin remodelers. An interesting area of future studies is to systemically identify MYSM1 partners and investigate the underlying molecular mechanisms for the selective gene targeting and transcription regulation by MYSM1. We realize that MYSM1 plays a broad role in the hematopoietic system and regulates the transcription of Ebf1, as well as other genes. Our preliminary ChIP-seq data identified over 1,000 genes that are associated with MYSM1 in B-cells (unpublished data). We observed a general reduction in absolute numbers of hematopoietic cells of all lineages in MYSM1-deficient mice, suggesting that MYSM1 may regulate HSC, in addition to its essential role in early B-cell lineage development. Thus, further studies are warranted to illustrate the biological roles and mechanisms for the spatiotemporal control of target genes by MYSM1 in HSCs and their progeny.
Experimental Procedures
Mice
Mysm1 targeted vector (L1L2_st0) was constructed by the EUCOMM consortium (http://www.sanger.ac.uk/htgt/report/project_gene_report?project_id=27041). The vector is composed of an FRT flanked splice acceptor (En2 SA), lacZ and neomycin and polyA sequence followed by a loxP site. An additional loxP site is inserted downstream of the targeted MYSM1 exon (E3). The Mysm1 mRNA truncation-first strategy was based on inserting a cassette into an intron of an intact target gene that produces a truncated mRNA at the RNA processing level. A splice acceptor (SA) in the cassette captures the RNA transcript and an efficient polyadenylation termination signal truncates the transcript, preventing the gene from being transcribed into mRNA downstream of the cassette site (Testa et al., 2004) (Supplementary Fig. S1a). Subsequent Cre expression results in the deletion of the floxed MYSM1 exon 3 by crossing with cre transgenic mice. The Mysm1 mRNA truncation-first floxed sperm in the C57BL/6J background (Mysm1_A04; Mysm1tm1a(ΔMP)Wtsi MGI#: 2444584) were provided by the KOMP Repository at UC Davis. In vitro fertilization, microinjection, chimera production, and the generation of Mysm1 mRNA truncation-first floxed mouse founders were carried out at the USC Transgenic Mouse Core Facility. In all experiments, wt littermates (+/+) were used for controls. Mice were maintained in a pathogen-free barrier facility, and all experiments were performed in accordance with the University of Southern California Institutional Animal Care and Use Committee.
Flow cytometric analyses and cell sorting
Sample preparation and cytometric analysis and sorting were performed as described previously (Sharabi et al., 2008; Song et al., 2008). Single-cell suspensions of bone marrow (BM), thymus, lymph nodes, and spleens were prepared and were first stained for 20 min at 4 °C with CD16/CD32 Fc-blocking antibody (2.4G2), unless indicated otherwise, in flow cytometry buffer, followed by incubation with a ‘cocktail’ of antibody conjugates. Data were collected on a FACSCanto II (BD) and were analyzed with FlowJo software (TreeStar). For cell progenitor population sorting, cells from BM were first depleted of mature hematopoietic cells using a lineage cell depletion kit (Miltenyi Biotec) and isolated by FACSAria cell sorter
Semiquantitative and quantitative RT-PCR
Semiquantitative and quantitative RT-PCR were performed as described previously (Sharabi et al., 2008). Total RNA from isolated cells was purified with RNeasy Microkit (Qiagen) according to the manufacturer’s instructions. The SuperScript III First-Strand Synthesis kit (Invitrogen) was used for reverse transcription. Serially diluted cDNA was used for semiquantitative PCR analysis. A SYBR Green PCR kit (BIO-RAD) was used for quantitative real-time PCR and results were quantified with an ICycler IQ (BIO-RAD). Sequences of primer pairs are in Supplementary Tables (1-3).
Lentivirus and retrovirus production and transduction
Recombinant lentiviral vectors were produced and transduced as described in our previous publications (Shen et al., 2004). Retroviruses were produced by transient transfection of 293T cells with plasmids that encode viral proteins (pCL-10A1) and a specific gene expression vector (pMIG-EBF1 or pMIG) as described (Pongubala et al., 2008).
Bone marrow transplantation
Unfractionated BM cells (2×106) from wt and CD45.2-deficient mice were transplanted into lethally irradiated (9.5 Gy) wild type C57BL/6 (CD45.1) mice through retro-orbital injection. PE conjugated CD45.2 and FITC conjugated CD45.1 (BD Bioscience) were used to determine the donor or recipient derived population by flow cytometry analysis (Sharabi et al., 2008). Peripheral blood was analyzed every week after transplantation.
Chromatin Immunoprecipitation
Chromatin was immunoprecipitated according to the manufacturer’s instruction (Cell Signaling) (Sharabi et al., 2008). Briefly, single cell suspensions were crosslinked with 1% (vol/vol) formaldehyde. Chromatin was isolated, digested by mung bean nuclease (MNase), sheared by sonication, and immunoprecipitated with antibodies. Immunoprecipitated DNA was washed and eluted according to the manufacturer's instructions. Eluted DNA and sheared input material was analyzed by PCR or ligated to an adaptor and amplified by PCR according to the manufacturer's protocol (Illumina). For sequential-ChIP experiments, complexes from initial anti-MYSM1 ChIP were eluted, diluted and then re-immunoprecipitated with anti-Brm (610390) or anti-BRG1 (07-478) antibody (Zhou et al., 2008).
Yeast two-hybrid screen
The Matchmaker2 Gal4 yeast two-hybrid system (Clontech, Mountain View, CA) was used to screen for MYSM1 binding partners in the Y190 yeast strain with MYSM1 and its domain cloned into the pGBKT7 vector in frame to the Gal4 binding domain and a human leukocyte matchmaker cDNA library cloned into the pACT2 vector in frame with the Gal4 transactivation domain (Clontech). Library screening and recovery of plasmids will be performed according to manufacturer’s instructions and was described previously (Lee et al., 2009).
Statistics
Groups of three to eight mice were used for statistical analysis. P values were calculated with Student's t-test.
Supplementary Material
Acknowledgments
We thank Bangxing Hong, Michelle Chong, Martina Gatzka, Sung-Hyung Lee and other members of the Chen lab for valuable assistance and suggestions. We thank Nancy Wu, Robert Maxson, Omid Akbari, Ping Wang, Michael Kahn, Judd Rice, Michael Stallcup, Peter Jones, and other colleagues at USC, Stephen Nutt, Biao Zheng, Harinder Singh and Kay Medina for providing valuable reagents and help. This work was supported by grants from the National Institute of Health (R01CA090427, AI084811, CA116677 and AI068472 to SYC, and CA100841 and AI08185 to XFH) and Leukemia & Lymphoma Society SCOR Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions X.X.J. Q.N. designed and performed experiments, and analyzed data; T.W. YC, PY, LW, L.J. V.N, H.W, and X.F.H. performed experiments and analyzed data; HRL and JJ performed yeast two-hybrid screening and provided reagents; MM provided reagents and analyzed data; S.Y.C. designed experiments, supervised the research and analyzed data. S.Y.C. and X.X.J. wrote the manuscript.
References
- Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- Cai SY, Babbitt RW, Marchesi VT. A mutant deubiquitinating enzyme (Ubp-M) associates with mitotic chromosomes and blocks cell division. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2828–2833. doi: 10.1073/pnas.96.6.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: Annotating Chromatin. Annual Review of Genetics. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Yi, Zhang Y. Role of Bmi-1 and Ring1A in H2A Ubiquitylation and Hox Gene Silencing. Molecular Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud J, Wakao R, Tang Y, Endoh M, Appanah R, Nesterova T, Silva J, Otte A, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Goldknopf IL, Taylor CW, Baum RM, Yeoman LC, Olson MO, Prestayko AW, Busch H. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. Journal of Biological Chemistry. 1975;250:7182–7187. [PubMed] [Google Scholar]
- Greig KT, de Graaf CA, Murphy JM, Carpinelli MR, Pang SHM, Frampton J, Kile BT, Hilton DJ, Nutt SL. Critical roles for c-Myb in lymphoid priming and early B-cell development. Blood. 2010;115:2796–2805. doi: 10.1182/blood-2009-08-239210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Wang B, Borde M, Nardone J, Maika S, Allred L, Tucker PW, Rao A. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. 2006;7:819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
- Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 Locus Marks the Earliest Lymphocyte Progenitors in Bone Marrow. Immunity. 2002;17:117–130. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes & Development. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Hashimshony T, Sawai CM, Pongubala JMR, Skok JA, Aifantis I, Singh H. Regulation of Immunoglobulin Light-Chain Recombination by the Transcription Factor IRF-4 and the Attenuation of Interleukin-7 Signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Fukuyama T, Biesen MA, Boussouar F, Tong C, de Pauw A, Murray PJ, van Deursen JMA, Brindle PK. Conditional Knockout Mice Reveal Distinct Functions for the Global Transcriptional Coactivators CBP and p300 in T-Cell Development. Mol Cell Biol. 2006;26:789–809. doi: 10.1128/MCB.26.3.789-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YW, Koo BK, Jeong HW, Yoon MJ, Song R, Shin J, Jeong DC, Kim SH, Kong YY. Defective Notch activation in microenvironment leads to myeloproliferative disease. Blood. 2008;112:4628–4638. doi: 10.1182/blood-2008-03-148999. [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, Chang H, Zhou FC, Gao SJ, Liang C, et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009;11:1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Leid M, Rothenberg EV. An Early T Cell Lineage Commitment Checkpoint Dependent on the Transcription Factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Turetsky A, Trinh L, Lu R. IFN Regulatory Factor 4 and 8 Promote Ig Light Chain {kappa} Locus Activation in Pre-B Cell Development. J Immunol. 2006;177:7898–7904. doi: 10.4049/jimmunol.177.11.7898. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M, et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di-and trimethylation. Genes & Development. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Kee BL. The Transcriptional Regulation of B Cell Lineage Commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Oguro H, Yuan J, Ichikawa H, Ikawa T, Yamazaki S, Kawamoto H, Nakauchi H, Iwama A. Poised Lineage Specification in Multipotential Hematopoietic Stem and Progenitor Cells by the Polycomb Protein Bmi1. Cell Stem Cell. 2010;6:279–286. doi: 10.1016/j.stem.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Osipovich O, Milley Cobb R, Oestreich KJ, Pierce S, Ferrier P, Oltz EM. Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Tcrb genes. Nat Immunol. 2007;8:809–816. doi: 10.1038/ni1481. [DOI] [PubMed] [Google Scholar]
- Pongubala JMR, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19− early B cell precursors. The Journal of Experimental Medicine. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys[thinsp]63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi AB, Aldrich M, Sosic D, Olson EN, Friedman AD, Lee SH, Chen SY. Twist-2 Controls Myeloid Lineage Development and Function. PLoS Biol. 2008;6:e316. doi: 10.1371/journal.pbio.0060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Evel-Kabler K, Strube R, Chen SY. Silencing of SOCS1 enhances antigen presentation by dendritic cells and antigen-specific anti-tumor immunity. Nat Biotech. 2004;22:1546–1553. doi: 10.1038/nbt1035. [DOI] [PubMed] [Google Scholar]
- Song XT, Kabler KE, Shen L, Rollins L, Huang XF, Chen SY. A20 is an antigen presentation attenuator, and its inhibition overcomes regulatory T cell-mediated suppression. Nat Med. 2008;14:258–265. doi: 10.1038/nm1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa G, Schaft J, Hoeven Fvd, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- Tsapogas P, Zandi S, Åhsberg J, Zetterblad J, Welinder E, Jönsson JI, Månsson R, Qian H, Sigvardsson M. IL-7 mediates Ebf-1–dependent lineage restriction in early lymphoid progenitors. Blood. 2011;118:1283–1290. doi: 10.1182/blood-2011-01-332189. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004a;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical Recruitment of Polycomb Group Silencing Complexes. Molecular Cell. 2004b;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Matsuda T, Watanabe S, et al. Structural and functional differences of SWIRM domain subtypes. J Mol Biol. 2007;369:222 –238. doi: 10.1016/j.jmb.2007.03.027. [DOI] [PubMed] [Google Scholar]
- Zandi S, Mansson R, Tsapogas P, Zetterblad J, Bryder D, Sigvardsson M. EBF1 Is Essential for B-Lineage Priming and Establishment of a Transcription Factor Network in Common Lymphoid Progenitors. The Journal of Immunology. 2008;181:3364–3372. doi: 10.4049/jimmunol.181.5.3364. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA Module Mediates Histone H2A and H2B Deubiquitination, Coactivates Nuclear Receptors, and Counteracts Heterochromatin Silencing. Molecular Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A Monoubiquitination Represses Transcription by Inhibiting RNA Polymerase II Transcriptional Elongation. Molecular Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A Histone H2A Deubiquitinase Complex Coordinating Histone Acetylation and H1 Dissociation in Transcriptional Regulation. Molecular Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453:306–313. doi: 10.1038/nature07038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


