Abstract
Gene silencing is essential for regulating cell fate in eukaryotes. Altered chromatin architectures contribute to maintaining the silenced state in a variety of species. The silent information regulator (SIR) proteins regulate mating type in S. cerevisiae. One of these proteins, Sir3, interacts directly with the nucleosome to help generate silenced domains. We determined the crystal structure of a complex of the yeast Sir3 BAH domain and the nucleosome core particle at 3.0 Å resolution. We see multiple molecular interactions between the protein surfaces of the nucleosome and the BAH domain that explain numerous genetic mutations. These interactions are accompanied by structural rearrangements in both the nucleosome and the BAH domain. The structure explains how covalent modifications on H4 K16 and H3 K79 regulate formation of a silencing complex that contains the nucleosome as a central component.
Eukaryotic cells normally carry the complete set of genes needed to specify every cell type. Establishment of a specific cell fate requires the silencing of genes whose expression would disrupt that fate. Several diverse families of protein complexes maintain silencing, however the mechanisms involved are similar in Saccharomyces cerevisiae (S. cerevisiae) and in multicellular eukaryotes (1). Regulation of mating type loci S. cerevisiae in serves as a paradigm for silencing. Yeast growing as haploids can adopt two mating types, a and α. The genes that are expressed at the MAT loci determine cell fate, while genes specifying the opposite fate can be found at the silent HMLα or HMRa loci (1, 2). The silent information regulator (Sir) proteins are essential for silencing of HMLα or HMRa and as well as telomeres and the rDNA loci (1, 2).
The Sir proteins create domains of silenced chromatin. A long-standing hypothesis is that these proteins form specific repressive architectures that involve the basic unit of chromatin, the nucleosome. In support of this hypothesis, the SIR complex or Sir3 alone can compact nucleosomal arrays in vitro (3-5). The involvement of nucleosomes in the mechanism of silencing was first indicated by the observation that yeast could not silence HMLα and HMRa when they contained a mutated form of histone H4 with a deletion of the N terminal tail (6). Subsequently, specific point mutations that impacted silencing were found in the N-terminal tails and in the globular portions of core histones (7-14) and deacetylation of histone H4 was identified as a hallmark of silenced regions (15). Reporter gene expression, restriction enzyme accessibility and MNase susceptibility were used to show that domains of silenced chromatin created by the SIR complex are several kb in length (16-21).
Several aspects of the extensive body of work on Sir3 interactions with nucleosomes are especially relevant to the structural work described here. Silencing requires de-acetylation of histone H4 lysine 16 (H4K16); we describe the atomic contacts in the Sir3 binding pocket for H4K16. We also describe contacts with H3K79, whose methylation has the potential to modulate silencing. Many of the mutations in histones that impact silencing lie in the ‘LRS’ (‘Loss of rDNA Silencing’)(11, 12) domain of the nucleosome core and we describe numerous contacts between that region and Sir3. Mutations that impact silencing have been found both at the N-terminus as well as at the C-teminal part of Sir3 (22). Most of these mutations are clustered in the ’bromo associated homology’ (BAH) domain that is found in the N-terminus of Sir3 (23-26). Here we use a mutation in Sir3 (D205N) that confers increased binding to nucleosomes in vitro. Expression of the BAH D205N domain fused to LexA partially restores silencing of mating type loci in sir3 null background. This domain is able, therefore, to combine with Sir2 and Sir4 to cause partial silencing when it is attached to an ectopic dimerization domain (27).
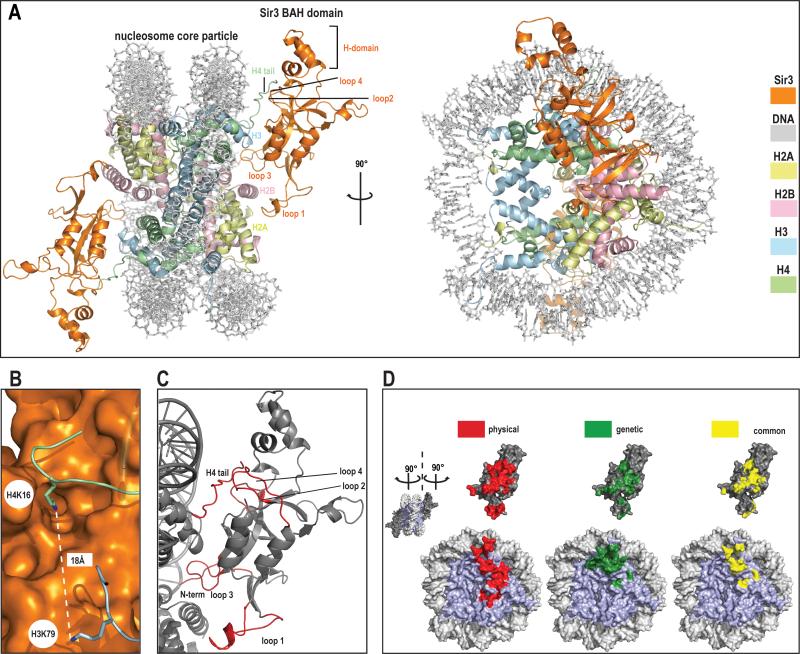
We report the crystal structure of the complex of the hypermorphic D205N Sir3 BAH domain (here after BAHSir3) and the nucleosome core particle at 3.0 Å resolution. Details of complex reconstitution, crystallization, data collection and refinement can be found in SOM (28). The BAH domain interacts extensively with each of the four core histones and consequently the solvent- accessible surface area buried between BAHSir3 and the nucleosome is large (1750 Å2, probe radius 1.4 Å). The structure shows a pseudo-two-fold symmetry, similar to that seen with the RCC1-nucleosome complex (29), in that BAHSir3 interacts in a similar manner with each of the two opposite faces of the nucleosome (Fig. 1). We observe 30 residues of BAHSir3 making contacts predominately with the core histones rather than nucleosomal DNA, suggesting that this protein-protein interface is critical to silencing.
Figure 1. Crystal structure of Sir3 BAH domain with nucleosome core particle.
a) General overview of the structure.
Two different views of the complex; front view and view rotated by 90° around y axis. The structure is color coded (BAH domain is depicted in orange, H2A in yellow, H2B in light pink, H3 in blue, H4 in green and DNA in light grey).
b) Histone H4 K16 and histone H3 K79.
Both residues that are critical in the regulation of SIR complex mediated silencing are shown in this figure. Histone H4 K16 is depicted as stick in green, histone H3 K79 in blue, the BAH domain surface is rendered in orange.
c) Folding transitions in the complex.
Both the nucleosome and BAH domain are depicted in grey and regions that get folded upon interaction are shown in red.
d) Correlation between structural and genetic contacts.
‘Open book’ view of the complex. NCP surface is shown on the bottom and BAH domain on top of the figure. Surfaces colored in red represent physical contacts as seen in the structure. Surfaces colored in green represent residues both in the NCP and the BAH domains where mutation has been shown to impact silencing. Yellow surfaces represent the overlay of physical (structure-derived) and genetic contacts.
Interactions with the core histones are mediated through five regions on the surface of BAHSir3. These regions map well to contacts inferred from genetic screens (see Figs. 1d and 2b for a summary). The BAH domain interacts with the H4 tail, which becomes folded upon binding, and the regions of histones H3 and H4 that make up the LRS domain. In addition, BAHSir3 contacts histone H2B at a position adjacent to the LRS surface and the H2A/H2B acidic patch. Of the histone residues contacted by BAHSir3 only one residue (H4V21) varies between the X. laevis histones used here and yeast histones (Fig. 4b and S3). Both of the histone residues that can be covalently modified and participate in the regulation of silencing (7-9, 30) (H3K79 and H4K16) are ordered in the structure (Fig. 1b and see below.)
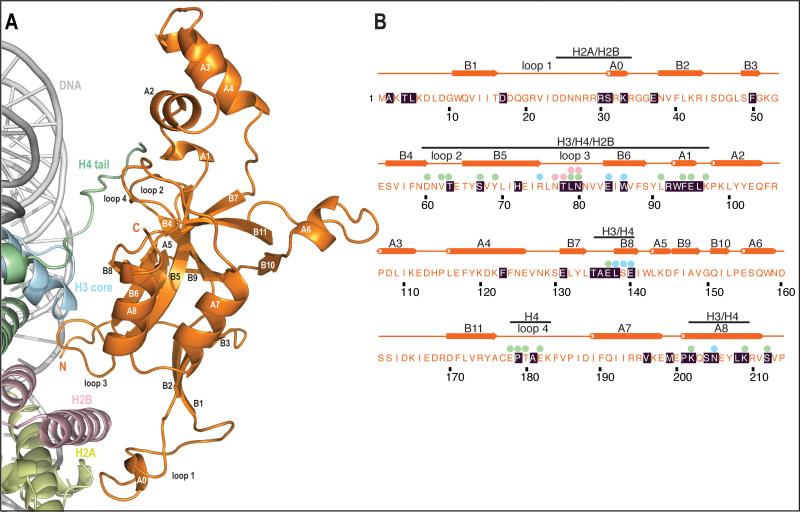
Figure 2. Overview of interactions in the complex.
a) Overview of interactions.
Same view as in Fig1a (front) BAH domain is depicted in orange, H2A in yellow, H2B in light pink, H3 in blue, H4 in green and DNA in light grey. Secondary structure elements are also depicted here. Secondary structure was assigned using KSDSSP.
b) Primary and secondary structure of D205N BAH.
Residues with black shading were mapped previously in genetic screens. Spheres above the sequence show histone interactions (within 4.1 Å) that are ordered and visible in the electron density. Colors of spheres represent which histone makes interaction with this residue of BAH domain. Bars above secondary structure inform which histone is interacting with this particular region of BAH domain. There are no spheres over residues in loop1 (residues 17-37) as this region is poorly ordered.
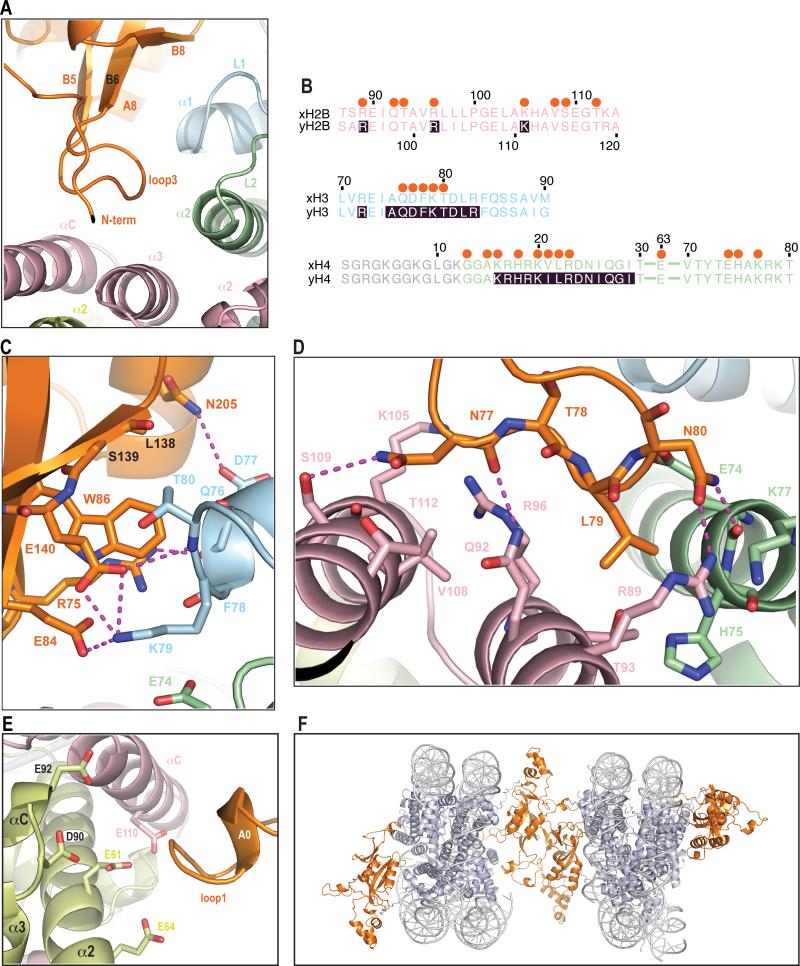
Figure 4. Interactions of BAH domain with a NCP body.
a) General view of interactions in this region.
Folded loop 3 and the β-strands that interact with regions of histones H3, H4 and H2B are shown.
b) Sequence alignment of regions of Xenopus and yeast histones (color coded the same as structure) that interact with BAH domain. Shaded residues were described in previous genetic screens. Orange spheres above the sequence depict which residues interact with BAH domain. The region of histone H4 that is disordered in the structure is depicted in grey.
c) Detailed interactions of BAH with H3.
A magnified view of a top part of panel a. Magenta dashes connect residues forming potential hydrogen bonds. Five LRS (Q76, D77, F78, K79 and T80) residues in helix α1 and loop L1 of H3 that contact the BAH domain in the structure. BAH W86 is within 4 Å of the H3Q76 carbonyl, T80 side chain and K79 Cα. There is a potential hydrogen bond between the H3D77 side chain carbonyl and the BAH N205 side chain amide. K79 could potentially form 3 hydrogen bonds with the BAH domain, one to the side chain of E84 and two to the E140 side chain. K79 conformation is further stabilized by van der Waals interactions with BAH W86 and H4E74. T80 side chain interacts with main chain of L138 and S139. BAH R75 forms polar interactions, one of which is a potential hydrogen bond with main chain of H3 residues D77 and F78. Additionally a hydrogen bond might also be formed between the BAH E140 side chain carbonyl and main chain amide of H3 T80.
d) Detailed interactions of BAH with H4 and H2B.
F78. Additionally a hydrogen bond might also be formed between the BAH
A magnified view of a bottom part of panel a. Magenta dashes connect residues forming potential hydrogen bonds. Two residues at the tip of loop 3 (L79 and N80) interact with histones H4 and histone H2B. They make van der Waals contacts with histone H4 residues E74, H75 and K77. Additionally the BAH N80 side-chain could form hydrogen bonds with main chain carbonyl of H4 E74 and side chain of H2B R89. L79 interacts with 3 H2B residues in helix a3 (R89, T93 and Q92). BAH N77 and T78 main chain carbonyls make charged interactions and a potential hydrogen bond with side-chains of H2B residues R96 and Q92, respectively. The side-chain of BAH N77 can additionally interact with four residues of H2B located in helix αC.
e) Interaction of the BAH domain with the acidic patch, same view as in Fig.1 (front).
A positively charged BAH patch (residues 28-34) is in close proximity to acidic residues E61, E64, D90 and E92 of H2A as well as residue E110 of H2B.
f) Crystal packing interaction.
Interactions between BAHSir3 and the nucleosome are established through flexible regions, which fold upon interaction (Fig. 2). Both the structure of the BAH domain and the nucleosome core particle (NCP) alone were determined previously(27, 31, 32), allowing comparison to the structure of the complex described here. One striking transition that accompanies assembly of the complex is folding and ordering of the histone H4 tail through extended interactions with loops 2 and 4 of BAHSir3 (Fig. 1c). Residues in flexible loops 1 and 3 of BAHSir3 are completely disordered in a free BAH domain structure but become ordered and partially ordered, respectively, upon binding the core region of the nucleosome. Additionally, the N-terminus of BAHSir3, which is in the vicinity of nucleosomal DNA (Fig. 1c), changes conformation upon binding the nucleosome. We conclude that BAHSir3 forms contacts with a large area of the histone octamer and that regions of the nucleosome and BAHSir3 become ordered upon this interaction.
Mutagenesis of the BAH domain of Sir3 has identified forty amino acid residues that impact silencing (Fig. 2b)(23-26). BAHSir3 contains at least 28 residues that form interactions (less than 4.1Å-distance) with a nucleosome. Of these, 17 were identified in genetic screens. Similarly, at least 30 mutations that impact silencing have been found in core histones (6, 10-14, 23, 25, 33, 34) and the structure provides an atomic description for 14 of these residues (Fig. 1d; red depicts physical contacts, green genetic contacts, yellow overlap).
Many of these mutations map to complementary electrostatic interactions in the interface between histones and BAHSir3. In several instances mutations that increase silencing increase the attractive charge in the interface between histones and BAH domain, emphasizing the importance of this type of interaction to the creation of a silenced chromatin state. The extensive correlation between mutations and molecular contacts indicates that the crystal structure reflects contacts important to biological function. We present the details of these contacts, and how they might explain both the genetic analysis and the role for covalent modification of histones in silencing, by starting with the H4 tail region and then moving through the body of the nucleosome to the acidic patch in histone H2A and H2B.
The demonstration that the N-terminus of histone H4 is critical for silencing in yeast was one of the initial findings indicating the importance of nucleosomes in transcriptional regulation. Deletions and mutations of the N-terminus of H4 (4-29) relieve silencing at HMLα and HMRa but do not impact growth of yeast (6). The charge of H4 residues 16-19 (a basic patch) was shown to be essential for silencing as mutations that sustain the positive charge maintained repression whereas mutations to glycine or glutamine abolished repression(7-9).
The histone H4 tail becomes ordered through residue G13 due to stabilizing interactions with BAHSir3. The H4 tail region interacts with loops 2 and 4, strand B5 and helix A1 of BAHSir3. Each of these structural features contains residues whose mutation generates a silencing phenotype (Fig. 2). Additionally, one residue in BAHSir3 located between strands B7 and B8 participates in this interaction (Fig. 3b). Binding interactions are largely electrostatic between the positively charged histone H4 tail and the negatively charged surface of th residues in BAH domain (Fig. 3c). Sixteen residues in BAHSir3 interact with H4 tail residues 13-23, primarily through their side-chains (Fig. 3b, c, Fig. 4b, SOM).
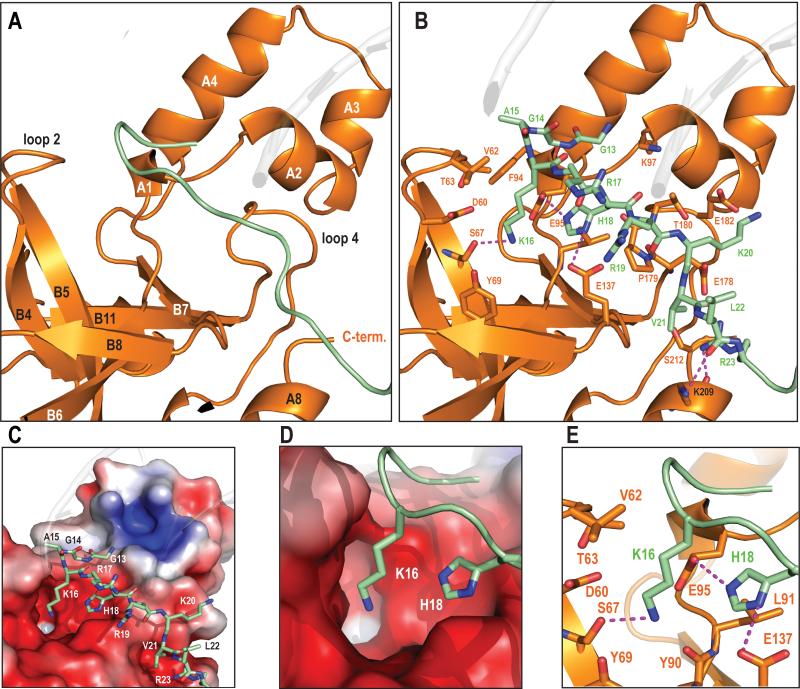
Figure 3. Overview of H4 tail interactions.
a) General view of BAH structural elements that interact with histone H4. BAH surface depicted here in orange interacts with H4 tail in green. The interaction surface is in between 2 domains of BAH, the helical H-domain and the β-sheet and that loops 2 and 4 play a crucial role in this interaction.
b) Detailed view of the H4 tail interface.
Same view as Fig. 3a. All H4 tail residues (13-23) are shown as sticks whereas in BAH only residues that make contacts are depicted as sticks. Magenta dashes connect residues forming potential hydrogen bonds (≤3.5Å). There are 6 possible hydrogen bonds in this interface; K16 forms one, H18 two, the R23 side chain two and L22 main chain carbonyl forms one. Other H4 residues that could participate in these polar interactions are K20 (with E182), and R23 (E178 HB and S212). G13, A15, V21 and R19 all make vander Waals interactions with numerous BAH residues (K97, F94, V62, T63, E95, L91, P179, T180, S212, and E178). Side-chain density for the majority of the H4 tail residues is visible (see SOM), the exceptions being side-chains of R17 and R19, which are apparently more flexible and display weak side-chain density.
c) Charge complementarity of the interface.
Basic histone H4 tail interaction with a negatively charged BAH domain surface. APBS-calculated electrostatics (−5kT to 5kT). Red surface represents negative and blue positive charge respectively.
d) Close up view of H4K16 and H4H18 binding in the charged pocket.
e) K16 binding pocket in BAH.
Detailed view of K16 and H18 side chain interactions. The K16 ε-amino group interacts with polar or negatively charged side chains of the BAH domain (D60, Y69, E95 and S67). K16 appears to form a hydrogen bond with S67 (3.1Å) and potentially a weak electrostatic interaction with the Y90 main chain carbonyl. Methyl groups of V62 and T63 could stabilize the alkyl chain of K16. Side-chain carbonyls of E137 and E95 and the main- chain carbonyl of P179 can form hydrogen bonds and an electrostatic interaction with the imidazole moiety of H18, respectively. H18 is additionally coordinated through van der Waals contacts.
An essential role for H4K16 in silencing has been demonstrated by mutational analyses, by ChIP and co-immunoprecipitation studies, and by biochemical studies showing acetylation of this residue disrupts Sir3 binding, (9, 35). (5, 36-42). A negatively charged binding pocket of BAHSir3 accommodates the side chains of H4K16 and H4H18 (Fig. 3d). Specificity for H4K16 in the unmodified state is achieved primarily by hydrogen bonding and electrostatic interactions between the ε-amino group of H4K16 and several polar or negatively charged side chains of BAH (Fig. 3e). Five of the BAH residues involved in contacts with K16 and H18 were identified in genetic screens. Of the potential electrostatic contacts that the BAH domain makes with histone residues 13-23 of the H4 N terminal tail, the majority are with K16 and H18. Acetylation of K16 could potentially disrupt most of the electrostatic contacts in this pocket (see Figs. 3d, e) and is therefore expected to significantly decrease the affinity of Sir3 for the nucleosome, concordant with previous studies, which infer a 1000-fold impact of acetylation (41).
The ‘LRS’ domain in the body of the nucleosome has been shown to be critically important for Sir3 dependent silencing at telomeres and at mating type loci (11, 12). A systematic mutagenesis study demonstrated that residues 72-83 of histone H3 and 78-81 of histone H4 are important for silencing (25). The BAH domain makes extensive interactions with a surface of the nucleosome body that includes portions of histone H3, H4, and H2B and that extends from the base of the H4 tail to an H2A region (Fig. 2a). The LRS surface is composed of helix α1 and loop L1 of histone H3, helix α2 and loop L2 of histone H4 and helices α3 and αC of of histone H2B. The LRS interacting region of BAHSir3 consists of loop 3, which becomes folded in the structure, as well as strands B6 and B8 and helix A8. There are five LRS residues (Q76, D77, F78, K79 and T80) in helix α1 and loop L1 of H3 that contact loop 3 and strands B6 and B8 of BAHSir3. All five of these H3 residues were identified in the slr screen (25) (Fig. 4b). BAH residues contacting histone H3 are located on both the sides of loop 3 and in strands B6 and B8. Most of the residues in the BAH domain that interact with H3 in the LRS region have been identified as regulating silencing in genetic screens (Figs. 2b and 4c). Many additional contacts are seen between BAHSir3 and other amino acids in the LRS (see Fig. 4). The strong correlation between the genetics and the physical interactions support the importance of the contacts between the BAH domain and the LRS surface in generating silencing.
We were interested in understanding how the structural contacts made by D205N might lead to a hypermorphic phenotype. We see a potential hydrogen bond between the H3D77 side chain carbonyl and the BAH N205 side chain amide (Fig. 4c). In WT BAHSir3 the interaction between D205 and D77 would be a repulsive interaction, thereby explaining why the affinity of BAHSir3 is increased by mutation to a neutral amino acid that can create hydrogen bonding in BAH D205N. Interestingly, mutations in H3D77 have also been shown to impact silencing (25). Mutations D77N and D77G would either increase binding to BAH D205 or remove repulsion, respectively, creating interactions similar to those seen in BAH D205N with the WT histone (Fig. 4c). Repulsive interactions have been proposed to limit binding affinity of WT Sir3 to the nucleosome, and this appears to be an important aspect of regulation. The BAH D205N mutation, which has increased binding affinity, causes increased telomeric silencing in some mutant backgrounds (9, 23, 25, 43, 44) but instead causes decreased silencing in a wild-type background (25), perhaps due to increased affinity impairing function.
Methylation of H3K79 by Dot1p has been implicated in regulating silencing (30, 45, 46). This methylation event, which occurs in the LRS region of the body of the nucleosome, has been shown to decrease binding by Sir3 in vitro(41) and has been proposed to modulate silencing in vivo by preventing localization of Sir3 to non-silenced regions(30). H3K79 could potentially form 3 hydrogen bonds with BAHSir3, one to the side chain of E84 and two to the side chain of E140. H3K79 conformation is further stabilized by van der Waals interactions with BAH W86 and H4E74 (Fig. 4c). Methylation of H3K79 would increase the cationic radius and the hydrophobicity of this residue. Progressive methylation would decrease the potential of H3K79 to form hydrogen bonds and trimethylation would ablate hydrogen bonding. This could potentially result in a decreased affinity of BAHSir3 for the nucleosome.
It is remarkable that at least 16 H4 and H2B residues in the LRS and adjacent regions have the potential to interact with only five residues of BAHSir3. Mutation of four of these amino acids (T78, L79, N80, K202) was shown to impact silencing in genetic screens, indicating the importance of this interface (Figs. 2b and 4d). In a manner similar to reciprocal mutations in BAH D205 and H3D77, the LRS mutations can be suppressed by a gain-of-function mutation BAH L79I, also identified in the slr screen. This mutation has the potential to increase van der Waals contacts with the BAH domain, elucidating a possible molecular mechanism for this genetic observation.
The acidic surface of histones H2A and H2B is a nucleosome interaction surface for proteins such as herpesvirus LANA (47) and RCC1 (29). A crystal packing interaction between a basic region of histone H4 tail and the acidic patch on adjacent nucleosome is observed in the crystal lattice of the Xenopus nucleosome core particle (32). The BAH domain apparently also makes contacts here as evidenced by electron density adjacent to the acidic patch; this density is poor and not continuous, but can only be accounted for by residues 17-37 of BAHSir3 (Fig. 4e). This region of the BAH domain is disordered in the apo structure (27). The density could be roughly modeled to locate the positively charged region of residues 28-34 of BAHSir3 as being close to the acidic residues of H2A and H2B. Mutations of these residues in the BAH domain were shown to impact silencing (24, 25). It is possible that in context of the full protein this interaction is stabilized and important for the overall affinity of Sir3 to nucleosome.
How might the interface between the nucleosome and BAHSir3 be integrated in a larger structure containing full length Sir3 to compact long regions of chromatin? The Sir3 protein has features not studied here that contribute to silencing, including acetylation of the N-terminus and dimerization determined by C- terminal regions (22, 48-51). In addition, interactions involving other proteins, especially Sir4, might be important, although overexpression of Sir3 alone can increase the size of the silent domain, implicating Sir3 as a fundamental architectural protein in establishing these extended domains (52, 53). In order to understand Sir3 oligomerization we will need to determine structures of full-length Sir3 with nucleosome arrays. Even in light of these caveats, there are features of the crystal packing of the BAHSir3-NCP structure that suggest a possible contribution of the BAH domain to nucleosome compaction.
Adjacent nucleosomes in the crystal lattice are bridged by dimerization of the BAH domain (Figs. 4f and S4). Interestingly, this dimer interface was also seen in the asymmetric unit of the apo BAH domain crystal lattice (27). To assess whether dimerization is solely a crystal packing phenomenon, we used sedimentation velocity analytical ultracentrifugation to determine if the BAH domain dimerizes in solution. Analysis of the weight average sedimentation coefficient for the BAH domain shows the presence of a weak self-association process, with a dimerization constant of ~2mM (Fig. S5). This weak interaction is expected to be insufficient by itself to promote compacted structures, but might contribute in the context of the full-length Sir3 protein, which has additional self-association interfaces, and linked nucleosomes, which would increase the effective relative concentration of each half of this BAH homodimer interface.
The complex visualized here is anticipated to be one of the central components for establishment of the silent state of chromatin in yeast. The BAH domain of Sir3 binds to an extensive histone surface within the nucleosome, causing structural transitions in both BAHSir3 and the H4 tail of the nucleosome. The correlation between mutations that impact silencing by Sir3 and amino acids that form physical contacts between BAHSir3 and the nucleosome show that this structure is important in the generation of silencing. The significance of a broad contiguous face in the interaction is underscored by our finding that mutations initially isolated as suppressors of H4 tail mutations, such as D205N, enhance interactions in the body of the nucleosome that are physically distant from the tail interactions. Numerous previous studies have implicated nucleosomes as being important for regulation via either their physical location on the genome relative to regulatory sites or their covalent modification to specify docking of regulatory complexes. We extend these examples by describing an extensive interface between a regulatory factor and the core histones of the nucleosome, thereby showing how the nucleosome can be a direct component of regulation.
It is instructive to note how covalent modification of histones impacts formation of this complex. Both acetylation of H4K16 and methylation of H3K79 are expected to disrupt several interactions that contribute to the BAHSir3-NCP interface. Acetylation of K16 is the more significant of these modifications in vivo, and would disrupt a larger number of molecular interactions based on the structure. Thus, with this complex, covalent modification of histones does not create a docking interface but rather has the potential to disrupt contacts and thereby cause a significant change in the energetics of interaction.
Supplementary Material
Acknowledgments
This work was supported by grant GM043901 from the NIH (to REK). K-J.A. was supported in part by a fellowship from the Human Frontier Science Program. We thank the staff at Beamlines 24-IDC/E at Argonne National Laboratory, especially K. Rajashankar and F. Murphy for excellent assistance with data collection. We thank T. Schwartz for use of the high-throughput crystallization facility as well as helpful discussions and critical reading of the manuscript. We thank R. Sternglanz for constructs of Sir3 BAH domain. We thank F. Winston for critical reading of the manuscript. We thank S. Jenni and D. Kostrewa for helpful discussions, S.Tan, and K. Luger for help with technical aspects of forming nucleosomes, J.Cochrane, S. Bowman, S. Miller, M. Simon and K. Bouazoune for critical reading of the manuscript, and members of the Kingston laboratory for helpful discussions. Coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with accession code 3TU4.
References
- 1.Rusche LN, Kirchmaier AL, Rine J. Annu Rev Biochem. 2003;72:481. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 2.Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 3.McBryant SJ, Krause C, Woodcock CL, Hansen JC. Mol Cell Biol. 2008 Jun;28:3563. doi: 10.1128/MCB.01389-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martino F, et al. Mol Cell. 2009 Feb 13;33:323. doi: 10.1016/j.molcel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Johnson A, et al. Mol Cell. 2009 Sep 24;35:769. doi: 10.1016/j.molcel.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kayne PS, et al. Cell. 1988 Oct 7;55:27. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- 7.Megee PC, Morgan BA, Mittman BA, Smith MM. Science. 1990 Feb 16;247:841. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- 8.Park EC, Szostak JW. Mol Cell Biol. 1990 Sep;10:4932. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson LM, Fisher-Adams G, Grunstein M. EMBO J. 1992 Jun;11:2201. doi: 10.1002/j.1460-2075.1992.tb05279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson LM, Kayne PS, Kahn ES, Grunstein M. Proc Natl Acad Sci U S A. 1990 Aug;87:6286. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD. Nat Genet. 2002 Oct;32:273. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- 12.Thompson JS, Snow ML, Giles S, McPherson LE, Grunstein M. Genetics. 2003 Jan;163:447. doi: 10.1093/genetics/163.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyriss MN, Jin Y, Gallegos IJ, Sanford JA, Wyrick JJ. Mol Cell Biol. 2010 Jul;30:3503. doi: 10.1128/MCB.00290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai J, Hyland EM, Norris A, Boeke JD. Genetics. 2010 Nov;186:813. doi: 10.1534/genetics.110.118489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Mol Cell Biol. 1996 Aug;16:4349. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahoney DJ, Broach JR. Mol Cell Biol. 1989 Nov;9:4621. doi: 10.1128/mcb.9.11.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loo S, Rine J. Science. 1994 Jun 17;264:1768. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- 18.Schnell R, Rine J. Mol Cell Biol. 1986 Feb;6:494. doi: 10.1128/mcb.6.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sussel L, Shore D. Proc Natl Acad Sci U S A. 1991 Sep 1;88:7749. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasmyth KA. Cell. 1982 Sep;30:567. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- 21.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Cell. 1990 Nov 16;63:751. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 22.Ehrentraut S, et al. Genes Dev. 2011 Sep 1;25:1835. doi: 10.1101/gad.17175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampath V, et al. Mol Cell Biol. 2009 May;29:2532. doi: 10.1128/MCB.01682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone EM, Reifsnyder C, McVey M, Gazo B, Pillus L. Genetics. 2000 Jun;155:509. doi: 10.1093/genetics/155.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norris A, Bianchet MA, Boeke JD. PLoS Genet. 2008 Dec;4:e1000301. doi: 10.1371/journal.pgen.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchberger JR, et al. Mol Cell Biol. 2008 Nov;28:6903. doi: 10.1128/MCB.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connelly JJ, et al. Mol Cell Biol. 2006 Apr;26:3256. doi: 10.1128/MCB.26.8.3256-3265.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Materials and methods are available as supporting material on Science Online [Google Scholar]
- 29.Makde RD, England JR, Yennawar HP, Tan S. Nature. 2010 Sep 30;467:562. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Leeuwen F, Gafken PR, Gottschling DE. Cell. 2002 Jun 14;109:745. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 31.Hou Z, Danzer JR, Fox CA, Keck JL. Protein Sci. 2006 May;15:1182. doi: 10.1110/ps.052061006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997 Sep 18;389:251. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 33.Yu Q, Olsen L, Zhang X, Boeke JD, Bi X. Genetics. 2011 Jun;188:291. doi: 10.1534/genetics.111.127886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fry CJ, Norris A, Cosgrove M, Boeke JD, Peterson CL. Mol Cell Biol. 2006 Dec;26:9045. doi: 10.1128/MCB.00248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aparicio OM, Billington BL, Gottschling DE. Cell. 1991 Sep 20;66:1279. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 36.Oppikofer M, et al. EMBO J. Jun;10:2011. [Google Scholar]
- 37.Hoppe GJ, et al. Mol Cell Biol. 2002 Jun;22:4167. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo K, Vega-Palas MA, Grunstein M. Genes Dev. 2002 Jun 15;16:1528. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rusche LN, Kirchmaier AL, Rine J. Mol Biol Cell. 2002 Jul;13:2207. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanny JC, Kirkpatrick DS, Gerber SA, Gygi SP, Moazed D. Mol Cell Biol. 2004 Aug;24:6931. doi: 10.1128/MCB.24.16.6931-6946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Mol Cell. 2007 Dec 28;28:1015. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Cell. 1995 Feb 24;80:583. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 43.Park Y, Hanish J, Lustig AJ. Genetics. 1998 Nov;150:977. doi: 10.1093/genetics/150.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C, Lustig AJ. Genetics. 1996 May;143:81. doi: 10.1093/genetics/143.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singer MS, et al. Genetics. 1998 Oct;150:613. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Proc Natl Acad Sci U S A. 2003 Feb 18;100:1820. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbera AJ, et al. Science. 2006 Feb 10;311:856. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 48.Liaw H, Lustig AJ. Mol Cell Biol. 2006 Oct;26:7616. doi: 10.1128/MCB.01082-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Cell. 2005 May 20;121:515. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 50.King DA, et al. J Biol Chem. 2006 Jul 21;281:20107. doi: 10.1074/jbc.M512588200. [DOI] [PubMed] [Google Scholar]
- 51.McBryant SJ, Krause C, Hansen JC. Biochemistry. 2006 Dec 26;45:15941. doi: 10.1021/bi061693k. [DOI] [PubMed] [Google Scholar]
- 52.Renauld H, et al. Genes Dev. 1993 Jul;7:1133. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 53.Hecht A, Strahl-Bolsinger S, Grunstein M. Nature. 1996 Sep 5;383:92. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 54.Weiss M. Journal of Applied Crystallography. 2001;34:130. [Google Scholar]
- 55.Connelly JJ, et al. Mol Cell Biol. 2006 Apr;26:3256. doi: 10.1128/MCB.26.8.3256-3265.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luger K, Rechsteiner TJ, Richmond TJ. Methods Enzymol. 1999;304:3. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 57.Leslie AGW. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992 [Google Scholar]
- 58.Collaborative Computational Project Number 4 Acta Crystallographica Section D. 1994;50:760. [Google Scholar]
- 59.McCoy AJ, et al. Journal of Applied Crystallography. 2007;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. J Mol Biol. 2002 Jun 21;319:1097. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 61.Bricogne EBG, Brandl M, et al. Global Phasing Ltd. , Cambridge; United Kingdom: 2008. [Google Scholar]
- 62.Adams PD, et al. Acta Crystallographica Section D. 2002;58:1948. doi: 10.1107/s0108767301016658. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Acta Crystallographica Section D. 2004;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Chen VB, et al. Acta Crystallographica Section D. 2010;66:12. [Google Scholar]
- 65.Laskowski RA, MacArthur MW, Moss DS, Thornton J. J. Appl. Crystallogr. 1993:283. [Google Scholar]
- 66.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc Natl Acad Sci U S A. 2001 Aug 28;98:10037. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kabsch W, Sander C. Biopolymers. 1983 Dec;22:2577. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 68.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Bioinformatics. 2009 May 1;25:1189. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.