Abstract
The human RPSA [ribosomal protein SA; also known as LamR1(laminin receptor 1)] belongs to the ribosome but is also a membrane receptor for laminin, growth factors, prion, pathogens and the anticarcinogen EGCG (epigallocatechin-gallate). It contributes to the crossing of the blood–brain barrier by neurotropic viruses and bacteria, and is a biomarker of metastasis. RPSA includes an N-terminal domain, which is folded and homologous to the prokaryotic RPS2, and a C-terminal extension, which is intrinsically disordered and conserved in vertebrates. We used recombinant derivatives of RPSA and its N- and C-domains to quantify its interactions with ligands by in-vitro immunochemical and spectrofluorimetric methods. Both N- and C-domains bound laminin with KD (dissociation constants) of 300 nM. Heparin bound only to the N-domain and competed for binding to laminin with the negatively charged C-domain, which therefore mimicked heparin. EGCG bound only to the N-domain with a KD of 100 nM. Domain 3 of the envelope protein from yellow fever virus and serotypes-1 and -2 of dengue virus bound preferentially to the C-domain whereas that from West Nile virus bound only to the N-domain. Our quantitative in-vitro approach should help clarify the mechanisms of action of RPSA, and ultimately fight against cancer and infectious agents.
Keywords: cancer, epigallocatechin-gallate (EGCG), flavivirus, heparin, intrinsically disordered protein, laminin receptor 1 (LamR1)
Abbreviations: 37LRP, 37 kDa laminin receptor precursor; 67LR, 67 kDa laminin receptor, an alternative name for LamR1; DENV, dengue virus; ED1, ED2 and ED3, domains 1, 2 and 3 of the flaviviral envelope protein E; EGCG, epigallocatechin gallate; LamR1, laminin receptor 1 (an alternative name for RPSA); mAb, monoclonal antibody; pNP, p-nitrophenol; pNPP, pNP phosphate; RPS2, ribosomal protein S2; RPSA, ribosomal protein SA; SINV, Sindbis virus; VEEV, Venezualian equine encephalitis virus; WNV, West-Nile virus; YFV, yellow fever virus
INTRODUCTION
The human 40S RPSA (ribosomal protein SA) is a multiform, multilocus and multifunctional protein. It has many alternative names, including LamR1 (laminin receptor 1). The amino acid sequence of RPSA includes 295 residues and corresponds to an MMth (theoretical molecular mass) of 32854 Da [1]. However, RPSA appears as polypeptides of MMapp (apparent molecular masses) of 37 and 67 kDa in immunoblots of cellular extracts. The 37 kDa form [37LRP (37 kDa laminin receptor precursor)] is a precursor of the 67 kDa form [67LR (67 kDa laminin receptor)] [2,3]. RPSA is not glycosylated; it is acylated in position Ser2 by fatty acids and this acylation is involved in the conversion of 37LRP into 67LR [4,5]; it is phosphorylated in position Tyr139 [6].
RPSA can be sub-divided in two main domains: an N-domain comprising residues 1–209 and a C-domain comprising residues 210–295. Sequence analyses have shown that the N-domain of RPSA is homologous to the RPS2 (ribosomal proteins S2) of prokaryotes. It contains a palindromic L173MWWML178 sequence that is conserved in all metazoans. Its C-domain is highly conserved in vertebrates. The amino acid sequence of RPSA is 98% identical in all mammals. These phylogenetic analyses have suggested that RPSA is a ribosomal protein that has acquired the function of laminin receptor during evolution [7,8].
The crystal structure of a recombinant N-domain of RPSA has been determined at 2.15 Å (1 Å=0.1 nm) resolution from crystals grown at 17°C and found to be similar indeed to those of prokaryotic RPS2 [9]. However, RPSA has multiple folding states: a recombinant C-domain is intrinsically disordered and a recombinant N-domain is mainly in an intermediate state of folding at 37°C, which is the body temperature. Such an intermediate state also exists for the full-length RPSA [10].
RPSA is associated with DNA and some histones in the nucleus, with the pre-ribosome in the nucleolus, with the 40S subunit of ribosomes in the cytoplasm, and with the cellular membrane (reviewed in [8,10]). RPSA is a membrane receptor for laminin, which is a component of the extracellular matrix, and for growth factors. As such, it has a role in tumour invasion and aggressiveness [8,10–12]. The 67LR form of RPSA is a membrane receptor for EGCG [(−)-epigallocatechin-3-gallate], which is a major constituent of green tea and has many health related effects. EGCG inhibits cancer cell growth, induces apoptosis of myeloma and leukaemia cells, has anti-allergic and anti-inflammatory actions, modulates the action of insulin, which is a key factor in stimulating fat cell mitogenesis and adipogenesis, and it inhibits the expression of tissue factor, which is an important trigger of arterial thrombosis [13]. There is evidence that the 37LRP form of RPSA interacts with heparan sulfates, which are other components of the extracellular matrix, and that this interaction is involved in its activity as a receptor for laminin and some pathogens [12,14–16]. RPSA is involved in signal transduction within the cell, in particular as part of its activities as a receptor for growth factors and EGCG [13,17].
RPSA is also a membrane receptor for toxins, prion, neurotropic viruses, and bacteria, of which it could promote adherence to the blood–brain barrier [16,18–23]. In particular, human RPSA is a potential cellular receptor for several flaviviruses, including DENV (dengue virus) and TBEV (tick-borne encephalitis virus) [19,20,24,25], and for several alphaviruses including VEEV (Venezualian equine encephalitis virus) and SINV (Sindbis virus) [9,18,26]. The envelope protein of flaviviruses includes three ectodomains, ED1, ED2 and ED3 and a transmembrane region. The ED3 domain is thought to contain the primary site of interaction between the virion and its cell receptors [27].
Previously, we have constructed recombinant plasmids for the expression of RPSA and its main domains in Escherichia coli, and their purification to homogeneity. RPSA-(2–295) corresponds to the full-length protein; RPSA-(2–209), exactly to exons 2–5 of the RPSA gene; RPSA-(2–220), to the protein domain that has been crystallized; RPSA-(210–295), exactly to exons 6–7; and RPSA-(225–295), to the C-terminal acid domain of the protein. RPSA-(2–209) also corresponds to the ribosomal domain of RPSA and includes all the residues that are visible in the crystal structure of RPSA-(2–220), i.e. residues 9–205. RPSA-(210–295) also corresponds to the domain of RPSA that is conserved in vertebrates. RPSA-(225–295) includes all five repetitions of an E/D–W–S/T motif, 13 negative charges and no positive charge [10].
Here, we used quantitative and semi-quantitative methods to measure the parameters of interaction between RPSA and several ligands, i.e. laminin, heparin, EGCG and the ED3 domain of flaviruses. We then used the recombinant derivatives of RPSA to map the interaction sites either in the N-domain or in the C-domain of RPSA. The results showed that the folded N-domain and intrinsically disordered C-domain of RPSA have both idiosyncratic and shared receptor functions. They shed light on the molecular mechanisms of these functions.
EXPERIMENTAL
Bacterial strains, reagents and buffers
The E. coli strains BL21 (F−, ompT), BLR (as BL21 but Δ(srl-recA)306::Tn10 (TetR)) (Novagen); NEB-Express (F−, lon, ompT) and NEB Express Iq (as NEB-Express but with miniF-lacIq) (New England Biolabs) have been described. Strain NZ1 was constructed by introducing the MiniF-lacIq of NEB Express Iq into BLR by chemical transformation. PBS, Tween 20, pNPP (4-nitrophenyl phosphate), heparin sodium salt from porcine intestinal mucosa, the mouse mAb (monoclonal antibody) LAM-89 to laminin, the conjugate between alkaline phosphatase and a goat antibody to mouse IgGs (Fc-specific), and N-acetyl-L-tryptophanamide were purchased from Sigma-Aldrich; laminin from mouse EHS (Engelbreth-Holm-Swarm) sarcoma and BSA fraction V from Roche Applied Science; the mouse mAb T320.11 to heparin and heparan-sulfates from Merck Millipore; Maxisorp ELISA plates from Nunc. The molecular mass of heparin was equal to 18000 Da. Buffer A contained 0.05% Tween 20 in PBS; buffer B, 3% BSA in PBS; Buffer C, 1% BSA in PBS; buffer D, 0.1 M diethanolamine, 10 mM MgCl2, pH 9.8; buffer E, 0.05% Tween 20, 1% BSA in PBS.
Recombinant proteins
The recombinant derivatives of RPSA, the method for their production and the method for their purification to homogeneity have been described [10]. The nomenclature of these derivatives indicates the segment of the RPSA sequence that they include. The RPSA residues are numbered from the initiator methionine. We used the murine rather than the human RPSA for regulatory reasons. The two proteins differ by only one residue, Asp293 in man and Glu293 in mouse. All the RPSA derivatives carry a His-tag extension at their N-terminus. In addition, RPSA-(2–295) carries a Strep-tag extension at its C-terminus. The construction of plasmids encoding the H6–PhoA and H6–ED3–PhoA hybrids, the production of the hybrids in the periplasm of E. coli and their purification through a His-tag were performed as described, except that NZ1 was used as a host strain [28,29]. NZ1 was recA to avoid recombination between the plasmidic and endogeneous phoA genes and ompT to avoid a proteolytic degradation of the recombinant hybrids. The origin of the viral ED3 domains and the corresponding segment of the envelope protein are indicated in Table 1. The protein concentrations were measured by absorbance spectrometry. The molar absorption coefficients and MMth values were computed from the amino acid sequences with subprogram PepStat of the EMBOSS software suite [30].
Table 1. Viral origins of the H6–ED3–PhoA hybrids.
The last column gives the residues of the viral glycoprotein E present in the H6–ED3–PhoA hybrids. The ED3 domains of WNV and JEV have three extra residues. The codons in the recombinant genes were synonymous but not necessarily identical with those in the original viral genomes.
| Virus | Strain | GenBank® accession no. | gpE residues |
|---|---|---|---|
| DENV1 | FGA/89 | AF226687 | 295–400 |
| DENV2 | Jamaica/N.1409 | M20558 | 295–400 |
| DENV3 | PaH881/88 | AF349753 | 293–398 |
| DENV4 | ThD4-0113-76 | AY618949 | 295–400 |
| YFV | 17D vaccine strain | X03700 | 293–398 |
| YFV | Asibi | AY640589 | 293–398 |
| WNV | IS-98-STD | AF481864 | 298–406 |
| JEV | SA14 | U14163 | 297–405 |
Enzymic activity of alkaline phosphatase
The rates for the formation of pNP (p-nitrophenol) from pNPP (2.7 mM initial concentration) by the H6–PhoA and H6–ED3–PhoA hybrids (10 nM in monomer) were measured at 25°C in buffer D. The variation of A405nm was measured during the first 400 s of reaction with a Jasco V-630 thermostated double-beam spectrophotometer. The rates were determined by linear regression of the data points and expressed relative to the rate for H6-PhoA.
Indirect ELISA
The indirect ELISA were performed in 96-well microtitre plates essentially as described in [31]. The wells were filled with a solution of the RPSA derivative under study (0.1 to 2.0 μg/ml in 100 μl of PBS) and the reaction of coating by adsorption run overnight at 4°C. The wells were emptied, washed with buffer A (three times) and PBS (twice). They were blocked with buffer B (400 μl) and then washed as above. An aliquot of the other interactant (100 μl in buffer C) was added to the wells, the reaction of capture was run at 25°C during either 1 h for laminin and heparin or 2 h for the (H6–ED3–PhoA)2 dimers, and then the wells were washed as above. The captured (H6–ED3–PhoA)2 or (H6–PhoA)2 dimers were detected by adding 5 mM pNPP in buffer D and monitoring the formation of pNP with A405nm. The captured laminin molecules were detected by the successive additions of the mouse mAb LAM-89 to laminin, a conjugate between alkaline phosphatase and a goat antibody to the mouse IgGs, and 5 mM pNPP as above. The captured heparin molecules were detected by the successive addition of the mouse mAb T320.11 to heparin/heparan sulfates, the conjugate between alkaline phosphatase and the goat antibody to mouse IgGs and 5 mM pNPP as above. The antibodies were diluted in buffer E before use.
Determination of a dissociation constant KD by competition ELISA
The dissociation constants at equilibrium in solution, KD, between laminin and RPSA derivatives were measured by a competition ELISA [32]. The assay was performed at 25°C in buffer C. Laminin at a constant concentration (2–7 μg/ml, 2.3–8.2 nM depending on the other partner) and the RPSA derivative at varying concentrations were first mixed together in solution and the reaction of binding was run for 20 h until equilibrium. The concentration of free laminin was then measured by an indirect ELISA using microtitre plates whose wells had been coated with the same RPSA derivative, as described in the previous paragraph. The density of coating was adjusted to capture less than 10% of the free laminin molecules during 1 h at 25°C and thus avoid shifting the binding equilibrium.
Determination of an inhibition constant Ki by competition ELISA
The inhibition constant at equilibrium in solution, Ki, of heparin for the interaction between RPSA-(225–295) and laminin was measured by a competition ELISA exactly as described for the determination of KD, except that laminin at a constant concentration and heparin at varying concentrations were first mixed together in solution. The concentration of free laminin was measured by an indirect ELISA in which RPSA-(225–295) was immobilized.
Determination of a dissociation constant KD by spectrofluorimetry
A quantitative analysis of the interaction between EGCG and RPSA derivatives at 25°C was performed by fluorimetric titration. The protein concentration was fixed and equal to 10 μg/ml, corresponding to 0.28 μM for RPSA-(2–295), 0.39 μM for RPSA-(2–220); 0.41 μM for RPSA-(2–209), 0.9 μM for RPSA-(210–295) and 1.09 μM for RPSA-(225–295). The protein was incubated with various concentrations of EGCG (0–400 μM) in PBS for 16 h to reach equilibrium. Control reactions were prepared by replacing the protein with 4 μM N-acetyl-L-tryptophanamide or buffer. The fluorescence measurements were carried out with a Jasco FP-6300 spectrofluorimeter, equipped with a thermostated cell holder. The molecules were excited at 278 nm and their emission spectrum recorded in the interval 310–360 nm. The slit width was equal to 2.5 nm for excitation and 5.0 nm for emission. The fluorescence signal of EGCG alone was measured in a control experiment and subtracted from the global signal of the binding mixtures to give the specific fluorescence signal of the protein or tryptophanamide.
Analysis of the experimental data
The raw data of the competition ELISAs and fluorescence titrations were processed essentially as described [33,34]. Let us consider the equilibrium of association and dissociation between a ligand L and a protein P to form the complex L:P. If [L]0 is the total concentration of L in the reaction; [P]0, the total concentration of P; [P], the concentration of the free molecules of P; [P:L], the concentration of complex; and KD, the dissociation constant of the complex L:P; then, the laws of mass action and conservation imply that [P] and [P:L] are solutions of the quadratic equations:
| (1) |
| (2) |
whose useful solutions are:
| (3) |
| (4) |
The indirect ELISA for measuring [P] was performed in conditions such that:
| (5) |
where A is the absorbance signal at 405 nm, and A0 and A∞ are the values of A at zero and saturating concentrations of L. The values of KD, A0 and A∞ were determined by fitting eqn (5), in which [P] is given by eqn (3), to the experimental values of A.
The spectrofluorimetric assay for measuring [P:L] was performed in conditions such that:
| (6) |
where F is the fluorescence intensity of the protein, and F0 and F∞ are the values of F at zero and saturating concentration of L. The values of ΔF∞/F0, [P]0 and KD were determined by fitting eqn (6), in which [P:L] is given by eqn (4), to the experimental values of ΔF/F0.
To evaluate the contribution of light absorption by ligand L to the variation of F in the experiments of fluorescence titration by L, we used N-acetyl-L-tryptophanamide, which is a close analogue of a Trp residue and generally does not bind L. The molar coefficient ∊ for the absorption of F by L was obtained by fitting a Beer's equation to the experimental values FW for tryptophanamide:
| (7) |
where δFW,∞ is an optional constant, usually less than 0.01FW,0, that allows for any residual fluorescence signal at high concentration of L. The values of ∊ and δFW,∞ that we obtained with tryptophanamide, were used to calculate a corrected value Fc of F for protein P, according to eqn (8):
| (8) |
Eqn (6) was then fitted to the Fc values as described above.
The concentration K1/2 for half-capture of ligand L by the immobilized protein P in an indirect ELISA was obtained by fitting either a Michaelis–Menten equation or a sigmoid equation to the experimental values of A405nm:
| (9) |
| (10) |
Similarly, we used eqn (9) to obtain the concentration EC50 at which the capture of ligand L1 by protein P was half-enhanced by ligand L2.
The results for the capture of a H6–ED3–PhoA hybrid by an immobilized RPSA derivative in an indirect ELISA were expressed as follows. Let us consider the A405nm signal in the ELISA as a function of the concentration of hybrid; s, the slope of the A405nm signal; v the rate of formation of pNP from pNPP by the same hybrid; s0 and v0, the corresponding parameters for the H6–PhoA control hybrid. The relative strength Sr of the interaction was expressed by the following ratio, where s and s0 were measured in the same experiment:
| (11) |
The free energy of dissociation between L and P is equal to ΔG=−RT ln(KD) and the variation of free energy on mutation of P is equal to ΔΔG=ΔG(wt)–ΔG(mut), where wt and mut refer to wild-type and mutant molecules of P. The S.E. (standard error) on ΔG and ΔΔG were calculated through the formula:
| (12) |
| (13) |
All the curve fits were performed with Kaleidagraph (Synergy Software).
RESULTS
Laminin binding
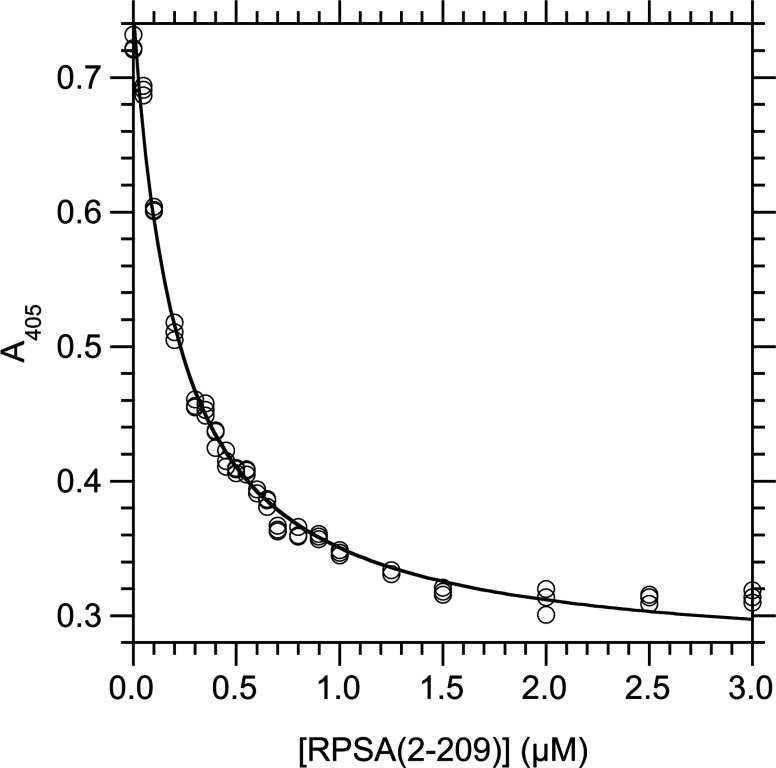
We determined the values of the KD in solution, at equilibrium and 25°C, between laminin and derivatives of RPSA by competition ELISA assays in which the concentration of the RPSA derivative varied (Figure 1). We analysed the experimental data with a 1:1 model of interaction (see the Experimental section). The best fittings were obtained for the RPSA-(2–209) and RPSA-(210–295) domains. They bound laminin with similar KD values, 273±37 nM and 374±54 nM respectively. Therefore both N- and C-domains bound laminin with similar affinities. Full-length RPSA bound laminin with a KD value of 666±33 nM (Table 1).
Figure 1. Determination of KD at 25°C in solution for the interaction between RPSA-(2–209) and laminin, by competition ELISA.
Laminin (5 μg/ml, 5.5 nM) and RPSA-(2–209) were first incubated for 20 h at 25°C in solution (buffer C) until the binding reaction reached equilibrium. The concentration of free laminin was then measured by an indirect ELISA in which RPSA-(2–209) (2 μg/ml, 81 nM) was immobilized in the wells of a microtitre plate and the captured laminin was revealed with a specific antibody. The total concentration of RPSA-(2–209) in the binding reaction is given along the x-axis; the A405 signal, which is linearly related to the concentration of free laminin in the binding reaction, is given along the y-axis. The curve was obtained by fitting eqn (5) to the experimental data (see the Experimental section). Totally, 21 concentrations of RPSA-(2–209) were used and each data point was done in triplicate.
Heparin binding
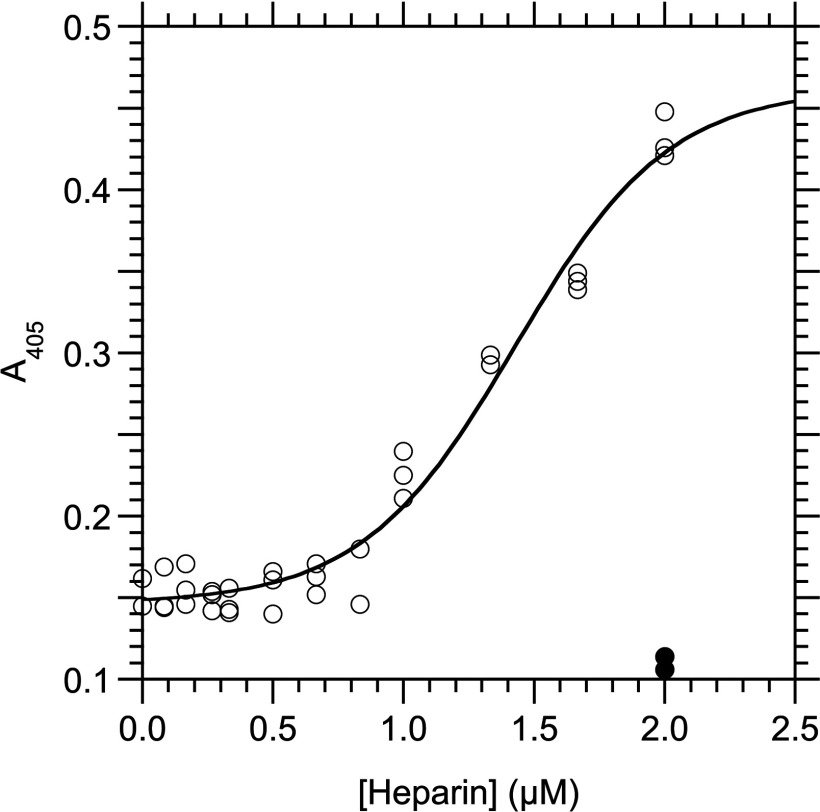
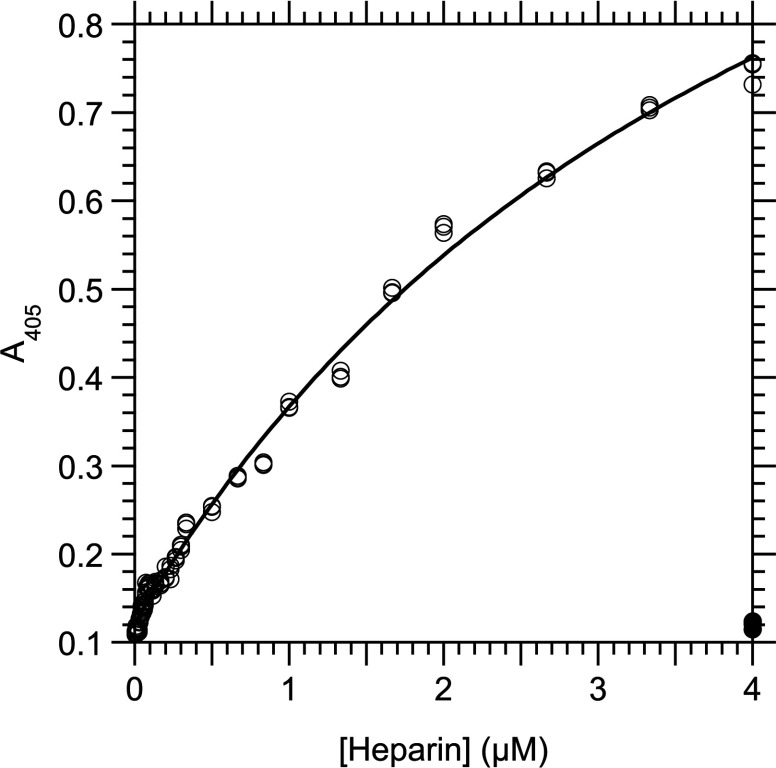
We analysed the interaction between heparin and derivatives of RPSA at 25°C by indirect ELISA assays in which the proteins were immobilized on microtitre plates. The positively charged N-domains, RPSA-(2–209) and RPSA-(2–220), bound heparin with K1/2 equal to 1.43±0.06 μM and 4.36±0.28 μM, respectively (Figures 2 and 3; Table 1). Neither the negatively charged C-domains nor the full-length RPSA bound heparin significantly (results not shown).
Figure 2. Interaction between RPSA-(2–209) and heparin as measured by an indirect ELISA at 25°C.
RPSA-(2–209) (2 μg/ml, 81 nM) was immobilized in the wells of a microtitre plate and used to capture heparin in buffer C. The total concentration of heparin in the capture reaction is given along the x-axis and the A405 signal along the y-axis. The curve was obtained by fitting eqn (10) to the experimental data. Open circles, with coating of RPSA-(2–209); closed circles, without coating of RPSA-(2–209) but with 2.0 μM heparin. Only part of the data points below 0.3 μM heparin have been represented for clarity.
Figure 3. Interaction between RPSA-(2–220) and heparin as measured by an indirect ELISA at 25°C.
RPSA-(2–220) (1 μg/ml, 39 nM) was immobilized in the wells of a microtitre plate and used to capture heparin in buffer C. The total concentration of heparin in the capture reaction is given along the x-axis and the A405 signal along the y-axis. The curve was obtained by fitting eqn (9) to the experimental data. Open circles, with coating of RPSA-(2–220); closed circles, without coating of RPSA-(2–220) but with 4.0 μM heparin.
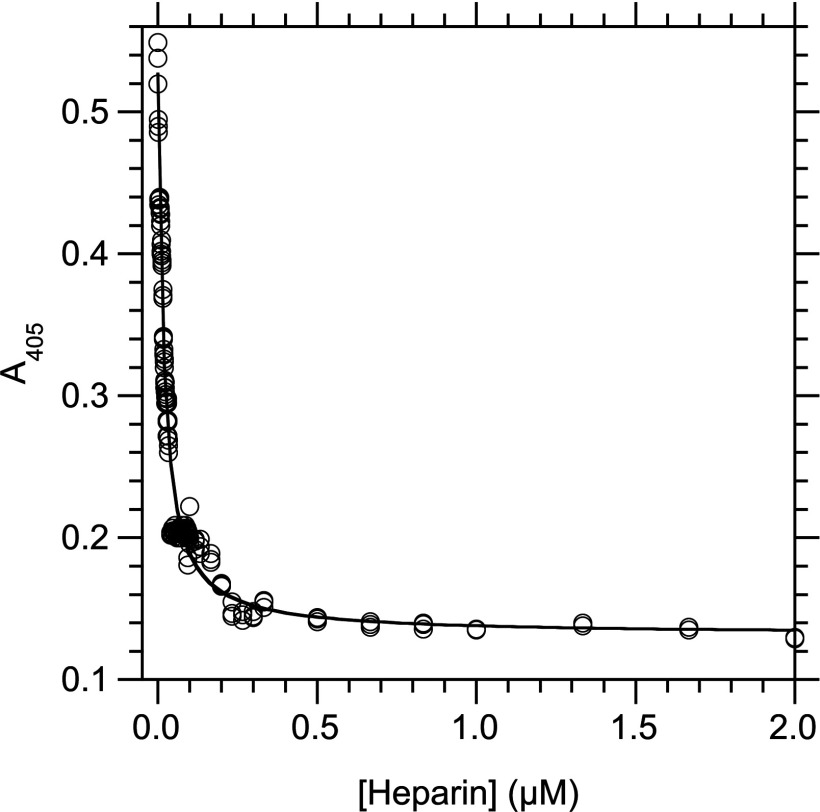
We observed that heparin increased the capture of laminin by immobilized RPSA-(2–220) in an indirect ELISA, with an EC50 equal to 35±4 nM of heparin. A control in the absence of immobilized RPSA-(2–220) showed that this enhancement was not due to a non-specific interaction between heparin and the blocking BSA (results not shown). In constrast, heparin strongly inhibited the capture of laminin by the immobilized RPSA-(225–295) domain in an indirect ELISA, with a constant of inhibition (Ki) equal to 16.1±0.7 nM (Figure 4). As the C-domain did not bind heparin (see above), these results could not be interpreted as a competition between heparin and laminin for the same binding site on RPSA-(225–295). Rather, they showed that the C-domain and heparin competed for interactions with the same site on laminin.
Figure 4. Determination of Ki at 25°C for the inhibition of the interaction between RPSA-(225–295) and laminin by heparin.
Laminin (5 μg/ml, 5.5 nM) and heparin were first incubated for 20 h at 25°C in solution (buffer C) until the binding reaction reached equilibrium. The concentration of free laminin was then measured by an indirect ELISA in which RPSA-(225–295) (0.15 μg/ml, 16 nM) was immobilized in the wells of a microtitre plate and the captured laminin was revealed with a specific antibody. The total concentration of heparin in the binding reaction is given along the x-axis; the A405 signal, which is linearly related to the concentration of free laminin in the binding reaction, is given along the y-axis. The curve was obtained by fitting eqn (5) to the experimental data. Totally, 52 concentrations of heparin were used and each data point was done in triplicate.
EGCG binding
The quenching of a protein fluorescence by the binding of a ligand is widely used to determine their dissociation constant [35]. We observed that the interaction between EGCG and the RPSA derivatives strongly decreased the intrinsic fluorescence intensity F of the latter, upon excitation at 278 nm (results not shown). EGCG does not fluoresce but it absorbs light between 210 and 320 nm with a maximum at 274 nm. Therefore the decrease in F could be due either to an absorption of the excitation light by EGCG or to a quenching of the protein fluorescence by the binding of EGCG. To determine the contribution of the light absorption, we titrated the fluorescence intensity FW of N-acetyl-L-tryptophanamide, a close analogue of a Trp residue, with increasing concentrations of EGCG. The variation of FW as a function of the EGCG concentration could be satisfactorily represented by Beer's law, with a correlation coefficient RP of 0.9996. The corresponding molar coefficient of absorption ∊ was equal to 0.0260±0.0004 μM−1 in our experimental conditions (λex=278 nm, 326 nm ≤ λem ≤350 nm, 25°C) and corresponded to a concentration of half-absorption ([EGCG]1/2) equal to 27 μM.
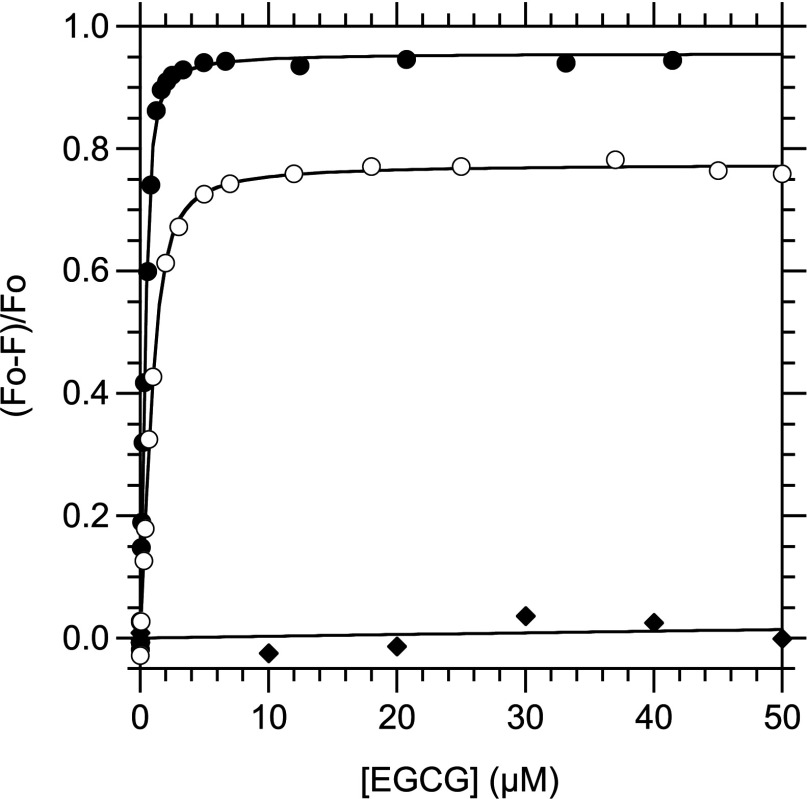
Similarly, the variation of F as a function of the EGCG concentration could be satisfactorily represented by Beer's law for RPSA-(210–295) and RPSA-(225–295), with RP values of 0.99 and [EGCG]1/2 values of 48 and 43 μM, respectively, consistent with that for tryptophanamide (results not shown). These results showed that the C-domains of RPSA did not bind EGCG. In contrast, fitting Beer's law to the experimental values of F for RPSA-(2–209), RPSA-(2–220) and RPSA-(2–295) resulted in [EGCG]1/2 values that were equal to 0.44, 0.61 and 2.1 μM, respectively, and inconsistent with a simple absorption of light by EGCG. Therefore to analyse the fluorescence profiles of these three last RPSA derivatives, we first corrected the measured values of F to take into account the absorption of light by EGCG, by using a value of ∊ that was measured for tryptophanamide in the same experimental conditions. We then fitted the equation of a 1:1 model of binding equilibrium to the corrected values Fc. This method allowed us to determine the dissociation constant KD between EGCG and each of RPSA-(2–209), RPSA-(2–220) and RPSA-(2–295) in solution, at equilibrium and 25°C with precision (Figure 5 and Table 2). The results showed that the binding site for EGCG was located fully within the N-domain of RPSA.
Figure 5. Determination of KD at 25°C in solution for the interaction between EGCG and RPSA derivatives, by spectrofluorimetry.
The binding reaction was performed at 25°C in PBS with a fixed concentration of the RPSA derivative (10 μg/ml) or N-acetyl-L-tryptophanamide (4 μM) and variable concentrations of EGCG. F, fluorescence intensity of the reaction mixture with λex=278 nm; F0, value of F in the absence of EGCG; closed circles, RPSA-(2–209) and λem=330 nm; open circles, RPSA-(2–295) and λem=327 nm; closed diamonds, N-acetyl-L-tryptophanamide and λem=330 nm. The graph gives the relative variation (F0–F)/F0 of F as a function of the total concentration in EGCG. The curves correspond to the fitting of eqn (6) to the experimental data. All the F values were corrected for the absorbance of light by EGCG through eqn (8) (see the Experimental section).
Table 2. Parameters of interaction between RPSA derivatives and ligands at 25°C.
The dissociation constants KD were measured at equilibrium in solution either by spectrofluorimetry for EGCG or by competition ELISA for laminin. The parameters K1/2 of half-saturation were measured by indirect ELISA. Each entry gives the mean parameter and associated S.E. in several independent experiments. The buffers are described in the Experimental section. na, not applicable; ≫, much greater than.
| Domain | pI | Charge | KD(EGCG) (nM) | KD(Laminin) (nM) | K1/2(Heparin) (μM) |
|---|---|---|---|---|---|
| 2–295 | 5.2 | −7.5 | 399±101 | 666±33 | ≫4 |
| 2–220 | 7.4 | 4 | 130±37 | 691±98 | 4.36±0.28 |
| 2–209 | 8.3 | 6 | 99±7 | 273±37 | 1.43±0.06 |
| 210–295 | 4.2 | −10 | na | 374±54 | ≫2 |
| 225–295 | 4.1 | −9 | na | 344±50 | ≫2 |
Contribution of Trp residues to folding and binding
To determine which Trp residues of RPSA were sensitive to the binding of EGCG and locate approximately its binding site, we changed each of the four Trp residues of RPSA-(2–209) into Ala by mutagenesis at the genetic level. We could produce and purify the W176A and W195A mutant proteins in sufficient quantities for their study. In contrast, we could not produce the W55A and W175A mutant proteins in significant amounts. These results suggested that residues Trp55 and Trp175 are essential for the folding or stability of RPSA, unlike Trp176 and Trp195. All four Trp residues are buried in the crystal structure of the N-domain of RPSA [9]. However, Trp55 and Trp175 are deeply buried within the protein core of RPSA whereas Trp176 and Trp195 belong to a more dynamic region that can become exposed to the solvent [10,36].
The interaction with EGCG decreased the fluorescence intensity of RPSA-(2–209) to similar extents for the W176A and W195A mutant derivatives as for the parental protein (results not shown). These decreases enabled us to determine the corresponding KD values at 25°C, 186±42 nM and 196±54 nM, respectively. From these KD values, we calculated that the W176A and W195A changes decreased the free energy of interaction (ΔΔG) between RPSA-(2–209) and EGCG by 0.4±0.1 kcal·mol−1, i.e. less than the energy of a Van der Waals bond. Thus, residues Trp176 and Trp195 did not contribute to the energy of interaction between EGCG and RPSA.
Flavivirus binding
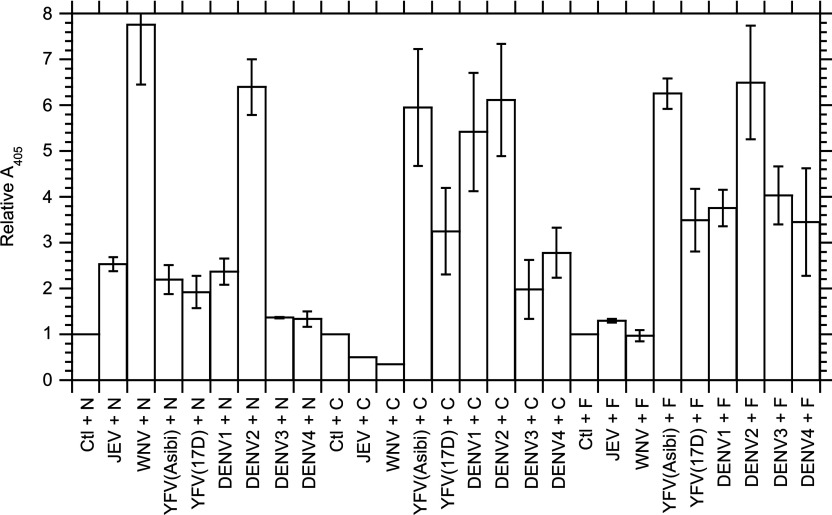
We tested the existence of interactions between RPSA and the ED3 domain from several pathogenic flaviviruses with an in-vitro immunochemical assay. This assay involved the immobilization of RPSA-(2–295), RPSA-(2–220) or RPSA-(225–295) on a solid support and the capture of dimeric hybrids (H6–ED3–PhoA)2 between an hexahistidine, the ED3 domain and alkaline phosphatase. We expressed the responses of the (H6–ED3–PhoA)2 hybrids relative to the unspecific response of a (H6–PhoA)2 hybrid in the same experiment (Figure 6). Moreover, we corrected the responses for the slight differences in enzymatic rates for the formation of pNP from pNPP between the (H6–ED3–PhoA)2 and (H6–PhoA)2 hybrids (see the Experimental section). The results showed a specific interaction between RPSA and the ED3 domain from YFV (yellow fever virus). The interaction was stronger for the wild-type strain YFV(Asibi) than for the derived vaccinal strain YFV(17D). It was restricted to the C-domain for both YFV strains. The results also showed specific interactions between RPSA and the ED3 domains from DENV1 and DENV2. The interaction was observed with both N- and C-domains of RPSA for DENV2, whereas it was restricted to the C-domain for DENV1. The interactions between RPSA and the ED3 domains from DENV3 and DENV4 were hardly significant. We found a specific interaction between the ED3 domain of WNV (West-Nile virus) and the N-domain of RPSA but no interaction with either the full-length RPSA or its C-domain. Finally, we did not find any interaction between the ED3 domain of the JEV (Japanese encephalitis virus) and RPSA or its domains (Figure 7).
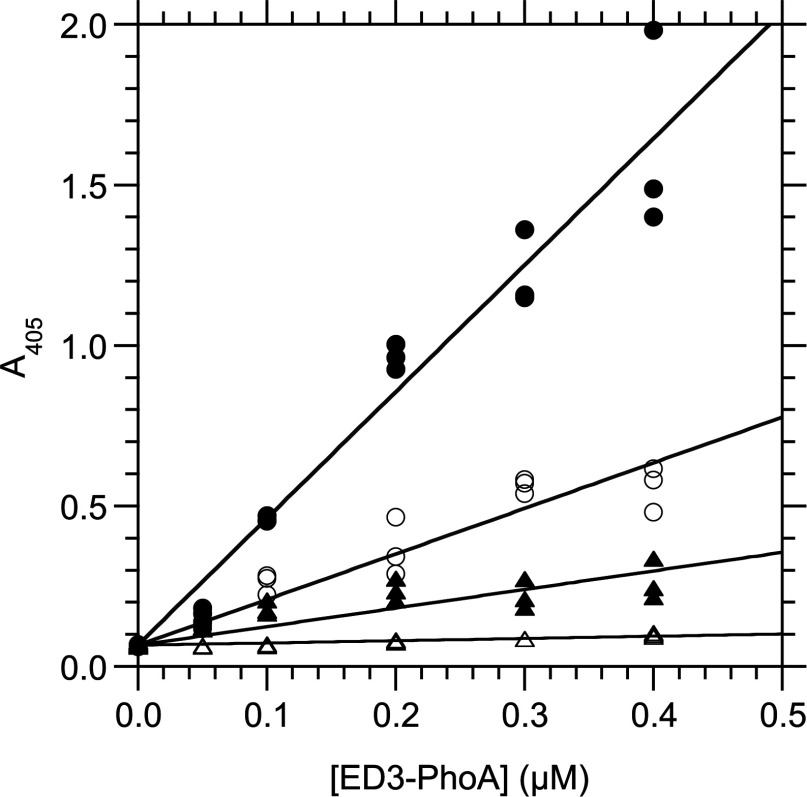
Figure 6. Interaction between RPSA-(225–295) and (H6–ED3–PhoA)2 dimers as measured by an indirect ELISA at 25°C.
RPSA-(225–295) (1 μg/ml, 110 nM) was immobilized in the wells of a microtitre plate and used to capture (H6–ED3–PhoA)2 dimers in PBS. The total concentration of H6–ED3–PhoA monomer in the capture reaction is given along the x-axis and the A405 signal along the y-axis. Linear equations were fitted to the experimental data and their slopes recorded. Closed circles, H6–ED3–PhoA hybrid for YFV(Asibi); empty circles, for YFV(17D); closed triangles, for DENV2; empty triangles, H6–PhoA alone without ED3 domain.
Figure 7. Comparison of the interactions between the ED3 domains of flaviviruses and RPSA derivatives at 25°C.
These interactions were measured by indirect ELISAs between (H6–ED3–PhoA)2 dimeric hybrids and immobilized RPSA derivatives as exemplified in Figure 6. The interactants are given along the x-axis: The (H6–ED3–PhoA)2 dimers are represented by the acronym of the virus from which they derived; Ctl, (H6–PhoA)2 control without any ED3 domain; N, RPSA-(2–220); C, RPSA-(225–295); F, RPSA-(2–295). The relative strength Sr of the interaction is given along the y-axis; it was calculated by eqn (11) as the relative slope of the linear dose–response curve in the indirect ELISA, taking the slope equal to 1 for the (H6–PhoA)2 control. Each column gives the mean and associated S.E.M. in at least three independent experiments.
DISCUSSION
Precise measurements of the parameters of interactions between RPSA and its ligands are important to understand the mechanism of these interactions, and design better ligands or inhibitors. Here, we used quantitative or semi-quantitative assays and RPSA derivatives whose structure or folding state had been carefully characterized in vitro [9,10].
Both N- and C-domains bind laminin
Our results showed that both N- and C-domains of RPSA contributed to the binding of laminin with similar KDs. It remains to be determined whether they bind to the same site or different sites on laminin. Our results were consistent with published data that conclude that the N-domain is sufficient for laminin binding [9,37] and with those, based on synthetic peptides, that show that the C-domain is involved in laminin binding [15,38,39]. The KD values that we obtained for the full-length RPSA and its N-domain, were similar to those reported in recent studies (Table 3) [9,11,12,37,40]. No KD value was available for the C-domain before the present study. We found that RPSA-(210–295) and RPSA-(225–295) bound laminin with nearly identical KDs. Therefore residues 210–224 appear dispensable for the interaction between the isolated C-domain and laminin.
Table 3. Dissociation constants KD between laminin and various forms of RPSA, as reported in the literature.
Cell membr., cell membranes; SPRadio-L, solid phase radio-ligand binding assay; SPR, surface plasmon resonance; kd/ka, the value of KD was calculated as the ratio of the rate constants for dissociation and association; Ind. ELISA, indirect ELISA; na, not applicable; nr, not reported.
| Form | Source | Folding | Assay | T (°C) | KD (nM) | Reference |
|---|---|---|---|---|---|---|
| 67LR | Cell membrane | Denatured | SPRadio-L | 4 | 2 | [11] |
| 67LR | Cell membrane | Denatured | SPRadio-L | 20 | 2 | [40] |
| 67LR | Cell shedding | Native | SPR, kd/ka | nr | 320 | [12] |
| 37LRP | E. coli | Renatured | SPR, kd/ka | nr | 410 | [12] |
| 1–220 | E. coli | Native | Ind. ELISA | 37 | 1700 | [9] |
| 1–295 | E. coli | Native | Ind. ELISA | 37 | 700 | [9] |
| 161–180 | Synthetic | na | SPRadio-L | 4 | 52 | [37] |
Previously, we have shown that the C-domain of RPSA is intrinsically disordered [10]. Therefore laminin could interact both with the N-domain of RPSA, which has a defined structure, and with its C-domain, which is intrinsically disordered. Such a result was not surprising since laminin binds heparin and heparan sulfates with high affinities, even though they have internal mobilities at the level of their iduronate residues [41]. Likely, the C-domain of RPSA and laminin interact through an induced-fit mechanism. As the isolated C-domain is intrinsically disordered, our results explained why RPSA binds laminin in experiments where its folding state is unknown because of denaturing treatments during its preparation, its immobilisation or the regeneration of the binding surface (Table 3).
Relations between heparin and laminin binding
We showed that heparin was captured by the N-domains, RPSA-(2–209) and RPSA-(2–220), in indirect ELISA assays, with K1/2 values in the low micromolar range and that it did not bind the C-domains significantly. These findings were consistent with the following properties: (i) heparin is a polyanion and can mimic nucleic acids; (ii) the N-domain of RPSA is positively charged and binds the 18S ribosomal RNA; and (iii) the C-domain is highly negatively charged.
Heparin was captured by the immobilized RPSA-(2–220) domain with a K1/2 value of 4.4 μM, and enhanced the capture of laminin by RPSA-(2–220) with and an EC50 value of 35 nM. Two different mechanims are consistent with the finding that EC50 was 130-fold smaller than K1/2: (i) laminin and heparin had different binding sites on RPSA-(2–220) and they cooperated for binding; (ii) heparin enhanced the capture of laminin by bridging several molecules of laminin together. The former mechanism is consistent with the report that 37LRP binds laminin and heparan sulfates in a non-exclusive manner [12]. The latter mechanism is favoured by the closeness between the value of EC50 (35 nM) that we found, and the values of KD that have been reported for the interaction between heparin and laminin (see below) [42].
We found that heparin strongly inhibited the capture of laminin by the immobilized RPSA-(225–295) domain and that this inhibition was due to a competition between heparin and RPSA-(225–295) for binding to the same site on laminin. Thus, the acidic C-domain of RPSA appeared as a molecular mimic of heparin. The method that we used to measure the constant of inhibition Ki, can be considered as a method to measure the dissociation constant KD between heparin and laminin since heparin did not bind RPSA-(225–295) and the indirect ELISA step of the competition ELISA method measured only the relative concentration of free laminin. The Ki value that we obtained, 16.1 nM, was consistent with the KD values, 50 nM and 130 nM, that have been reported for the interaction between laminin and heparin, which involves two sites of different strengths [42]. There is at least one previous example of a protein that mimics heparin, binds laminin, behaves as a basement membrane protein and binds prion, i.e. acetylcholine esterase [43]. RPSA-(225–295) could simply behave as a negatively charged polyelectrolyte, as heparin and heparan sulfates [44]. Its five repeated motifs of sequence E/D–W–S/T could also mimic more closely the structures of heparin or nucleic acids.
It has been shown previously that peptide G, i.e. the synthetic peptide RPSA-(161–180), binds heparin and proposed that peptide G binds laminin via heparin or heparan sulfates. In particular, it has been assumed that the basal level of peptide G binding to laminin, in the absence of added heparin, is due to residual heparan sulfates in the preparations of laminin [14,15]. We found a KD value of 0.273 μM for the interaction between laminin and RPSA-(2–209), and a K1/2 value of 1.43 μM for the capture of heparin by RPSA-(2–209). Likewise, we found KD values of 0.374 and 0.666 μM for the interactions between laminin and either RPSA-(210–295) or RPSA-(2–295) respectively, whereas heparin did not bind to these two RPSA derivatives. Therefore the affinity of RPSA for laminin was greater than its affinity for heparin and it is unlikely that RPSA or its domains bind laminin through low concentrations of heparan sulfates in the laminin preparations.
The binding site for EGCG is fully included within the N-domain
A precise measurement of the KD value between RPSA and EGCG is important for comparison with the physiological concentration of EGCG after drinking green tea. Here, we showed that RPSA-(2–295) bound EGCG with a KD value of 399 nM at 25°C. We also showed that RPSA-(2–209), whose atomic structure is known, bound EGCG with a KD value of 99 nM and that RPSA-(210–295) did not bind EGCG. Thus, the binding site for EGCG was fully included within the N-domain of RPSA. The KD values that we measured were compatible with the concentrations of EGCG that are found in plasma samples 1 h after the drinking of green tea, i.e. between 0.1 and 0.6 μM [45].
The KD value (99 nM at 25°C) for the interaction between EGCG and RPSA-(2–209) that we measured by spectrofluorimetry, was 2.5-fold higher than the KD value (39.9 nM at 25°C) for the interaction between EGCG and the 67LR form of RPSA, purified from human cancer cells, that has been calculated as the ratio kd/ka of the rate constants for dissociation and association, measured by surface plasmon resonance [46]. However, KD measured at equilibrium in solution as here, is generally not equal to KD calculated as kd/ka from kinetics of interaction measured at the interface between a solid phase and a liquid phase [47].
We found that EGCG strongly quenched the intrinsic fluorescence of the RPSA derivatives. Such a quenching has been reported previously for the interaction between EGCG and either bovine liver dihydrofolate reductase or human serum albumin [48,49]. Here additionally, we introduced a correction for the absorbance of light by EGCG to obtain reliable KD values. We found that the residue changes W176A and W195A of RPSA-(2–209) did not affect significantly the affinity between RPSA-(2–209) and EGCG. Therefore EGCG did not interact with residues Trp176 and Tpr195. Recently, it has been shown that a synthetic peptide, corresponding to residues RPSA-(161–170), decreases both interaction between human cancer cells and EGCG, and inhibition of their growth by EGCG. It has also been reported that the RPSA-(161–170) peptide and EGCG form a non-covalent complex at high concentrations [50].
The interaction between EGCG and RPSA could be studied further with the derivatives of RPSA and spectrofluorimetric method that we describe here. This method is precise and easily implemented. It could be used to study and compare the interaction between RPSA or mutant derivatives on the one hand, and polyphenols or peptides on the other hand.
Both N- and C-domains could serve as viral receptors
The wild-type strain (Asibi) and vaccine strain (17D) of YFV have five differences of residues between their ED3 domains [51]. Our results showed that RPSA could be a receptor for YFV. They suggested that the five differences of residues in the ED3 domains of YFV(17D) could attenuate the virus by weakening its interaction with RPSA. Our results confirmed that RPSA could be a receptor or co-receptor for some serotypes of DENV, in particular DENV1 and DENV2 [19,20]. There was no relation between the net charge or pI of the flaviviral ED3 domains and their ability to bind the N- or C-domain of RPSA (Table 1 and Figure 7). Therefore their binding to the RPSA derivatives was not purely electrostatic. The lack of detectable interaction between the ED3 domain from some flaviviruses and either the N-domain or the C-domain or both further showed that the observed interactions were specific. The ED3 domain from YFV interacted mainly with the acid C-domain of RPSA whereas the ED3 domain from WNV interacted only with its N-domain. As regards alphaviruses, it has been reported that the C-domain of RPSA includes a binding site for VEEV and that its N-domain includes a binding site for SINV [9,26]. Therefore our results and previous data indicate that both N- and C-domains of RPSA could serve as viral receptors.
Are the specific but weak interactions that we observed in vitro between RPSA and the ED3 domain of flaviviruses, significant in vivo? The envelope protein and therefore its ED3 domain are present in 180 copies on the surface of the flaviviral particles [27]. The RPSA molecules are clustered in membrane rafts at the cell surface and the infection by some flaviviruses depends on these membrane rafts [52,53]. Therefore the weak interactions that we observed in vitro, could be reinforced in vivo by an extensive effect of avidity, in which a single viral particle would interact simultaneously with several molecules of RPSA on the cell surface.
The (ED3–PhoA)2 dimers displayed the ED3 domain in two copies and we relied on an avidity effect to enhance their interaction with an immobilized receptor. They could be useful to further analyse the weak interactions between the ED3 domain of flaviviruses and RPSA, to analyse the enhancing or inhibiting effects of additional molecules on interaction, e.g. glycosaminoglycans, and to map the binding sites by mutagenesis. For example, such a dimeric hybrid has enabled us to show that the ED3 domain from WNV interacts directly with the NKp44 receptor of natural killer NK-cells [29].
Interactions between the N- and C-domains
We have shown previously that RPSA-(2–209) and RPSA-(225–295) interact weakly together and assumed that this interaction could be significant in the context of the full-length RPSA molecule where their effective concentrations would be high [10]. Several observations of the present study were consistent with this assumption. The KD value was 4-fold higher for the interaction of EGCG with the full-length RPSA-(2–295) than for its interaction with the N-domain. The ED3 domain from WNV bound the N-domain of RPSA but not the full-length protein. Therefore the C-domain may mask binding sites in the N-domain, either sterically or through its negative contribution to the electrostatic field of RPSA.
Furthermore, the N-domain of RPSA bound heparin whereas neither the C-domain nor the full-length RPSA protein bound it. This result indicated that the binding site of heparin in the N-domain was masked by the C-domain. The acidic domain of RPSA and heparin competed for binding to laminin (see the Results section). They may also compete for binding to the N-domain. This would explain why the full-length RPSA did not bind heparin, unlike the N-domain. One role of the C-domain could be to shield the binding site of the 18S RNA on RPSA in the absence of the former, or to prevent the binding of unrelated nucleic acids, e.g. tRNAs, by competing with them. The neutralization of the basic N-domain by the acidic C-domain could also help to control the residence of RPSA in the nucleus or nucleolus, where the ribosome matures [54].
We showed that there were several binding sites for laminin in RPSA, at least one in RPSA-(2–209) and one in RPSA-(225–295). We did not explore the importance of the binding site that had been identified previously between residues 205 and 229 of RPSA using synthetic peptides, because these residues are located at the junction between the N- and C-domains [15,38]. The existence of several binding sites in RPSA for the same ligand complicates the characterization of its interactions. For example, the KD value was 2-fold higher for the interaction between laminin and the full-length RPSA-(2–295) than for the interaction between laminin and either its N- or C-domain. This higher value of KD is difficult to interpret in the absence of information about the binding sites of the N- and C-domains on the surface of laminin. The interaction between RPSA-(2–295) and laminin might not conform to a 1:1 model.
Conformation and function
The binding activities of RPSA that we characterized in this study, except heparin binding, had initially been observed for the 67LR form of RPSA. Our results showed that these binding activities were also present in RPSA-(2–295), the 37LRP recombinant form of RPSA, or even in sub-domains. That both the 37LRP and 67LR forms share the same functions suggests that their structures or folding states are rather similar and that the maturation of 37LRP into 67LR is not necessary for many functions of RPSA. As mentioned earlier, the C-domain of RPSA is in an intrinsically disordered state of folding, i.e. in a continuum of different conformations [10]. Its flexibility could provide a range of finely tuned affinities for different proteins in different circumstances. At 17°C, the temperature at which the protein crystals were grown, 97% of the RPSA-(2–209) molecules are in their native, most stable state. At 25°C, the temperature at which we measured the interactions between RPSA and ligands, 91% of its molecules are still in the native state. At 37°C, the temperature of the human body, only 33% of the RPSA-(2–209) molecules are in a native state and 62% of the molecules are in an intermediate state of folding [10]. Therefore RPSA may be present in multiple exchanging folding states in vivo and this plasticity complicates the characterization of its interactions and functions.
Conclusions
We have shown that the N- and C-domains harboured both idiosynchratic and shared functions (Table 4) and that the C-domain mimicked heparin. The robust methods of expression, purification and measurement of interactions in vitro that we report here for RPSA and its domains, should allow one to assay additional ligands or inhibitors, and identify precisely their binding sites by mutagenesis. They could be applied to RPSA from other organisms, in particular mosquitoes and ticks that are the vectors of many pathogenic flaviviruses and alphaviruses. Ultimately, the results of such studies will serve applications against cancer and the various pathogens that interact with RPSA.
Table 4. The N- and C-domains of RPSA have both idiosyncratic and shared functions.
| Function | N-domain | C-domain | Full-length |
|---|---|---|---|
| Laminin binding | + | + | + |
| Heparin binding | + | − | − |
| Heparin mimic | − | + | − |
| EGCG binding | + | − | + |
| ED3 binding | + | + | + |
AUTHOR CONTRIBUTION
Nora Zidane, Mohamed Ould-Abeih and Isabelle Petit-Topin mainly studied the interactions with the ED3 domains of flaviviruses, with EGCG and with laminin and heparin respectively. Hugues Bedouelle designed and co-ordinated the study and was responsible for the final versions of the data analysis and paper.
ACKNOWLEDGEMENTS
We thank Elodie Brient-Litzler for advice and Nicole Guiso for her constant interest.
FUNDING
This work was supported by the Pasteur-Weizmann Council and Fondation ARC (grants to M.B.O.-A.) and by ANR [grant number 2010-INTB-1601-03 (to H.B.)].
References
- 1.Jackers P., Minoletti F., Belotti D., Clausse N., Sozzi G., Sobel M. E., Castronovo V. Isolation from a multigene family of the active human gene of the metastasis-associated multifunctional protein 37LRP/p40 at chromosome 3p21.3. Oncogene. 1996;13:495–503. [PubMed] [Google Scholar]
- 2.Liotta L. A., Horan Hand P., Rao C. N., Bryant G., Barsky S. H., Schlom J. Monoclonal antibodies to the human laminin receptor recognize structurally distinct sites. Exp. Cell Res. 1985;156:117–126. doi: 10.1016/0014-4827(85)90266-6. [DOI] [PubMed] [Google Scholar]
- 3.Castronovo V., Claysmith A. P., Barker K. T., Cioce V., Krutzsch H. C., Sobel M. E. Biosynthesis of the 67 kDa high affinity laminin receptor. Biochem. Biophys. Res. Commun. 1991;177:177–183. doi: 10.1016/0006-291x(91)91965-f. [DOI] [PubMed] [Google Scholar]
- 4.Landowski T. H., Dratz E. A., Starkey J. R. Studies of the structure of the metastasis-associated 67 kDa laminin binding protein: fatty acid acylation and evidence supporting dimerization of the 32 kDa gene product to form the mature protein. Biochemistry. 1995;34:11276–11287. doi: 10.1021/bi00035a037. [DOI] [PubMed] [Google Scholar]
- 5.Buto S., Tagliabue E., Ardini E., Magnifico A., Ghirelli C., van den Brule F., Castronovo V., Colnaghi M. I., Sobel M. E., Menard S. Formation of the 67-kDa laminin receptor by acylation of the precursor. J. Cell. Biochem. 1998;69:244–251. doi: 10.1002/(sici)1097-4644(19980601)69:3<244::aid-jcb2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 6.Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 7.Ardini E., Pesole G., Tagliabue E., Magnifico A., Castronovo V., Sobel M. E., Colnaghi M. I., Menard S. The 67-kDa laminin receptor originated from a ribosomal protein that acquired a dual function during evolution. Mol. Biol. Evol. 1998;15:1017–1025. doi: 10.1093/oxfordjournals.molbev.a026000. [DOI] [PubMed] [Google Scholar]
- 8.Nelson J., McFerran N. V., Pivato G., Chambers E., Doherty C., Steele D., Timson D. J. The 67 kDa laminin receptor: structure, function and role in disease. Biosci. Rep. 2008;28:33–48. doi: 10.1042/BSR20070004. [DOI] [PubMed] [Google Scholar]
- 9.Jamieson K. V., Wu J., Hubbard S. R., Meruelo D. Crystal structure of the human laminin receptor precursor. J. Biol. Chem. 2008;283:3002–3005. doi: 10.1074/jbc.C700206200. [DOI] [PubMed] [Google Scholar]
- 10.Ould-Abeih M. B., Petit-Topin I., Zidane N., Baron B., Bedouelle H. Multiple folding states and disorder of ribosomal protein SA, a membrane receptor for laminin, anticarcinogens, and pathogens. Biochemistry. 2012;51:4807–4821. doi: 10.1021/bi300335r. [DOI] [PubMed] [Google Scholar]
- 11.Rao N. C., Barsky S. H., Terranova V. P., Liotta L. A. Isolation of a tumor cell laminin receptor. Biochem. Biophys. Res. Commun. 1983;111:804–808. doi: 10.1016/0006-291x(83)91370-0. [DOI] [PubMed] [Google Scholar]
- 12.Fatehullah A., Doherty C., Pivato G., Allen G., Devine L., Nelson J., Timson D. J. Interactions of the 67 kDa laminin receptor and its precursor with laminin. Biosci. Rep. 2010;30:73–79. doi: 10.1042/BSR20090023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachibana H. Green tea polyphenol sensing. Proc. Japan. Acad. B, Phys. Biol. Sci. 2011;87:66–80. doi: 10.2183/pjab.87.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo N. H., Krutzsch H. C., Vogel T., Roberts D. D. Interactions of a laminin-binding peptide from a 33-kDa protein related to the 67-kDa laminin receptor with laminin and melanoma cells are heparin-dependent. J. Biol. Chem. 1992;267:17743–17747. [PubMed] [Google Scholar]
- 15.Kazmin D. A., Hoyt T. R., Taubner L., Teintze M., Starkey J. R. Phage display mapping for peptide 11 sensitive sequences binding to laminin-1. J. Mol. Biol. 2000;298:431–445. doi: 10.1006/jmbi.2000.3680. [DOI] [PubMed] [Google Scholar]
- 16.Hundt C., Peyrin J. M., Haik S., Gauczynski S., Leucht C., Rieger R., Riley M. L., Deslys J. P., Dormont D., Lasmezas C. I., Weiss S. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO J. 2001;20:5876–5886. doi: 10.1093/emboj/20.21.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K., Li L., Kozlowski K., Suh H. S., Cao W., Ballermann B. J. The protein phosphatase-1 targeting subunit TIMAP regulates LAMR1 phosphorylation. Biochem. Biophys. Res. Commun. 2005;338:1327–1334. doi: 10.1016/j.bbrc.2005.10.089. [DOI] [PubMed] [Google Scholar]
- 18.Wang K. S., Kuhn R. J., Strauss E. G., Ou S., Strauss J. H. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J. Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thepparit C., Smith D. R. Serotype-specific entry of dengue virus into liver cells: identification of the 37-kilodalton/67-kilodalton high-affinity laminin receptor as a dengue virus serotype 1 receptor. J. Virol. 2004;78:12647–12656. doi: 10.1128/JVI.78.22.12647-12656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tio P. H., Jong W. W., Cardosa M. J. Two dimensional VOPBA reveals laminin receptor (LAMR1) interaction with dengue virus serotypes 1, 2 and 3. Virol. J. 2005;2:25. doi: 10.1186/1743-422X-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akache B., Grimm D., Pandey K., Yant S. R., Xu H., Kay M. A. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J. Virol. 2006;80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim K. J., Chung J. W., Kim K. S. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. J. Biol. Chem. 2005;280:1360–1368. doi: 10.1074/jbc.M410176200. [DOI] [PubMed] [Google Scholar]
- 23.Orihuela C. J., Mahdavi J., Thornton J., Mann B., Wooldridge K. G., Abouseada N., Oldfield N. J., Self T., Ala’Aldeen D. A., Tuomanen E. I. Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J. Clin. Invest. 2009;119:1638–1646. doi: 10.1172/JCI36759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Protopopova E. V., Konavalova S. N., Loktev V. B. Isolation of a cellular receptor for tick-borne encephalitis virus using anti-idiotypic antibodies. Vopr. Virusol. 1997;42:264–268. [PubMed] [Google Scholar]
- 25.Sorokin A. V., Mikhailov A. M., Kachko A. V., Protopopova E. V., Konovalova S. N., Andrianova M. E., Netesov S. V., Kornev A. N., Loktev V. B. Human recombinant laminin-binding protein: isolation, purification, and crystallization. Biochemistry (Mosc). 2000;65:546–553. [PubMed] [Google Scholar]
- 26.Malygin A. A., Bondarenko E. I., Ivanisenko V. A., Protopopova E. V., Karpova G. G., Loktev V. B. C-terminal fragment of human laminin-binding protein contains a receptor domain for venezuelan equine encephalitis and tick-borne encephalitis viruses. Biochemistry (Mosc). 2009;74:1328–1336. doi: 10.1134/s0006297909120050. [DOI] [PubMed] [Google Scholar]
- 27.Mukhopadhyay S., Kuhn R. J., Rossmann M. G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 28.Bedouelle H., Brient-Litzler E., Dussart P., Despres P., Bremand L. Method for the diagnosis or the screening of an arbovirus infection, reagents useful in said method and their applications. Patent publication Nos: EP2003144, WO2008152528, US2010291586 (A1) CNRS, Institut Pasteur; 2008 date of priority, 15/06/2007; applicants:
- 29.Hershkovitz O., Rosental B., Rosenberg L. A., Navarro-Sanchez M. E., Jivov S., Zilka A., Gershoni-Yahalom O., Brient-Litzler E., Bedouelle H., Ho J. W., et al. NKp44 receptor mediates interaction of the envelope glycoproteins from the West Nile and dengue viruses with NK cells. J. Immunol. 2009;183:2610–2621. doi: 10.4049/jimmunol.0802806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice P., Longden I., Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 31.Harlow E., Lane D. Antibodies: A Laboratory Manual. NY: Cold Spring Harbor Laboratory, Cold Spring Harbor; 1988. [Google Scholar]
- 32.Friguet B., Chaffotte A. F., Djavadi-Ohaniance L., Goldberg M. E. Measurements of the true affinity constant in solution of antigen–antibody complexes by enzyme-linked immunosorbent assay. J. Immunol. Methods. 1985;77:305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 33.Rondard P., Goldberg M. E., Bedouelle H. Mutational analysis of an antigenic peptide shows recognition in a loop conformation. Biochemistry. 1997;36:8962–8968. doi: 10.1021/bi9625096. [DOI] [PubMed] [Google Scholar]
- 34.Renard M., Belkadi L., Bedouelle H. Deriving topological constraints from functional data for the design of reagentles fluorescent immunosensors. J. Mol. Biol. 2003;326:167–175. doi: 10.1016/s0022-2836(02)01334-7. [DOI] [PubMed] [Google Scholar]
- 35.Miranda F. F., Brient-Litzler E., Zidane N., Pecorari F., Bedouelle H. Reagentless fluorescent biosensors from artificial families of antigen binding proteins. Biosens. Bioelectron. 2011;26:4184–4190. doi: 10.1016/j.bios.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 36.Di Giovanni C., Grottesi A., Lavecchia A. Conformational switch of a flexible loop in human laminin receptor determines laminin-1 interaction. Eur. Biophys. J. 2012;41:353–358. doi: 10.1007/s00249-012-0793-9. [DOI] [PubMed] [Google Scholar]
- 37.Castronovo V., Taraboletti G., Sobel M. E. Functional domains of the 67–kDa laminin receptor precursor. J. Biol. Chem. 1991;266:20440–20446. [PubMed] [Google Scholar]
- 38.Landowski T. H., Uthayakumar S., Starkey J. R. Control pathways of the 67 kDa laminin binding protein: surface expression and activity of a new ligand binding domain. Clin. Exp. Metastasis. 1995;13:357–372. doi: 10.1007/BF00121912. [DOI] [PubMed] [Google Scholar]
- 39.Wewer U. M., Taraboletti G., Sobel M. E., Albrechtsen R., Liotta L. A. Role of laminin receptor in tumor cell migration. Cancer Res. 1987;47:5691–5698. [PubMed] [Google Scholar]
- 40.Malinoff H. L., Wicha M. S. Isolation of a cell surface receptor protein for laminin from murine fibrosarcoma cells. J. Cell. Biol. 1983;96:1475–1479. doi: 10.1083/jcb.96.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulloy B., Forster M. J. Conformation and dynamics of heparin and heparan sulfate. Glycobiology. 2000;10:1147–1156. doi: 10.1093/glycob/10.11.1147. [DOI] [PubMed] [Google Scholar]
- 42.Skubitz A. P., McCarthy J. B., Charonis A. S., Furcht L. T. Localization of three distinct heparin-binding domains of laminin by monoclonal antibodies. J. Biol. Chem. 1988;263:4861–4868. [PubMed] [Google Scholar]
- 43.Johnson G., Swart C., Moore S. W. Interaction of acetylcholinesterase with the G4 domain of the laminin alpha1-chain. Biochem. J. 2008;411:507–514. doi: 10.1042/BJ20071404. [DOI] [PubMed] [Google Scholar]
- 44.Seyrek E., Dubin P. Glycosaminoglycans as polyelectrolytes. Adv. Colloid Interface Sci. 2010;158:119–129. doi: 10.1016/j.cis.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Lee M. J., Wang Z. Y., Li H., Chen L., Sun Y., Gobbo S., Balentine D. A., Yang C. S. Analysis of plasma and urinary tea polyphenols in human subjects. Cancer Epidemiol. Biomarkers Prev. 1995;4:393–399. [PubMed] [Google Scholar]
- 46.Tachibana H., Koga K., Fujimura Y., Yamada K. A receptor for green tea polyphenol EGCG. Nat. Struct. Mol. Biol. 2004;11:380–381. doi: 10.1038/nsmb743. [DOI] [PubMed] [Google Scholar]
- 47.Bedouelle H., Belkadi L., England P., Guijarro J. I., Lisova O., Urvoas A., Delepierre M., Thullier P. Diversity and junction residues as hotspots of binding energy in an antibody neutralizing the dengue virus. FEBS J. 2006;273:34–46. doi: 10.1111/j.1742-4658.2005.05045.x. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez-del-Campo L., Saez-Ayala M., Chazarra S., Cabezas-Herrera J., Rodriguez-Lopez J. N. Binding of natural and synthetic polyphenols to human dihydrofolate reductase. Int. J. Mol. Sci. 2009;10:5398–5410. doi: 10.3390/ijms10125398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maiti T. K., Ghosh K. S., Dasgupta S. Interaction of (−)-epigallocatechin-3-gallate with human serum albumin: fluorescence, fourier transform infrared, circular dichroism, and docking studies. Proteins. 2006;64:355–362. doi: 10.1002/prot.20995. [DOI] [PubMed] [Google Scholar]
- 50.Fujimura Y., Sumida M., Sugihara K., Tsukamoto S., Yamada K., Tachibana H. Green tea polyphenol EGCG sensing motif on the 67-kDa laminin receptor. PLoS ONE. 2012;7:e37942. doi: 10.1371/journal.pone.0037942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hahn C. S., Dalrymple J. M., Strauss J. H., Rice C. M. Comparison of the virulent Asibi strain of yellow fever virus with the 17D vaccine strain derived from it. Proc. Natl. Acad. Sci. U.S.A. 1987;84:2019–2023. doi: 10.1073/pnas.84.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujimura Y., Yamada K., Tachibana H. A lipid raft-associated 67kDa laminin receptor mediates suppressive effect of epigallocatechin-3-O-gallate on FcepsilonRI expression. Biochem. Biophys. Res. Commun. 2005;336:674–681. doi: 10.1016/j.bbrc.2005.08.146. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi T., Suzuki T. Function of membrane rafts in viral lifecycles and host cellular response. Biochem. Res. Int. 2011;2011:245090. doi: 10.1155/2011/245090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houmani J. L., Ruf I. K. Clusters of basic amino acids contribute to RNA binding and nucleolar localization of ribosomal protein L22. PLoS ONE. 2009;4:e5306. doi: 10.1371/journal.pone.0005306. [DOI] [PMC free article] [PubMed] [Google Scholar]