Abstract
Epigenetic mechanisms are thought to help regulate the unique transcription program that is established in germ cell development. During the germline cycle of many organisms, the epigenome undergoes waves of extensive resetting events, while a part of epigenetic modification remains faithful to specific loci. Little is known about the mechanisms underlying these events, how loci are selected for, or avoid, reprogramming, or even why these events are required. In particular, although the significance of genomic imprinting phenomena involving DNA methylation in mammals is now well accepted, the role of histone modification as a transgenerational epigenetic mechanism has been the subject of debate. Such epigenetic mechanisms may help regulate transcription programs and / or the pluripotent status conferred on germ cells, and contribute to germ line continuity across generations. Recent studies provide new evidence for heritability of histone modifications through germ line cells and its potential effects on transcription regulation both in the soma and germ line of subsequent generations. Unraveling transgenerational epigenetic mechanisms involving highly conserved histone modifications in elegant model systems will accelerate the generation of new paradigms and inspire research in a wide variety of fields, including basic developmental studies and clinical stem cell research.
Keywords: chromatin, epigenetics, germ cell, histone modification, transcription
Introduction
The term epigenome refers to the profile of epigenetic modification across any genome, i.e., the genome-wide patterns of DNA and histone modifications within any nucleus. It is becoming clear that the epigenome is a crucial component of the information that is passed through cell division and even across generations besides the genome.
Extensive reprogramming of the epigenome is a conserved feature of germ cell specification in many organisms (Seydoux & Braun 2006; Surani et al. 2007; Sasaki & Matsui 2008). Since August Weismann proposed the theory of germline continuity more than a century ago, the mechanism(s) ensuring germline “immortality” across generations has long been unclear. Recent studies have suggested that this transgenerational continuity can involve epigenetic regulation that includes histone modifications and DNA methylation (Surani et al. 2007; Katz et al. 2009). However, it is still largely unknown how epigenetic modifications regulated in one generation can directly ensure appropriate control of the specialized gene expression program in germ cells of the next generation. To explore these potential generational effects, genetic, cytological, and epigenomic analyses have been performed in many model systems, and information about this is being accumulated. Some elegant studies regarding these epigenetic mechanisms have been conducted using Caenorhabditis elegans as a model system.
This review will mainly describe some previous discoveries in genetic studies using C. elegans, in which epigenetic regulators required for germline immortality were elegantly identified, and will focus on “erasure” and maintenance of heritable histone modifications as epigenetic memory, both of which appears to have an impact on germ line continuity.
Epigenetic regulators required for germ-line continuity
The MES proteins
The maternal effect sterile (mes) genes were identified in screens for “grandchildless” mutants in C. elegans 20 years ago (Capowski et al. 1991). The mes mutations cause maternal-effect sterility, the result of degeneration of the germ line in the F2 generation. The phenotype has always been very interesting, since MES proteins (maternal in origin) are last detected in the primordial germ cells (PGCs) of the F1 mutant animals, yet the PGCs produce approximately 1000 functional descendant germ cells. The germ line only fails when specification is attempted in the absence of maternal MES protein in the following generation, that is, in the PGCs of F2 mutants. The emerging model is that the MES proteins help specify the chromatin organization (e.g., chromatin modification status) that the PGCs inherit, and that participates in specifying the germline transcription program in the nascent germ cells. Indeed, MES proteins are homologues of factors identified as critical epigenetic components. MES-2, MES-3 and MES-6 compose a complex resembling the Polycomb Repressive Complex PRC2, which is highly conserved among multicellular organisms and is involved in chromatin based repression via the histone H3 lysine 27 (H3K27) methyltransferase activity of MES-2 (Paulsen et al. 1995; Xu et al. 2001; Bender et al. 2004). MES-4 is a histone H3 lysine 36 (H3K36) methyltransferase that is homologous to mouse NSD1 (Bender et al. 2006) and likely functions with a unique set of partners that have not yet been identified. The MES-mediated histone modifications are detectable in all stages of germline development, while a subset of active chromatin marks are erased and almost undetectable in the PGCs (Fig. 1; discussed below). These MES factors have been shown to play important roles in X chromosome silencing that is observed in adult germ cells, although MES-4’s role in this process is presumably indirect, as MES-4 associates predominantly with autosomal chromatin (Fong et al. 2002; Bender et al. 2006). In the embryo, the maternally-provided MES proteins are initially found in all cells, but they become restricted to the PGCs in later stages (Fong et al. 2002). The function of MES proteins in the embryonic germ line, however, remains unclear. Interestingly, among the mes mutants, mes-4 mutants display the most severe PGC proliferation defects (Capowski et al. 1991). We recently discovered that MES-4 contributes to transgenerational-transcription repression in the PGCs (Furuhashi et al. unpubl. data, 2009). Intriguingly, MES-4 has also been shown to be required for ectopic expression of germ line genes in somatic cells lacking the NuRD (Nucleosome Remodeling and Deacetylase) complex component MEP-1, suggesting that there is a context-dependent function for MES-4-mediated H3K36 methylation (Unhavaithaya et al. 2002).
Fig. 1.
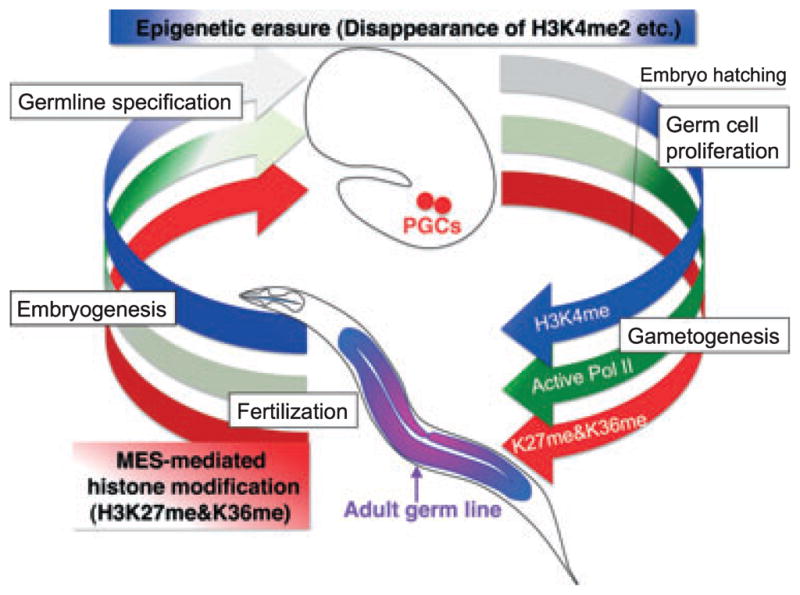
Epigenetic erasure/maintenance in C. elegans germline cycle. Epigenetic reprogramming involving widespread changes in chromatin is a conserved feature of germ cell specification in many organisms (Schaner et al. 2003; Seki et al. 2005, 2007; Hajkova et al. 2008). In C. elegans germline, a subset of conserved epigenetic marks of “active” chromatin, such as H3K4me2, disappears specifically from PGC chromatin soon after the birth of these cells and remains extremely low until hatching. MES-mediated histone modifications, H3K27me and H3K36me, are detectable during the germline cycle (Bender et al. 2004, 2006).
MRG-1
Mutations in the mrg-1 gene cause a germline degeneration phenotype that is very similar to that observed in mes mutants (Takasaki et al. 2007). MRG-1 is the C. elegans orthologue of MRG15, a mammalian chromodomain protein related to the mortality factor MORF4, which has been shown to be required for cell proliferation and embryo survival (Tominaga et al. 2005a, b). The yeast homologue Eaf3 has been shown to recognize and bind H3K36me. and is required for preventing transcriptional initiation from cryptic promoters within gene bodies (Carrozza et al. 2005; Joshi & Struhl 2005; Keogh et al. 2005). MRG-1 is required for the repression of genes that are also mis-regulated in mes-4 mutant animals, and like MES-4 is concentrated on autosomes and excluded from the X chromosome (Takasaki et al. 2007). Furthermore, PGCs lacking MRG-1 show a severe proliferation defect and degeneration similar to the phenotypes observed in mes-4 mutant PGCs, implying that MRG-1 may be a “reader”, but perhaps even an “interpreter” of the MES-4 dependent H3K36me. MRG-1 homologues in other systems associate with both histone acetyltransferase (HAT, also called KAT) and histone deacetylase / demethylase (HDAC/ KDM) complexes (Carrozza et al. 2005; Joshi & Struhl 2005; Keogh et al. 2005; Morillon et al. 2005; Martin et al. 2006; Taverna et al. 2006; Hayakawa et al. 2007; Larschan et al. 2007; Moshkin et al. 2009). This protein may thus function as “reader / interpreter” for both transcription activation and repression. This bifunctional property might contribute to the context dependent function of MES-4-mediated H3K36me described above.
SPR-5 / LSD1
A recent study demonstrated that mutants of spr-5, the C. elegans orthologue of the H3K4me2 demethylase LSD1 (also called KDM1), exhibit progressive sterility over approximately 20–30 generations (Katz et al. 2009). This sterility was shown to correlate with the misregulation of a specific gene set, which is expressed in spermatogenesis, and aberrant transgenerational accumulation of H3K4me2. Details of the exciting discovery are described / discussed below.
Resetting histone modification between generations
H3K4 methylation is a well-characterized, transcription-coupled histone modification that can provide an epigenetic memory for regions of active transcription (Li et al. 2007; Shilatifard 2008; Muramoto et al. 2010). For example, H3K4 methylation by Trithorax in Drosophila is critical for the maintenance of a transcriptionally active state through multiple cell divisions, even in the absence of activating transcription factors (Ringrose & Paro 2004). Ng & Gurdon (2008) recently reported that endoderm genes are inappropriately expressed in Xenopus embryos derived from the transfer of differentiated endoderm nuclei. This effect was correlated with lysine 4 of the histone variant H3.3 that is incorporated during endodermal transcription, implying that the complete reprogramming of a somatic nucleus may require efficient erasure of epigenetic information at H3K4 (Ng & Gurdon 2008). The natural target of erasure mechanisms operating in oocytes during somatic cell nuclear transfer experiments are likely to be epigenetic information arriving in gametes. Inappropriate propagation of epigenetic information acquired during gametogenesis to the next generation could result in the misregulation of gamete- and meiosis-specific genes in the zygote. Therefore, resetting of H3K4me2, and other histone marks, that is acquired during gametogenesis may be required to prevent the inappropriate transmission of epigenetic memory across generations.
DNA methylation does not occur in C. elegans, and thus all epigenetic information on chromatin is presumably encoded in histone modifications. In C. elegans, a number of conserved epigenetic marks of “active” chromatin, such as H3K4me2, dramatically erased from the C. elegans PGCs soon after their birth (Schaner et al. 2003; Fig. 2). This event might be analogous to the epigenetic erasure observed in the PGCs of other organisms (Seki et al. 2005, 2007; Hajkova et al. 2008). One of the candidates that could be involved in such an active epigenetic erasure is histone demethylase activity. Katz et al. (2009) tested whether C. elegans homologues of LSD1, which catalyzes H3K4me demethylation and is a component of CoREST transcriptional repressor complex (Shi et al. 2004), were involved in the H3K4me2 erasure in the worm PGCs. Mutations of all three LSD1 orthologs, alone or in combination, did not initially show apparent defects in the PGC erasure process, indicating the existence of LSD1-independent mechanism(s). However, it was noticed that as lines carrying mutations in one of the orthologs, spr-5, were passaged, they produced fewer progeny, that the progeny were often sterile, and that the frequency of sterility increased with each passage. Subsequent careful generational analyses revealed the “germ line mortality” phenotype in the spr-5 mutant, a phenotype in which the germline becomes progressively dysfunctional in successive generations. In contrast to previously identified mutants displaying this phenotype, no evidence of accumulating genetic defects was obtained during the generational analyses. Instead, Katz et al. observed that the above phenotypes could be fully rescued by transient exposure to SPR-5 activity, suggesting that the underlying defects were of an epigenetic nature. Expression-profiling analyses comparing samples from multiple generations showed striking and coordinated expression changes in genes expressed during spermatogenesis. The expression of these genes was gradually elevated in early generations and peaked just before the generations that showed a sharp decrease in fertility. At this time point, the expression of these genes was also markedly decreased. However, what was particularly intriguing was that the level of H3K4me2 in the promoter regions of these genes continued to rise. The above observations suggest that aberrant accumulation of epigenetic marks can persist, and even inappropriately accumulate, in successive generations independently of increases in transcription. Moreover, in the later generations, Katz et al. observed an increased failure to efficiently erase H3K4me2 from the mutant PGCs. Collectively, these data suggest that SPR-5 / LSD1 is required for removing epigenetic information acquired from the parental germ line, and that defects in the erasure mechanism leads to aberrant accumulation of the epigenetic memory that becomes increasingly difficult to erase in the PGCs (via the SPR-5 independent mechanisms) in successive generations. The transgenerational accumulation of H3K4me2 leads to defects in gametogenesis in the worms and abolishes germ line immortality. However, many questions remain to be answered. For instance, it is not clear when histone demethylase is required in the germ line cycle. SPR-5 protein is observed in both adult germ cells and in the early embryo, and may be functional in either or both stages. It is also unclear why defects in gametogenesis appear after many generations. The SPR-5 / LSD1-independent mechanism that erases H3K4me in the PGCs is presumed to help “mop up” H3K4me2 erasure that is not removed in spr-5 mutants, but this is not certain. Inefficiencies in this mechanism may normally and stochastically increase the persistence of H3K4me2 in successive generations, and the loss of SPR-5 function may reveal this stochastisticity. The exact targets of either erasure mechanism are still unknown. Regardless of these current uncertainties, recent studies have shown that histone demethylases are critical for the maintenance of fertility in other organisms (Di Stefano et al. 2007; Okada et al. 2007; Rudolph et al. 2007), suggesting that there is a conserved requirement for “epigenetic erasers” to maintain germline continuity.
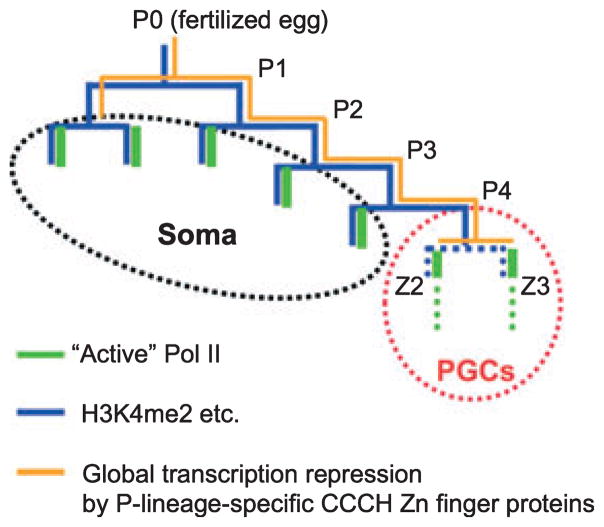
Fig. 2.
Transcription status and histone modification dynamics in C. elegans early embryonic blastomeres. During early embryogenesis, the asymmetric divisions of P-lineage cells (P0 to P3) produces germ line precursors and somatic cells. Subsequently, the last P cell (P4) finally makes two germline-committed cells, Z2 and Z3, and they are arrested at G2 phase during the rest of embryogenesis (Fukuyama et al. 2006). Throughout P-lineage, CCCH zinc finger proteins, OMA-1/2 and PIE-1, are sequentially acting to maintain transcriptional quiescence independently of the chromatin environment (orange line: Seydoux et al. 1996; Seydoux and Dunn, 1997; Guven-Ozkan et al. 2008). However, soon after the symmetric cell division of P4, PIE-1 is degraded quickly and “Active Pol II” suddenly appears in the PGCs (green). H3K4me2 (blue) are initially present in both the transcriptionally quiescent P-lineage and their somatic sisters where transcription is activated. H3K4me2 in P-lineage is maintained by unidentified mechanism, but in the PGCs the level of H3K4me2 begin to decrease (indicated by dotted blue lines), and becomes almost undetectable soon after the birth of Z2/Z3 (Schaner et al. 2003). Our recent study revealed that the appearance of “active Pol II” in the PGCs is actually transient, and that MES-4 activity is important for the Pol II repression (indicated by dotted green lines) (Furuhashi et al. unpubl. data, 2009).
Heritability of histone modification across generations
The mechanisms for stability and maintenance of epigenetic information, particularly those involving histone modifications, have been extensively investigated in several model systems. Recent discoveries that modifications such as H3K4me and H3K27me can be inherited through gametes raise new questions about whether the transmitted epigenetic modifications play critical roles in the development of the next generation and, what mechanisms ensure the stable inheritance of these modifications (Hammoud et al. 2009). Although the maintenance DNA methyltransferase activity of Dnmt1 in mammals has been well characterized, histone methyltransferase activities predominantly responsible for maintenance, but not de novo, methylation have not been described. Existence of such machinery in germ line may be expected to ensure faithful maintenance of the histone modification pattern across generations.
Recently, molecular mechanisms underlying the maintenance of some epigenetic marks after DNA/ chromatin replication have been identified. For example, DNA methylation is maintained via the unique recognition of hemi-methylated CpG sequence by the SRA domain of UHRF1, which recruits Dnmt1 to the DNA (Bostick et al. 2007; Sharif et al. 2007; Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008). Similarly, the PRC2 complex, which catalyzes H3K27me3, specifically binds to pre-existing H3K27me3, and this binding appears to be critical for the heritable transmission of this modification (Hansen et al. 2008; Margueron et al. 2009). As described above, MES proteins in C. elegans are the homologues of the PRC2 components and the NSD-type H3K36 methyltransferases and are critical for germline immortality across generations. Further investigation on the potential transgenerational nature of the MES-mediated epigenetic marks and its maintenance mechanism would provide new insights into the epigenetic/ transgenerational transcription regulation as a critical component in development.
Conclusions
The model system C. elegans, which allows a combination of traditional genetic screens, cytological, and next generation genome-wide analyses, has made significant contributions to the field of germ cell research. The studies discussed here are of overlapping interest to scientists involved in both traditional chromatin research, because of its impact on the debate surrounding the “heritability” of histone modifications and the histone code, as well as the wide variety of scientists interested in epigenetic regulation of developmental programs and the establishment and maintenance of the germ line.
An important question that is still largely unclear is exactly how any molecular mechanism determines what information is to be reset and what is to be maintained between generations. Future studies focusing on this will allow us to gain better understanding of the molecular basis of germ line continuity.
Acknowledgments
I would like to thank David J. Katz, Susan Strome, Shoichiro Kurata and members of the Kelly, Strome and Kurata labs for their support and helpful discussions. This review was written partially based on the studies supported by grants from the National Institute of Health to W.G.K. H.F. is supported by Grant-in-Aid for Young Scientists (Start-up).
References
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- Bender LB, Cao R, Zhang Y, Strome S. The MES-2 / MES-3 / MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol. 2004;14:1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
- Bender LB, Suh J, Carroll CR, Fong Y, Fingerman IM, Briggs SD, Cao R, Zhang Y, Reinke V, Strome S. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133:3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Capowski EE, Martin P, Garvin C, Strome S. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics. 1991;129:1061–1072. doi: 10.1093/genetics/129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Di Stefano L, Ji JY, Moon NS, Herr A, Dyson N. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr Biol. 2007;17:808–812. doi: 10.1016/j.cub.2007.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y, Bender L, Wang W, Strome S. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science. 2002;296:2235–2238. doi: 10.1126/science.1070790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama M, Rougvie AE, Rothman JH. C. elegans DAF-18 / PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr Biol. 2006;16:773–779. doi: 10.1016/j.cub.2006.02.073. [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T, Nishi Y, Robertson SM, Lin R. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell. 2008;135:149–160. doi: 10.1016/j.cell.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Ohtani Y, Hayakawa N, Shinmyozu K, Saito M, Ishikawa F, Nakayama J. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12:811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DG, Grimes DE, Baetz K, Howe L. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol Cell Biol. 2006;26:3018–3028. doi: 10.1128/MCB.26.8.3018-3028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell. 2009;35:782–793. doi: 10.1016/j.molcel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Muramoto T, Müller I, Thomas G, Melvin A, Chubb JR. Methylation of H3K4 is required for inheritance of active transcriptional states. Curr Biol. 2010;20:397–406. doi: 10.1016/j.cub.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- Paulsen JE, Capowski EE, Strome S. Phenotypic and molecular analysis of mes-3, a maternal-effect gene required for proliferation and viability of the germ line in C. elegans. Genetics. 1995;141:1383–1398. doi: 10.1093/genetics/141.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schäfer C, Phalke S, Walther M, Schmidt A, Jenuwein T, Reuter G. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Schaner CE, Deshpande G, Schedl PD, Kelly WG. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev Cell. 2003;5:747–757. doi: 10.1016/s1534-5807(03)00327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Molecular implementation & physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Takasaki T, Liu Z, Habara Y, Nishiwaki K, Nakayama J, Inoue K, Sakamoto H, Strome S. MRG-1, an autosome-associated protein, silences X-linked genes and protects germline immortality in Caenorhabditis elegans. Development. 2007;134:757–767. doi: 10.1242/dev.02771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga K, Kirtane B, Jackson JG, Ikeno Y, Ikeda T, Hawks C, Smith JR, Matzuk MM, Pereira-Smith OM. MRG15 regulates embryonic development and cell proliferation. Mol Cell Biol. 2005a;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga K, Matzuk MM, Pereira-Smith OM. MrgX is not essential for cell growth and development in the mouse. Mol Cell Biol. 2005b;25:4873–4880. doi: 10.1128/MCB.25.12.4873-4880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- Xu L, Fong Y, Strome S. The Caenorhabditis elegans maternal-effect sterile proteins, MES-2, MES-3, and MES-6, are associated in a complex in embryos. Proc Natl Acad Sci USA. 2001;98:5061–5066. doi: 10.1073/pnas.081016198. [DOI] [PMC free article] [PubMed] [Google Scholar]