Abstract
SUMMARY
A hexanucleotide GGGGCC repeat expansion in the noncoding region of the C9ORF72 gene is the most common genetic abnormality in familial and sporadic amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). The function of the C9ORF72 protein is unknown, as is the mechanism by which the repeat expansion could cause disease. Induced pluripotent stem cell (iPSC)-differentiated neurons from C9ORF72 ALS patients revealed disease-specific (1) intranuclear GGGGCCexp RNA foci, (2) dysregulated gene expression, (3) sequestration of GGGGCCexp RNA binding protein ADARB2, and (4) susceptibility to excitotoxicity. These pathological and pathogenic characteristics were confirmed in ALS brain and were mitigated with antisense oligonucleotide (ASO) therapeutics to the C9ORF72 transcript or repeat expansion despite the presence of repeat-associated non-ATG translation (RAN) products. These data indicate a toxic RNA gain-of-function mechanism as a cause of C9ORF72 ALS and provide candidate antisense therapeutics and candidate human pharmacodynamic markers for therapy.
INTRODUCTION
A recently discovered hexanucleotide ‘‘GGGGCC’’ repeat’’ expansion in the noncoding region of the C9ORF72 gene has been found in at least 8% of sporadic ALS (sALS) and FTD cases and more than 40% of familial ALS (FALS) and FTD cases (DeJesus-Hernandez et al., 2011; Majounie et al., 2012; Renton et al., 2011). Since the discovery of pathogenic repeat expansions as a mechanism of disease in the 1990s, the list of neurodegenerative and neuromuscular disorders characterized by unstable repeat expansions has grown to over 20 (Brouwer et al., 2009; Pearson et al., 2005; Todd and Paulson, 2010). Repeat expansions are classified as coding or noncoding according to their gene location, and the disease-causing mechanisms include protein gain-of-function (Huntington’s disease, HD), protein loss-of-function (FRAXA, FRDA), toxic RNA gain-of-function (DM1&2) (for reviews, see Brouwer et al., 2009; Gatchel and Zoghbi, 2005; Todd and Paulson, 2010), and non-ATG-initiated translation (RAN) peptides (Mori et al., 2013b) (Ash et al., 2013). The repeat expansion in DM1 alters activities of RNA binding proteins (RBPs), including muscleblind-like 1 (MBLN1) (Fardaei et al., 2002; Grammatikakis et al., 2011; Miller et al., 2000). MBLN1 is sequestered in the nucleus by the repeat-containing RNA resulting in the formation of a pathogenic protein:RNA complex that, when visualized by RNA fluorescent in situ hybridization, form an intranuclear RNA foci, which leads to a loss of protein activity and reduces alternative splicing of other genes (Kanadia et al., 2003, 2006). Notably, intranuclear GGGGCC RNA foci have also been found in the motor cortex and spinal cord of C9ORF72 ALS/FTD patients (DeJesus-Hernandez et al., 2011), suggesting that, like myotonic dystrophy, RNA toxicity plays a role in C9ORF72 neurodegeneration.
To understand the pathogenesis of the C9ORF72 expansion and to develop possible therapeutics, we generated a collection of C9ORF72 ALS induced pluripotent stem cells (iPSCs) and differentiated them into neurons (iPSNs). Using this model system, we discovered intranuclear C9ORF72 repeat-containing RNA fociin all tested human C9ORF72 iPSN cell lines. Furthermore, we identified several protein binding partners for the expanded GGGGCC RNA (GGGGCCexp) and confirmed that the RNA binding protein ADARB2 interacts with nuclear GGGGCC RNA foci. In addition, we discovered aberrantly expressed genes in C9ORF72 cells and determined that C9ORF72 ALS iPSNs are highly susceptible to glutamate-mediated excitotoxicity. To validate the use of this iPSC model, we confirmed these expanded C9ORF72-related phenotypes in postmortem human ALS CNS tissue. Finally, iPSN treatment with novel antisense oligonucleotides (ASOs) that target the GGGGCCexp RNA sequence but do not lower C9ORF72 RNA levels mitigate all toxic phenotypes. Although RAN proteins, translated from the mutant GGGGCC expansion, are present in these iPSNs, they do not appear to contribute to the observed acute neurotoxicity. Taken together, these data support the theory that the generation of toxic RNA plays a major role in C9ORF72 ALS and that specifically targeted antisense can effectively prevent neurotoxicity. These iPSC studies, through the development of pathological readouts, support the development of a novel antisense therapy to treat neurodegeneration due to the C9ORF72 repeat expansion.
RESULTS
C9ORF72 iPSCs Exhibit the GGGGCC Repeat Expansion and Have Reduced C9ORF72 RNA Levels
iPSC models are useful for studying human disease pathogenesis and could serve as a powerful human and allele-specific tool to evaluate therapeutics. To study the pathology of the C9ORF72 repeat expansion, we isolated fibroblasts from unrelated C9ORF72 ALS patients whose repeat expansion was confirmed by repeat-primed PCR (Renton et al., 2011) and Southern blot analysis (Figures 1A and 1B; for demographic information on all cell lines see Table S1 available online), reprogrammed them to TRA-1–60+ iPSCs (Dimos et al., 2008), and differentiated them to Tuj-1+ iPS-derived neurons (Figure S1A). iPSC lines were generated from fibroblasts reprogramed using Sox2, Oct4, Klf4, and c-Myc encoding vectors (data not shown). All iPSC lines were validated via strict quality control profiling including expression of pluripotency markers as well as normal karyotyping (data not shown). The iPSN cultures are composed of a heterogeneous neuronal cell population, of which about 30%–40% stained positive for motor neuron marker HB9 (Figure S1B). It is widely known that not only motor neurons, but also cortical neurons, interneurons, and glia are pathologically injured in ALS (Morrison et al., 1998; Kang et al., 2010; Reis et al., 2011), which is why studies were carried out using a mixed neuronal cell population. Southern blot analysis revealed that the GGGGCC expansion is maintained in all lines after reprogramming and differentiation from fibroblast to iPSC neurons or astroglia (Figure 1A) with little or minor changes in expansion size, which is likely to be reflected by clonal selection of fibroblasts. No expansion size instability was observed after increasing cell passage numbers in vitro (>50; Figure 1B).
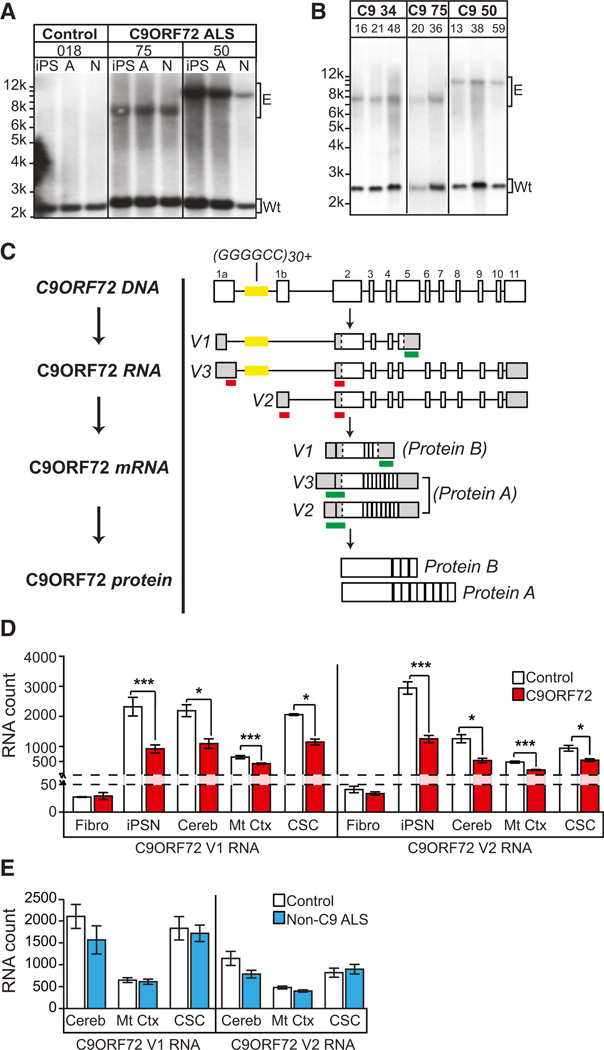
Figure 1. C9ORF72 ALS iPSNs Exhibit GGGGCCexp Repeat Allele and Reduced C9ORF72.
(A) Southern blot of C9ORF72 ALS fibroblasts and subsequent iPSCs for lines used in this study. All C9ORF72 lines contained normal length allele (Wt) and an expanded ‘‘GGGGCC’’ repeat allele (E) that is maintained during differentiation to iPSN (N) or astrocytes (A).
(B) Repeat length of iPSCs was maintained over several iPSC cell passage numbers (ranging from p13 to p59).
(C) Schematic of the C9ORF72 gene, location of the repeat sequence (yellow), validated RNA variants, and location of the nanostring probesets. For detection of nanostring probe, each individual probe (red) must bind in tandem for detection (green). Probes to C9ORF72 V1 will detect pre-mRNA and mRNA; probes to V2 and V3 will only detect C9ORF72 mRNA.
(D) C9ORF72 ALS iPSNs and CNS tissue exhibit reduced C9ORF72 RNA levels (n for C9ORF72 versus Control: n = (fibroblast, Fibro) 5 versus 5; (iPSN) 3 versus 3; (cerebellum, Cereb) 4 versus 6; (motor cortex, Mt Ctx) 10 versus 6; (cervical spinal cord, CSC) 4 versus 6).
(E) Non-C9ORF72 ALS patients do not show attenuated levels of C9ORF72 RNA.
(*p < 0.05; ***p < 0.001). See also Figures S1 and S2; Tables S1, S2, and S3.
Earlier studies have shown that ALS and FTD patients exhibit decreased C9ORF72 RNA levels in patient tissue as measured by real-time PCR (Ciura et al., 2013; DeJesus-Hernandez et al., 2011; Gijselinck et al., 2012). Therefore, to determine whether patient fibroblasts and iPSNs exhibit similar in vivo C9ORF72 RNA variant expression patterns, we quantified C9ORF72 RNA levels in C9ORF72 ALS fibroblasts, in iPSNs, and in human CNS regions from multiple unrelated patients (Table S2) using the highly sensitive, probe-based nanostring RNA detection system (probe sequences in Table S3). This method is ideal for screening human tissue due to the lack of any nucleotide amplification step. There are three validated mRNA products transcribed from the C9ORF72 gene, C9ORF72 variant 1, 2, and 3 (NM_145005.5, NM_018325.3, and NM_001256054.1, respectively) with variant 1 and 3 containing open reading frames (ORFs) upstream of the expanded GGGGCC repeat. To detect the levels of each C9ORF72 RNA variant and to compare each variant between samples we generated two 50-mer nanostring probes for each transcript that target one of the C9ORF72 variants. In order for the RNA variant to be detected, each 50-mer probe must bind in tandem (schematic shown in Figure 1C). Similar to previous studies, C9ORF72 V1 and V2 RNA levels were attenuated by ∼50% in C9ORF72 ALS CNS tissue (n = 5–10) compared to healthy controls (n = 4–10) (Figure 1D). Interestingly, iPSNs (n = 3) exhibited a similar expression pattern with an approximate 50% reduction in C9ORF72 V1 and V2 RNA levels, while patient fibroblasts showed low C9ORF72 expression with no obvious differences between the control and C9ORF72 ALS lines (although slight differences may be confounded by the lower absolute RNA counts) (Figure 1D). In human CNS tissue, the highest quantity of C9ORF72 RNA is found in the cerebellum and cervical spinal cord. Importantly, in non-C9ORF72 ALS tissue, C9ORF72 RNA levels were comparable to healthy control tissue (Figure 1E). While we tested for the presence of C9ORF72 V3 RNA using probes targeting the spliced mRNA sequence (Figure 1C), it could only be detected slightly above background in iPSNs and cervical spinal cord tissue with no obvious expression differences between control and C9ORF72 ALS cells/tissue (Figure S2A). However, probes targeting C9ORF72 intron 7, which detect V2 and V3 pre-mRNA, exhibited a statistically significant reduction of this RNA sequence in C9ORF72 ALS compared to control and non-C9ORF72 ALS tissue, suggesting that the prespliced levels of C9ORF72 V2 and V3 RNA are also reduced in endogenous tissue (Figure S2B). Whether these changes in RNA correlate with endogenous protein are uncertain, as current antibodies to C9ORF72 protein are not suitable for specific quantification (data not shown). While the function of the C9ORF72 protein is unknown, a recent study in zebrafish has shown that C9ORF72 deficiency via ASO knockdown results in locomotor deficits and abnormal growth and morphology of motor axons (Ciura et al., 2013). Further studies are required to determine whether a C9ORF72 deficit in human ALS/FTD contributes to the disease phenotype.
C9ORF72 ALS iPSCs Exhibit Toxic RNA Foci
We detected intranuclear GGGGCC RNA foci in C9ORF72 ALS patient tissue, iPSNs, and fibroblasts by RNA fluorescent in situ hybridization (RNA FISH), similar to previous reports of postmortem CNS tissue (DeJesus-Hernandez et al., 2011), employing fluorescently labeled locked nucleic acid (LNA) oligonucleotide probes targeting the expanded repeat (Figures 2A and S3A). This pathology was not present in non-C9ORF72 control cells or C9ORF72 cells labeled with scrambled FISH probes (Figures 2B and S3A). Specifically, we observed that, across five C9ORF72 fibroblast and four C9ORF72 iPSN lines, approximately 25% and 35% of cells contained intranuclear GGGGCC RNA foci, respectively (Figures 2B, 2C, and S3A). When compared to fibroblasts, the higher percentage of RNA foci containing cells and the higher number of foci/cell in the iPSN might be explained by the higher C9ORF72 expression levels (Figure 1D and 2C). It is important to note that the high G:C content of the C9ORF72 expanded RNA poses technical challenges for RNA FISH probe targeting; as such, it is possible that we are only visualizing a fraction of the actual RNA foci present in the cultures. Nevertheless, this same approach has successfully identified nuclear GC repeats in fragile X diseased tissue (Sellier et al., 2013). Specificity of the FISH probes was confirmed by treatment of cells with RNase A and DNase I, which reduced and maintained the RNA foci respectively, strongly suggesting that these intranuclear inclusions are comprised of ‘‘GGGGCC’’ RNA and not DNA (Figures 2B and 2C).
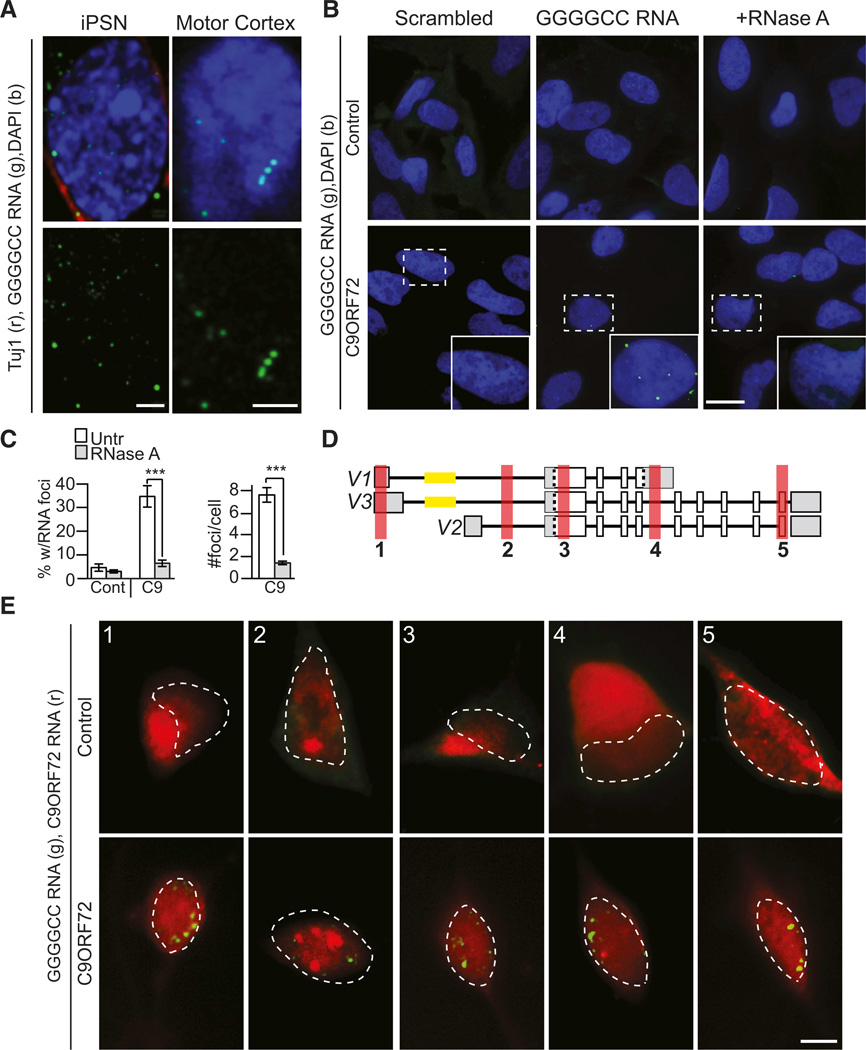
Figure 2. C9ORF72 ALS iPSN Exhibit Pathological Nuclear GGGGCCexp RNA Foci.
(A) Single optical plane (∼0.3 µm) of C9ORF72 iPS-differentiated neurons processed for GGGGCC RNA FISH exhibit clear intranuclear foci (left) similar to those found in C9ORF72 ALS motor cortex (right).
(B and C) GGGGCC probe is specific to C9ORF72 RNA foci and foci are RNase A sensitive (n = 30 fields of view).
(D) Schematic of RNA FISH probe location relative to C9ORF72 RNA variants.
(E) Probes to exonic and intronic regions upstream and downstream of the repeat (red) do not form nuclear foci, do not consistently overlap with GGGGCC RNA foci (green), and exhibit similar localization in control and C9ORF72 ALS iPSN lines. Dotted line outlines cell nucleus.
Data in (C) indicate mean ± SEM (p < 0.001). Scale bar = 2 mm for (A), 20 mm for (B), and 5 mm for (E). See also Figure S3A.
The C9ORF72 mutation resembles the DM2 mutation, which is caused by a long CTTG tract in the first intron of the ZNF9 gene that generates a toxic CCUGexp RNA (Lee and Cooper, 2009; Udd and Krahe, 2012). In DM2, the intranuclear RNA foci are comprised of the expanded RNA repeat and not the surrounding intronic or exonic regions (Margolis et al., 2006). Therefore, utilizing dual RNA FISH methodologies, we investigated the composition of the C9ORF72 GGGGCC RNA foci in the iPSNs (Margolis et al., 2006). To achieve this, we probed control and C9ORF72 iPSNs with a 5’digoxigenin-labeled LNA probe to the GGGGCC repeat and a 5’FAM-labeled LNA probe to sequences upstream (probe 1) or downstream (probes 2–5) of the expanded repeat targeting either C9ORF72 exons or introns (Figure 2D). This approach allowed us to determine whether the full C9ORF72 transcript is sequestered into nuclear GGGGCC RNA foci. We did not observe nuclear foci when visualizing FISH probes that target these sequences. Moreover, the staining pattern of 5’FAM-labeled LNA probes in control non-C9ORF72 iPSNs was similar to C9ORF72 iPSN staining (Figure 2E, upper panel) and differed from the GGGGCC targeting probes in the C9ORF72 iPSN (Figure 2E, lower panel), suggesting that C9ORF72 RNA exonic or intronic sequences upstream or downstream of the repeat are not primary components of the nuclear RNA foci (Figure 2E).
C9ORF72 ALS iPSCs Exhibit RAN Translation Pathology
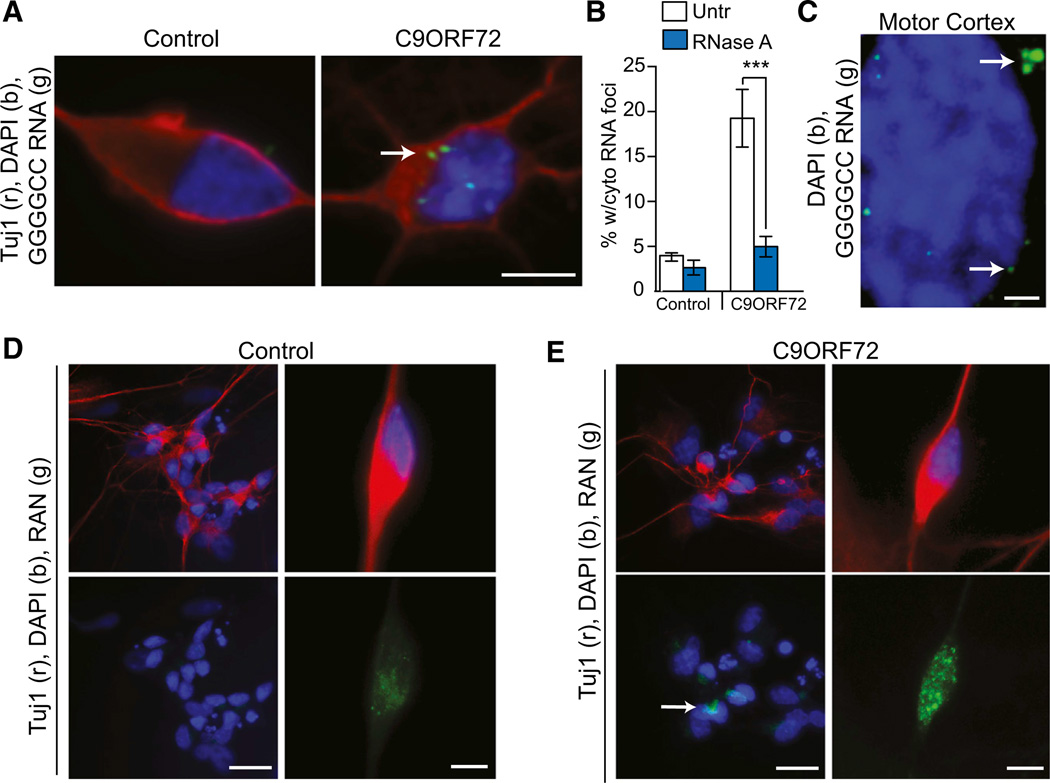
The percentage of cells with cytoplasmic foci in C9ORF72 cells was significantly higher than in control fibroblasts and iPSNs, as was the number of cytoplasmic GGGGCC foci per cell (Figures 3A, 3B, and S3B). Moreover, similar cytoplasmic foci could be found in C9ORF72 ALS postmortem motor cortex (Figure 3C). The presence of these cytoplasmic RNA foci suggested that the expanded GGGGCC RNA may undergo non-ATG-initiated translation (RAN) (Ash et al., 2013; Mori et al., 2013b) resulting in the accumulation of high-molecular-weight cytoplasmic dipeptide protein products, namely, poly-(Gly-Ala), poly-(Gly-Pro), and poly-(Gly-Arg) (Ash et al., 2013), a process similar to microsatellite RNA products in DM1 and spinocerebellar ataxia type 8 (SCA8) (Zu et al., 2011). Utilizing an antibody that was raised against the poly-(Gly-Ala), poly-(Gly-Pro), and poly-(Gly-Arg) RAN protein products but preferentially detects the poly-(Gly-Pro) RAN product in C9ORF72 human tissue (Ash et al., 2013), we tested for the presence of cytoplasmic RAN protein in C9ORF72 iPSNs. Immunofluorescent staining of the C9ORF72 iPSNs revealed cytoplasmic poly-(Gly-Pro) RAN protein in C9ORF72 ALS iPSNs with only light background staining in some non-ALS control iPSNs (Figures 3D and 3E) (Ash et al., 2013), thus matching the pathology of C9ORF72 postmortem patient CNS tissue.
Figure 3. C9RF72 ALS Cells Show Cytoplasmic RAN Translation Peptides.
(A) C9ORF72 iPSN cells contain cytoplasmic GGGGCC RNA foci, while control iPSNs do not.
(B) Quantification of cytoplasmic RNA foci reveals that cytoplasmic foci are RNase A sensitive.
(C) Single-optical plane (∼0.3 µm) image of C9ORF72 ALS motor cortex also shows cytoplasmic GGGGCC RNA foci similar to iPSNs.
(D) RAN protein shown by IF using C9RANT antibody that preferentially detects the poly-(Gly-Pro) RAN product in C9ORF72 iPSNs but not control iPSNs.
***p < 0.001. Scale bar = 5µm for (A), (D), and (E), right panels; 1 mm for (C), and 20 µm for (D) and (E), left panels. See also Figure S3B.
RNA Binding Protein ADARB2 Interacts with GGGGCC RNA
In noncoding repeat expansion disorders, pathogenesis may be due to the accumulation of expanded repeat-containing RNA transcripts that sequester RNA binding proteins (RBPs) (Eche-verria and Cooper, 2012). The presence of intranuclear RNA foci in C9ORF72 ALS cells suggests that the expanded GGGGCC RNA might also sequester RBPs. The identification of such GGGGCCexp RBPs may prove critical for understanding the mechanisms of C9ORF72-mediated neurodegeneration and could further be important for the identification of candidate therapies.
Previously, proteome arrays have been successfully utilized to identify protein-binding partners for long noncoding RNAs (Rapicavoli et al., 2011). We utilized this unbiased, in vitro high-throughput methodology to investigate potential GGGGCC RNA interactors. A 5’Cy5-labeled GGGGCC × 6.5 RNA was synthesized and hybridized to a proteome array containing nearly two-thirds of the annotated human proteome as yeast-expressed, full-length ORFs with N-terminal GST-His × 6 fusion proteins (a total of 16,368 full-length human proteins repeated 2–3 times per chip) (Jeong et al., 2012). A 5’Cy5-labeled scrambled RNA of the same G:C content as the 5’Cy5-labeled GGGGCC × 6.5 RNA was used as a negative control. For each RNA sequence, three proteome arrays were hybridized in parallel as technical replicates. Using this method, we identified 19 ORFs that consistently exhibited high affinity for the GGGGCC × 6.5 RNA as compared to the scrambled RNA determined via the ΔZ-score (GGGGCC × 6.5 RNA Z score – G:C scrambled RNA Z score) (Table S4; for a complete list of all binding proteins and their respective Z score for each RNA see Table S5). Notably, the GGGGCC RNA has been shown to form a G-quadruplex structure (Fratta et al., 2012; Reddy et al., 2013) and our G:C scrambled negative probe is predicted to form the same structure. Therefore, we very conservatively screened for binding partners to the repeat sequence, excluding any hits that would nonspecifically bind the G-quadruplex structure. In addition, the proteome array will identify protein interactors independent of their respective cellular abundance unlike standard RNA affinity assays from cell lysates and protein identification via mass spectrometry. From the 19 GGGGCC × 6.5 interactor candidates, we chose ADARB2, a known RBP, to study its role in C9ORF72 pathology in greater detail.
Simultaneous RNA FISH and RBP immunofluorescence (RNA FISH-IF) studies in C9ORF72 iPSNs revealed that ADARB2 protein colocalizes with the nuclear GGGGCC RNA foci (Figure 4A), with unchanged levels of mRNA (Figure S4A). In addition, RBP:RNA coimmunoprecipitation (RNA co-IP) studies allowed us to isolate C9ORF72 RNA from the RNA co-IP using primers to exon 1a and the intronic region 5’ of the GGGGCC expanded repeat (Figures 4B and S4B), indicating that ADARB2 interacts with endogenous C9ORF72 RNA in living cells. Finally, we performed an electrophoretic gel shift assay (EMSA) with recombinant ADARB2 purified from E. coli (Figures S4C and S4D). Titrating ADARB2 clearly shows depletion of free RNA and shift to slower mobility or a well shift, the latter of which is presumably due to multimerization of the protein:RNA complexes (Figures S4D and S4E). Taken together, these data indicate that both biochemically and in living cells, ADARB2 protein interacts with C9ORF72 RNA and has a high binding affinity for the GGGGCC repeat RNA sequence, which could be useful as a readout to monitor C9ORF72-specific drug efficacy.
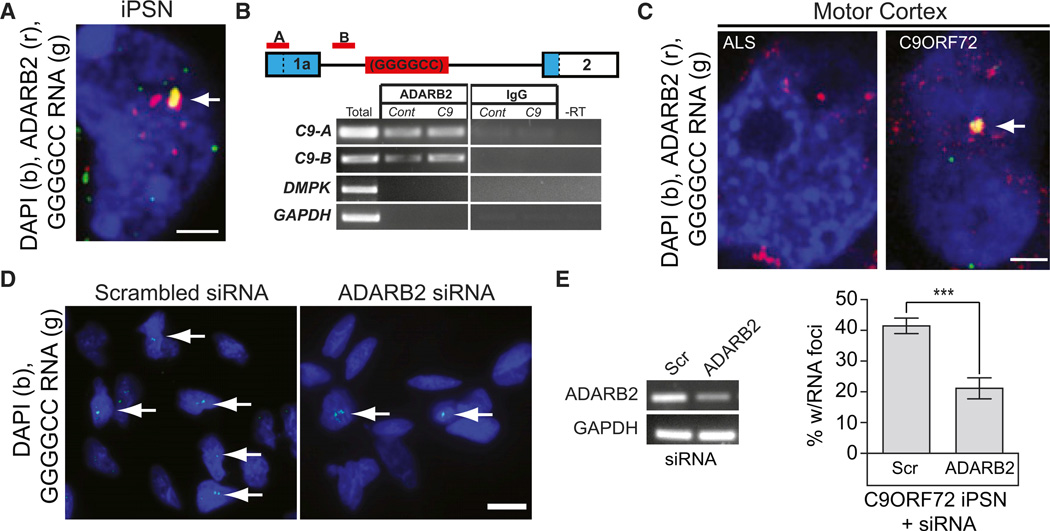
Figure 4. ADARB2 Protein Binds to the GGGGCCexp of C9ORF72 RNA.
(A) Single optical plane (0.34 µm) colocalization of GGGGCC RNA foci with ADARB2 protein signal in C9ORF72 iPSN.
(B) ADARB2 co-IP with bound RNA was performed; RNA was isolated from ADARB2 protein co-IP in control and C9ORF72 iPSNs. RT-PCR of the co-IP RNA using two primer sets (red) upstream of the C9ORF72 GGGGCC repeat (schematic, blue = UTR) indicates that ADARB2 binds to the C9ORF72 RNA in both control and C9ORF72 iPSNs. Amplification of DMPK and GAPDH were used to test for immunoprecipitation specificity (lower panel).
(C) Single optical plane (∼0.3µm) image demonstrating colocalization of GGGGCC RNA foci and ADARB2 in C9ORF72 patient postmortem motor cortex tissue, layers 3–5.
(D and E) siRNA knockdown of ADARB2 results in a significant reduction in the percent of iPSNs with nuclear RNA foci (arrows). Data in (E) indicate mean ±SEM (***p < 0.001).
Scale = 2 mm for (A) and (C), and 15 mm for (D). See also Figure S4 and S5; Tables S4 and S5.
To determine whether these in vitro observations are recapitulated in vivo, we examined the colocalization of ADARB2 protein to GGGGCC RNA foci in human postmortem C9ORF72 patient tissue. RNA FISH-IF confirmed that ADARB2 colocalizes with GGGGCC RNA foci in motor cortex of C9ORF72 ALS patients, while there is no nuclear accumulation or colocalization in non-C9 ALS tissue (Figure 4C).
Since ADARB2 appears to interact with endogenous C9ORF72 RNA through the GGGGCC repeat sequence, we examined whether ADARB2 is required for RNA foci formation, similar to the MBNL1 requirement for foci formation in DM1 and DM2 (Lee and Cooper, 2009; Udd and Krahe, 2012). To test this, we treated iPSNs with siRNA against ADARB2 and performed RNA FISH for the nuclear GGGGCC RNA foci. siRNA-mediated knockdown of ADARB2 resulted in a statistically significant 48.99% reduction in the number of iPSNs with RNA foci (Figures 4D and 4E). These studies suggest that an interaction between ADARB2 and the C9ORF72 RNA expansion plays a role in the formation or maintenance of the RNA foci in vitro, supporting the hypothesis that interactions of RBPs with the GGGGCC repeat may play a role in GGGGCCexp RNA toxicity. Moreover, we observed that ADARB2 appeared to statistically accumulate in the nucleus of C9ORF72 iPSN by immunostaining (Figures S5A and S5B) and this was recapitulated in C9ORF72 ALS postmortem tissue (Figures S5C and S5D).
Recent studies performing in vitro pull down assays have implicated other RBPs as potential GGGGCCexp RNA binding partners, including hnRNPA3 (Mori et al., 2013a) and Pur α (Xu et al., 2013). While these RBPs were not included on the prote-ome arrays used in this study, we performed immunohistochemical analyses of multiple RBPS, including P62, hnRNPA1, hnRNPA1B2, FUS, P62, and Pur a (Figure S5E). We also included TDP43, a well-characterized RNA binding protein in ALS pathology and pathophysiology. Macroscopic immunostaining suggests that unlike for ADARB2, none of the other RBPs formed distinct nuclear aggregates in the C9ORF72 iPSC neuron cultures when compared to control iPSNs.
Transcriptome of C9ORF72 Cells and Tissue
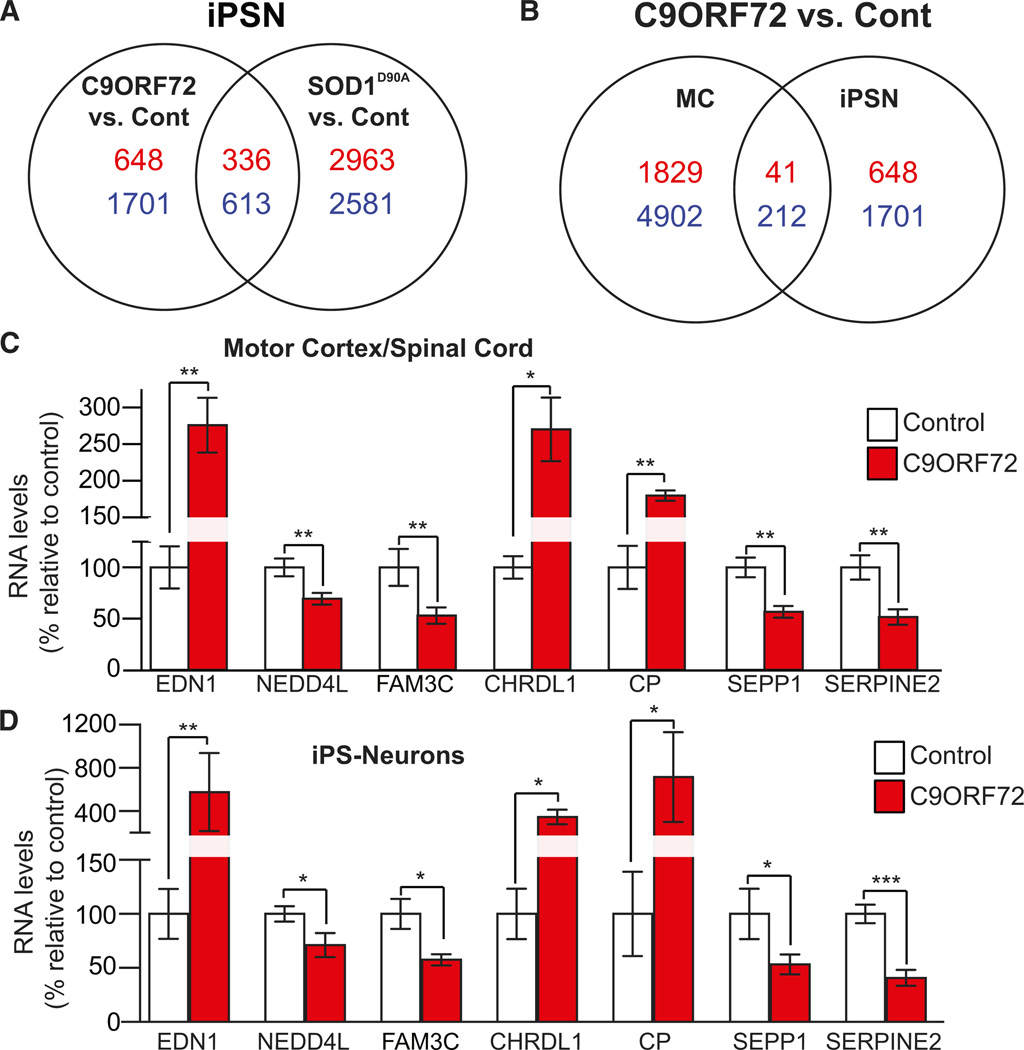
Sequestration of RBPs and the presence of nuclear foci suggest that the expansion mutation may alter the cellular transcriptome, which could provide yet another readout for therapeutic intervention. Using five C9ORF72 ALS fibroblast lines, we identified unique gene expressions changes (p < 0.05) when compared to healthy controls, and accounted for significantly altered genes from SOD1mut fibroblasts (Figures S6A and S6B; Tables S6 and S7). Similarly, we found that, using four iPSN lines a unique population of genes were dysregulated as compared to control, again subtracting the aberrantly expressing genes from SOD D90A iPSN lines (Figures 5A and S6C; Table S8). iPSNs that carry a SOD1D90A mutation exhibited a large number of dys-regulated genes when compared to control cells, although a subset of expression abnormalities were common between C9ORF72 and SOD1D90A iPSNs (Figures 5A and S6C; Tables S9 and S10). Taken together, these data indicate that the C9ORF72 transcriptome is different from the SOD1mut transcriptome in both fibroblasts and iPSNs. This can be visualized when comparing the expression levels of statistically significant genes in C9ORF72 iPSNs to that of SOD1D90A iPSNs (Figure S6D).
Figure 5. C9ORF72 ALS-Specific Gene Expression.
(A) Venn diagrams comparing the differentially expressed, statistically significant genes in C9ORF72 iPSNs versus control iPSNs and SOD1D90A iPSN versus control iPSNs indicate vast differences in global gene expression. Red indicates upregulated genes; blue indicates down-regulated genes.
(B) Gene expression comparison of common, statistically significant gene expression in C9ORF72 iPSNs versus control (negating any genes significant in SOD1D90A versus control iPSNs) and C9ORF72 human motor cortex tissue (versus healthy control motor cortex). p < 0.05.
(C and D) Targeted gene expression analysis of aberrantly expressed secreted genes, chosen by microarray results, in human motor cortex and spinal cord revealed dysregulation of seven genes that were also found to be similarly dysregulated in C9ORF72 iPSNs (versus control). Data in (C) and (D) indicate mean ± SEM.
*p < 0.05; **p < 0.01; ***p < 0.001. See also Figure S6 and S7; Tables S6–S16.
To evaluate whether cultured iPSNs recapitulate the C9ORF72 ALS human brain transcriptome and might there fore be used ultimately to evaluate future therapeutics, we next examined any commonalities between C9ORF72 iPS-derived neurons and postmortem motor cortex(n=3) (Figure 5B). We identified a large number of aberrantly expressed genes(p<0.05)in C9ORF72 ALS motor cortex (compared to control) ofwhich a subset overlapped with genes aberrantly expressed in C9ORF72 iPSNs, including those expressed concordantly (Figures 5B and S6E–S6F and Tables S11 and S12). When comparing C9ORF72 fibroblasts to C9ORF72 iPSN and motor cortex, fewer genes were found to be common suggesting that these cell types are not very similar (Figures S6E and S6F and Tables S13 and S14). Only a population of altered genes is shared between the postmortem C9ORF72 human motor cortex and the C9ORF72 iPSNs, most likely due to the cellular heterogeneity of the human motor cortex as compared to a neuron-enriched iPSN culture system. Interestingly, all C9ORF72 cell and tissue gene arrays consistently showed a larger number of downregulated genes than upregulated genes, which was not observed in the SOD1mut samples (Table S15).
With the goal of identifying genes that might be utilized as therapeutic biomarkers, we selected genes that exhibited altered expression in C9ORF72 iPSNs, fibroblasts, or human motor cortex via exon microarray. We specifically selected genes coding for proteins that are expressed in the CNS and predicted to be secreted. In choosing these, we hoped that their levels could be quantified in patient cerebrospinal fluid (CSF) as this would allow for monitoring during antisense treatment, similar to what we recently reported for human SOD1 antisense therapy (Reddy et al., 2013). We selected 26 target genes (Table S16) and tested their expression in C9ORF72 autopsied CNS tissue against non-ALS control tissue using nanostring gene expression methodologies. Sixteen of the target genes tested were also aberrantly expressed in C9ORF72 ALS patient tissue (Figures 5C, S7A, and S7B), of which seven showed the same direction of dysregulation (up or down) when compared to iPSNs (Figure 5D). These genes could be potential candidates for the future development of a pharmacodynamic biomarker to monitor C9ORF72 therapy in human CSF and/or blood.
C9ORF72 ALS iPSC Neurons Are Highly Susceptible to Glutamate-Mediated Excitotoxicity
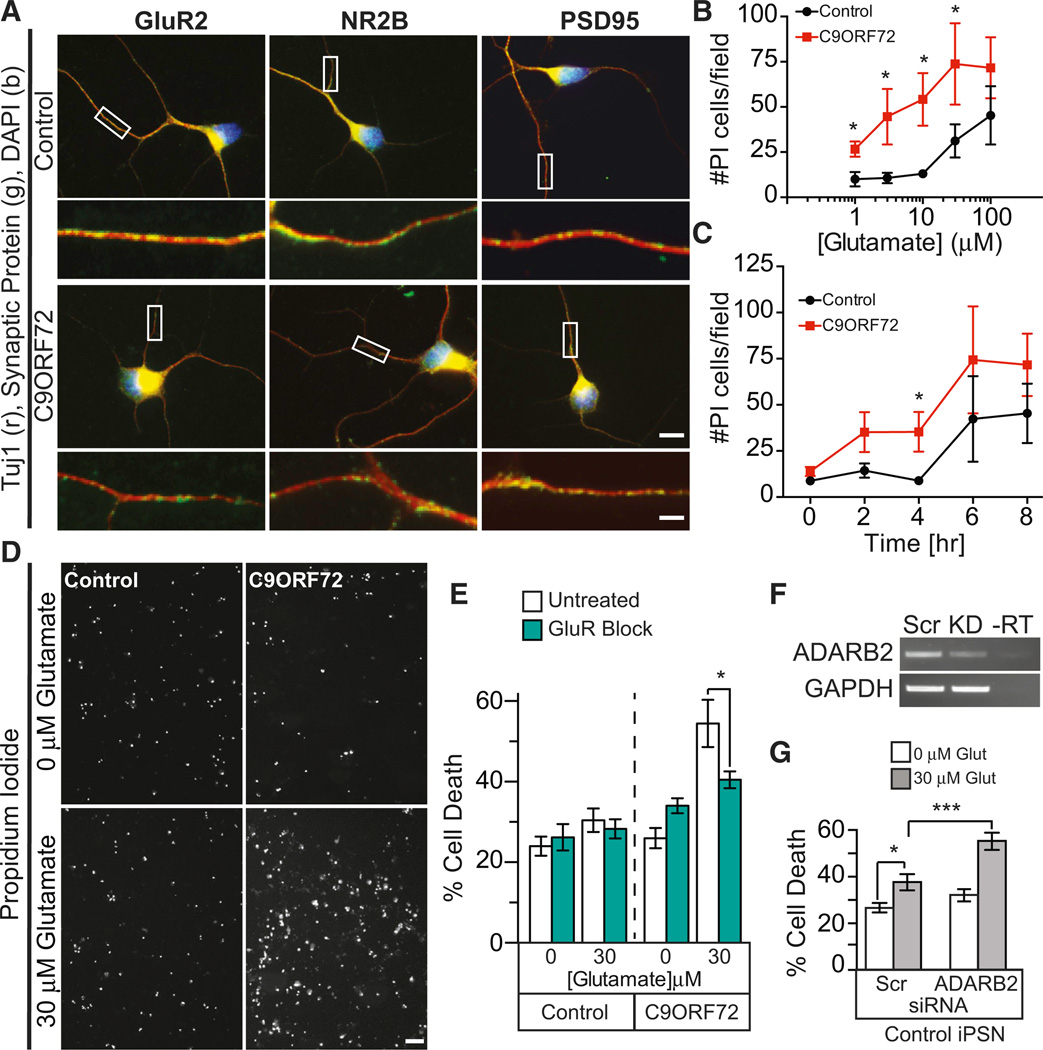
Glutamate toxicity has been shown to play a major role in ALS as sporadic ALS and C9ORF72 patients exhibit a loss of astroglial glutamate transporter 1 (GLT-1/EAAT2), which buffers synaptic glutamate thus preventing excitotoxicity (Lin et al., 1998; Roth-stein et al., 1995; Renton et al., 2011). Notably, C9ORF72 iPSNs express glutamate receptors (GluR2), NMDA receptors (NR2B), and postsynaptic marker protein postsynaptic density protein-95 (PSD95), comparable to control iPSNs, thus suggesting that these cells form functional synapses and are capable of responding to glutamate-induced excitotoxicity (Figure 6A).
Figure 6. C9ORF72 ALS iPSC Neurons Are Highly Susceptible to Glutamate Toxicity.
(A) Immunofluorescent staining of control and C9ORF72 iPSN cultures show expression of glutamate receptors GluR2, NR2B, and post-synaptic density protein PSD95 at comparable levels as determined by qualitative analysis. Box indicates region of high magnification seen below each image (scale = 10 mm [top] and 2.5 mm [bottom]).
(B) Dose response curve of control and C9ORF72 iPSC neurons revealed that C9ORF72 iPSNs are highly susceptible to glutamate excitotoxicity at 1, 3, 10, and 30 µM concentrations after 8 hr of treatment by popidium iodide staining.
(C) Glutamate-induced excitotoxicity of C9ORF72 iPSNs shows statistically significant cell death after 4 hr of 30 µM glutamate treatment when compared to control iPSNs.
(D) Representative image of propidium iodide staining of control and C9ORF72 ALS iPSN after 4 hr of 0 and 30 µM glutamate treatment; note the increased prodium iodide signal in the C9ORF72 iPSNs as compared to the control iPSNs.
(E) Blocking glutamate receptors prevents glutamate-induced C9ORF72 iPSN cell death (4 hr, 30 µM glutamate).
(F and G) Knockdown of ADARB2 via siRNA treatment resulted in a statistically significant increased susceptibility to glutamate-induced excitotoxicity in control non-C9ORF72 iPSNs at 4 hr, 30 µM glutamate treatment.
Data in (B), (C), (E), and (G) indicate mean ± SEM (*p < 0.05; ***p < 0.001). See also Figure S8.
To determine whether the C9ORF72 mutation leads to an altered physiology in iPSC neurons, we explored sensitivity to glutamate excitotoxicity. Glutamate treatment resulted in a dose- and time-dependent increase of cell death in control and C9ORF72 iPSN cultures as determined by cellular propidium iodide (PI) uptake (Sattler et al., 1997; Sattler et al., 1999) (Figures 6B–6D and S8). Notably, C9ORF72 ALS iPSNs are almost 100-fold more sensitive to glutamate treatment, as toxicity at 3 µM glutamate in healthy control iPSNs was comparable to death at 100 µM glutamate in C9ORF72 iPSNs (Figures 6B and S8). Glutamate-induced cell death was blocked by inhibitors of glutamate receptors and calcium channels (MK-801, 10 µM; CNQX, 10 µM; nimodipine, 2 µM; Sattler et al., 1999), confirming that C9ORF72 iPSN cell death was gluta-mate-receptor dependent (Figure 6E).
To test whether the sequestration of ADARB2 plays a role in the observed glutamate susceptibility, we knocked down ADARB2 levels in healthy control iPSNs via siRNA treatment (Figure 6F) and investigated their susceptibility to glutamate-mediated cell death. Interestingly, a loss of about 50% of ADARB2 RNA significantly enhances control iPSC neurons’ susceptibility to glutamate (30 µM at 4 hr) to levels similar to those observed in C9ORF72 iPSNs (Figures 6F and 6G). This suggests that a partial loss of ADARB2 via sequestration to GGGGCC RNA might play a role in C9ORF72-mediated RNA toxicity.
Antisense Oligonucleotides Rescue C9ORF72 Toxic Phenotype
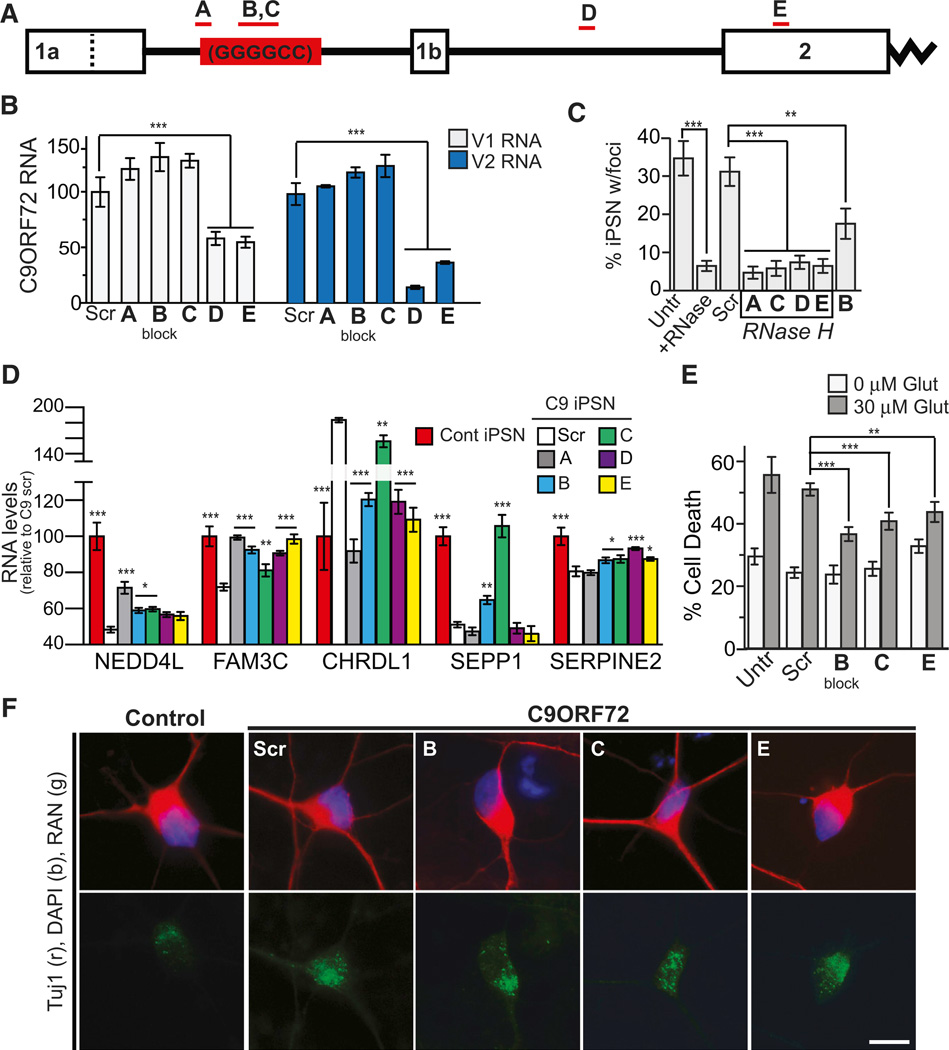
If an RNA-dominant pathogenic mechanism exists for C9ORF72 ALS, it could be a candidate for antisense oligonucleotide therapeutics. For example, in DM1, (CAG)7 ASOs have successfully been utilized to reverse expanded RNA toxicity in vitro as well as to normalize in vivo aberrant RNA splicing in transgenic models by reducing toxic (CUG)n RNA (Mulders et al., 2009). Similarly, (CAG)25 ASOs (PMOs), which bind, but not degrade, the (CUG)n RNA, block MBNL1 interaction with the toxic RNA and rescue the animal model disease phenotype (Wheeler et al., 2009). In the DM1 mouse nuclear-retained RNA species are highly susceptible to ASOs that work through the RNase H mechanism (Osborne et al., 2009; Wheeler et al., 2012), resulting in loss of RNA foci and reversal of myotonia and spliceopathies (Wheeler et al., 2012). Based upon these findings we screened over 250 chimeric MOE/DNA RNase H active ASOs (Gapmers) (Figure S9A) designed to bind to multiple regions of the C9ORF72 transcript as well as to the GGGGCCexp RNA, a selection of which are included in this study (Figures 7A and S9B). In addition, we studied an ASO that binds the GGGGCCexp RNA repeat to block any RBP interaction but does not degrade the transcript (Figure S9B).
Figure 7. Antisense Oligonucleotides Rescue RNA Toxicity Observed in C9ORF72 iPSC Neurons.
(A) Map of the C9ORF72 gene indicating the ASOs utilized in this study. ASO B is endonuclease resistant and designed to block the repeat while the remaining are gapmers, which mediate RNA degradation through an RNase H-based mechanism. Dotted line indicates end of V1 Exon 1A sequence.
(B) ASOs that target the repeat sequence do not statistically alter C9ORF72RNA levels in C9ORF72 iPSNs, while ASOs downstream of the repeat significantly reduce the levels of C9ORF72 variants 1 and 2.
(C) All ASOs significantly reduced the number of nuclear GGGGCC RNA foci observed in C9ORF72 iPSNs to levels comparable to the RNase A-treated cells while treatment with a nontargeting scrambled probe did not affect the percent of cells with nuclear foci.
(D) ASO treatment was able to rescue the aberrant expression of NEDD4IL, FAM3C, CHRDL1, SEPP1, and SERPINE2 in a statistically significant manner.
(E) ASO B, C, and E rescued the increased susceptibility of C9ORF72 iPSNs to glutamate exposure (30 µM glutamate, 5 hr).
(F) ASO treatment for 3 days does not dramatically reduce the levels of cytoplasmic RAN proteins.
Data in (B), (C), and (E) indicate mean ± SEM; data in (D) indicates mean ± SD (*p < 0.05; **p < 0.01; ***p < 0.001). See also Figure S9.
We tested the efficacy of ASOs using C9ORF72 ALS patient fibroblast lines as well as three patient iPSN lines. Our results indicate that ASOs targeting the GGGGCCexp (ASO: A–C) do not reduce C9ORF72 V1 or V2 RNA levels in fibroblasts or iPSNs regardless of their intended effect (block or RNase H activation) (Figures 7A, 7B, and S9C). In contrast, gapmer ASOs targeting the intronic region downstream ofthe repeat or exon 2 effectively reduced C9ORF72 V1 and V2 RNA levels in C9ORF72 patient fibroblasts and iPSNs when compared to cells treated with a non-targeting ASO (Figures 7B and S9C).
RNase H-mediated C9ORF72 knockdown significantly reduced both the percentage of cells that contain GGGGCCexp RNA foci (Figures 7C and S9D) and the number of foci per cell in both fibroblasts and iPSN cultures regardless of its target location (expansion, intronic region, or coding sequence) or effect on C9ORF72 RNA levels (Figures 7C and S9D–S9F). This suggests that ASO protection against RNA toxicity can be obtained without reducing C9ORF72 RNA levels, which could be important for future therapeutic development.
If ADARB2 colocalization with RNA foci (Figures 4A and 4C) is due to protein:GGGGCCexp RNA interaction, then ASOs that reduce RNA foci might also attenuate the observed ADARB2 nuclear accumulation observed in C9ORF72 ALS iPSN and motor cortex. Indeed, all ASOs tested reduced the nuclear ADARB2 protein signals in C9ORF72 iPSNs as determined by immunostaining (Figures S9G and S9H).
ASO treatment also normalized the dysregulated gene expression of our candidate biomarker genes NEDD4L, FAM3C, CHRDL1, SEPP1, and SERPINE2 in C9ORF72 iPSNs (Figure 7D) independent of the ASO target region, hence independent of C9ORF72 RNA knockdown. Thus, these genes could indeed serve as potential biomarkers to monitor ASO therapy efficacy. Importantly, genes that were not altered between control and C9ORF72 iPSNs did not change when treated with ASOs, suggesting that ASO treatment does not have untoward effects on general gene transcription (Figure S9I).
Finally, we tested whether ASO treatment might rescue the susceptibility to the glutamate excitotoxicity observed in C9ORF72 iPSNs. iPSNs treated with ASOs targeting the repeat sequence or targeting C9ORF72 exon 2 were exposed to 30 µM glutamate for 4 hr and monitored for cell death. ASOs targeting the repeat, without altering C9ORF72 RNA levels, significantly rescued the glutamate toxicity phenotype by up to 30%. ASOs targeting exon 2, which reduced C9ORF72 mRNA levels, still significantly protected the iPSN by 16% (Figures 7E and S9J). This suggests that the loss of C9ORF72 mRNA and subsequent loss of C9ORF72 protein do not play a role in the observed vulnerability to glutamate, but instead implies that RNA toxicity causes C9ORF72 cells to be highly sensitive to excitotoxicity. This is further supported by the fact that RAN products were still detected in the C9ORF72 iPSNs after ASO treatment, through either immunocytochemistry or protein blotting, despitearescue of the described phenotypes, including glutamate toxicity (Figures 7F and S9K). Notably, whether the ASO altered a population of newly synthesized RAN or other RAN products that are not detected with the present antibodies is not known.
DISCUSSION
Taken together, the current studies provide evidence that RNA toxicity plays a key role in C9ORF72 ALS based on the molecular, biochemical, and functional studies described here. Specifically, we have (1) demonstrated that patient fibroblasts and iPSNs contain intranuclear GGGGCC RNA foci similar to those found in vivo (DeJesus-Hernandez et al., 2011), (2) identified numerous proteins that interact with the C9ORF72 GGGGCCexp RNA, (3) confirmed interaction of ADARB2 with the RNA expansion in vitro and in vivo, (4) described atypical gene expression in C9ORF72 ALS tissue and cell lines that match C9ORF72 CNS patient tissue, and (5) determined that C9ORF72 iPSC neurons are highly susceptible to glutamate toxicity. Most importantly, by using these various pathological and physiological readouts in human iPSC neurons, we were able to identify antisense oligonucleotides that can abrogate C9ORF72 RNA expansion-dependent pathology, RNA binding protein aggregation, aberrant gene expression, and neurotoxicity. Furthermore, ASO that selectively blocked the hexanucleotide expansion without lowering C9ORF72 RNA levels could minimize pathology and toxicity (Figure 8). Notably, iPSCs derived from ALS patients appear to accurately recapitulate the pathological and genomic abnormalities found in the C9ORF72 ALS brain. Modeling this expansion mutation in animals can be particularly challenging in part due to the fact that the vast majority of human disease is caused by very large numbers of G:C-rich repeats that prove difficult to artificially express. Therefore, human C9ORF72 iPSCs provide an exceptionally valuable tool to investigate ALS/FTD pathophysiological pathways and therapy development and could also be used to validate future in vivo disease models.
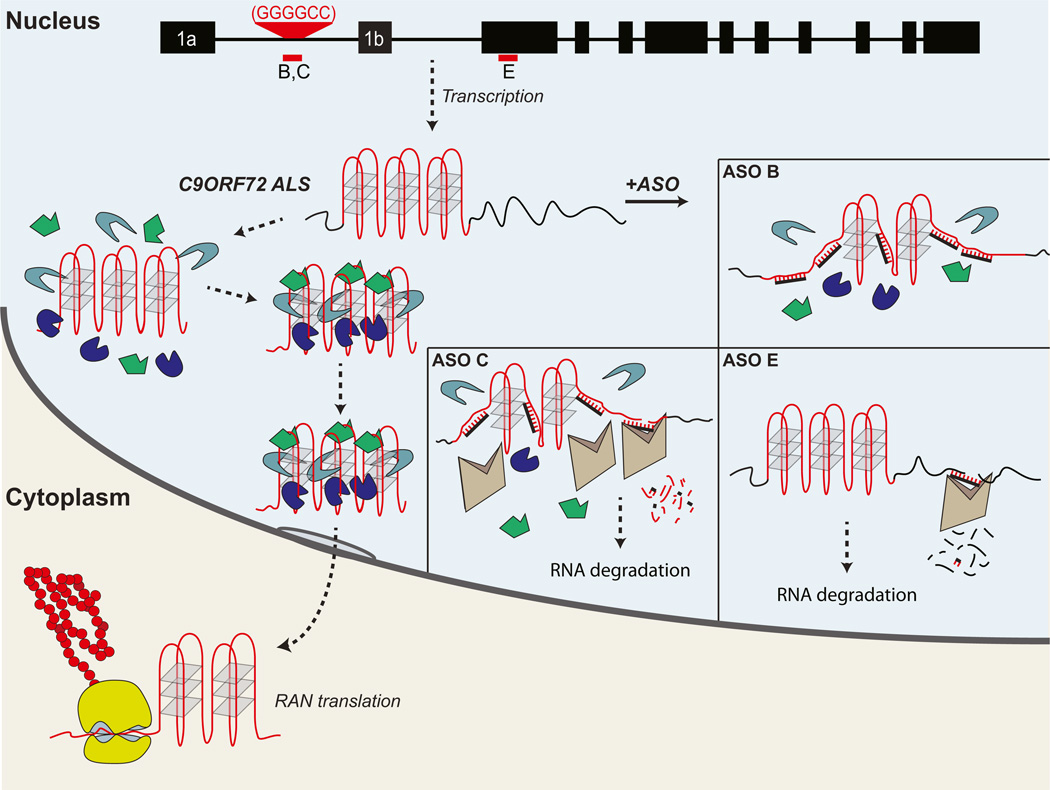
Figure 8. Pathogenesis Resulting from the C9ORF72 Noncoding GGGGCC Expansion in ALS/FTD.
Top. Representation of the human C9ORF72 gene, showing hexanucleotide expansion (red) and location of various antisense oligonucleotide (ASO) probes (B, C, E). Hexanucleotide expansion is located between the noncoding exons 1a and 1b. The expanded RNA is predicted to form a series of G-quartet tertiary structures. At least three different pathophysiological pathways may results from the mutation: (1) RNA toxicity resulting from excessive binding of various different RNA binding proteins (green, blue, gray polygons) leading to sequestration of these RBPs, and loss of their activity resulting in anomalous downstream events; (2) export of the expansion to cytoplasm with repeat-associated translation (RAN) (in the sense or antisense directions) and the formation of excessive high-molecular-weight cytoplasmic peptides (red), which may be toxic or protective; and (3) loss of C9ORF72 mRNA leading to loss of protein function (not shown). ASOs may disrupt C9ORF72 RNA toxicity by several pathways. ASO B: an endonuclease-resistant oligonucleotide that is predicted to disrupt the expansion, interfering with the G-quartet structure and preventing RBP interaction/sequestration. ASO C; a gapmer ASO that binds to the expansion targeting it RNase H-mediated cleavage and subsequent RNA degradation by nuclear endogenous nucleases, since this ASO targets the repeat it will also disrupt the repeat sequence structure prior to cleavage and prevent RBP sequestration. ASO E: a gapmer that targets the C9ORF72 RNA coding sequence marking the RNA for RNase H cleavage, thus resulting in the degradation of wild-type and the expanded C9ORF72 RNA; this will further reduce the already attenuated levels of cellular C9ORF72 RNA. Notably, ASOs B and C do not reduce cellular C9ORF72 RNA.
ASOs designed to activate RNase H-mediated C9ORF72 RNA degradation, or that block the toxic GGGGCCexp RNA, rescued all of the described pathogenic phenotypes. Interestingly, ASOs downstream of the repeat were able to rescue some of the observed toxic phenotypes (e.g., nuclear foci), despite the fact that our data indicate that RNA far downstream of the repeat where ASOs D and E target are not sequestered into C9ORF72 intranuclear GGGGCC RNA foci. It is possible that the ASOs might degrade the RNA very early during transcription and prior to any splicing events and foci formation as previously described (Rigo et al., 2012).
The role of RNA binding proteins in C9ORF72 RNA pathology as described in this study is supported by recent findings of other RBPs binding to the GGGGCCexp repeat. Various in vitro pulldown assays with different sources of cell lysates, such as human kidney cell lysates and mouse CNS lysates, have identified commonly known RBPs such as hnRNP (e.g., hnRNPA3) proteins and Pur a as interactors to the GGGGCCexp repeat (Xu et al., 2013; Mori et al., 2013a; Almeida et al., 2013). We tested some of these interactors in our iPSC culture model but were not able to confirm their nuclear sequestration, similar to what has been shown using C9ORF72 FTD patient-derived iPSCs (Almeida et al., 2013). Except for Pur a, none of the previously described RBPs have been yet shown to have functional implications for C9ORF72 RNA toxicity; hence, their role in GGGGCC repeat disease remains unclear. Importantly, traditional pulldown methods to determine protein:RNA interactors from cell lysates followed by mass spectrometry protein identification are limited in that they are biased toward the most abundant proteins of the cell lysates. Proteome arrays, as performed here, are advantageous for nonbiased interactor screening since protein abundance will not dictate the identification of candidate binding partners.
Overall, the current study strongly implicates aberrant RNA metabolism in the pathogenesis of C9ORF72 disease. More than a decade ago, aberrant astroglial RNA metabolism was first described in ALS (Lin et al., 1998) and some of those patients were subsequently found to carry the C9ORF72 mutation (Renton et al., 2011). In order to better understand the mechanism of toxicity associated with the C9ORF72 mutation, we chose to study in more detail ADARB2 based on its known function as a RNA editing regulator. ADAR proteins are a family of CNS-enriched adenosine deaminases that mediate A-to-I editing of RNAs such as the (Q/R) site of the GluR2 AMPA receptor (Hideyama et al., 2012). Loss of (Q/R) editing results in Ca2+-permeable GluR2 receptors that increase neuronal excitation and cause motor neuron death in a conditional knockout model (Hideyama et al., 2010). Moreover, improper editing via the ADAR family has been implicated as a contributor to sporadic ALS (Hideyama et al., 2012). While the function of ADARB2 in editing is unclear, it is capable of competitively binding RNAs to regulate ADAR family editing in vitro (Chen et al., 2000). The human and iPSC data in the current study suggest that ADARB2 could play a role in human C9ORF72 ALS by interacting with the C9ORF72 RNA foci. One hypothesis is that the interaction of this RBP with the GGGGCCexp RNA might result in its loss of function. Additional studies to investigate the downstream effects of the loss of ADARB2 on biological processes such as RNA editing would be important to further characterize the roles of this protein in CNS cytotoxicity. While we have only thus far investigated one RBP interactor identified in the present screen (Table S4), further studies are required to determine whether any of the other RBPs might interact with the GGGGCCexp RNA in vivo. Moreover, a GC-rich scrambled RNA sequence was used, which probably forms a G-quadruplex similar to the GGGGCC × 6.5 RNA (Fratta et al., 2012; Reddy et al., 2013), thereby enriching for structure and sequence-specific protein interactors. However, the stringency of this analysis might have inadvertently excluded RBPs whose binding to the GGGGCC RNA was not apparent since it also showed an affinity to the GC rich scrambled RNA structure. Finally, it is possible that multiple RBPs bind to the expansion, leading to either additive or synergistic effects, implicating various downstream pathways.
RAN protein has been reported in C9ORF72 tissue and hypothesized tocontribute to disease toxicity. Our studies document that C9ORF72 iPSC neurons have at least one RAN protein and thus recapitulate the pathology seen in human C9ORF72 postmortem brain. However, despite the mitigation of excitotoxicity, large reductions in RNA foci and RBP aggregation and the normalization of various genes by the ASO therapy, RAN peptide can still be detected after ASO treatment. This would suggest that the detected accumulated cytoplasmic peptide was not a major contributor to neurotoxicity in our culture model. Although it is important to note that it is possible that our ASO treatment could rescue the formation of newly synthesized RAN peptides and that longer ASO treatment would eventually show a reduction of RAN products. Furthermore, there could be other RAN peptides such as, e.g., antisense RAN peptides, contributing to the observed phenotypes, but that were not detected with the present antibodies, since our antibody preferentially detects the poly-(Gly-Pro) RAN product.
While C9ORF72 protein exhibits structural similarity to the DENN protein family, the function of the C9ORF72 protein remains unclear (Levine et al., 2013). DENN proteins are GTP-GDP exchange factors (GEFs) for Rab GTPases, thus implicating the C9ORF72 protein in vesicular trafficking (Zhang et al., 2012). C9ORF72 RNA levels are reduced in patients that contain the expanded allele and a recent study has suggested that C9ORF72 haploinsufficiency causes neurodegeneration in a zebrafish model (Ciura et al., 2013). However, a subset of ASOs utilized in the current study very effectively reduce C9ORF72 RNA levels by more than 50% (D, E) in the human iPSNs and rather than being toxic, these ASOs actually abrogate toxicity associated with the endogenous C9ORF72 mutation. This strongly suggests that loss of C9ORF72 is not a major cause of C9ORF72 ALS pathology and toxicity seen in iPSNs. In support of this, a recent pathoclinical study revealed that a homozygous mutation, which are generally far more severe for loss of function disorders, in C9ORF72 mutation individuals had a clinical phenotype similar to heterozygous C9ORF72 mutations, suggesting that C9ORF72 loss of function was not pathogenic (Fratta et al., 2013).
EXPERIMENTAL PROCEDURES
Fibroblast Collection, Reprogramming, and iPS Differentiation to Neurons
Patient fibroblasts were collected at Johns Hopkins Hospital with patient consent (IRB protocol: NA_00021979) or by Dr. Pentti Tienari at the Helsinki University Central Hospital (Table S1).
Collection of Human Autopsied Tissue
Human autopsied tissue, collected with Institutional Review Board and ethics approval, used for these data are described in detail in Table S2.
Antisense Oligonucleotide Treatment
Modified 2’-methoxyethyl (MOE)/DNA ASOs were generated by Isis Pharmaceuticals and the 2’ O-methyl RNA (OME)/DNA ASOs were designed by C.J.D.
RNA FISH, Immunofluorescence, and RAN Detection
RNA fluorescent in situ hybridization (FISH) of fibroblasts and iPSNs was performed as previously described (Donnelly et al., 2011) with modification. Human CNS tissue (see Table S2 for tissue used) was fixed in 4% PFA and cryoprotected in 30% sucrose. Primary antibody was applied in the following dilutions: ADARB2, 1:100 (Sigma HPA031333) and TDP-43, 1:500 (Proteintech, 10782-2-AP), Pur a, 1:50 (Lifespan), P62, 1:100 (Abcam), hnRNPA1, 1:500 (gift from G. Dreyfuss), hnRNPA1B2, 1:500 (Santa Cruz), and FUS, 1:500 (Sigma). RAN protein products were detected in iPSNs, using the C9RANT antibody that preferentially detects the poly-(Gly-Pro) RAN product (Ash et al., 2013). For dot blot analysis of RAN protein products, human tissue was processed as previously described using urea fractionation (Ash et al., 2013).
Proteome Array to Identify GGGGCC RNA Interactors
Proteome array was performed as previously described (Jeong et al., 2012; Rapicavoli et al., 2011) by hybridizing either a 5’-Cy5-GGGGCC6.5 HPLC purified RNA or a 5’-Cy5-GGGCGGGGCGGCGCGGGGGCGGGGCGGCGCGG GGGCGGGG scrambled RNA as a control (IDT DNA).
Nanostring Gene Expression Analysis
RNA was quantified using a probe codeset (Table S3). RNA quantification was performed with 100–200 ng RNA on an nCounter Analysis System per the manufacturer’s protocol. RNA counts were normalized using the nCounter program (Nanostring) and either GAPDH + GUSB for fibroblasts or GAPDH + GUSB + OAZ1 endogenous controls for iPSN and human tissue.
RNA Isolation and Microarray Analysis
RNA was isolated from cell cultures using the RNeasy Kit with on-column DNase treatment (QIAGEN). Total RNA was labeled and hybridized to the human 1.0 ST Exon Array (Affymetrix) at the Johns Hopkins Deep Sequencing and Microarray Core Facility following the manufacturer’s instructions. Microarray raw data were analyzed using the Partek Genomics Suite Software (Partek). See the Accession Numbers section below for microarray data.
Glutamate-Induced Excitotoxicity
For testing C9ORF72 iPSN sensitivity to glutamate-induced excitotoxicity, healthy control and C9ORF72 iPSNs were treated with various concentrations of L-glutamate (1, 3, 10, 30, 100 µM) for 2–8 hr. At the appropriate time point, cells were incubated with 1 µM propidium iodide and 1 µM calcein AM (Invitrogen) for 30 min to visualize dead and live cells, respectively. For ASO rescue experiments, iPSNs were treated with ASOs for 72 hr and then treated with L-glutamate prior to dead/live cell quantification.
Protein Purification and EMSA Binding Analysis
ADARB2 fusion protein was expressed and purified from Rosetta II cells following gateway expression system (Invitrogen) and GSTrap HP column purification. Increasing concentrations of purified protein was incubated with 10 nM Cy5-labeled RNA. The fraction of RNA shifted, due to ADARB2 binding, was densitometrically quantified in ImageJ (NIH).
Microscopy and Image Analysis
Z-stack images were taken on a Zeiss Axioimager with the Apotome tool or a Zeiss LSM510-meta single-point laser scanning confocal microscope matched exposure times or laser settings and normalized within their respective experiment.
Statistical Analysis
Statistical analysis was performed using the Student’s t test or one-way analysis of variance with the Turkey’s or Dunnet’s post-hoc test and the Prism 6 software (GraphPad Software, Inc.).
Additional details are provided in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by grants from NIH (J.D.R.), P2ALS (J.D.R.), Muscular Dystrophy Association (J.D.R.), The Judith and Jean Pape Adams Charitable Foundation (R.S.), ALS Association (R.S.), the Johns Hopkins Brain Science Institute, The Ansari ALS Center for Cell Therapy and Regeneration Research at Johns Hopkins, The Alzheimer Drug Discovery Foundation and the Association for Frontotemporal Degeneration, The Finnish Academy, The Sigrid Juselius Foundation, the Helsinki University Central Hospital, Robert Packard Center for ALS Research, Maryland Stem Cell Research Fund (C.J.D.), Intramural Research Programs of the US National Institutes of Health (NIH) (B.T.), and National Institute on Aging (B.T.). We would like to thank the Johns Hopkins Deep Sequencing and Microarray Core for the valuable insight on high-throughput experimental design and analysis. Dr. Phillip Wong provided data analysis and interpretation. Dr. Lyle Ostrow provided human tissue demographics. Additional technical and reagent support was graciously provided by Meredith Davitt, Uma Balasubramanian, Conover Talbot Jr., Dr. Tania Gendron, and Dr. Jean-Phillipe Richard. J.D.R, R.S., C.J.D., F.R., and C.F.B. have patents pending on antisense therapeutics and associated genetic biomarkers. B.T. has patents pending for the diagnostic and therapeutic uses of the C9ORF72 hexanucleotide repeat expansion. The remaining authors have no competing financial interests.
Footnotes
ACCESSION NUMBERS
Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-1925, E-MTAB-1926, and E-MTAB-1927.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, nine figures, and sixteen tables and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2013.10.015.
AUTHOR CONTRIBUTIONS
C.J.D designed, performed, and analyzed the experiments and the OME ASO; maintained, treated, and characterized the C9ORF72 fibroblasts and iPSNs; and wrote the manuscript. P.Z. optimized and performed Southern blot methodologies and characterized repeat lengths of the cell and tissue used in these data sets. J.T.P. aided in iPSN differentiation, iPSN characterization, and siRNA and RNA-FISH experiments. A.R.H. and J.W. generated and purified ADARB2 protein and performed the EMSA experiments. N.A.M. analyzed and categorized microarray data. S.V. maintained C9ORF72 cell lines, aided in RNA isolation, tested antibody specificity, and optimized and performed the RNA-coIP experiments. E.L.D. differentiated and maintained all iPSCs cells to the neuronal stage and quantified iPSC differentiation efficacy. E.M.P. performed the proteome array assay. B.H. performed imaging analysis for proteome array hits. D.M.F. aided in blinded imaging and gene expression analysis. N.M. and P.T. provided fibroblast or iPSC lines. B.T. screened human tissue for C9ORF72 expanded repeat and provided critical input for experimental design and strategies. F.R. and C.F.B. designed and provided MOE ASOs and performed initial ASO screening as well as providing input on experimental ASO strategies. S.B. provided the proteome array and aided in analysis and interpretation. R.S. and J.D.R. oversaw project development, experimental design, data interpretation, and manuscript writing.
REFERENCES
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer JR, Willemsen R, Oostra BA. Microsatellite repeat instability and neurological disease. Bioessays. 2009;31:71–83. doi: 10.1002/bies.080122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciura S, Lattante S, Le Ber I, Latouche M, Tostivint H, Brice A, Kabashi E. Loss of function of C9orf72 causes motor deficits in a zebrafish model of Amyotrophic Lateral Sclerosis. Ann. Neurol. 2013 May 30; doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p–linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, et al. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30:4665–4677. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria GV, Cooper TA. RNA-binding proteins inmicrosatel-lite expansion disorders: mediators of RNA toxicity. Brain Res. 2012;1462:100–111. doi: 10.1016/j.brainres.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, Isaacs AM. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Poulter M, Lashley T, Rohrer JD, Polke JM, Beck J, Ryan N, Hensman D, Mizielinska S, Waite AJ, et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 2013;126:401–409. doi: 10.1007/s00401-013-1147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchel JR, and Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- Grammatikakis I, Goo YH, Echeverria GV, Cooper TA. Identification of MBNL1 and MBNL3 domains required for splicing activation and repression. Nucleic Acids Res. 2011;39:2769–2780. doi: 10.1093/nar/gkq1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideyama T, Yamashita T, Suzuki T, Tsuji S, Higuchi M, Seeburg PH, Takahashi R, Misawa H, Kwak S. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J. Neurosci. 2010;30:11917–11925. doi: 10.1523/JNEUROSCI.2021-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideyama T, Yamashita T, Aizawa H, Tsuji S, Kakita A, Takahashi H, Kwak S. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol. Dis. 2012;45:1121–1128. doi: 10.1016/j.nbd.2011.12.033. [DOI] [PubMed] [Google Scholar]
- Jeong JS, Jiang L, Albino E, Marrero J, Rho HS, Hu J, Hu S, Vera C, Bayron-Poueymiroy D, Rivera-Pacheco ZA, et al. Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol. Cell. Proteomics. 2012;11:O111–016253. doi: 10.1074/mcp.O111.016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68:668–681. doi: 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem. Soc. Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29:499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CL, Bristol LA, Jin L, Dykes-Hoberg M, Crawford T, Clawson L, Rothstein JD. Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron. 1998;20:589–602. doi: 10.1016/s0896-6273(00)80997-6. [DOI] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chiò A, Restagno G, Nicolaou N, Simon-Sanchez J, et al. Chromosome 9-ALS/FTD Consortium; French research network on FTLD/FTLD/ALS; ITALSGEN Consortium. Lancet Neurol. 2012;11:323–330. Frequency of the C9orf72 hexanu-cleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. [Google Scholar]
- Margolis JM, Schoser BG, Moseley ML, Day JW, Ranum LP. DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum. Mol. Genet. 2006;15:1808–1815. doi: 10.1093/hmg/ddl103. [DOI] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Lammich S, Mackenzie IR, Forné I, Zilow S, Kretzschmar H, Edbauer D, Janssens J, Kleinberger G, Cruts M, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013a;125:413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013b;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Morrison BM, Janssen WG, Gordon JW, Morrison JH. Time course of neuropathology in the spinal cord of G86R superoxide dismutase transgenic mice. J. Comp. Neurol. 1998;391:64–77. doi: 10.1002/(sici)1096-9861(19980202)391:1<64::aid-cne6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Mulders SA, van den Broek WJ, Wheeler TM, Croes HJ, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, Thornton CA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne RJ, Lin X, Welle S, Sobczak K, O’Rourke JR, Swanson MS, Thornton CA. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum. Mol. Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long non-coding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K, Zamiri B, Stanley SY, Macgregor RB, Jr, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 2013;288:9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B, Gullane P, Irish J, Jurisica I, Kamel-Reid S. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 2011;11:46. doi: 10.1186/1472-6750-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. ITALSGEN Consortium. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo F, Hua Y, Chun SJ, Prakash TP, Krainer AR, Bennett CF. Synthetic oligonucleotides recruit ILF2/3 to RNA transcripts to modulate splicing. Nat. Chem. Biol. 2012;8:555–561. doi: 10.1038/nchembio.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein JD, Van Kammen M, Levey AI, Martin LJ, Kuncl RW. Selective loss of glial glutamate transporter GLT-1 in amyotrophic lateral sclerosis. Ann. Neurol. 1995;38:73–84. doi: 10.1002/ana.410380114. [DOI] [PubMed] [Google Scholar]
- Sattler R, Charlton MP, Hafner M, Tymianski M. Determination of the time course and extent of neurotoxicity at defined temperatures in cultured neurons using a modified multiwell plate fluorescence scanner. J. Cereb. Blood Flow Metab. 1997;17:455–463. doi: 10.1097/00004647-199704000-00011. [DOI] [PubMed] [Google Scholar]
- Sattler R, Xiong Z, Lu WY, Hafner M, MacDonald JF, Tymianski M. Specific coupling of NMDA receptor activation to nitric oxide neurotoxicity by PSD-95 protein. Science. 1999;284:1845–1848. doi: 10.1126/science.284.5421.1845. [DOI] [PubMed] [Google Scholar]
- Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, Alunni V, Moine H, Thibault C, Page A, et al. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–880. doi: 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udd B, Krahe R. The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol. 2012;11:891–905. doi: 10.1016/S1474-4422(12)70204-1. [DOI] [PubMed] [Google Scholar]
- Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, Jin P. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc. Natl. Acad. Sci. USA. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, He F, Aravind L. Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front Genet. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.