Abstract
Objective
To develop RNA splicing biomarkers of disease severity and therapeutic response in myotonic dystrophy type 1 (DM1) and type 2 (DM2).
Methods
In a discovery cohort we used microarrays to perform global analysis of alternative splicing in DM1 and DM2. The newly identified splicing changes were combined with previous data to create a panel of 50 putative splicing defects. In a validation cohort of 50 DM1 subjects we measured the strength of ankle dorsiflexion (ADF) and then obtained a needle biopsy of tibialis anterior (TA) to analyze splice events in muscle RNA. The specificity of DM-associated splicing defects was assessed in disease controls. The CTG expansion size in muscle tissue was determined by Southern blot. The reversibility of splicing defects was assessed in transgenic mice by using antisense oligonucleotides (ASOs) to reduce levels of toxic RNA.
Results
Forty-two splicing defects were confirmed in TA muscle in the validation cohort. Among these, 20 events showed graded changes that correlated with ADF weakness. Five other splice events were strongly affected in DM1 subjects with normal ADF strength. Comparison to disease controls and mouse models indicated that splicing changes were DM-specific, mainly attributable to MBNL1 sequestration, and reversible in mice by targeted knockdown of toxic RNA. Splicing defects and weakness were not correlated with CTG expansion size in muscle tissue.
Interpretation
Alternative splicing changes in skeletal muscle may serve as biomarkers of disease severity and therapeutic response in myotonic dystrophy.
Introduction
DM1 is a dominantly inherited neuromuscular disorder resulting from expansion of a CTG repeat in the 3′ untranslated region of DM protein kinase (DMPK).1 DM2 results from a similar expansion of CCTG repeats in the first intron of ZNF9/CNBP.2 Both mutations give rise to toxic RNAs that contain repetitive segments – an expanded CUG repeat in DM1 and a CCUG repeat in DM2.3
Abnormal regulation of alternative splicing is a molecular hallmark of DM.4 The mutant RNA with expanded repeats is retained in nuclear foci, causing changes of alternative splicing for a specific group of transcripts (reviewed by Orengo and Cooper5). One mechanism for splicing misregulation is that proteins in the Muscleblind-like (MBNL) family are sequestered in nuclear foci of CUG- or CCUG-repeats.6 Expression of expanded CUG repeats (CUGexp) also triggers upregulation of splicing factor CELF1, which further compounds the problem with splicing regulation.7
Agents that promote CUGexp degradation or inhibit CUGexp-MBNL interaction have shown beneficial effects in animal models of DM1.8–13 As these treatments advance to clinical trials, there is a pressing need for biomarkers to assess disease progression and therapeutic response. Splicing defects served this purpose during the preclinical phases of drug development. However, splicing changes have not been comprehensively studied in DM1 and DM2, and connections between alternative splicing and functional impairment have not been established. We therefore used microarrays to perform global analysis of alternative splicing in DM1 and DM2. We then studied an independent cohort to confirm splicing defects and determine which splice events are associated with muscle weakness.
Subjects and Methods
Research subjects and muscle testing
Participants were recruited through the National Registry of Myotonic Dystrophy and Facioscapulohumeral Dystrophy (FSHD) Patients and Family Members.14 The studies were approved by the University of Rochester Research Subjects Review Board. Subjects with DM1 or DM2 were ambulatory adults with proven CTG or CCTG expansions. Nonambulant individuals and patients with congenital or childhood onset of DM1 were excluded to eliminate confounding effects of muscle disuse or maldevelopment. Subjects were recruited in two prospective, nonoverlapping cohorts. The discovery cohort and disease controls are described in Supplementary Table 1. The validation cohort consisted of 50 DM1 subjects (mean age 47, range 18 to 69, male:female 18:32) and 8 healthy controls (n = 8, mean age 26, range 20 to 37, male:female 5:3). Strength of ankle dorsiflexion (ADF) and hand grip was determined by standardized manual muscle testing and quantitative myometry as previously described.15, 16 Manual testing was expressed as Medical Research Council (MRC) grades 5, 4+, 4, 4−, 3, and 2. Quantitative testing was expressed as the percentage of the predicted strength in healthy individuals of the same age, sex, and height.15 Manual and quantitative testing were correlated (r = 0.86, P < 0.001) and showed a broad spectrum of ADF weakness in the validation cohort (Supplementary Fig 1). On the day following the strength testing, at 10 to 11 AM, after a standardized meal, each subject underwent a needle biopsy of TA muscle as previously described.17 The biopsy procedure was well tolerated in all subjects (Supplementary Fig 2).
RNA and DNA analysis
Reverse transcriptase (RT)-PCR analysis of alternative splicing was performed as previously described.18 Procedures for RNA, microarray, and DNA analysis are described in Supplementary Methods.
Histological analysis
Fluorescence in situ hybridization (FISH, for CUGexp RNA) and immunofluorescence (IF) was performed on frozen sections using CAG-repeat probe, antibody A2764 for MBNL1, and antibody F1.652 for embryonic myosin (DSHB, Iowa City, IA) as previously described.18, 19
Mouse models
HSALR transgenic and Mbnl1 knockout mice were previously described. 20,21 To determine the effects of CUGexp knockdown on splicing outcomes, HSALR mice were treated with subcutaneous injection of antisense oligonucleotide (ASO) 445236 (25 mg/kg twice weekly for four weeks) or saline (n = 4 per group) as previously described.10 ASO 445236 was a gift from Dr. Frank Bennett at Isis Pharmaceuticals. Adr (Clcn1 null, non-dystrophic myotonia), mdx (dystrophin null), and strain-appropriate control mice were obtained from Jackson Laboratories.
Statistical analysis
Associations of splicing with age, strength, CTG expansion, and MYH3 expression were described using Pearson’s correlation coefficients. Exploratory multiple regression analyses were performed to examine candidate sets of splice events associated with ADF weakness (see Supplementary Methods). The significance of splicing differences between groups (DM with full mutations or protomutations vs. disease controls vs. healthy controls) or ASO- vs. saline-treated mice was determined using two-sample t tests.
Results
Transcriptome-wide discovery of splicing defects in DM1 and DM2
Previously we studied splicing changes in vastus lateralis (VL) because knee extensors are functionally important and accessible for needle biopsy.18 However, VL is less involved than distal muscles in DM1, which may affect its suitability for biomarker discovery. To address this question we examined three MBNL1-dependent splice events in VL biopsies from 16 subjects with DM1, 11 with DM2, and 3 healthy controls. The splice events were strongly affected in all subjects with DM2 but changes were less consistent in DM1 (Supplementary Fig 3), in line with previous observations that sequestration of MBNL1 in VL muscle was more pronounced in DM2 than DM1.18
These results suggested that VL biopsies were suitable for biomarker discovery in DM2 but not in DM1. We next examined 27 postmortem muscles from 7 individuals with DM1. Eight samples (4 biceps, 2 quadriceps, 1 tibialis anterior, 1 diaphragm) showed good RNA integrity and major defects of SERCA1/ATP2A1 and Insulin Receptor (INSR) alternative splicing (not shown). These samples and seven DM2 VL biopsies were carried forward into microarray analysis, along with VL biopsies from eight healthy subjects and 8 disease controls (FSHD). The age and sex distributions were similar across groups (Supplementary Table 1). The RNA was analyzed on Human Exon 1.0 ST arrays. The arrays contained 5.4 million probes mapping to 1.1 million exons from 35 thousand RefSeq mRNAs.22 The overall pattern of altered gene expression was strikingly similar in DM1 and DM2 (Supplementary Fig 4), consistent with a shared RNA-dominant disease mechanism.
To identify misregulated splice events we used signal intensity from each probe set to estimate expression level for each exon. We then normalized the expression level to other exons in the same transcript, and tested for differences of exon inclusion between groups. Through empiric testing of different selection criteria we developed a filtering procedure (Supplementary Methods) to identify exons that showed differential inclusion in DM1 and DM2 compared to healthy and disease controls. The selection criteria identified 438 candidate exons belonging to 322 genes (Supplementary Table 4). The criteria captured > 50% of previously described DM-associated splicing defects (Supplementary Table 5). We then chose 109 candidate exons for initial confirmation by RT-PCR, using the same RNA samples that were analyzed on the arrays (Supplementary Table 6). The results were supportive of misregulated splicing for 73 (67%) candidates and 10 positive controls (exons with previously documented splicing misregulation), indicating that our selection criteria were moderately sensitive and specific.
Validation of splicing defects in a different muscle and independent cohort
Next we used biopsy samples from a different muscle (TA) to confirm splicing defects in DM1, selecting novel 31 events that appeared to show large effects. TA was used for this analysis because it is preferentially affected and its function (ankle dorsiflexion, ADF) can be directly assessed. Comparing TA samples from 5 DM1 subjects with 5 healthy controls we provisionally confirmed splicing misregulation for all 31 events (Supplementary Table 7, nominal P < 0.05 for each confirmed event). Based on these results and previous studies we selected a group of 50 putative splicing defects for further validation in a larger cohort (the 5 initial subjects + 45 additional DM1 subjects, vs. 8 healthy controls). Initially we focused on subjects who clearly displayed ADF weakness (MRC score 4+ or below, quantitative myometry < 80% of predicted, n = 33), on the assumption that clinically affected muscles were more likely to show splicing misregulation. This analysis reconfirmed the splicing defects for 42 of 50 splice events (nominal P < 0.001 for DM vs. healthy controls, Bonferroni-corrected P < 0.05, Table 1). Notably, the splicing in TA was much more severely affected than in VL in the discovery cohort (Supplementary Fig 5), suggesting that splicing misregulation may contribute to selective patterns of muscle involvement. There was no association of splicing outcome with age or gender.
Table 1.
Effects of DM1 on the regulation of alternative splicing in TA muscle. For each splice event, the fraction of splice products that included or skipped the alternative exon(s) was compared for eight healthy controls and 33 DM1 subjects with ADF weakness (MRC score of 4+ or below, “weak DM1”). Splice events are listed in descending order of effect size (percentage splicing shift) in the weak DM1 vs. healthy subjects. The correlation of splicing outcome with weakness is calculated for all DM1 subjects (weak or not weak, n = 50). Splice events that show early transition (misregulated splicing in TA with normal strength) are underlined. Downward arrows indicate that inclusion of the exon is reduced in DM1, upward arrows indicates that exon inclusion is increased. Splicing shift is the absolute difference of exon inclusion (or exclusion) in DM1 vs. controls, as calculated from signal intensity of RT-PCR products: exon inclusion (or exclusion) product ÷ sum of all products × 100%.
| Splice event | % splicing shift in weak DM1 vs. healthy, direction of shift (P value) | Correlation of splicing with weakness (P value) | Function of gene product (effect of splicing change) |
|---|---|---|---|
| SOS1 exon 25 | 82.6 ↓ (1.E-25) | 0.57 (1.5E-05) | receptor signaling (↓ signaling) |
| ATP2A1 exon 22 | 78.3 ↓ (3.E-09) | 0.65 (4.0E-07) | Ca++ reuptake |
| ALPK3 exon 2 | 71.9 ↑ (4.E-16) | 0.64 (6.6E-07) | signaling, myogenesis |
| NFIX exon 7 | 68.1 ↑ (1.E-15) | 0.68 (7.5E-08) | transcription factor, myogenesis |
| INSR exon 11 | 66.2 ↓ (6.E-26) | 0.44 (1.5E-03) | insulin signaling (Δ signaling) |
| CAPZB exon 8 | 66.1 ↓ (6.E-18) | 0.65 (2.5E-07) | assembly of actin filaments |
| ARFGAP2 exon 6 | 65.2 ↓ (5.E-25) | 0.36 (1.1E-02) | vesicle trafficking |
| PDLIM3 exon 5 | 65.0 ↓ (3.E-12) | 0.70 (1.5E-08) | Z disc |
| CACNA1S exon 29 | 64.3 ↓ (3.E-16) | 0.67 (1.3E-07) | excitation contraction coupling (↑ Ca++ entry) |
| CAMK2B exon 13 | 62.9 ↓ (3.E-31) | 0.44 (1.4E-03) | Signaling |
| VPS39 exon 3 | 62.4 ↓ (1.E-14) | 0.58 (8.3E-06) | vesicle trafficking, TGFbeta signaling |
| CLCN1 exon 7a | 58.5 ↑ (2.E-25) | 0.52 (1.3E-04) | chloride channel (loss of ion conductance) |
| LDB3 exon 11 | 53.0 ↑ (3.E-05) | 0.58 (1.1E-05) | Z disc |
| GFPT1 exon 9 | 50.6 ↓ (9.E-12) | 0.66 (2.3E-07) | protein glycosylation (↓ feedback inhibition) |
| DTNA (DB2) exons 11a, 12 | 47.5 ↑ (4.E-12) | 0.71 (1.0E-08) | membrane integrity, signaling (altered signaling) |
| IMPDH2 exon 9b | 44.3 ↓ (1.E-24) | 0.57 (1.5E-05) | nucleotide biosynthesis |
| MBNL1 exon 7 | 42.8 ↑ (2.E-09) | 0.68 (6.7E-08) | alternative splicing (↑ nuclear localization) |
| NRAP exon 12 | 42.0 ↓ (1.E-10) | 0.46 (7.5E-04) | myofibril assembly |
| ANK2 exon 21 | 41.6 ↓ (3.E-08) | 0.70 (1.2E-08) | membrane targeting |
| OPA1 exon 4b | 40.9 ↓ (3.E-25) | 0.60 (3.6E-06) | mitochondrial dynamics (↑ proteolytic cleavage of OPA1 protein) |
| RYR1 exon 70 | 38.4 ↓ (8.E-19) | 0.45 (9.1E-040) | Ca++ release (↓ open probability) |
| COPZ2 exon 9b | 33.6 ↑ (2.E-07) | 0.33 (2.0E-02) | vesicle trafficking |
| KIF13A exon 32 | 33.5 ↓ (7.E-06) | 0.64 (4.8E-07) | positioning of endosomes |
| TTN exon Mex5 | 31.5 ↑ (8.E-30) | 0.50 (2.1E-04) | cytoskeletal protein |
| PHKA1 exon 28 | 28.8 ↓ (4.E-08) | 0.63 (9.0E-07) | muscle glycogenesis |
| FHOD1 exon 11a | 25.8 ↓ (2.E-09) | 0.39 (5.5E-03) | actin organization |
| MBNL2 exon 7 | 24.8 ↑ (8.E-09) | 0.55 (3.2E-05) | alternative splicing |
| DTNA (DB1) exons 11a, 12 | 23.8 ↑ (3.E-07) | 0.64 (4.9E-07) | membrane integrity, signaling (Δ signaling) |
| DMD exon 78 | 18.0 ↓ (3.E-06) | 0.71 (9.6E-09) | membrane integrity |
| PHKA1 exon 19 | 17.6 ↑ (3.E-05) | 0.68 (5.1E-08) | muscle glycogenesis |
| MLF1 exon 3 | 15.6 ↓ (1.E-04) | 0.58 (1.1E-05) | oncoprotein |
| ABLIM2 exon 12 | 14.6 ↑ (2.E-07) | 0.65 (3.0E-07) | Z disc |
| UBE2D3 exon 10 | 14.2 ↓ (1.E-25) | 0.55 (2.9E-05) | Ubiquitination |
| BIN1 exon 11 | 14.0 ↓ (5.E-04) | 0.72 (4.5E-09) | T-tubule formation (↓ formation of T tubules) |
| LDB3 exon 5 | 13.6 ↑ (3.E-17) | 0.49 (2.6E-04) | Z disc |
| DMD exon 71 | 13.3 ↓ (3.E-04) | 0.62 (1.9E-06) | membrane integrity |
| TBC1D15 exon 10 | 13.1 ↓ (7.E-18) | 0.42 (2.6E-03) | intracellular trafficking |
| USP25 exons 19a,19b | 12.5 ↓ (1.E-05) | 0.45 (1.1E-03) | deubiquitination (↑ proteolysis of muscle proteins) |
| TXNL4A exon 4 | 12.5 ↓ (1.E-09) | 0.38 (6.9E-03) | PQBP1 binding protein |
| VEGFA exons 6a, 6b | 7.3 ↓ (2.E-15) | 0.45 (1.0E-03) | angiogenesis (altered diffusibility) |
| CAPN3 exon 16 | 6.4 ↓ (3.E-06) | 0.64 (5.0E-07) | intracellular protease (↓ protease activity) |
| ATP2A2 intron 19 | 3.8 ↓ (1.E-08) | 0.45 (1.1E-03) | Ca++ reuptake |
Associations of splicing changes with muscle weakness
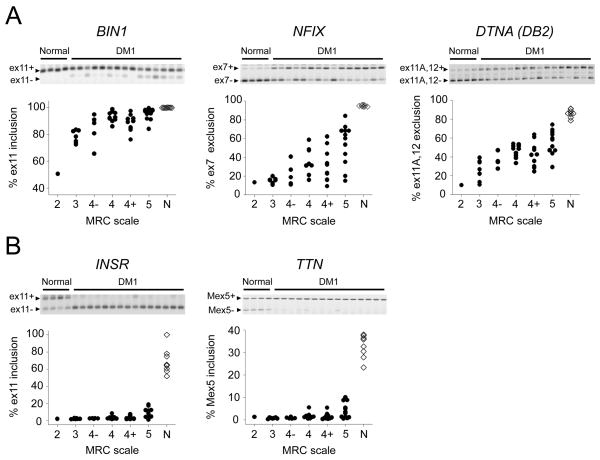
Next we examined splicing outcomes across the entire validation cohort (n = 50) to test for associations of splicing defects with muscle weakness. Twenty of the 42 DM1-affected splice events showed a correlation with TA function, using manual muscle testing to gauge the severity of ADF weakness (r > 0.6, nominal P < 10−5, Table 1; three examples are shown in Fig 1A). With one exception the same events were also correlated with ADF weakness assessed by quantitative myometry, and 10 events were also correlated with handgrip weakness (Supplementary Table 8). The relationship between ADF weakness and splicing outcome is shown for every splice event in Supplementary Data: All Splice Events.
Figure 1.
Relationships between alternative splicing in TA muscle and strength of ankle dorsiflexion in DM1. For each splice event the fractional inclusion or exclusion of the indicated exon is shown for 45 DM1 subjects with full mutations (> 100 CTG repeats) and 8 healthy controls. Representative gel images of RT-PCR products are shown for each splice event. The exon inclusion splice product is the top band and the exon exclusion product is the lower band. ADF strength on the side of the TA biopsy was determined by manual testing and expressed as an MRC score. (A) Three examples of splice events that showed correlation of splicing outcome with muscle weakness. (B) Examples of “early transition” events that are strongly affected even in TA muscles that exhibit normal strength.
Five splice events were strongly affected by DM1 (> 30% shift of exon inclusion) yet not associated with ADF weakness (r ≤ 0.5, Table 1). These events were markedly affected even when TA weakness was not clinically apparent (Fig 1B shows two examples). Events in this “early transition” group included INSR, titin (TTN), ryanodine receptor (RYR1), calcium/calmodulin-dependent protein kinase 2B (CAMK2B), and ARFGAP2, all of which rely on MBNL1 for normal splicing regulation (see below), indicating that sequestration of MBNL1 is an early molecular event in DM1.
Results of exploratory multiple regression analyses are presented in Supplementary Tables 9 and 10. Although the values of adjusted R2 presented here are optimistic (a more realistic assessment of model performance would have to be done using a new data set), the results suggest that models containing several splice events should provide substantial improvements over models that contain a single splice event. Some splice events such as MBNL1 exon 7, COPZ2, and LDB3 were consistently associated with ADF strength as measured by either quantitative myometry or manual muscle testing. Others appeared mainly in models for ADF strength measured by quantitative myometry (ALPK3, CAPZB, VPS39, and DTNA (DB2)) or manual muscle testing (NFIX, RYR1, USP25, and TXNL4A) (Supplementary Tables 9 and 10).
Splicing defects are not correlated with CTG expansion size in muscle or blood
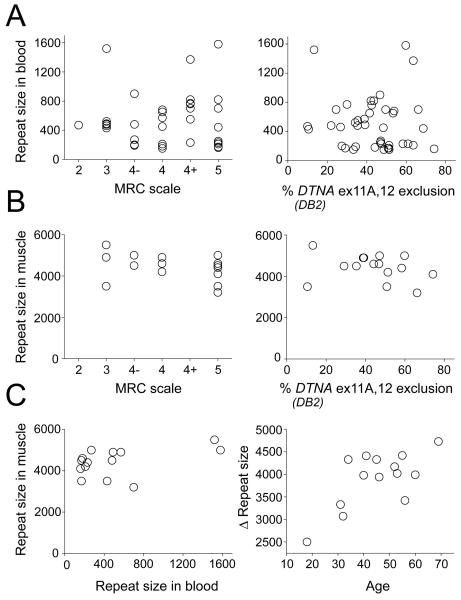
Among subjects who carried full DM1 mutations (> 100 CTG repeats, n = 45) there was no correlation of TA weakness or splicing misregulation with CTG expansion size in peripheral blood (Fig 2A). However, CTG expansions in leukocytes are smaller and not predictive of expansion size in skeletal muscle.23–25 We therefore examined CTG expansions in TA muscle, selecting subjects whose ADF strength was relatively preserved (n = 7, mean age 42, strength = 5) or significantly weak (n = 8, mean age 48, ADF strength of 4, 4−, or 3). However, once again there was no correlation of CTG expansion size with splicing outcomes or ADF strength (Fig 2B). In fact, all 15 subjects had muscle expansions that were very large and extremely heterogeneous, with modal expansion size greater than 3,000 repeats (Supplementary Fig 6). The CTG expansions in muscle also did not correlate with leukocyte expansions from the same subjects (Fig 2C). However, in line with a previous study,25 the difference of repeat size between leukocytes and muscle was correlated with age (r = 0.68, P < 0.01), suggesting an age-dependent process of somatic expansion that is more pronounced in muscle than hematopoietic cells.
Figure 2.
CTG expansion size is not associated with splicing misregulation or weakness in TA. (A) CTG expansion size in leukocytes is not correlated with ADF strength (left) or misregulated splicing of DTNA DB2 exons 11A and 12 (right). (B) CTG expansion size in TA muscle tissue is not correlated with ADF strength or DTNA splicing (right). (C) CTG expansion size in TA muscle tissue is not correlated with leukocytes (left). However, the difference of repeat size between muscle tissue and leukocytes (Δ repeat size) is correlated with age (right) (r = 0.68, P < 0.01)
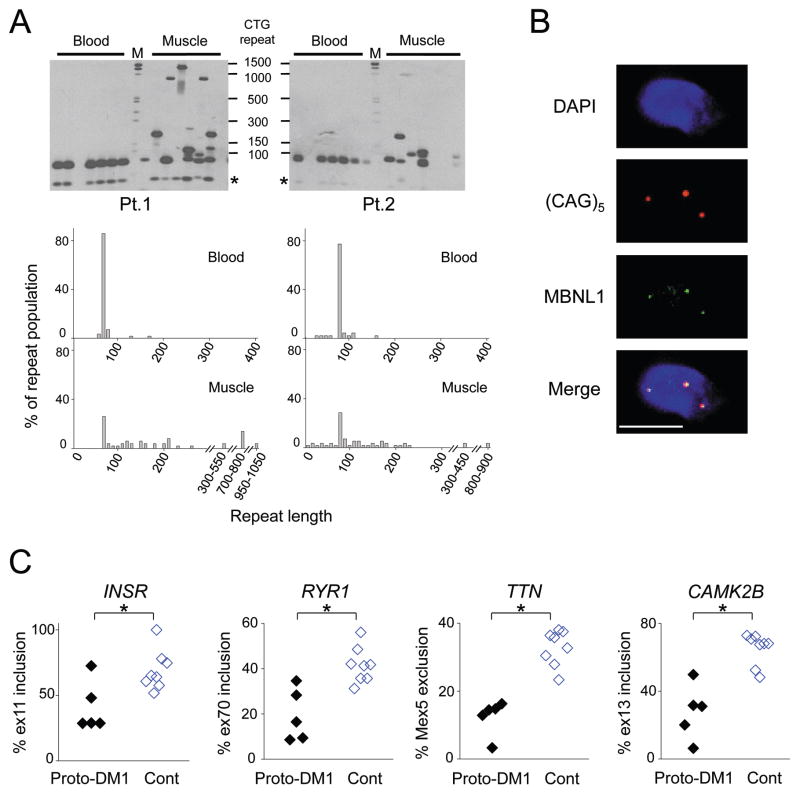
The DM cohort included 5 subjects (age 39 to 68 yrs) who carried small expansions of 80 to 90 CTG repeats (protomutations). These individuals were minimally symptomatic and they exhibited normal ADF strength. We used small-pool PCR to examine expanded alleles in two of these subjects. While protomutations were relatively stable in peripheral blood (14% and 23% unstable alleles, average size change of 4 and 7 repeats) they were remarkably unstable in skeletal muscle (74% and 71% unstable alleles, with average size change of 192 and 75 repeats, P < 0.001, chi-square for muscle vs. blood), producing muscle alleles with several hundred to more than 1,000 CTG repeats (Fig 3A). These repeat tracts were long enough to induce nuclear foci of CUGexp RNA, partial sequestration of MBNL1 (Fig 3B), and misregulated splicing for 11 of 42 DM1-affected splice events (Supplementary Table 8 P < 0.01), as shown for four early transition events in Fig 3C.
Figure 3.
Molecular features of the DM1 protomutation. (A) Tissue-specific somatic instability of the DM1 protomutation. (Top) Representative data from small-pool PCR + Southern blot analysis of CTG repeat length. DM1 expansions were amplified from genomic DNA at high dilution, so that each reaction contains one or several amplifiable alleles, then detected by Southern blot. * indicates normal DMPK allele. (Bottom) Histograms showing allelic distribution of CTG expansions in leukocytes or muscle from subjects with DM1 protomutations. CTG expansions are binned in groups spanning 10 repeats. More than 44 alleles were sized for each sample. M, molecular weight standard. (B) Fluorescence in situ hybridization and immunofluorescence of TA section showing colocalization of MBNL1 protein (green) with CUGexp foci (red) in a myonucleus (blue) of a subject who carries a DM1 protomutation. Bar = 5 μm. (C) Alternative splicing of early transition events is misregulated in subjects with DM1 protomutations (Proto-DM1, CTG expansion size of 80 to 90 repeats), compared to healthy controls (Cont). * P<0.05, t test.
Disease specificity of splicing defects
Splicing changes in DM1 may recapitulate patterns of RNA processing that normally occur during fetal muscle development.18, 26 Mouse studies have shown that non-DM dystrophies can impact alternative splicing, presumably due to post-natal myogenesis during muscle regeneration. RNA toxicity, however, had a more profound effect, in terms of the number and extent of the splicing changes.27,18, 28 We examined the expression of embryonic myosin heavy chain (MYH3), a sensitive marker of muscle regeneration or denervation.29 MYH3 mRNA was variably elevated in DM1 TA, and associated with splicing changes for 35 of 50 splice events (r > 0.6 and P < 10−5, Supplementary Fig 7A, Supplementary Table 8). However, examination of tissue sections indicated that MYH3 protein was confined to a small fraction of the muscle fibers (Supplementary Fig 7B), consistent with observations that regenerating fibers are infrequent in DM1 and therefore unlikely to account for the large splicing effects that we observed. To further assess disease specificity and the possible contribution of muscle regeneration we compared 10 splice events in DM1 with other muscle disorders (listed in Supplementary Table 1) and healthy controls, using TA samples for all analyses. Although some disease controls exhibited minor splicing changes, the defects in DM1 were more severe and consistent for each event (Supplementary Fig 8), in line with previous observations that misregulated alternative splicing is relatively specific for DM.30–33
Cross-correlation of splicing abnormalities
Previous work has shown that alternative splicing of CAPZB, FXR1, and ANK2 are regulated by CELF1 but not MBNL1.26 We found that CAPZB was misregulated in all subjects with full mutations, whereas effects on FXR1 were limited to severely affected subjects (Supplementary Fig 9A). We also compared 5 splice events regulated by MBNL1 but not CELF1 (CLCN1, TTN, MBNL1 exon 7, MBNL2 exon 7, and PDLIM3 exon 5).18, 26, 34 CLCN1 splicing was affected in nearly all subjects, whereas MBNL1 and MBNL2 mis-splicing were mainly present in the severely affected subjects (Supplementary Fig 9B). These results indicate that some splice events are more sensitive to RNA toxicity than others, even when regulated by the same splicing factor. Notably, some events were highly correlated even though they were regulated by different splicing factors (Supplementary Fig 9C).
Molecular basis of splicing defects
Next we determined which splice events show conservation in humans and mice. Out of 68 DM1-affected exons, 55 (81%) showed developmentally regulated alternative splicing in wild-type mice, defined as a shift of splicing between embryonic day 18 and postnatal day 20 (Supplementary Table 11). We then used mouse models to compare the effects of MBNL1 ablation, CUGexp expression, chronic dystrophy (dystrophin deficiency), or non-dystrophic myotonia (chloride channelopathy) on the regulation of these events. Twenty nine of the 55 events (53%) showed DM-like splicing changes in Mbnl1 knockout mice, and, with one exception, a similar pattern in CUGexp-expressing mice (Supplementary Table 11), consistent with previous observations that splicing defects in CUGexp-expressing and Mbnl1 knockout mice were highly concordant.18, 28 These results suggest that MBNL1 sequestration contributes to most of the splicing changes that we observed in DM1 patients. However, at least twelve of the human DM1 splicing defects were not recapitulated in any of the mouse models.
Reversibility of splicing defects in transgenic mice
Targeted knockdown of toxic RNA is being pursued as a therapeutic strategy for DM1. Levels of CUGexp RNA in transgenic mice were reduced by systemic administration of ASOs, thus correcting several of the splicing defects that are characteristic of DM1.10 To determine whether reversibility is a general feature of DM-associated spliceopathy we examined nine events that showed parallel changes in DM1 patients and CUGexp-expressing mice. In all cases the splicing defects in muscle were fully corrected after eight subcutaneous injections of the ASO (Supplementary Fig 10).
Discussion
Misregulated alternative splicing is a fundamental molecular feature of DM1,4 affecting many genes involved in muscle homeostasis and function.18, 30–33, 35–37 The current study enlarges the number of known DM-affected splice events and provides the first detailed assessment of splicing changes in relation to functional impairment. We identified a group of splice events that are affected even before there is evidence of muscle weakness – the earliest transcriptomic changes that are presently known in DM1. Beyond this phase emerges a spreading hierarchy of splicing changes whose number and extent are proportional to disease severity. The organization of the hierarchy appears to reflect: (1) differences among splicing factors in their sensitivity to RNA toxicity; and (2) differences among exons in their response to perturbations of their cognate splicing factors. The earliest changes involve a group of MBNL1-dependent exons, which fits with observations that MBNL proteins have greater CUGexp binding affinity than other RNA binding proteins.6, 38 These “early transition” changes are apparent even in protomutation carriers and individuals with minimal DM1 symptoms. These events are MBNL1-dependent but they are normally regulated in heterozygous Mbnl1 knockout mice (data not shown), suggesting that protomutation carriers already have > 50% sequestration of MBNL1 protein. With increasing disease severity there are changes of other MBNL1- and CELF1-dependent splice events, and it is possible that other splicing factors are also sequestered or indirectly affected through the activation of regeneration or stress response pathways. Since DM-associated splicing defects resemble splicing patterns that normally occur during muscle development, we expect that any process involving acute widespread muscle necrosis and regeneration has potential to trigger transient re-expression of fetal splice products.27 However, previous work30–33 and the current study indicate that major splicing defects are relatively specific to DM1 and DM2 among chronic neuromuscular disorders. It is noteworthy that our array analysis identified hundreds of candidate splicing defects in DM but failed to uncover a single splicing change in FSHD. Also, we failed to identify any splicing changes in DM1 that did not also occur in DM2.
Our study indicates that alternative splice events have good potential to function as biomarkers of DM severity and therapeutic response: (1) the analytical precision is good; (2) the mechanism for splicing misregulation is well defined and directly connected to the disease process (RNA toxicity) and therapeutic goal (reduction of toxic RNA and release of sequestered proteins); (3) many splicing defects are correlated with muscle weakness and some are directly implicated in symptoms of DM1;33, 35, 39 and (4) in mouse models the splicing defects are fully reversible by RNA targeted therapy. The main drawback is that tissue sampling is required. However, the analysis can be performed on small tissue samples that are easily obtained by a minimally invasive biopsy procedure. Currently we are examining the feasibility and test-retest reliability of serial sampling for splicing analysis.
Expanded CTG repeats are unstable in muscle fibers but the clinical consequences of this process are uncertain.40 It is unclear whether the growth of expanded repeats over time determines the onset or progression of DM1, or whether individual differences of somatic instability may contribute to the enormous clinical variability of DM1. We observed subjects with protomutations who carry hundreds to >1,000 CTG repeats in muscle tissue, yet their muscle strength remained intact. This suggests that growth of the repeat into a larger size range (> 2,000 repeats) is required to develop a progressive myotonic myopathy. We also found that adults with full mutations displayed huge CTG expansions in TA muscle, regardless of whether their contractile function was markedly affected or relatively preserved. This suggests that some individuals tolerate large expansions better than others, and raises the possibility that DM1 severity is modulated by other factors, such as modifier genes, sequence interruptions in the CTG repeat tract,41 epigenetic changes at the DM1 locus,42 or physical activity. It is also possible that muscle deficits are not cell autonomous, but depend partly on RNA toxicity in other cells, such as motor neurons.43, 44 A limitation of our study, however, is that resolution and sizing of large CTG fragments on Southern blots is not precise. Alternative technologies to confirm expansion size are needed but are not currently available.
A precise molecular explanation for DM-associated muscle weakness and wasting remains elusive. Previous studies have indicated that mis-splicing of BIN1 exon 11 (BIN1ex11) may contribute to muscle weakness in DM1.33 BIN1ex11 splicing is required for normal formation of transverse tubules,45 which are critical structures for excitation-contraction coupling (ECC). Reduction of BIN1ex11 inclusion below 80% was sufficient to induce T tubule abnormalities and muscle weakness in mice.33 We found that mis-splicing of BIN1ex11 ranked first among splicing defects that correlated with ADF weakness. However, effects on BIN1 splicing were not large, and only 14% of our cohort had BIN1ex11 inclusion levels below 80%. It is possible, however, that BIN1ex11 inclusion varies among fibers, falling below 80% in some fibers or domains, or that mis-splicing of other ECC components, such as ryanodine receptor or CACNA1S (dihydropyridine receptor), may potentiate the effects of BIN1ex11 skipping. Another candidate for involvement in muscle weakness is dystrobrevin (DTNA). Mis-splicing of DTNA exons 11A and 12 ranked third for correlation with weakness (r = 0.7) and is known to affect signaling pathways that influence muscle growth.32 In addition, the largest effect on alternative splicing that we observed was an increase in the skipping of SOS1 exon 25, a splicing outcome that inhibits signaling pathways involved in muscle hypertrophy.46, 47 Healthy subjects included exon 25 in 99% of SOS1 transcripts, whereas the average inclusion rate in DM1 was only 16%.
The selection of optimal splicing biomarkers for future studies will depend on the specific requirements of study design. If muscle samples are obtained before and after an intervention, it is reasonable to examine splice events that correlate with muscle weakness, on the assumption that correction of these defects may predict subsequent potential for functional improvement (as was the case for myotonia rescue in transgenic mice).8–10, 12 In this regard, our exploratory multiple regression analyses indicate that a panel containing several splice events is likely superior to a single event for predicting TA function. Alternatively, by examining “early transition” events it may be possible to assess therapeutic response in a single post-intervention sample, because these events are maximally affected in nearly all subjects at baseline. However, it appears that these events are quite sensitive to RNA toxicity, so that their correction may require a stronger therapeutic effect (although three such events were fully corrected by ASO treatment in mice). Current data also suggest that different splice events may report on distinct mechanisms, such as MBNL1 sequestration or upregulation of CELF1. We therefore propose that a panel of splicing biomarkers may prove optimal for gauging therapeutic effects in clinical trials. However, further validation and reliability studies are needed to guide the selection of splice events that are most suitable for this purpose.
Supplementary Material
Acknowledgments
The authors thank Kirti Bhatt, Sarah Leistman, Linda Richardson, and Bharati Shah for technical assistance. ASO 445236 was kindly provided by Dr. Frank Bennett at Isis Pharmaceuticals. This work comes from the University of Rochester Paul D. Wellstone Muscular Dystrophy Cooperative Research Center (NIH/NS048843) with support from the National Institute of Health (AR049077, AR48143, AR055947, UL1RR024160-06S4, KO8NS064293); the University of Rochester CTSI/Clinical Research Center (UL1RR024160 and UL1RR024160-06S4), the Saunders Family Fund, Run America, the Muscular Dystrophy Association [M.N.]; and postdoctoral fellowships [to M.N.] from the Cell Science Research Foundation and the Uehara Memorial Foundation.
Footnotes
Potential Conflicts of Interest
The authors have no conflicts of interest to disclose.
References
- 1.Brook JD, McCurrach ME, Harley HG, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 2.Liquori CL, Ricker K, Moseley ML, et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–7. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 3.Udd B, Krahe R. The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol. 2012;11:891–905. doi: 10.1016/S1474-4422(12)70204-1. [DOI] [PubMed] [Google Scholar]
- 4.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–41. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 5.Orengo JP, Cooper TA. Alternative splicing in disease. Adv Exp Med Biol. 2007;623:212–23. doi: 10.1007/978-0-387-77374-2_13. [DOI] [PubMed] [Google Scholar]
- 6.Miller JW, Urbinati CR, Teng-umnuay P, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–48. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler TM, Sobczak K, Lueck JD, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–9. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18551–6. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler TM, Leger AJ, Pandey SK, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–5. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulders SA, van den Broek WJ, Wheeler TM, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13915–20. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkesh R, Childs-Disney JL, Nakamori M, et al. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. J Am Chem Soc. 2012;134:4731–42. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francois V, Klein AF, Beley C, et al. Selective silencing of mutated mRNAs in DM1 by using modified hU7-snRNAs. Nat Struct Mol Biol. 2011;18:85–7. doi: 10.1038/nsmb.1958. [DOI] [PubMed] [Google Scholar]
- 14.Hilbert JE, Kissel JT, Luebbe EA, et al. If you build a rare disease registry, will they enroll and will they use it? Methods and data from the National Registry of Myotonic Dystrophy (DM) and Facioscapulohumeral Muscular Dystrophy (FSHD) Contemp Clin Trials. 2012;33:302–11. doi: 10.1016/j.cct.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Group F-DS. A prospective, quantitative study of the natural history of facioscapulohumeral muscular dystrophy (FSHD): implications for therapeutic trials. Neurology. 1997;48:38–46. doi: 10.1212/wnl.48.1.38. [DOI] [PubMed] [Google Scholar]
- 16.Moxley RT, III, Logigian EL, Martens WB, et al. Computerized hand grip myometry reliably measures myotonia and muscle strength in myotonic dystrophy (DM1) Muscle and Nerve. 2007;36:320–8. doi: 10.1002/mus.20822. [DOI] [PubMed] [Google Scholar]
- 17.Edwards R, Young A, Wiles M. Needle biopsy of skeletal muscle in the diagnosis of myopathy and the clinical study of muscle function and repair. N Engl J Med. 1980;302:261–71. doi: 10.1056/NEJM198001313020504. [DOI] [PubMed] [Google Scholar]
- 18.Lin X, Miller JW, Mankodi A, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Human molecular genetics. 2006;15:2087–97. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 19.Mankodi A, Urbinati CR, Yuan QP, et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Human molecular genetics. 2001;10:2165–70. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 20.Mankodi A, Logigian E, Callahan L, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–73. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 21.Kanadia RN, Johnstone KA, Mankodi A, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–80. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 22.Gardina PJ, Clark TA, Shimada B, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMCGenomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashizawa T, Dubel JR, Harati Y. Somatic instability of CTG repeat in myotonic dystrophy. Neurology. 1993;43:2674–8. doi: 10.1212/wnl.43.12.2674. [DOI] [PubMed] [Google Scholar]
- 24.Thornton CA, Johnson K, Moxley RT., 3rd Myotonic dystrophy patients have larger CTG expansions in skeletal muscle than in leukocytes. Ann Neurol. 1994;35:104–7. doi: 10.1002/ana.410350116. [DOI] [PubMed] [Google Scholar]
- 25.Zatz M, Passos-Bueno MR, Cerqueira A, Marie SK, Vainzof M, Pavanello RC. Analysis of the CTG repeat in skeletal muscle of young and adult myotonic dystrophy patients: when does the expansion occur? Human molecular genetics. 1995;4:401–6. doi: 10.1093/hmg/4.3.401. [DOI] [PubMed] [Google Scholar]
- 26.Kalsotra A, Xiao X, Ward AJ, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20333–8. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orengo JP, Ward AJ, Cooper TA. Alternative splicing dysregulation secondary to skeletal muscle regeneration. Ann Neurol. 2011;69:681–90. doi: 10.1002/ana.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du H, Cline MS, Osborne RJ, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiaffino S, Reggiani C. Myosin isoforms in mammalian skeletal muscle. J Appl Physiol. 1994;77:493–501. doi: 10.1152/jappl.1994.77.2.493. [DOI] [PubMed] [Google Scholar]
- 30.Kimura T, Nakamori M, Lueck JD, et al. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Human Molecular Genetics. 2005;14:2189–200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 31.Nakamori M, Kimura T, Fujimura H, Takahashi MP, Sakoda S. Altered mRNA splicing of dystrophin in type 1 myotonic dystrophy. Muscle Nerve. 2007;36:251–7. doi: 10.1002/mus.20809. [DOI] [PubMed] [Google Scholar]
- 32.Nakamori M, Kimura T, Kubota T, et al. Aberrantly spliced alpha-dystrobrevin alters alpha-syntrophin binding in myotonic dystrophy type 1. Neurology. 2008;70:677–85. doi: 10.1212/01.wnl.0000302174.08951.cf. [DOI] [PubMed] [Google Scholar]
- 33.Fugier C, Klein AF, Hammer C, et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med. 2011;17:720–5. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 34.Ward AJ, Rimer M, Killian JM, Dowling JJ, Cooper TA. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Human molecular genetics. 2010;19:3614–22. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–7. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 36.Charlet-B N, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 37.Mankodi A, Takahashi MP, Jiang H, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 38.Yuan Y, Compton SA, Sobczak K, et al. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–86. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler TM, Lueck JD, Swanson M, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–7. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–70. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 41.Braida C, Stefanatos RK, Adam B, et al. Variant CCG and GGC repeats within the CTG expansion dramatically modify mutational dynamics and likely contribute toward unusual symptoms in some myotonic dystrophy type 1 patients. Human molecular genetics. 2010;19:1399–412. doi: 10.1093/hmg/ddq015. [DOI] [PubMed] [Google Scholar]
- 42.Lopez Castel A, Nakamori M, Tome S, et al. Expanded CTG repeat demarcates a boundary for abnormal CpG methylation in myotonic dystrophy patient tissues. Human molecular genetics. 2011;20:1–15. doi: 10.1093/hmg/ddq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wheeler TM, Krym MC, Thornton CA. Ribonuclear foci at the neuromuscular junction in myotonic dystrophy type 1. Neuromuscul Disord. 2007;17:242–7. doi: 10.1016/j.nmd.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panaite PA, Gantelet E, Kraftsik R, Gourdon G, Kuntzer T, Barakat-Walter I. Myotonic dystrophy transgenic mice exhibit pathologic abnormalities in diaphragm neuromuscular junctions and phrenic nerves. J Neuropathol Exp Neurol. 2008;67:763–72. doi: 10.1097/NEN.0b013e318180ec64. [DOI] [PubMed] [Google Scholar]
- 45.Lee E, Marcucci M, Daniell L, et al. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 2002;297:1193–6. doi: 10.1126/science.1071362. [DOI] [PubMed] [Google Scholar]
- 46.Rojas JM, Subleski M, Coque JJ, et al. Isoform-specific insertion near the Grb2-binding domain modulates the intrinsic guanine nucleotide exchange activity of hSos1. Oncogene. 1999;18:1651–61. doi: 10.1038/sj.onc.1202483. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y, Jiang D, Thomason DB, Jarrett HW. Laminin-induced activation of Rac1 and JNKp46 is initiated by Src family kinases and mimics the effects of skeletal muscle contraction. Biochemistry. 2007;46:14907–16. doi: 10.1021/bi701384k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.