Abstract
Protein misfolding and deposition underlie an increasing number of debilitating human disorders. We have shown that model proteins unrelated to disease, such as the Src homology 3 (SH3) domain of the p58α subunit of bovine phosphatidyl-inositol-3′-kinase (PI3-SH3), can be converted in vitro into assemblies with structural and cytotoxic properties similar to those of pathological aggregates. By contrast, homologous proteins, such as α-spectrin-SH3, lack the capability of forming amyloid fibrils at a measurable rate under any of the conditions we have so far examined. However, transplanting a small sequence stretch (6 aa) from PI3-SH3 to α-spectrin-SH3, comprising residues of the diverging turn and adjacent RT loop, creates an amyloidogenic protein closely similar in its behavior to the original PI3-SH3. Analysis of specific PI3-SH3 mutants further confirms the involvement of this region in conferring amyloidogenic properties to this domain. Moreover, the inclusion in this stretch of two consensus residues favored in SH3 sequences substantially inhibits aggregation. These findings show that short specific amino acid stretches can act as mediators or facilitators in the incorporation of globular proteins into amyloid structures, and they support the suggestion that natural protein sequences have evolved in part to code for structural characteristics other than those included in the native fold, such as avoidance of aggregation.
Keywords: protein misfolding, protein aggregation, protein evolution
The spontaneous conversion of soluble proteins or protein fragments into aggregates and amyloid fibrils is a challenging problem in biological and medical sciences. An increasing body of evidence supports the anomalous misassembly of proteins into insoluble deposits as the fundamental cause behind a growing number of debilitating human disorders, such as Alzheimer's disease and Parkinson's disease, type II diabetes, and the transmissible spongiform encephalopathies (1-4). Important clues to understand the molecular basis of amyloid diseases and, more generally, the biological significance of protein aggregation have emerged recently from observations made in proteins unrelated to human disease, which have been found to convert in vitro into aggregates with structural and cytotoxic properties indistinguishable from those exhibited by amyloid assemblies associated with pathological conditions (5-14).
The Src homology 3 (SH3) domain of the p58α subunit of phosphatidyl-inositol-3′-kinase (PI3-SH3) is one of the best characterized examples of a small globular protein unrelated to any known pathological condition that can form amyloid fibrils in vitro (5, 15, 16). Aggregated species obtained from this protein have been found to be cytotoxic when added to cell cultures (13). This observation suggests that the protein aggregates underlying different human disorders could show similar mechanisms of cytotoxicity and more generally that during evolution nature had to develop strategies to avoid protein misassembly to preserve the viability of living organisms (11, 13). It also suggests that toxicity is not restricted to assemblies formed by a handful of polypeptide sequences and that, indeed, many more misfolded proteins or peptides could be behind human disorders for which the causes are so far largely unknown. This suggestion seems to be supported by a growing number of pathologies and physiological conditions for which a correlation with protein misfolding and deposition has been reported (17-20). Interestingly, recent studies suggest the existence of common structural features between different cytotoxic protein assemblies, pointing perhaps to generic therapeutic strategies against amyloid disorders (21).
We have previously shown that the α-spectrin-SH3 (SPC-SH3) domain, which shares the same fold and 24% sequence identity with PI3-SH3, does not form amyloid fibrils under any conditions tested. Moreover, we have shown that the longer n-src loop, characteristic of PI3-SH3 and notably shorter in most of the SH3 domains, does not confer amyloidogenic properties to PI3-SH3, suggesting that differences in other regions of the amino acid sequence that form the consensus SH3 structural core are likely to be the origin of this different behavior (22). The choice of system and conditions used in this and previous work does not seek to mimic a physiological environment but to reproduce in vitro and with an amenable model those stages of aggregation that precede the formation of amyloid structures. Indeed, the study of PI3-SH3 has undoubtedly provided extremely valuable information about the structural properties of amyloid fibrils, the molecular mechanism of amyloid formation, and, most importantly, the cytotoxicity of protein assemblies, with significant implications for a better understanding of the pathological processes and the exploration of future therapeutic possibilities (5, 13, 15, 16, 23, 24).
Materials and Methods
Protein Expression and Sample Preparation. Wild-type and variant proteins were obtained as previously described (5, 16, 22). The wild-type proteins used in this study consist of residues 1-84 from the SH3 domain of bovine PI3-SH3 (plus a GS extension in the N terminus) and of residues 1-62 from the SH3 domain of chicken brain SPC-SH3. After purification, the proteins were dialyzed against 50 mM ammonium carbonate, freeze-dried, and then kept at -20°C until required. Unless otherwise indicated, amyloid formation was achieved as described (5, 16). Briefly, fibril samples were obtained by incubating protein samples at pH 2.0 (at room temperature and at a concentration of 10 mg/ml) for periods of time from several days to 1 month.
Biophysical Characterization of Proteins and Amyloid Fibril Formation. Circular dichroism spectra were recorded in a Jasco 710 spectropolarimeter (Easton, MD) at 25°C and at a protein concentration of 10 μM by averaging 20 scans per spectrum. Thioflavin-T binding assays and electron microscopy analysis were carried out as reported (9, 16, 25). Electron micrographs were obtained on H-7000 (Hitachi, Tokyo) and CM100 (Philips, Eindhoven, the Netherlands) transmission electron microscopes operating at 75 kV and 80 kV, respectively. Fourier transform infrared measurements were made as described (16, 23). Samples were prepared by dissolving directly the various proteins species in 2H2O at a protein concentration of 10 mg/ml in 2H2O p2Hc 2.0 (corrected for isotope effect). Second derivatives of the amide I band spectra were used to determine the frequencies at which the different spectral components were located.
Quantification of Protein Deposition and Cytotoxicity Tests. Protein deposition was quantified by centrifuging 20-μl aliquots of protein samples at 300,000 × g for 30 min. The amount of protein present in both the supernatant and pellet was estimated by resuspending the pellet in 20 μl of 20 mM phosphate buffer, pH 2.0, and diluting both supernatant and resuspended pellet in three volumes of 8 M Gnd·HCl. Four additional volumes of 6 M Gnd·HCl were then added to the mixture, which was then incubated for 30 min at room temperature. As reported before, PI3-SH3 amyloid fibrils and protein aggregates dissolve in the presence of 6 M Gnd·HCl (23). The fraction of aggregated protein was calculated as the ratio between the amounts of protein remaining in the pellet and supernatant. Unless otherwise stated the experiments were carried out in triplicate to minimize errors during sample handling. Cell culture and viability tests were performed as reported (13).
Solid-State NMR. Uniformly 15N-labeled fibril samples for NMR were obtained as described above. Fibrillar material was sedimented at 150,000 × g for 24 h by using a Beckman ultracentrifuge. The pellet (≈40 μl) was transferred into a 4-mm magic-angle spinning solid-state NMR rotor. Solid-state NMR experiments were carried out on a 500-MHz widebore spectrometer (Bruker, Billerica, MA) at a magic-angle spinning rotation frequency of 10 kHz by using a commercial triple-resonance probe. Typically, 2,000 scans were accumulated for each 1D spectrum. All spectra were referenced by using external 15NH4Cl, setting the ammonium peak to be 35.9 ppm.
Results
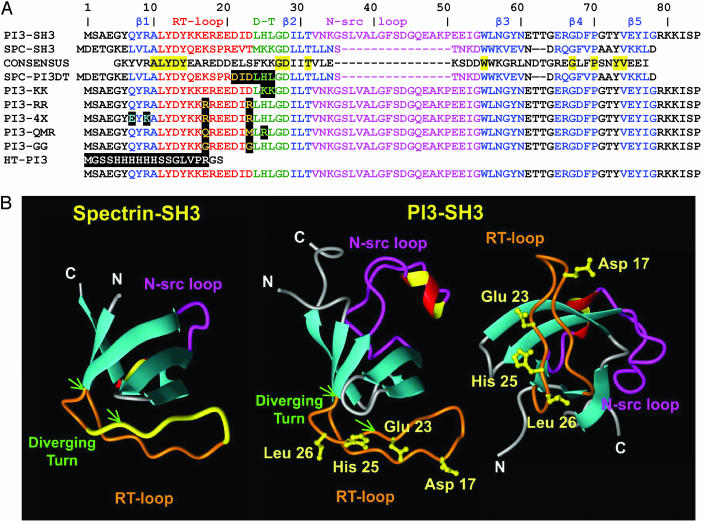
Sequence Conservation in the Diverging Turn Region of SH3 Domains. The diverging turn is a member of a particular subset of type II β-turns and forms part of the folding nucleus of SH3 domains (26-28). Examination of the smart (Simple Modular Architecture Research Tool, http://smart.embl-heidelberg.de) alignment (29, 30) for SH3 domains and the consensus sequence obtained through such alignment (a similar consensus has been reported in refs. 31 and 32) reveal that the diverging turn of PI3-SH3 shows a number of unusual features (Fig. 1A). First, the position that is equivalent to residue 26 [unless otherwise stated, residue numbering corresponds to that of the PI3-SH3 sequence (see Fig. 2A)] in PI3-SH3 is occupied by a charged residue in >58% of the SH3 sequences. In 40% of the cases, it is a basic residue, with preference for Lys (30%). By contrast, position 26 is occupied in PI3-SH3 by Leu, which appears exposed to solvent in the native structure of the domain. This finding is extremely unusual, given that only <10% of SH3 sequences show a hydrophobic residue at this position. The adjacent position 25 is usually occupied by a basic residue in 40% of the sequences, usually Lys (30%). The canonical sequence for these two positions in the SH3 family is in fact Lys-25-Lys-26, as in SPC-SH3 (Fig. 1A), a protein domain that does not form amyloid fibrils at low pH under any of the conditions we have so far explored (22). In addition, it is interesting that the PI3-SH3 sequence spanning residues 21-26 (Asp-Ile-Asp-Leu-His-Leu) has a binary polar/nonpolar pattern shared by <3% of SH3 sequences and absent in both the consensus and SPC-SH3 sequences (Fig. 1A). Interestingly, comparison of the sequences of both domains reveals that the diverging turn of the PI3-SH3 domain (residues 24-28) has a net charge of +1 at low pH, whereas the corresponding region of SPC-SH3 has a net charge of +2 at acidic pH (Fig. 1A) and a significantly reduced hydrophobic character when compared with PI3-SH3. Indeed, recent studies have shown that the balance of charges and the hydrophobic character of a given sequence can play an important role in driving amyloid fibril formation and that an increase of the net charge in an amyloidogenic polypeptide can severely interfere with its ability to self-assemble (14, 33-35). Finally, the atypical alternating polar/nonpolar pattern in PI3-SH3 (that is absent in SPC-SH3) could also contribute to the observed differences in amyloid formation exhibited by both domains, as suggested by recent studies showing a high aggregation propensity for these sequence patterns (36, 37).
Fig. 1.
Proteins used in this study. (A) Sequences of SPC-SH3, PI3-SH3, and the variants used in this study. Numbers correspond to the PI3-SH3 sequence. Conserved residues between SPC-SH3 and PI3-SH3 are highlighted in yellow. Data Submission Software Package and the Protein Data Bank files corresponding to PI3-SH3 (1pnj) and SPC-SH3 (1shg) were used to locate secondary structure motifs (60): β-strands appear in blue, and RT loops, diverging turns (D-T), and n-src loops appear in red, green, and purple, respectively. Highlighted in black are positions substituted in the respective mutants. (B) Three-dimensional structures of SPC-SH3 and PI3-SH3. The location of the RT loop and diverging turn is indicated in orange in both proteins, and the n-src loop is shown in magenta. The diverging turn is indicated with two green arrows in both the SPC-SH3 and PI3-SH3 structures. The fragment colored yellow in SPC-SH3 is the stretch of amino acids replaced by those of PI3-SH3 to generate the mutant SPC-PI3DT. The side chains of the residues of the RT loop and the diverging turn that were mutated in PI3-SH3 protein are colored yellow.
Fig. 2.
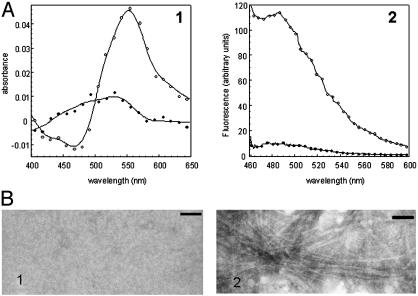
Amyloid fibril formation by wild-type SPC-SH3 and mutant SPC-PI3DT. Samples at concentrations of 10 mg/ml were incubated for 30 d at pH 2.0. (A) Binding of Congo red (Left) and Thioflavin-T (Right) by wild-type SPC-SH3 (•) and SPC-PI3DT mutant (○) samples. The contribution of the dyes were subtracted from the spectra. (B) Electron micrographs of 1:10 diluted samples of wild-type SPC-SH3 (Left) and the SPC-PI3DT mutant (Right). (Scale bar, 100 nm.)
Analysis of SH3 Domains with Chimeric Diverging Turns. To find out whether this region could play a critical role in the formation of amyloid fibrils by PI3-SH3, we created a chimeric SH3 domain (SPC-PI3DT) by replacing residues 22-27 of SPC-SH3, corresponding to the diverging turn and neighboring RT loop residues of the protein, with the homologous residues 21-26 from PI3-SH3 (Fig. 1). Incubation of the SPC-PI3DT mutant under the same conditions used to study wild-type PI3-SH3 (pH 2.0 at 10 mg/ml for 30 d) results in the ready formation of a gel, with properties resembling those of samples of PI3-SH3 known to contain amyloid fibrils (5, 16). SPC-PI3DT samples obtained in this way are able to bind both Congo red and thioflavin-T (Fig. 2A), and transmission electron microscopy analysis reveals the presence of abundant amyloid fibrils (Fig. 2B). These results show clearly that a short amino acid sequence is sufficient to increase dramatically the amyloidogenicity of a protein. Because replacement of the other regions of these two proteins (i.e., the n-src loop of PI3-SH3 with that of SPC-SH3) does not alter significantly the amyloid formation properties of the domain, (22), the diverging turn and neighboring RT loop amino acids appear to have a rather specific role in influencing PI3-SH3 aggregation.
To investigate further the significance of the amino acid composition of this region of the PI3-SH3 domain for amyloid formation at low pH, we analyzed a series of PI3 mutants designed to modify specific residues of the diverging turn as well as neighboring residues corresponding to the RT loop of the protein (Fig. 1). The idea behind this approach was to test whether simple sequence modifications in this particular region of the protein could alter its ability to form fibrils as we had observed for the chimeric mutant SPC-PI3DT. The first mutant we analyzed had the amino acids His-25 and Leu-26 of PI3-SH3 replaced by two Lys residues, as found in SPC-SH3 and in the majority of known SH3 domains, to create the chimeric mutant PI3-KK.
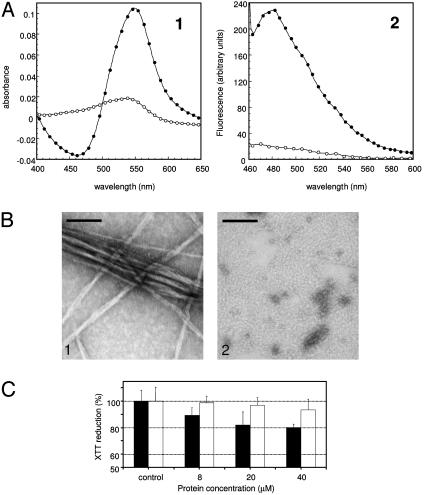
Both PI3-SH3 and PI3-KK were incubated at pH 2.0 for 30 d at a protein concentration of 10 mg/ml and then analyzed for features typical of amyloid structures. The wild-type PI3-SH3 protein readily formed a gel, whereas the mutant protein showed no indication of aggregation during the entire experiment. Moreover, although a red shift of the maximum in the absorbance spectra of Congo red and an enhancement of the fluorescence of thioflavin-T occurs when the dyes are added to PI3-SH3 samples (Fig. 3A), no significant effects were observed for PI3-KK at the end of the 30-d incubation period. Transmission electron microscopy analysis confirms the absence of ordered amyloid fibrils in solutions containing the mutant protein (Fig. 3B), whereas long, straight, and un-branched fibrils are evident in solutions of wild-type PI3-SH3 (Fig. 3B). Thus, introducing the consensus KK sequence of SH3 family at position 25-26 of PI3-SH3 by itself inhibits dramatically the propensity of the protein to form amyloid structures. Given that PI3-SH3 and PI3-KK are both denatured to a similar extent at pH 2.0 (see the supporting information, which is published on the PNAS web site), these differences cannot be related to protein stability, but rather support the importance of specific regions of the sequence in facilitating fibril formation from the denatured state.
Fig. 3.
Amyloid fibril formation by wild-type PI3-SH3 and PI3-KK mutant. Samples were prepared as in Fig. 2. (A) Binding of Congo red (Left) and Thioflavin-T (Right) by wild-type PI3-SH3 (•) and PI3-KK mutant (○) samples. The contribution of the dyes were subtracted from the spectra. (B) Electron micrographs of 1:10 diluted samples of wild-type PI3-SH3 (Left) and PI3-KK mutant (Right). (Scale bar, 100 nm.) (C) Cell viability in the presence of PI3-SH3 and PI3-KK proteins preincubated in 50 mM acetate buffer, pH 5.5, containing 25% (vol/vol) trifluoroethanol (13) at the protein concentrations indicated. Black bars refer to wild-type PI3-SH3, and white bars refer to the PI3-KK mutant. Values are normalized to those of control cells treated with medium alone.
PI3-KK was also incubated in the presence of 25% trifluoroethanol in 50 mM acetate buffer at pH 5.5, in which the wild-type protein PI3-SH3 rapidly self-assembles to generate granular aggregates that are toxic to cells in culture (13). Under these conditions, PI3-KK aggregates to a lower extent than the PI3-SH3 wild-type protein, as indicated by the differences in turbidity at 340 nm (1.18 a.u. for the wild-type solution vs. 0.26 a.u. for the PI3-KK solution) and transmission electron microscopy analysis of both samples (data not shown). Solutions containing wild-type PI3-SH3 aggregates seem to have a higher cytotoxic effect on NIH 3T3 cells than those containing PI3-KK assemblies (Fig. 3C). This observation can be attributed to the much lower aggregation propensity exhibited by the PI3-KK mutant under these conditions. It is interesting to note that mutations in the diverging turn that inhibit the formation of mature amyloid fibrils also interfere with the generation of cytotoxic species. This observation is consistent with the existence of common elements in the pathways followed by PI3-SH3 and presumably other proteins to form different types of assemblies, ranging from cytotoxic granular aggregates to well defined amyloid fibrils.
Effect of PI3-SH3 RT Loop and Diverging Turn Sequence Character on the Amyloidogenic Properties Exhibited by the Domain. It has been described previously in some SH3 domains a high helical propensity in the region between strands 1 and 2, including the RT loop and diverging turn (38). Interestingly, studies in other polypeptides (9, 33, 39) have shown that an increase in helical propensity can reduce the propensity of proteins to form amyloid fibrils. To analyze in detail the involvement of different factors (charges, helical propensity, etc.) that could favor or inhibit the formation of amyloid fibrils by PI3-SH3, we designed an additional set of mutants (Fig. 1 and Table 1). The mutational variants were as follows. (i) E17R/D23R (PI3-RR), which incorporates two additional positive charges in the RT loop region of the molecule and is predicted to have a slightly enhanced helical propensity; (ii) Q7E/R9K/E17R/D23R (PI3-4X), which incorporates the same two additional charges incorporated in PI3-RR but also has a substantially enhanced helical propensity; (iii) E17Q/D23M/H25R (PI3-QMR), which has the same net charge at low pH as the wild-type protein but is predicted to have a significantly enhanced helical propensity; (iv) E17G/D23G (PI3-GG), which has again the same charge at low pH as the wild-type protein but is predicted to have a decreased helical propensity and has Gly substitutions that are incorporated to decrease the β-sheet propensity (40); and (v) His-tagged PI3-SH3 (HT-PI3), which has a wild-type PI3-SH3 domain with a His-tag attached to its N terminus and exhibits an increase in charge of +7, enabling the effect of perturbations by charges distantly located from the RT loop/diverging turn region to be probed.
Table 1. Aggregation properties exhibited by PI3-SH3 mutants.
| Mutant | Substitutions | Helix, % (pH 2.0)* | ΔCharge (pH 2.0) | Aggregated protein, %† | β-Structure FTIR‡ | Fibrils EM§ |
|---|---|---|---|---|---|---|
| PI3-SH3 | Wild-type domain | 2.77 | 0 | 92.3 ± 1.5 | + (β) | ++ |
| PI3-KK | H25K/L26K | 3.16 | +1 | n.t. | n.t. | − |
| PI3-RR | E17R/D23R | 4.21 | +2 | 1.5 ± 1.5 | − (r.c.) | − |
| PI3-4X | Q7E/R9K/E17R/D23R | 6.33 | +2 | 1.0 ± 0.4 | − (r.c.) | − |
| PI3-QMR | E17Q/D23M/H25R | 5.57 | 0 | 49.7 ± 1.6 | + (β) | + |
| PI3-GG | E17G/D23G | 1.91 | 0 | 2.4 ± 0.7 | − (r.c.) | − |
| HT-PI3 | His-tag in N terminus | 2.77 | +7 | 88.5 ± 0.6 | + (β) | ++ |
n.t., not tested.
The percentage of helix was estimated by using agadir and corresponds to residues 1-30 of the sequence under the following conditions: 278°K; ionic strength, 0.1 M; pH 2.0 (61-63).
The fraction of aggregated protein was determined as described in Materials and Methods. Protein concentrations were 1 mM, and the samples were incubated at pH 2.0 and 35°C for 10 days. No significant changes were observed at longer times of incubation.
FTIR, Fourier transform infrared spectroscopy. +, presence of characteristic β-sheet-related bands (β) appearing at 1610-1620 cm−1 as reported previously (16, 23). −, random-coil (r.c.) spectrum with a main broad band appearing at 1640 cm−1.
EM, electron microscopy. ++, abundant fibrils in samples. +, presence of fibrils in samples. −, absence of fibrils in samples.
Amyloid fibril formation by the wild-type and mutant PI3-SH3 sequences was evaluated by using techniques described above coupled with Fourier transform infrared spectroscopy as reported previously (16, 23). The amount of aggregated protein was determined by sample fractionation by using sedimentation at 300,000 × g (see Materials and Methods). PI3-RR and PI3-4X, both having an increase in net charge of +2 and enhanced helical propensities relative to the wild-type PI3-SH3 protein, remain soluble under conditions in which the latter forms amyloid fibrils; no significant amount of protein could be detected in the pelletable fraction of the samples, and no fibrils could be observed by electron microscopy (Table 1). In addition, the Fourier transform infrared spectra of these samples resemble those of disordered structures with bands having maxima close to 1640 cm-1 (see Table 1).
By contrast, the PI3-QMR mutant, which shows no variation in net charge but is predicted to have an enhanced helical propensity compared to the wild-type sequence, seems to aggregate into amyloid fibrils, although to a lesser extent than the wild-type protein, as revealed by sedimentation data (49.7% ± 1.6% of aggregated protein for PI3-QMR versus 92.3% ± 1.5% for the wild-type protein) (Table 1). By contrast, samples of the PI3-GG mutant, which has the same net charge as the wild-type protein and is predicted to have a slightly decreased helical propensity in its denatured state, showed no significant evidence for aggregation under these conditions (Table 1).
In addition, HT-PI3, in which seven additional charges have been placed at the N-terminal region of the protein by means of a His-tag, still aggregates after incubation at low pH (Table 1), showing that the simple addition of charges at the terminus of the protein does not interfere significantly with its amyloidogenic properties. Despite the ability of HT-PI3 to form amyloid fibrils, the presence of a His-tag does affect the morphology of the fibrils as observed in electron micrographs of wild-type PI3-SH3 and HT-PI3 samples, (see the supporting information). Thus, although the basic interactions involved in the early stages of aggregation appear largely unaffected by this His-tag, the detailed arrangement of protofilaments within the fibril seems to be significantly perturbed.
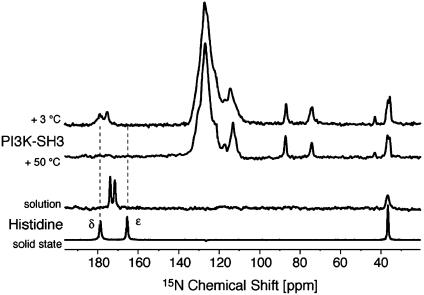
Solid-State NMR Spectra of PI3-SH3 Amyloid Fibrils. PI3-SH3 amyloid samples, prepared from uniformly 15N-labeled protein, were examined by 1D solid-state 15N NMR. At low temperatures, the spectra are broad, as can be seen most clearly in the resolved side-chain resonances of His (185.0 and 181.5 ppm), Arg (≈92.1, 79.8, and 48.9 ppm) and Lys (43.0 ppm) residues (Fig. 4). At higher temperatures the linewidths of the majority of the signals become narrower, although the resonances corresponding to His-25 (situated in the diverging turn), broaden again at a temperature of 30°C, disappearing above 50°C (Fig. 4). This behavior is fully reversible and suggests that that the environment surrounding His-25 is significantly different from that of the other side chains whose resonances can be observed. We have compared 15N chemical-shift values from histidine alone in solution and in a microcrystalline state (Fig. 4). The fact that both His-25 Nε and Nδ chemical shifts appear significantly downfield shifted in the 15N spectra of PI3-SH3 amyloid fibrils can be attributed (by analogy with the spectrum of microcrystalline histidine monohydrochloride) to the involvement of Hδ in an intermolecular hydrogen bond (41). Given that His-25 is exposed in the native structure of PI3-SH3 (42), these observations suggest its interaction with other residues within the amyloid structure, which is consistent with the conformational rearrangement of the PI3-SH3 domain required for amyloid formation (5, 15, 16).
Fig. 4.
Solid-state NMR and 15N NMR spectra of PI3-SH3. One-dimensional spectra of PI3-SH3 amyloid fibrils recorded at different temperatures are shown. The spectra are compared with that of 15N-histidine monohydrochloride in solution at low pH and in the crystalline state.
Discussion
Short Stretches of Amino Acids Modulate the Amyloidogenic Behavior of Globular Proteins. In previous studies we have shown that virtually the entire PI3-SH3 protein seems to be incorporated into the amyloid structure, which is relatively dense to electrons and highly resistant to proteinase digestion (15, 16, 23). The present observations, therefore, cannot be simply explained in terms of the involvement of the RT loop and adjacent diverging turn of PI3-SH3 in the formation of the core structure of the amyloid fibril, but it rather suggests that this particular region confers enhanced amyloidogenic properties to PI3-SH3 (Fig. 2). Moreover, these observations imply that a key stretch of amino acids can act as a particularly effective “mediator” or “facilitator” in the conversion of an entire protein into amyloid structures by expediting the formation of intermolecular assemblies and the incorporation of additional polypeptide regions into these complexes. Therefore, the removal or addition of such a region of the sequence has dramatic effects in the ability of a particular polypeptide to misfold into amyloid aggregates and, therefore, confirms the importance of specific residues in controlling the structural fate of a polypeptide chain.
Both amyloidogenic and nonamyloidogenic variants of PI3-SH3 are denatured at pH 2.0, indicating that the ability of this region to confer amyloidogenicity on PI3-SH3 is not simply related to changes in protein stability but rather results from the intrinsic ability of this region to promote aggregation. Preliminary results with octapeptides expanding this region support this conclusion (S. Ventura, unpublished results). The present work also confirms the involvement charge as a key factor modulating amyloid formation (14, 34). Moreover, mutations inhibiting aggregation PI3-SH3 aggregation (with the exception of E17G/D23G) also decrease the hydrophobicity at the site of the mutation, whereas those that increase the aggregation potential of SPC-SH3 cause the hydrophobicity of the corresponding region of the protein to increase. This observation supports the conclusion that, in addition to charge (14), the hydrophobicity of the polypeptide chain is a fundamental factor in the amyloidogenic potential of an unstructured polypeptide chain (33). Finally, mutations aimed at increasing helical propensity reduce aggregation, although to a lower extent. It is interesting to note that the observations made in the PI3-KK and the PI3-QMR mutants, both of which include substitutions affecting His-25 (in PI3-KK to Lys and in PI3-QMR to Arg), suggest that the presence of His-25 by itself is not sufficient to confer amyloidogenic properties on the PI3-SH3 protein. Rather, it is more likely that the charge and hydrophobic balance of the RT loop and diverging turn in the various nonamyloidogenic mutants could have a similar effect in preventing aggregation, even if it originates from substitutions in a variety of neighboring residues. From these results we can therefore conclude that the RT loop and diverging turn of PI3-SH3 are specific regions that influence its amyloidogenic behavior. Moreover, the alteration of the charge and hydrophobic balance in this particular region of the PI3-SH3 protein perturbs dramatically its ability to form amyloid fibrils, whereas modifications, insertions, or deletions at the N terminus (HT-PI3 mutant) or in the n-src loop of the protein, do not show such effects (22). The present study suggests that the propensity of a given protein to aggregate and form amyloid fibrils under certain conditions may, to a large extent, be determined by the properties of specific regions of its sequence. This observation is in agreement with data for both pathological and nonpathological polypeptides, which suggest the existence of “hotspots” for protein aggregation (24, 33, 43-49).
We have recently reported that a simple combination of factors that characterize the physicochemical properties of a given polypeptide sequence can rationalize to a very significant extent the changes observed in aggregation propensity that result from individual residue substitutions in unstructured peptides and proteins (50). Although this approach has not yet been developed in such detail for multiple amino acid substitutions as those included in this work, the application of the algorithm derived from the single mutations to the sequences used in this study qualitatively predicts a substantial decrease in aggregation rate for PI3-KK, PI3-RR, and PI3-4X mutants when compared with the wild-type PI3-SH3 sequence. This prediction is in good agreement with the experimental observations reported in this paper. The observed behavior of the PI3-GG mutant, however, cannot be rationalized on the same basis, which suggests that additional elements could interfere with the early stages of aggregation or with the packing of the polypeptide chain within the amyloid structure. The disruption of sequence patterns prone to aggregation might be the reason behind the observations made for this particular mutant. Indeed, the PI3-GG mutant interrupts the binary alternating polar/nonpolar pattern formed by residues 21-26, which some authors have shown to be relevant in defining the aggregation propensity exhibited by polypeptides (36, 37). Interestingly, the use of predictive algorithms (based on modifications of the above mentioned equations) (50) that include the effect of amino acid patterns on amyloid formation (51) unequivocally predict a dramatic decrease in the aggregation propensity exhibited by the PI3-GG mutant compared with that of PI3-SH3, which is in remarkable agreement with the experimental results.
Evolution of Polypeptide Sequences and Selection Against Amyloid Formation. Replacement of residues 22-27 in SPC-SH3 by those corresponding to the same region in the PI3-SH3 sequence increases dramatically the aggregation propensity exhibited by the former protein, suggesting that the residues in the diverging turn and RT loop of the SH3 domains could play a protective role against protein aggregation. Substitution of residues 25 and 26 of the PI3-SH3 sequence by those that are most highly represented in SH3 domains (lysine in both equivalent positions) significantly reduces the propensity of PI3-SH3 to aggregate, indicating that a significant degree of protection could occur from these two residues. These interpretations support the idea that proteins have evolved not only to maintain their required stability and functionality but also to avoid aggregation and amyloid formation (24, 34, 35, 52-57). This goal could be achieved by eliminating altogether the sequence patterns susceptible to promoting aggregation (53). Other strategies to prevent uncontrolled self-assembly in nature appear to include the sequestering of amyloidogenic sequences inside the globular core of the protein (33), the creation of structural or sequence motifs able to cap regions of the protein prone to aggregation (55), the stabilization of local interactions that might prevent aggregation-prone regions from forming intermolecular interactions (57), and the selection of residues able to silence aggregation through their insertion within aggregation-prone regions or in flanking segments (55, 58). In the case of the SH3 domain family, in the majority of the known sequences, positions 25 and 26 are occupied by positively charged residues, which we have shown are able to reduce substantially the amyloidogenic behavior exhibited by PI3-SH3.
Finally, the finding that short sequence stretches can generate high amyloidogenicity and mediate the conversion of an entire globular protein into amyloid fibrils has a number of interesting practical consequences. For example, knowledge of characteristics that promote or inhibit the amyloidogenic behavior of a given sequence will provide key information for the design of new or modified proteins having enhanced stability and resistance to aggregation (9, 14, 24, 33, 35). In addition, this observation suggests that the design of molecules able to “screen” or “cap” such aggregation-prone regions or even to promote intrachain interactions that might inhibit aggregation could be successfully exploited as a strategy for tackling deleterious disorders linked to protein deposition, such as Alzheimer's or Parkinson's disease (19, 24, 59).
Supplementary Material
Acknowledgments
We thank C. E. MacPhee, D. Hall, and O. S. Hen for valuable insights. This work was supported by the Ministerio de Ciencia y Tecnología, Spain, the Centre de Referència en Biotecnologia (Generalitat de Catalunya, Spain) and a Program Grant from the Wellcome Trust (to J.Z. and C.M.D.). S.V. is supported by a Ramón y Cajal project awarded by the Ministerio de Ciencia y Tecnología and cofinanced by the Universitat Autonoma de Barcelona. J.Z. acknowledges support from the Leverhulme Trust.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SH3, Src homology 3; PI3-SH3, SH3 domain of the p58α subunit of bovine phosphatidyl-inositol-3′-kinase; SPC-SH3, SH3 domain of chicken α-spectrin; HT-PI3, His-tagged PI3-SH3.
References
- 1.Tan, S. Y. & Pepys, M. B. (1994) Histopathology 25, 403-414. [DOI] [PubMed] [Google Scholar]
- 2.Kelly, J. W. (1998) Curr. Opin. Struct. Biol. 8, 101-106. [DOI] [PubMed] [Google Scholar]
- 3.Rochet, J. C. & Lansbury, P. T., Jr. (2000) Curr. Opin. Struct. Biol. 10, 60-68. [DOI] [PubMed] [Google Scholar]
- 4.Dobson, C. M. (2001) Philos. Trans. R. Soc. London 356, 133-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guijarro, J. I., Sunde, M., Jones, J. A., Campbell, I. D. & Dobson, C. M. (1998) Proc. Natl. Acad. Sci. USA 95, 4224-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litvinovich, S. V., Brew, S. A., Aota, S., Akiyama, S. K., Haudenschild, C. & Ingham, K. C. (1998) J. Mol. Biol. 280, 245-258. [DOI] [PubMed] [Google Scholar]
- 7.Chiti, F., Webster, P., Taddei, N., Clark, A., Stefani, M., Ramponi, G. & Dobson, C. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3590-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramirez-Alvarado, M., Merkel, J. S. & Regan, L. (2000) Proc. Natl. Acad. Sci. USA 97, 8979-8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villegas, V., Zurdo, J., Filimonov, V. V., Aviles, F. X., Dobson, C. M. & Serrano, L. (2000) Protein Sci. 9, 1700-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fandrich, M., Fletcher, M. A. & Dobson, C. (2001) Nature 410, 165-166. [DOI] [PubMed] [Google Scholar]
- 11.Dobson, C. M. (1999) Trends Biochem. Sci. 24, 329-332. [DOI] [PubMed] [Google Scholar]
- 12.Ellis, R. J. & Pinheiro, T. J. (2002) Nature 416, 483-484. [DOI] [PubMed] [Google Scholar]
- 13.Bucciantini, M., Giannoni, E., Chiti, F., Baroni, F., Formigli, L., Zurdo, J., Taddei, N., Ramponi, G., Dobson, C. M. & Stefani, M. (2002) Nature 416, 507-511. [DOI] [PubMed] [Google Scholar]
- 14.Lopez De La Paz, M., Goldie, K., Zurdo, J., Lacroix, E., Dobson, C. M., Hoenger, A. & Serrano, L. (2002) Proc. Natl. Acad. Sci. USA 99, 16052-16057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jimenez, J. L., Guijarro, J. I., Orlova, E., Zurdo, J., Dobson, C. M., Sunde, M. & Saibil, H. R. (1999) EMBO J. 18, 815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zurdo, J., Guijarro, J. I., Jimenez, J. L., Saibil, H. R. & Dobson, C. M. (2001) J. Mol. Biol. 311, 325-340. [DOI] [PubMed] [Google Scholar]
- 17.Haggqvist, B., Naslund, J., Sletten, K., Westermark, G. T., Mucchiano, G., Tjernberg, L. O., Nordstedt, C., Engstrom, U. & Westermark, P. (1999) Proc. Natl. Acad. Sci. USA 96, 8669-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picken, M. M. (2001) Arch. Pathol. Lab. Med. 125, 38-43. [DOI] [PubMed] [Google Scholar]
- 19.Sacchettini, J. C. & Kelly, J. W. (2002) Nat. Rev. Drug Discovery 1, 267-275. [DOI] [PubMed] [Google Scholar]
- 20.Sandilands, A., Hutcheson, A. M., Long, H. A., Prescott, A. R., Vrensen, G., Loster, J., Klopp, N., Lutz, R. B., Graw, J., Masaki, S., et al. (2002) EMBO J. 21, 6005-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kayed, R., Head, E., Thompson, J. L., McIntire, T. M., Milton, S. C., Cotman, C. W. & Glabe, C. G. (2003) Science 300, 486-489. [DOI] [PubMed] [Google Scholar]
- 22.Ventura, S., Lacroix, E. & Serrano, L. (2002) J. Mol. Biol. 322, 1147-1158. [DOI] [PubMed] [Google Scholar]
- 23.Zurdo, J., Guijarro, J. I. & Dobson, C. M. (2001) J. Am. Chem. Soc. 123, 8141-8142. [DOI] [PubMed] [Google Scholar]
- 24.Zurdo, J. (2004) Protein Pept. Lett., in press. [DOI] [PubMed]
- 25.Klunk, W. E., Pettegrew, J. W. & Abraham, D. J. (1989) J. Histochem. Cytochem. 37, 1273-1281. [DOI] [PubMed] [Google Scholar]
- 26.Grantcharova, V. P., Riddle, D. S. & Baker, D. (2000) Proc. Natl. Acad. Sci. USA 97, 7084-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gsponer, J. & Caflisch, A. (2002) Proc. Natl. Acad. Sci. USA 99, 6719-6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindorff-Larsen, K., Paci, E., Serrano, L., Dobson, C. M. & Vendruscolo, M. (2003) Biophys. J. 85, 1207-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz, J., Milpetz, F., Bork, P. & Ponting, C. P. (1998) Proc. Natl. Acad. Sci. USA 95, 5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ponting, C. P., Schultz, J., Milpetz, F. & Bork, P. (1999) Nucleic Acids Res. 27, 229-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larson, S. M. & Davidson, A. R. (2000) Protein Sci. 9, 2170-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larson, S. M. & Pande, V. S. (2003) J. Mol. Biol. 332, 275-286. [DOI] [PubMed] [Google Scholar]
- 33.Chiti, F., Taddei, N., Baroni, F., Capanni, C., Stefani, M., Ramponi, G. & Dobson, C. M. (2002) Nat. Struct. Biol. 9, 137-143. [DOI] [PubMed] [Google Scholar]
- 34.Chiti, F., Calamai, M., Taddei, N., Stefani, M., Ramponi, G. & Dobson, C. M. (2002) Proc. Natl. Acad. Sci. USA 99, Suppl. 4, 16419-16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez De La Paz, M. & Serrano, L. (2004) Proc. Natl. Acad. Sci. USA 101, 87-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West, M. W., Wang, W., Patterson, J., Mancias, J. D., Beasley, J. R. & Hecht, M. H. (1999) Proc. Natl. Acad. Sci. USA 96, 11211-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, W. & Hecht, M. H. (2002) Proc. Natl. Acad. Sci. USA 99, 2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prieto, J., Wilmans, M., Jimenez, M. A., Rico, M. & Serrano, L. (1997) J. Mol. Biol. 268, 760-778. [DOI] [PubMed] [Google Scholar]
- 39.Kallberg, Y., Gustafsson, M., Persson, B., Thyberg, J. & Johansson, J. (2001) J. Biol. Chem. 276, 12945-12950. [DOI] [PubMed] [Google Scholar]
- 40.Minor, D. L., Jr., & Kim, P. S. (1994) Nature 367, 660-663. [DOI] [PubMed] [Google Scholar]
- 41.Fuess, H., Hohlwein, D. & Mason, S. A. (1977) Acta Crystallogr. B33, 654-659. [Google Scholar]
- 42.Booker, G. W., Gout, I., Downing, A. K., Driscoll, P. C., Boyd, J., Waterfield, M. D. & Campbell, I. D. (1993) Cell 73, 813-822. [DOI] [PubMed] [Google Scholar]
- 43.Mulkerrin, M. G. & Wetzel, R. (1989) Biochemistry 28, 6556-6561. [DOI] [PubMed] [Google Scholar]
- 44.Wetzel, R. (1994) Trends Biotechnol. 12, 193-198. [DOI] [PubMed] [Google Scholar]
- 45.Betts, S., Haase-Pettingell, C. & King, J. (1997) Adv. Protein Chem. 50, 243-264. [DOI] [PubMed] [Google Scholar]
- 46.Tenidis, K., Waldner, M., Bernhagen, J., Fischle, W., Bergmann, M., Weber, M., Merkle, M. L., Voelter, W., Brunner, H. & Kapurniotu, A. (2000) J. Mol. Biol. 295, 1055-1071. [DOI] [PubMed] [Google Scholar]
- 47.von Bergen, M., Friedhoff, P., Biernat, J., Heberle, J., Mandelkow, E. M. & Mandelkow, E. (2000) Proc. Natl. Acad. Sci. USA 97, 5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wurth, C., Guimard, N. K. & Hecht, M. H. (2002) J. Mol. Biol. 319, 1279-1290. [DOI] [PubMed] [Google Scholar]
- 49.Kammerer, R. A., Kostrewa, D., Zurdo, J., Detken, A., Garcia-Echeverria, C., Green, J. D., Muller, S. A., Meier, B. H., Winkler, F. K., Dobson, C. M. & Steinmetz, M. O. (2004) Proc. Natl. Acad. Sci. USA 101, 4435-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiti, F., Stefani, M., Taddei, N., Ramponi, G. & Dobson, C. M. (2003) Nature 424, 805-808. [DOI] [PubMed] [Google Scholar]
- 51.Dubay, K. F., Chiti, F., Zurdo, J., Dobson, C. M. & Vendruscolo, M. (2004) J. Mol. Biol., in press. [DOI] [PubMed]
- 52.Wood, S. J., Wetzel, R., Martin, J. D. & Hurle, M. R. (1995) Biochemistry 34, 724-730. [DOI] [PubMed] [Google Scholar]
- 53.Broome, B. M. & Hecht, M. H. (2000) J. Mol. Biol. 296, 961-968. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz, R., Istrail, S. & King, J. (2001) Protein Sci. 10, 1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richardson, J. S. & Richardson, D. C. (2002) Proc. Natl. Acad. Sci. USA 99, 2754-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobson, C. M. (2002) Nature 418, 729-730. [DOI] [PubMed] [Google Scholar]
- 57.Tahiri-Alaoui, A., Bouchard, M., Zurdo, J. & James, W. (2003) Protein Sci. 12, 600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otzen, D. E., Kristensen, O. & Oliveberg, M. (2000) Proc. Natl. Acad. Sci. USA 97, 9907-9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobson, C. M. (2003) Nat. Rev. Drug Discovery 2, 154-160. [DOI] [PubMed] [Google Scholar]
- 60.Kabsch, W. & Sander, C. (1983) Biopolymers 22, 2577-2637. [DOI] [PubMed] [Google Scholar]
- 61.Munoz, V. & Serrano, L. (1994) Nat. Struct. Biol. 1, 399-409. [DOI] [PubMed] [Google Scholar]
- 62.Munoz, V. & Serrano, L. (1997) Biopolymers 41, 495-509. [DOI] [PubMed] [Google Scholar]
- 63.Lacroix, E., Viguera, A. R. & Serrano, L. (1998) J. Mol. Biol. 284, 173-191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.