Abstract
Rapid advances in next-generation sequencing technology are revolutionizing approaches to genomic and epigenomic studies of skin. Deep sequencing of cutaneous malignancies reveals heavily mutagenized genomes with large numbers of low-prevalence mutations and multiple resistance mechanisms to targeted therapies. Next-generation sequencing approaches have already paid rich dividends in identifying the genetic causes of dermatologic disease, both in heritable mutations and the somatic aberrations that underlie cutaneous mosaicism. Although epigenetic alterations clearly influence tumorigenesis, pluripotent stem cell biology, and epidermal cell lineage decisions, labor and cost-intensive approaches long delayed a genome-scale perspective. New insights into epigenomic mechanisms in skin disease should arise from the accelerating assessment of histone modification, DNA methylation, and related gene expression signatures.
INTRODUCTION
No longer merely rumblings of a far-off herd, the forerunners of the DNA sequencing revolution are upon us, trampling familiar benchmarks. For the past 15 years, genomics has referred primarily to experiments probing genes or transcripts using high-density arrays of oligonucleotides. These technologies recognize millions of prespecified differences in DNA sequence and RNA abundance. Although arrays may theoretically tile across gigabases (Bertone et al., 2006), commercially available versions generally interrogate less than 2% of the full informational content of a human genome. Somatic or germline mutations occur virtually throughout the genome, rendering these technologies of limited value in detecting new genetic differences. As a consequence, some of the most basic genetic questions had remained unresolved: e.g., whether most human cancers arise from a small number of common mutations or many combinations of rarer aberrations.
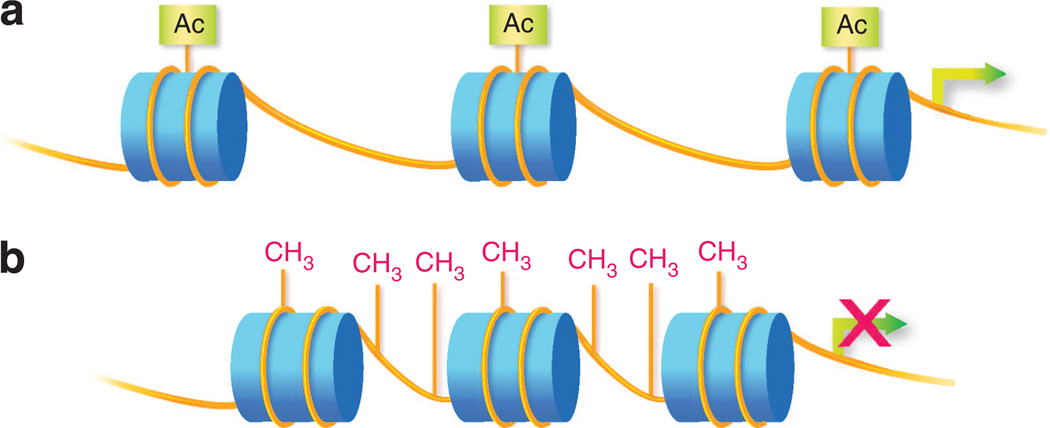
Heritable changes in gene expression may occur without alteration of DNA sequence, in the form of covalent histone and DNA modifications, collectively known as epigenetics. Histone proteins package DNA into nucleosomes and chromatin fibers. Chemical modifications to histones alter the recruitment of transcriptional machinery and modulate gene expression, a layer of regulation known as the “histone code” (Figure 1a; Jenuwein and Allis, 2001). DNA methylation also changes the binding and activity of transcriptional apparatus, often attenuating gene activity in skin and other tissues (Figure 1b). Until the past few years, genome-wide epigenetic studies relied heavily on microarray-based methods with limitations in coverage similar to that for mutational assessment (e.g., DNA methylation studies using arrays assessed less than 0.1% of cytosine phosphate guanine (CpG) dinucleotide methylation sites).
Figure 1. Epigenetic modifications.
(a) Histone modification. Histone proteins (depicted as blue cylinders) spool DNA (depicted as yellow lines) to form nucleosomes and chromatin. Posttranslational modifications (e.g., addition of acetyl, methyl, or ubiqituin groups) to the N-terminal tail regions of histones alter local chromatin conformation. Variability in chromatin condensation affects the accessibility of genes. Loosely condensed regions (euchromatin) are more actively expressed and tightly condensed regions (heterochromatin) are repressed. Depicted in the figure is histone acetylation, associated with loosening of local chromatin and more active gene expression. (b) DNA methylation. DNA methylation occurs through the addition of a methyl group to the C5 position of cytosine to form 5-methylcytosine, typically at cytosine phosphate guanine (CpG) dinucleotides. In promoter regions, DNA methylation silences genes by interfering with transcription factor binding and/or recruitment of methyl-CpG-binding proteins that recruit repressor complexes. DNA methylation is typically associated with tightly condensed chromatin.
With rapid advances in chemistries and imaging technology, the so-called “next-generation sequencing” methodologies (Ansorge, 2009) have reduced the expense in identifying all 6 billion nucleotides in a diploid genome to <$4,000, quite recently the bill for a few dozen genes (Figure 2a). Costs of high-resolution assessment of DNA methylation and histone modification have dropped proportionately. Bisulfite conversion of unmethylated cytosines to uracil before sequencing yields true single-nucleotide CpG resolution for <$4,000. Techniques combining enrichment of methylated genomic DNA by immunoprecipitation and methylation-sensitive enzyme digestion, followed by next-generation sequencing, interrogate ~78% of genomic CpGs for <$1,500 (Harris et al., 2010). Similarly, immunoprecipitation of chromatin DNA bound to specific histone modifications, characterized by next-generation sequencing (chromatin immunoprecipitation–sequencing), maps these marks to the genome for as little as $500. In an era of cheap analytics, the accessibility of cutaneous tissues represents a distinct advantage. Skin cancers represent a classic model for somatic mutation, as do rashes for epigenetic alteration. However, bargain whole-genome analyses tempt us to inspect and challenge even such basic classifications.
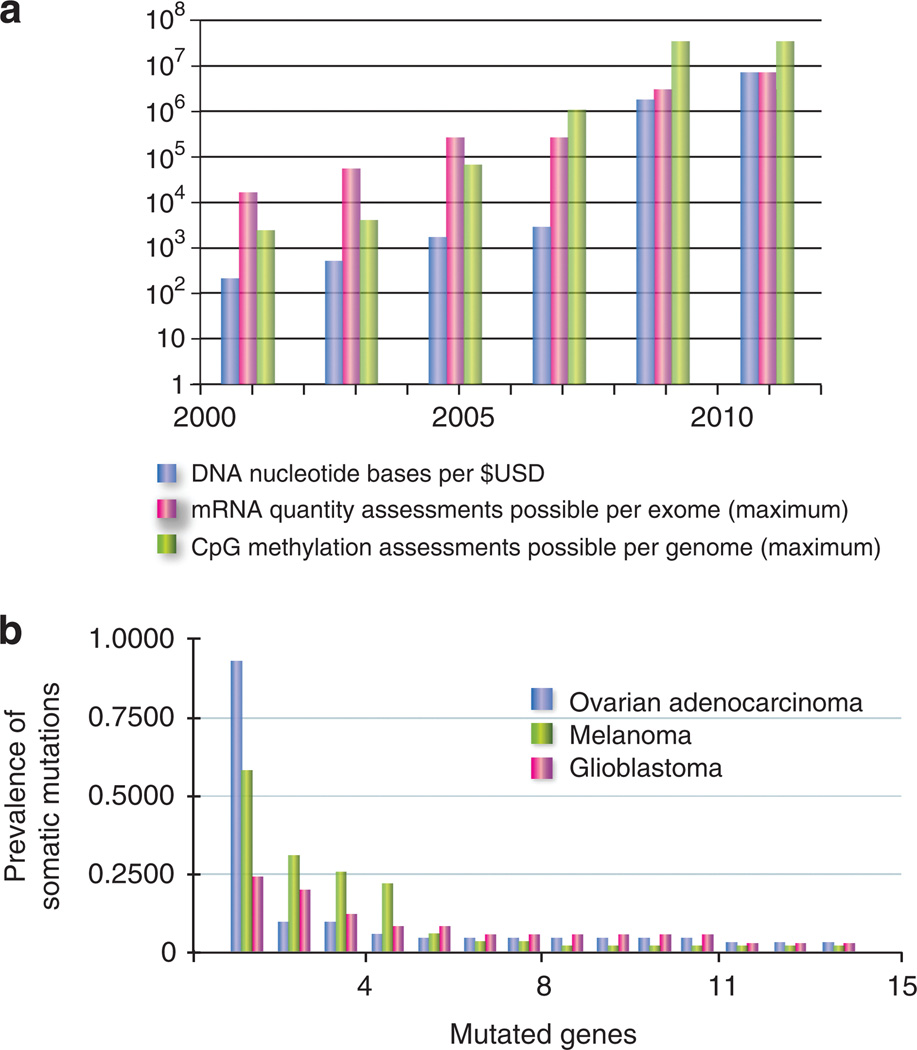
Figure 2. Rapidly expanding sequence profiles of cancer.
(a) For DNA, graph depicts the number of sequenced nucleotides per $1 USD (NHGRI 2011 data sheet, www.genome.gov/sequencingcosts/). For gene expression quantification, the number of discrete assessments across all coding regions is graphed. From 2001 to 2005, the standard Affymetrix (Santa Clara, CA) platform primarily assessed expression at the 3′ ends of transcripts, reaching a maximum of B50,000 transcripts and variants per array. By 2005, so-called ‘‘exon arrays’’ were introduced, carrying probes for each individual exon, interrogating expression of more than 250,000 exons. From 2009 to present, RNA quantification has increasingly been performed using sequencing (RNAseq), approximating all >6 million coding nucleotides in the human genome. For cytosine phosphate guanine (CpG) methylation, the primary advances represent new technologies making denser assessment feasible. Individual platforms, techniques, and genomic CpG coverage are reviewed in Laird (2010) and Fouse et al. (2010). (b) Histogram of genetic heterogeneity in cancer. On the basis of recent sequencing studies, for each cancer type, a histogram of the most commonly mutated genes are arrayed from most to least frequent (The Cancer Genome Atlas Research Network, 2008, 2011; Wei et al., 2011). All non-synonymous mutations per gene were binned; copy number changes are not displayed.
CANCER
Even curable cancers, defying fully predictable response to therapy, suggest highly individualized genetic etiology. In the age of Sanger sequencing, before 2008, fewer than 300 genes were reported to harbor mutations in any cancer (Futreal et al., 2004). In the past 3 years, more than 1,000 tumors representing about 50 classes of malignancy have been sequenced at large scale—either at the level of the whole genome, or simply for all known genes (about 50–70 million bases, or 2–3% of the genome). The catalog has thus grown by an order of magnitude (Stratton, 2011). To say a particular primary cancer has been “deep sequenced” now generally implies sufficient redundancy that changes at more than 90% of inspected nucleotides, and present in 10% or more cells of the population, can be detected reliably.
Tumor samples test the limitations of deep sequencing. As with comparative genomic hybridization and other sequencing approaches, contamination with stromal or inflammatory cells rapidly dampens signal, necessitating expensive over-sampling to detect mutations (Thomas et al., 2006). Read lengths are short, generally less than 150 bp. Therefore, multiple mutations in a single gene cannot be ascribed to the same or different chromosomes without subcloning large fragments. Short reads also make it difficult to distinguish genuine mutation in stromal cells from those in the malignancy. Any amplification approach targeting single or very small cell populations is hindered by diminished representation and allele loss (Frumkin et al., 2008).
Nevertheless, given decreasing costs, hundreds of thousands of tumors, as well as matching normal tissue for comparison, may be comprehensively sequenced by 2020. Most existing data originated from large collaborative efforts such as the International Cancer Genome Consortium (Hudson et al., 2010) and the Cancer Genome Atlas Project, but improving affordability has opened access to individual laboratories. As maturing chemistries and solid-state technology for sequencing (Mardis and Wilson, 2009; Metzker, 2010) and analytical approaches to mutation detection (Meyerson et al., 2010) are discussed elsewhere, we focus on takeaways for the cutaneous oncologist.
Somatic heterogeneity
Although some cancers appear genetically homogenous, generated from a few predominant aberrations, many appear driven by hundreds or even thousands of rare combinations of mutations. The latter profile—several highly prevalent tumor suppressors or oncogenes, but also a long “tail” of less frequent somatic changes—typifies cancers as diverse as pancreatic adenocarcinoma (Jones et al., 2008), glioblastoma (The Cancer Genome Atlas Research Network, 2008), and cutaneous melanoma (Wei et al., 2011) (Figure 2b).
Beyond the BRAF oncogene activated in more than half of primary melanomas arising in sun-damaged skin, sequencing and copy number analysis have revealed the ERBB4 and c-KIT signaling receptors as amplified or mutated in 20–30% of primary cancers (Prickett et al., 2009; Flaherty et al., 2010), as well as NRAS, at similar proportions (Hocker et al., 2008). For cancers harboring these activated oncogenes, targeted drugs, such as the Braf inhibitor vemurafenib and c-kit inhibitor imatinib, have produced striking instances of clinical response and extended survival (Carvajal et al., 2011; Chapman et al., 2011). Some novel, low-prevalence mutations, which may serve as driver oncogenes, appear to recur in melanoma, such as GRIN2A (glutamate receptor, ionotropic, N-methyl d10-aspartate 2A) (~5%; Wei et al., 2011), FLT1 (fms-like tyrosine kinase 1), and PTK2B (protein tyrosine kinase 2β) (~10%; Prickett et al., 2009). Outside these groups, no well-characterized activating mutation yet shows a prevalence greater than 5%. Similarly, initial exome sequencing of cutaneous squamous cell carcinomas has failed to identify recurrent oncogenic aberrations, even those, such as in Ras proteins, that commonly produce related cancers in mouse models (Kemp et al., 1993; Durinck et al., 2011).
Both melanoma and cutaneous squamous cell carcinomas tolerate extraordinarily mutated genomes, in cases approaching 100,000 individual base changes per cancer (Pleasance et al., 2010; Durinck et al., 2011). This frequency eclipses the several hundred (or fewer) aberrations typically found in solid cancers. Such high levels of somatic mutations, mainly generated by ultraviolet radiation, produce legions of socalled “passenger” changes, obscuring those actually contributing to malignancy. At observed mutation rates, 5% of average-sized genes (with ~1,100 base pairs of coding sequence) carry a passenger substitution altering amino acid sequence. Without very large sample sizes, or recurrence at structurally significant amino acids, low-prevalence driver candidates in skin require substantive functional validation.
Once understood, the biology of rare, recurrent aberrations should influence the development of rational therapies. In the case that dozens of distinct cellular pathways can each drive malignancy, although infrequently, extending traditional targeting strategies might take decades. Such a scenario would be complicated by lower economic incentive and patient advocacy for unusual genetic origins of cancer. On the other hand, if many somatic aberrations represent different points of activation for a few core signaling pathways, analytical methods detecting such convergence (Akavia et al., 2010) will prove critical in directing treatment. The recurrent upregulation (but not mutation) of proteins such as Polo-like 1 kinase in cutaneous squamous cell carcinomas may represent targetable instances of such common activated effector genes (Watt et al., 2011).
The high mutation rate in skin cancers may also generate synergistic activation of more oncogenic pathways than found in a corresponding visceral tumor. For example, disabling mutations in Notch receptors reach 75% prevalence in cutaneous squamous cell carcinomas (Wang et al., 2011), but occur in fewer than 30% of squamous cell carcinomas of the head, neck, and lung (Agrawal et al., 2011; Stransky et al., 2011). The cause of this disparity remains unknown, but we speculate that highly mutagenized cancers may activate additional pathways requiring combination strategies in targeted therapies.
Drug resistance
Large-scale cancer sequencing raises expectations for circumventing acquired drug resistance. The first targeted inhibitor of the oncogenic hedgehog signaling pathway, vismodegib, binds and inactivates the G protein–coupled receptor and oncogene Smoothened (Robarge et al., 2009). Multiple cancer types activate this pathway, including basal cell cancers, pancreatic cancers, and medulloblastoma (Theunissen and de Sauvage, 2009). Unfortunately, more than 70% of patients treated with systemic vismodegib develop resistance after 6 months. Comprehensive sequencing of hedgehog signaling proteins in these resistant cancers detected new mutations in Smoothened that reduce drug binding (Yauch et al., 2009).
Similarly, Braf inhibitors rapidly induce responses in even advanced BRAF-mutant melanomas, but resistance commonly (>50% of patients) presents within 6 months of initiation (Chapman et al., 2011). Rather than mutation of the drug target, as seen in receptors such as Smoothened or EGFR, vemurafenib-resistant melanomas appear to activate parallel signaling pathways, via upregulation of the plateletderived growth factor receptor-β, or reactivate MEK through CRAF and oncogenic Ras (Heidorn et al., 2010), MAP3K8 (Johannessen et al., 2010), or gain-of-function mutations in NRAS (Nazarian et al., 2010). The malignancy rapidly adopts new machinery to drive growth.
For many cancers, especially of the skin, tumor samples can be easily accessed before and after treatment. In vemurafenib-resistant melanoma, BRAF was exhaustively sequenced to rule out the possibility of new mutations (Nazarian et al., 2010). Profiling of mRNA transcript levels—now possible through sequencing-based quantification (RNA-seq)—distinguished two distinct forms of resistance. Finally, comprehensive sequencing of defined, candidate oncogenes identified the new mutations in NRAS. These steps, once the product of months of labor, can be completed in only a few weeks using deep sequencing.
As the number of targeted drugs grows, these analytical capabilities anticipate personalized second-line therapies for treatment-resistant melanomas, non-melanoma skin cancers, and cutaneous sarcomas. Crossover strategies have already shown measurable benefits for sorafenib directed at nonsmall cell lung cancers bearing activated forms of KRAS (Kim et al., 2011).
Epigenomics
Cancers show hypermethylation of CpG-rich regions (socalled “CpG islands”) in promoter regions of tumor suppressor genes, in a context of global DNA hypomethylation. Promoter hypermethylation effectively substitutes for mutational gene inactivation, e.g., leading to loss of tumor suppressor gene expression (Nakhasi et al., 1981). In contrast, hypomethylation may lead to oncogene activation, genomic instability and chromosomal rearrangements, and/or loss of parental imprinting (Feinberg et al., 2006). In the Sézary syndrome variant of cutaneous T-cell lymphoma, loss of Fas receptor expression, which in turn suppresses T-cell apoptosis, is believed pathogenic (Contassot and French, 2010). Recent reports suggest DNA hypermethylation as the most common cause of FAS inactivation, more so than gene mutation (Jones et al., 2010). In melanoma, at least 90 genes display aberrant DNA methylation, including genes whose loss-of-function is well established in melanoma pathogenesis, such as CDKN2A and PTEN (Sigalotti et al., 2010).
Similarly, dysregulation of histone modifications can enhance malignancy, presumably through perturbation of local chromatin structure, thereby altering gene expression. Among the more well-studied modifications are histone 3 lysine 4 trimethylation (H3K4me3, enriched at the transcription start sites of actively expressed genes; Santos-Rosa et al., 2002) and histone 3 lysine 27 trimethylation (H3K27me3, associated with gene silencing; Boyer et al., 2006). Gene rearrangement of MLL (mixed-lineage leukemia), a H3K4 methyltransferase, frequently occurs in leukemias. Mice engineered to express partially duplicated mixed-lineage leukemia show increased H3K4me3 levels and associated overexpression of leukemia-associated genes (Dorrance et al., 2006). Melanomas often overexpress EZH2 (enhancer of zeste homolog 2), a H3K27 methyltransferase (Bachmann et al., 2006), as well as SETDB1 (SET domain, bifurcated 1), a H3K9 methyltransferase. SETDB1, when overexpressed in combination with mutant Braf in zebrafish, accelerates melanoma development, although the end effector genes are not yet clear (Ceol et al., 2011).
DNA methylation profiles may provide biomarkers for detection of and prognostication in cancers. In dermatopathology, the discovery of epigenetic markers or profiles may augment the diagnostic capabilities of techniques currently in use (e.g., immunohistochemistry, comparative genomic hybridization, or fluorescence in situ hybridization). Recently, a multi-locus methylation profile discriminated nevi from melanomas (Conway et al., 2011). Gene hypermethylation may also refine prognosis (e.g., PTEN is hypermethylated in 60% of melanomas and associated with worse patient survival (Lahtz et al., 2009)).
In other instances, epigenetic changes predict treatment response. Hypermethylation and loss of expression of MGMT (O-6-methylguanine-DNA methyltransferase; a DNA repair enzyme) in glioblastoma multiforme (Costello et al., 1994) correlates with enhanced survival from alkylating agent chemotherapy (Hegi et al., 2005). Unfortunately, MGMT status in metastatic melanoma does not predict response to the commonly used alkylating agent, dacarbazine (Hassel et al., 2010).
Finally, modulating epigenetic activity can treat disease. Currently, the histone deacetylase inhibitors vorinostat and romidepsin are Food and Drug Administration approved for treatment of cutaneous T-cell lymphomas. Similarly, azacitidine and decitabine (DNA methyltransferase inhibitors) are approved for hematological malignancies (Boumber and Issa, 2011). The mechanisms underlying these agents are not understood completely. For example, histone deacetylase inhibitors not only block histone deacetylases but also increase acetylation of other structural proteins and transcription factors (Lane and Chabner, 2009).
Earlier, low-resolution studies of a mouse skin carcinogenesis model show global changes in epigenetic marks with early and progressive accumulation of DNA hypomethylation and loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 (Fraga et al., 2004, 2005). In addition, hypermethylation of tumor suppressor genes (e.g., MLH1, mutL homolog 1), VHL (Von Hippel–Lindau), and p16INK4a) often occurs early in tumorigenesis (Jones and Baylin, 2002). The mechanisms underlying both these specific and more global DNA methylation and histone modification alterations remain unclear (Feinberg and Tycko, 2004). The next 5 years should mark completion of many high-resolution epigenomes and genomes from normal, premalignant, and malignant tissues. These data should help determine which alterations affect disease and whether they result from mutations in epigenetic machinery, develop sporadically under selective pressure, and/or are secondary to other processes in tumorigenesis.
GENETIC DISEASE
Somatic mosaicism
Dermatologists are practiced skeptics of the dogma “one genome per individual”. Genetic diversity within individuals reveals itself routinely in skin: the mosaic presentations of X-linked conditions such as incontinentia pigmenti and McCune-Albright disease (Rieger et al., 1994); localized lesions of neurofibromatosis or Darier disease (Kehrer-Sawatzki and Cooper, 2008; Harboe et al., 2011); and even the abundant nevi harboring BRAF mutations (Lin et al., 2011). Most of these examples result from single-nucleotide changes. Simple mutation, given its potential to finely modify any gene at any position, may represent the most common source of somatic mosaicism. Indeed, deep sequencing has recently revealed that nucleotide substitutions generate the striking segmental presentation of the Proteus syndrome (Lindhurst et al., 2011).
Recently, cheaper sequencing identified a novel mechanism of somatic diversity. Ichthyosis en confetti, a rare defect in keratinization, is distinguished by macules of apparently normal skin. These islands represent true reversion of mutations in the keratin 10 gene through mitotic recombination of chromosome 17 (Choate et al., 2010). Such copy-conservative chromosomal duplication is well established, known as “isoparental disomy” in heritable genetics and “copy-neutral loss-of-heterozygosity” in cancer, but this discovery reveals an assiduous process in normal adult cells. Such somatic instability appears at least partially position specific (as in chromosome 17), as none of the other keratin-based ichthyosiform disorders demonstrate such common reversion. Sequence-dependent mechanisms that may underlie such chromosomal aberration have recently been proposed (De and Michor, 2011).
Chromosomal duplication conferring reversion may also correct inherited blistering disorders, albeit only focally. Some of these patches of reversion result from the types of chromosomal displacement described in ichthyosis en confetti, erasing dominant alleles encoding structural defects (Jonkman et al., 1997; Pasmooij et al., 2005). Surprisingly, single-nucleotide insertions and deletions may also, on occasion, precisely edit inherited mutations in collagens and laminins (Jonkman et al., 2003; McGrath, 2004).
These diverse examples imply that occult, localized genetic variation probably both enhances and ameliorates human pathology constantly (Cho, 2010). Detecting small populations of mutant cells demands the screening of exomes for aberrant populations less than 1 in 10,000 affordably and reliably. Continued gains in ultra-deep sequencing efficiency may surpass this threshold in the next 10 years.
Inherited disease
Traditional genome-wide association studies (GWAS) profile genetic polymorphism in individuals with and without heritable conditions such as psoriasis. Some of these single-nucleotide polymorphisms recurrently associate with disease; the chromosomal neighborhood of these markers is then scoured for potential causative genes. During the past 10 years, the improved ability to type markers and organize large-scale studies of affected populations have greatly expanded candidate loci contributing to common diseases such as diabetes, cardiovascular disease, and psoriasis (Stranger et al., 2011). However, many GWAS identified loci have not been linked definitively to causative genes, as markers often flank large regions spanning many genes. New, population-based approaches linking genotype to phenotype may accelerate the functional annotation and validation of such candidates (Dendrou et al., 2009).
In psoriasis, a large number of studies have confirmed the immunological loci HLAA, HLAB, and HLAC as the genomic regions most strongly associated with classic psoriasis vulgaris (Nair et al., 2006; Liu et al., 2008). Remarkably, it appears that the greatest genetic contributions to a common disease were identified decades in advance of unbiased genome-wide screening (Rimbaud et al., 1974). However, genome-wide association studies have expanded the list of candidate effectors to polymorphisms in the IL12/23 complex subunits (targets of highly specific new targeted drugs for psoriasis such as ustekinimab; Cargill et al., 2007; Zhang et al., 2009) and also new pathways such as defensinmediated immunity (Hollox et al., 2008) whose clinical significance is not yet clear.
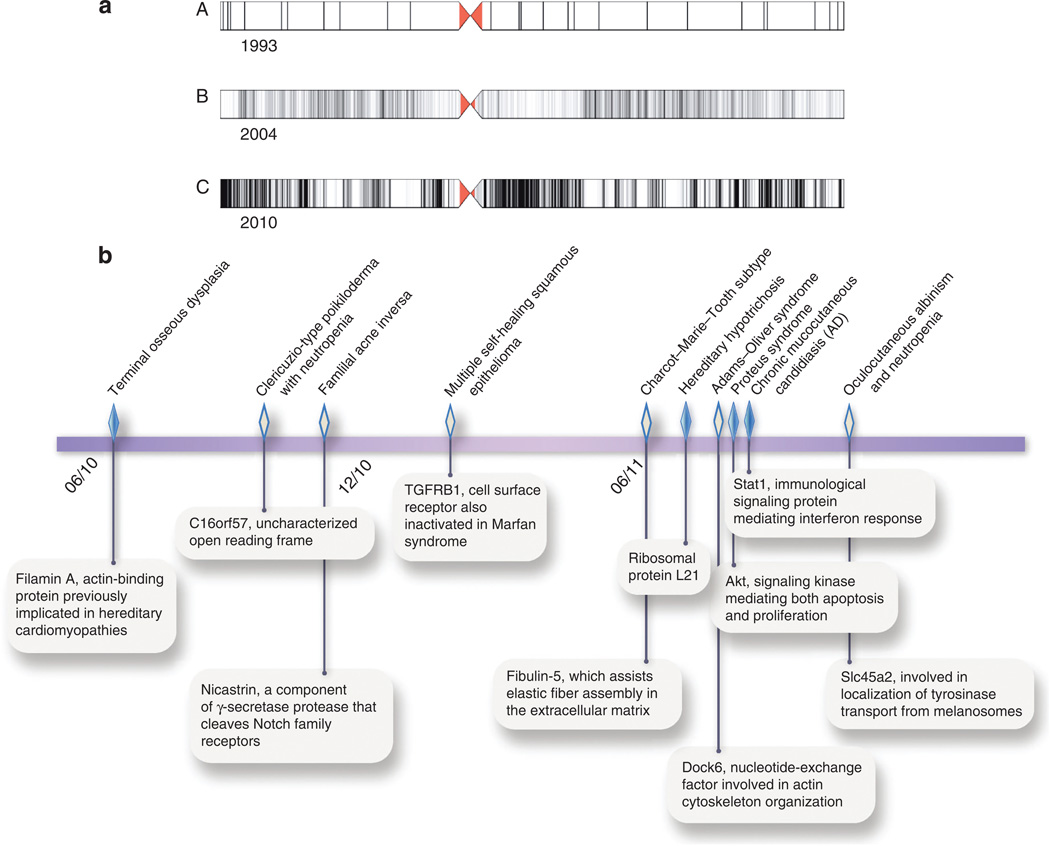
Exome sequencing seems poised to supplant array-based genome-wide association studies (Figure 3a). Deep sequencing retrieves not only portions of chromosomes associated with disease but also those nucleotide variants potentially altering protein sequence. If samples reveal particularly consequential changes to a gene, a small number of affected individuals may suffice to identify the causative gene. In only the past 12 months, exome sequencing has been used to identify novel causative mutations for conditions as diverse as oculocutaneous albinism with neutropenia (Cullinane et al., 2011), familial hypotrichosis (Zhou et al., 2011a), and hidradenitis suppurativa (Wang et al., 2010), often from limited kindreds (Figure 3b).
Figure 3. Accelerating gains in correlating genotype to phenotype.
(a) For chromosome 11, distribution of interrogated DNA sequence displayed for (A) microsatellite markers (1993), (B) single-nucleotide polymorphisms on Affymetrix 100K genotypic oligonucleotide array (2004), and (C) whole exome (2010). (b) Genetic bases of recent cutaneous phenotypes identified by exome sequencing Affymetrix, USA (Sun et al., 2010; Concolino et al., 2010; Wang et al., 2010; Goudie et al., 2011; Auer-Grumbach et al., 2011; Zhou et al., 2011a; Shaheen et al., 2011; Lindhurst et al., 2011; Liu et al., 2011; Cullinane et al., 2011). Closed diamonds show activating mutations; open diamonds represent loss-of-function mutations.
EPIGENETIC CONTROL OF CELL FATE
Although an individual’s stem cells are essentially genetically identical, regardless of tissue of origin, epigenetic mechanisms critically maintain the stem cell state and direct subsequent differentiation. Embryonic stem (ES) cells, derived from the inner cell mass of the blastocyst, self-renew indefinitely, and as pluripotent cells, give rise to all cell types of the body. Thus, these populations offer unique insight into early developmental biology and suggest possible therapeutic cell replacement. ES cells harbor such characteristic epigenetic features as “bivalent domains” in promoters of early developmental genes, where both activating H3K4me3 and repressive H3K27me3 histone modifications reside. Consequently, these genes may lie silent but responsive to developmental stimuli (Mikkelsen et al., 2007).
With cell fate determination, increased expression of lineage-specific genes associates with loss of H3K27me3 marks, whereas genes for other lineages are not expressed and maintain their bivalent marks, or lose H3K4me3 at their promoters (Mikkelsen et al., 2007; Pan et al., 2007). Similar mechanisms influence the maintenance and differentiation of skin progenitor cells. In proliferating human keratinocytes, H3K27me3 is enriched at the promoters of and presumably represses transcription of epidermal differentiation genes. During differentiation, a subset of these genes (e.g., keratin 1 and involucrin) lose repressive H3K27me3 marks with upregulation of their gene products (Sen et al., 2008).
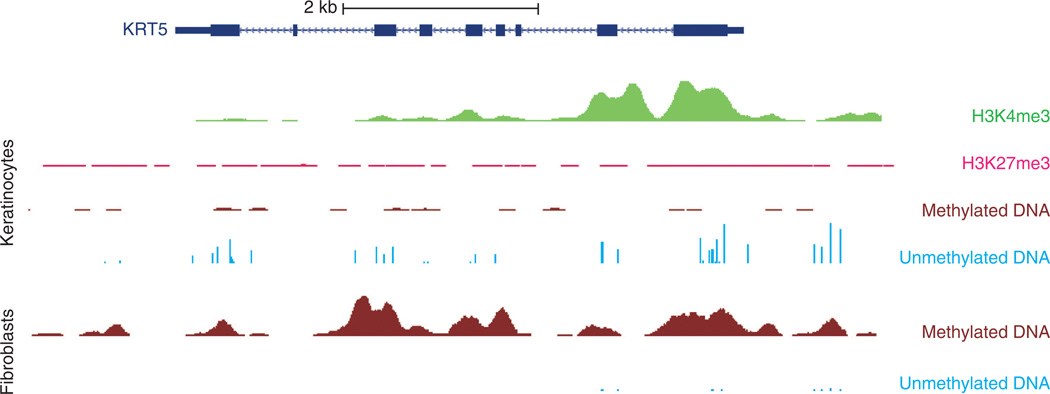
DNA methylation patterns also associate with differing states of cellular identity. In ES cells, genes associated with pluripotency tend to be hypomethylated, whereas those associated with differentiation tend to be hypermethylated and inactive (Huang and Fan, 2010). For somatic progenitor and stem cells, DNA methylation appears required for maintenance of cell renewal. Depletion of DNA methyltransferase-1 (an enzyme that maintains DNA methylation marks) in human epidermal progenitor cells leads to cell cycle arrest, premature differentiation, and loss of self-renewal. These changes correspond with loss of promoter DNA methylation at epidermal differentiation genes with differentiation (Sen et al., 2010). During differentiation, numerous genetic loci undergo changes in methylation, likely promoting lineage commitment by repressing genes related to other lineages and leaving lineage-specific genes hypomethylated and expressed (Huang and Fan, 2010). For example, during hematopoietic myeloid cell specification, genes important for myeloid development such as GADD45 α (growth arrest and DNA-damage-inducible α) and myeloperoxidase become hypomethylated and upregulated (Ji et al., 2010)). In skin cells, primary cultured KCs grown under low calcium basal conditions show DNA hypomethylation of the keratin 5 locus, whereas primary cultured fibroblasts show hypermethylation (Figure 4).
Figure 4. Histone modification and DNA methylation profile of keratin 5 (KRT5).
University of California, Santa Cruz (UCSC) genome browser snapshot encompassing an ~9 Kb segment of DNA spanning the KRT5 locus. The dark blue rectangles and lines depict the KRT5 gene structure. Profiled cells are primary cultured neonatal foreskin keratinocytes (KCs, top) and fibroblasts (bottom). These KCs express KRT5, whereas fibroblasts do not. Starting from the top, there are high levels of H3K4me3 histone modification signal (associated with active promoters) at the promoter in KCs (depicted in green). In pink, H3K27me3 signal (associated with gene repression) shows low levels in KCs. Depicted in brown are regions of methylated DNA (assessed by methylated DNA immunoprecipitation sequencing), with minimal signal in KCs and high levels in fibroblasts. Unmethylated DNA (assessed by methylation-sensitive restriction enzyme sequencing) is depicted with light blue vertical bars and shows a high number of sequencing peaks for KCs compared with a low number for fibroblasts. These data and other reference epigenomes are publicly available from the NIH Roadmap Epigenome Project website (http://vizhub.wustl.edu; Zhou et al., 2011b).
Given their similar roles in regulating gene expression programs, the significant interplay between histone modifications and DNA methylation is not surprising. Histone modifications appear more dynamic. DNA methylation functions as a more stable regulatory mechanism to lock in these expression programs (Cedar and Bergman, 2009). Additional layers of regulation are evident; e.g., the long noncoding RNA HOTAIR interacts with the Polycomb repressive complex 2 (which mediates histone H3 lysine 27 methylation) to help specify expression programs associated with fibroblast anatomic positional identity (Rinn et al., 2007).
Induced pluripotent stem cells (iPSCs) illustrate the importance of epigenetic mechanisms in maintaining cellular state, as somatic cells can be epigenetically reprogrammed back to an embryonic cell–like state through the expression of certain transcription factors (e.g., Oct 3/4, Sox2, krueppel-like factor 4, and c-Myc; Wu and Hochedlinger, 2011). Induced pluripotent stem cells embody features of true ES cells such as morphology, stem cell gene expression, and ability to differentiate into many cell lineages, but circumvent ethical issues associated with ES cells. Their potential clinical benefits include use in tissue regeneration, disease modeling, and gene therapy (Galach and Utikal, 2011). Cutaneous diseases may be among the earliest to benefit from the use of iPSCs through autologous replacement of diseased skin with genetically corrected skin. Mutations in the collagen VII gene cause recessive dystrophic epidermolysis bullosa, a cutaneous blistering disease with devastating clinical consequences. Recent reports of iPSC generation from these patients, differentiation into KCs and skin-like structures, and reengineering of these KCs to express functional collagen VII provide an early proof of principle (Itoh et al., 2011; Tolar et al., 2011).
Despite the great potential of iPSCs, similarity to ES cells may not be complete. Initial maps show hundreds of differentially methylated DNA regions between ES cells and iPSCs, which may partly represent an epigenetic memory from the original somatic cell type, with incomplete DNA methylation reprogramming (Lister et al., 2011). Observed, subtle differences in gene expression may share a similar cause (Ohi et al., 2011; Ghosh et al., 2010). These residual epigenetic marks may explain why low-passage iPSCs preferentially differentiate into lineages related to their original somatic cell type and restrict alternative cell lineages (Kim et al., 2010; Polo et al., 2010).
EPIGENETICS ON A GENOME SCALE
Different cell types, individuals, and disease states develop unique epigenomes. Methodological limitations of older DNA methylation studies constrained analysis to CpG islands and promoter regions, largely ignoring the remainder of the genome. Deep sequencing appears to be the likeliest path to expanding the catalog of differences distinguishing cell types (e.g., KCs and fibroblasts) and elucidating their significance. A number of collaborative efforts (e.g., the International Human Epigenome Consortium, Encyclopedia of DNA Elements (Encode), Blueprint and NIH Roadmap Epigenomics projects) seek to bring hundreds of such “reference epigenomes” to the community during the next 10 years.
This high-resolution cartography of genome-wide DNA methylation in normal tissue and tumors has begun. Early, surprising findings include underappreciated rates of non-CpG methylation (Lister et al., 2009), the intragenic and intergenic addresses (rather than 5′ promoter regions) of most differentially methylated CpG islands (Maunakea et al., 2010), and the fact that CpG island shores (low CpG density regions that lie up to 2 Kb from CpG islands) show highly variant methylation. The significant overlap of differentially methylated CpG island shore locations between different tissue types and normal tissue and cancers suggests dysregulated tissue-specific DNA methylation mechanisms in malignancy (Irizarry et al., 2009).
DNA methylation may not correlate only with decreased expression. Gene body methylation appears to regulate cell type–specific alternative promoters/transcripts and can increase gene expression (Laurent et al., 2010; Maunakea et al., 2010). New gene expression profiling techniques such as RNA-seq assess these tissue-specific alternative transcripts (observed in ~98% of multi-exon genes) and non-annotated regions at far higher resolution (Wang et al., 2008).
Genome-wide histone modification maps based on ChIP-seq should further dissect the machinery determining these marks and their regulatory roles in different cell types and development (e.g., H3K4me1 associates with active and poised enhancers, H3K4me3 with active promoters, and H3K36me3 with actively transcribed gene bodies). Given the complex interplay between DNA methylation, histone modification, and subsequent gene regulation, we anticipate discovery of new, coordinated changes in epigenetic patterns that drive cutaneous tumorigenesis, maintenance of stem cell renewal, and cell fate decision.
ACKNOWLEDGMENTS
We thank Zachary Sanborn for assistance with generating chromosomal illustrations. Both JBC and RJC are supported by the Career Development Awards from the Dermatology Foundation. RJC is also supported as a Samsung Biotechnology Scholar-in-Residence.
Abbreviations
- CpG
cytosine phosphate guanine
- ES
embryonic stem
- iPSC
induced pluripotent stem cell
- KC
keratinocyte
- KRT5
keratin 5
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
References
- Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akavia UD, Litvin O, Kim J, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Auer-Grumbach M, Weger M, Fink-Puches R, et al. Fibulin-5 mutations link inherited neuropathies, age-related macular degeneration and hyperelastic skin. Brain. 2011;134:1839–1852. doi: 10.1093/brain/awr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann IM, Halvorsen OJ, Collett K, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- Bertone P, Trifonov V, Rozowsky JS, et al. Design optimization methods for genomic DNA tiling arrays. Genome Res. 2006;16:271–281. doi: 10.1101/gr.4452906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumber Y, Issa J-PJ. Epigenetics in cancer: what’s the future? Oncology (Williston Park, NY) 2011;25:220–6. 228. [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Cargill M, Schrodi SJ, Chang M, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal RD, Antonescu CR, Wolchok JD, et al. KIT as a therapeutic target in metastatic melanoma. JAMA. 2011;305:2327–2334. doi: 10.1001/jama.2011.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ. Rethinking the prosaicism of mosaicism; is it in us? Exp Dermatol. 2010;19:1037–1039. doi: 10.1111/j.1600-0625.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- Choate KA, Lu Y, Zhou J, et al. Mitotic recombination in patients with ichthyosis causes reversion of dominant mutations in KRT10. Science. 2010;330:94–97. doi: 10.1126/science.1192280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concolino D, Roversi G, Muzzi GL, et al. Clericuzio-type poikiloderma with neutropenia syndrome in three sibs with mutations in the C16orf57 gene: delineation of the phenotype. Am J Med Genet. 2010;152A:2588–2594. doi: 10.1002/ajmg.a.33600. [DOI] [PubMed] [Google Scholar]
- Contassot E, French LE. Epigenetic causes of apoptosis resistance in cutaneous T-cell lymphomas. J Invest Dermatol. 2010;130:922–924. doi: 10.1038/jid.2009.427. [DOI] [PubMed] [Google Scholar]
- Conway K, Edmiston SN, Khondker ZS, et al. DNA-methylation profiling distinguishes malignant melanomas from benign nevi. Pigment Cell Melanoma Res. 2011;24:352–360. doi: 10.1111/j.1755-148X.2011.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JF, Futscher BW, Kvoes RA, et al. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. 1994;269:17228–17237. [PubMed] [Google Scholar]
- Cullinane AR, Vilboux T, O’Brien K, et al. Homozygosity mapping and whole-exome sequencing to detect SLC45A2 and G6PC3 mutations in a single patient with oculocutaneous albinism and neutropenia. J Invest Dermatol. 2011;131:2017–2025. doi: 10.1038/jid.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendrou CA, Plagnol V, Fung E, et al. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat Genet. 2009;41:1011–1015. doi: 10.1038/ng.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrance AM, Liu S, Yuan W, et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J Clin Invest. 2006;116:2707–2716. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Ho C, Wang NJ, et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 2011;1:137–143. doi: 10.1158/2159-8290.CD-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Hodi FS, Bastian BC. Mutation-driven drug development in melanoma. Curr Opin Oncol. 2010;22:178–183. doi: 10.1097/cco.0b013e32833888ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse S, Nagarajan R, Costello J. Genome-scale DNA methylation analysis. Epigenomics. 2010;2:105–117. doi: 10.2217/epi.09.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Herranz M, Espada J, et al. A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer Res. 2004;64:5527–5534. doi: 10.1158/0008-5472.CAN-03-4061. [DOI] [PubMed] [Google Scholar]
- Frumkin D, Wasserstrom A, Itzkovitz S, et al. Amplification of multiple genomic loci from single cells isolated by laser micro-dissection of tissues. BMC Biotechnol. 2008;8:17. doi: 10.1186/1472-6750-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galach M, Utikal J. From skin to the treatment of diseases—the possibilities of iPS cell research in dermatology. Exp Dermatol. 2011;20:523–528. doi: 10.1111/j.1600-0625.2011.01282.x. [DOI] [PubMed] [Google Scholar]
- Ghosh Z, Wilson KD, Wu Y, et al. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS ONE. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudie DR, D’Alessandro M, Merriman B, et al. Multiple self-healing squamous epithelioma is caused by a disease-specific spectrum of mutations in TGFBR1. Nat Genet. 2011;43:365–369. doi: 10.1038/ng.780. [DOI] [PubMed] [Google Scholar]
- Harboe TL, Willems P, Jespersgaard C, et al. Mosaicism in segmental darier disease: an in-depth molecular analysis quantifying proportions of mutated alleles in various tissues. Dermatology. 2011;222:292–296. doi: 10.1159/000328404. [DOI] [PubMed] [Google Scholar]
- Hassel JC, Sucker A, Edler L, et al. MGMT gene promoter methylation correlates with tolerance of temozolomide treatment in melanoma but not with clinical outcome. Br J Cancer. 2010;103:820–826. doi: 10.1038/sj.bjc.6605796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Heidorn SJ, Milagre C, Whittaker S, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocker TL, Singh MK, Tsao H. Melanoma genetics and therapeutics approaches in the 21st century: moving from the benchside to the bedside. J Invest Dermatol. 2008;128:2575–2595. doi: 10.1038/jid.2008.226. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Huffmeier U, Zeeuwen PLJM. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Fan G. DNA methylation in cell differentiation and reprogramming: an emerging systematic view. Regen Med. 2010;5:531–544. doi: 10.2217/rme.10.35. [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Anderson W, Artez A, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kiuru M, Cairo MS, et al. Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. Proc Natl Acad Sci USA. 2011;108:8797–8802. doi: 10.1073/pnas.1100332108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Ji H, Ehrlich LIR, Seita J, et al. A comprehensive methylome map of lineage commitment from hematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CL, Wain EM, Chu C-C. Downregulation of Fas gene expression in Sézary syndrome is associated with promoter hypermethylation. J Invest Dermatol. 2010;130:1116–1125. doi: 10.1038/jid.2009.301. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkman MF, Castellanos Nuijts M, van Essen AJ. Natural repair mechanisms in correcting pathogenic mutations in inherited skin disorders. Clin Exp Dermatol. 2003;28:625–631. doi: 10.1046/j.1365-2230.2003.01400.x. [DOI] [PubMed] [Google Scholar]
- Jonkman MF, Scheffer H, Stulp R, et al. Revertant mosaicism in epidermolysis bullosa caused by mitotic gene conversion. Cell. 1997;88:543–551. doi: 10.1016/s0092-8674(00)81894-2. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Cooper DN. Mosaicism in sporadic neurofibromatosis type 1: variations on a theme common to other hereditary cancer syndromes? J Med Genet. 2008;45:622–631. doi: 10.1136/jmg.2008.059329. [DOI] [PubMed] [Google Scholar]
- Kemp CJ, Donehower LA, Bradley A, et al. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74:813–822. doi: 10.1016/0092-8674(93)90461-x. [DOI] [PubMed] [Google Scholar]
- Kim ES, Herbst RS, Wistuba II, et al. The BATTLE Trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahtz C, Stranzenbach R, Fiedler E, et al. Methylation of PTEN as a prognostic factor in malignant melanoma of the skin. J Invest Dermatol. 2009;130:620–622. doi: 10.1038/jid.2009.226. [DOI] [PubMed] [Google Scholar]
- Laird P. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Gen. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Goto Y, Murata H, et al. Polyclonality of BRAF mutations in primary melanoma and the selection of mutant alleles during progression. Br J Cancer. 2011;104:464–468. doi: 10.1038/sj.bjc.6606072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhurst MJ, Sapp JC, Teer JK, et al. A mosaic activating mutation in AKT1 associated with the proteus syndrome. N Engl J Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Okada S, Kong X-F. Gain-of-function human STAT1. mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Helms C, Liao W, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4:e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Wilson RK. Cancer genome sequencing: a review. Hum Mol Genet. 2009;18:R163–R168. doi: 10.1093/hmg/ddp396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JA. Keratinocyte heal thyself: a new form of “natural gene therapy”. J Invest Dermatol. 2004;122:x–xi. doi: 10.1046/j.0022-202X.2003.22141.x. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RP, Stuart PE, Nistor I, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78:827–851. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhasi HL, Lynch KR, Dolan KP, et al. Covalent modification and repressed transcription of a gene in hepatoma cells. Proc Natl Acad Sci USA. 1981;78:834–837. doi: 10.1073/pnas.78.2.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Pasmooij AMG, Pas HH, Deviaene FCL. Multiple correcting COL17A1 mutations in patients with revertant mosaicism of epidermolysis bullosa. Am J Hum Genet. 2005;77:727–740. doi: 10.1086/497344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett TD, Agrawal NS, Wei X, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger E, Kofler R, Borkenstein M, et al. Melanotic macules following Blaschko’s lines in McCune-Albright syndrome. Br J Dermatol. 1994;130:215–220. doi: 10.1111/j.1365-2133.1994.tb02903.x. [DOI] [PubMed] [Google Scholar]
- Rimbaud P, Meynadier J, Guilhou JJ, et al. Immunological disorders and histocompatibility antigens in psoriasis. Ann Dermatol Syphiligr (Paris) 1974;101:359–374. [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robarge KD, Brunton SA, Castanedo GM, et al. GDC-0449-a potent inhibitor of the hedgehog pathway. Bioorg Med Chem Lett. 2009;19:5576–5581. doi: 10.1016/j.bmcl.2009.08.049. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, et al. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, et al. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Faqeih E, Sunker A, et al. Recessive mutations in DOCK6, encoding the guanidine nucleotide exchange factor DOCK6, lead to abnormal actin cytoskeleton organization and Adams-Oliver syndrome. Am J Hum Genet. 2011;89:328–333. doi: 10.1016/j.ajhg.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigalotti L, Covre A, Fratta E, et al. Epigenetics of human cutaneous melanoma: setting the stage for new therapeutic strategies. J Transl Med. 2010;8:56. doi: 10.1186/1479-5876-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011 doi: 10.1126/science.1208130. Available from: http://www.sciencemag.org/content/early/2011/07/27/science.1208130.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- Sun Y, Almomani R, Aten E, et al. Terminal osseous dysplasia is caused by a single recurrent mutation in the FLNA gene. Am J Hum Genet. 2010;87:146–153. doi: 10.1016/j.ajhg.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen J-W, de Sauvage FJ. Paracrine Hedgehog signaling in cancer. Cancer Res. 2009;69:6007–6010. doi: 10.1158/0008-5472.CAN-09-0756. [DOI] [PubMed] [Google Scholar]
- Thomas RK, Nickerson E, Simons JF, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- Tolar J, Xia L, Riddle MJ, et al. Induced pluripotent stem cells from individuals with recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2011;131:848–856. doi: 10.1038/jid.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yang W, Wen W, et al. γ-secretase gene mutations in familial acne inversa. Science. 2010;330:1065. doi: 10.1126/science.1196284. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NJ, Sanborn Z, Arnett KL, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci USA. 2011;108:17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt SA, Pourreyron C, Purdie K, et al. Integrative mRNA profiling comparing cultured primary cells with clinical samples reveals PLK1 and C20orf20 as therapeutic targets in cutaneous squamous cell carcinoma. Oncogene. 2011;30:4666–4677. doi: 10.1038/onc.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch RL, Dijkgraaf GJP, Alicke B, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-J, Huang W, Yang S, et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat Genet. 2009;41:205–210. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]
- Zhou C, Zang D, Jin Y, et al. Mutation in ribosomal protein L21 underlies hereditary hypotrichosis simplex. Hum Mutat. 2011a;32:710–714. doi: 10.1002/humu.21503. [DOI] [PubMed] [Google Scholar]
- Zhou X, Maricque B, Xie M, et al. The human epigenome browser at washington university. Nat Methods. 2011b;8:989–990. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]