Abstract
Hepatitis C virus (HCV) envelope glycoproteins E1/E2 can pseudotype retroviral particles and efficiently mediate entry into target cells. Using this experimental system, we determined HCV tropism for different cell types. Only primary hepatocytes and one hepatoma cell line were susceptible to HCV pseudovirus entry, which could be inhibited by sera from HCV-infected individuals. Furthermore, expression of the putative HCV receptor CD81 on nonpermissive human hepatic but not murine cells enabled HCV pseudovirus entry. Importantly, inhibition of viral entry by an anti-CD81 mAb occurred at a step following HCV attachment to target cells. Our results indicate that CD81 functions as a post-attachment entry coreceptor and that other cellular factors act in concert with CD81 to mediate HCV binding and entry into hepatocytes.
It is estimated that 170 million people worldwide are infected with the hepatitis C virus (HCV) and are at risk of developing chronic hepatitis or cirrhosis, the latter often leading to hepatocellular carcinoma (1, 2). In the past, difficulties with culturing the virus and expressing fusogenic envelope glycoproteins limited studies of HCV tropism and entry. RT-PCR- and electron microscopy-based approaches were relied on to demonstrate the presence of HCV RNA and proteins in primary hepatocytes and certain hepatoma cell lines (3-8). Furthermore, the existence of extrahepatic HCV reservoirs was suggested by the detection of viral RNA in serum and peripheral blood mononuclear cells (PBMC) (9-11). Recently, a major technical advance in the field has been the discovery that unmodified HCV envelope glycoproteins can pseudotype retroviral particles and mediate entry into target cells (12-15). This model seems to authentically replicate the early steps of the HCV life cycle, enabling detailed studies of HCV tropism and entry into target cells.
The cellular tropism of enveloped viruses is largely determined by selective interactions of viral envelope glycoproteins with specific cell-surface receptors. Entry generally proceeds by a cascade of coordinated events wherein virus binding to a host molecule triggers exposure of cryptic envelope glycoprotein domains that mediate downstream interactions and functions. We and others recently demonstrated that DC-SIGN (dendritic cell-specific ICAM-3 grabbing nonintegrin; CD209) and L-SIGN (DC-SIGNR, liver and lymph node specific; CD209L) function as HCV capture receptors but do not mediate viral entry into target cells (16, 17). Candidate HCV entry receptors include CD81, scavenger receptor class B type 1, low density lipoprotein receptor, and glycosaminoglycans (18-20). CD81 is the most extensively characterized putative HCV receptor. A number of groups have demonstrated that the soluble ectodomain of HCV envelope glycoprotein E2 binds specifically and with relatively high affinity (Kd ≈10-8 M) to human and chimpanzee CD81 (21-24). However, CD81 is widely expressed on human cells and therefore cannot account for the restricted tropism of HCV to hepatocytes and perhaps certain lymphocytes (25).
Here, we report that HCV E1/E2-pseudotyped retroviral particles enter into primary human hepatocytes as well as one human hepatoma cell line. In contrast to previous reports, no entry was observed into resting or stimulated human PBMC. Entry of HCV E1/E2-pseudotyped particles into target cells was inhibited by HCV RNA+/Ab+ human sera and an anti-CD81 mAb. Furthermore, expression of CD81 in a CD81-human hepatoma cell line rendered it permissive to HCV pseudovirus entry. A murine fibroblast cell line expressing CD81, however, remained resistant to HCV pseudovirus entry. Surprisingly, virus binding to CD81+ hepatic cells was not inhibited by an anti-CD81 mAb, which could still inhibit entry of prebound HCV pseudoviruses. Taken together, our results indicate that CD81 functions as an HCV entry coreceptor and that an unidentified primary receptor mediates virus attachment to hepatic cells.
Materials and Methods
Cells and Abs. All cell lines were cultured under standard conditions. NKNT3 hepatoma cells were provided by I. Fox (University of Nebraska Medical Center, Omaha), and Huh-7 hepatoma cells were provided by R. Chowdhurry (Albert Einstein College of Medicine). All of the other cell lines were purchased from the American Type Culture Collection. CD81+ derivatives of HepG2 and 3T3 were generated by transfection of a pcDNA3.1-CD81 expression construct, followed by selection in G418 (1 mg/ml). Individual clones were isolated on the basis of CD81 expression as determined by labeling with an anti-CD81 mAb, JS-81 (Pharmingen), followed by flow cytometry analyses. The Quantum Simply Cellular Microbead kit (Sigma) was used in a quantitative fluorescence-activated cell sorting assay (26) to estimate the number of CD81 molecules per cell for each line. Human PBMC were isolated and cultured by standard methods and used either immediately or stimulated with 10 μg/ml phytohemagglutinin for 72 h before use (27). Primary human hepatocytes were provided by S. Strom (University of Pittsburgh, Pittsburgh) through the National Institutes of Health Liver Tissue Procurement and Distribution System. Briefly, human livers are perfused with collagenase, and explanted hepatocytes are filtered and centrifuged. Purified hepatocytes are plated at 50-70% final confluency on collagen I-treated tissue culture plates in serum-free Williams' E medium (28). They are used for pseudotype infection 48 h postplating. The viability of primary hepatocytes at the time of infection is >85%. Anti-E1 mAb A4 and anti-E2 mAb H53 were provided by J. Dubuisson (Institut Pasteur, Lille, France) (29). Anti-E2 mAb 091b was purchased from Austral Biologicals. All of the mAbs used in the study are IgG1, and, where indicated, an isotype-matched nonspecific murine IgG1 was used. Sera from HCV RNA+/Ab+ and HCV- donors were collected by R. Klein at the Montefiore Medical Center (Bronx, NY) and by P. Dény (Université Paris, Paris).
Production of HIV Particles Pseudotyped with HCV Envelope Glycoproteins. 293T cells were calcium phosphate transfected with a 1:3 ratio of NLluc+env- reporter vector (30) and E1/E2 or E1/E2-p7 expression vectors, which are described in detail elsewhere (13). Cell culture supernatants were collected 48 h posttransfection and analyzed for p24 content by ELISA, as previously described (27). Target cells (104) were incubated overnight with 200 μl of supernatant containing pseudoviruses (≈100 ng/ml p24), then washed and placed in fresh medium for another 36 h. Luciferase activity was measured in cell lysates, as previously described (31). PBMC (5 × 106) were infected with 5 ml of supernatant containing pseudoviruses, and luciferase activity was measured in lysates of total cells 48 h postinfection. For inhibition of entry studies, mAbs (10 μg/ml) or sera (1:100) were added to target cells immediately before infection with pseudoviruses. Alternatively, target cells were preincubated with pseudoviruses or JS-81 (10 μg/ml) for 2 h at 4°C, washed three times with cold PBS, and incubated with JS-81 (10 μg/ml) or pseudoviruses, respectively, for another 2 h at 4°C. Control incubations were performed with a nonspecific, murine IgG1 (10 μg/ml) instead of JS-81. After washing with cold PBS, cells were placed at 37°C, and luciferase activity was measured in cell lysates 48 h later.
Virus-Binding Assay. HCV virion binding to cells was evaluated by RT-PCR as described previously (16). Cells were preincubated with mAbs (10 μg/ml) for 15 min at room temperature before addition of sera (10-20 μl) from HCV RNA+ donors. Cells were washed, and viral RNA was extracted by using a QIAmp Viral RNA Mini Spin kit (Qiagen, Valencia, CA) and quantified by an HCV Quantasure Plus assay at Laboratory Corporation of America (Research Triangle Park, NC).
Results
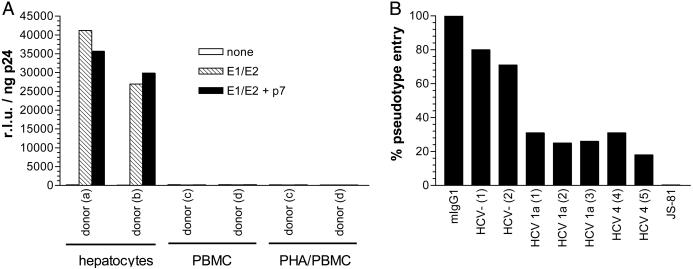
Entry of HCV Pseudoviruses into Primary Human Cells. HCV pseudoviruses were generated by coexpression of an envelope-negative, luciferase-expressing HIV-1 genome (NLluc+env-) and HCV E1/E2, with or without p7. E1/E2 were from the H77 subtype 1a isolate, which was derived from serum of an HCV-infected individual (32). Target cells were infected with pseudovirus-containing supernatants, and luciferase activity was measured 48 h postinfection and standardized for p24 (HIV-1 Gag) content. Primary human hepatocytes were consistently infected by HCV pseudoviruses, and no significant variability in viral entry levels was observed between the two donors (Fig. 1a). Primary human PBMC, however, were completely resistant to HCV pseudovirus entry, irrespective of whether they were resting or stimulated with phytohemagglutinin for 3 days before infection (Fig. 1a). No differences in pseudovirus entry patterns were observed when E1/E2 were coexpressed with p7, indicating that in this system at least, p7 does not influence viral fusion. Furthermore, HCV pseudovirus entry into primary hepatocytes was consistently inhibited by ≈70% with HCV RNA+/Ab+ sera (1:100) from subtype 1a- or 4-infected individuals, but not by sera from HCV- individuals (Fig. 1b). Finally, anti-CD81 mAb JS-81 (10 μg/ml) completely inhibited pseudovirus entry into primary hepatocytes (Fig. 1b). We note that hepatocytes as well as resting and stimulated PBMC express similar levels of CD81 (data not shown).
Fig. 1.
HCV pseudovirus entry into human primary cells. (A) Primary hepatocytes (≈104) from two healthy, HCV- donors were infected with NLluc+env- viruses (≈100 ng/ml p24) pseudotyped with no envelope glycoproteins, E1/E2, or E1/E2 coexpressed with p7. Similarly, PBMC (5 × 106) from healthy, HCV- donors were either infected or phytohemagglutinin-stimulated then infected with HCV pseudotypes. Luciferase activities (relative light units, r.l.u.) were measured 48 h postinfection and standardized for p24 content. Results are from a representative experiment. Entry into primary hepatocytes varied by ±100%. (B) Alternatively, primary hepatocytes were premixed with control murine IgG1 (10 μg/ml), sera (1:100) from two HCV-, two HCV subtype 1a+ and two HCV subtype 4+ donors, or anti-CD81 mAb JS-81 (10 μg/ml) immediately followed by infection with NLluc+env- viruses (≈100 ng/ml p24) pseudotyped with E1/E2. The % pseudovirus entry was calculated by (r.l.u. in the presence of inhibitor)/(r.l.u. in the absence of inhibitor) × 100%. Results are from a representative experiment with an assay error of ±25%.
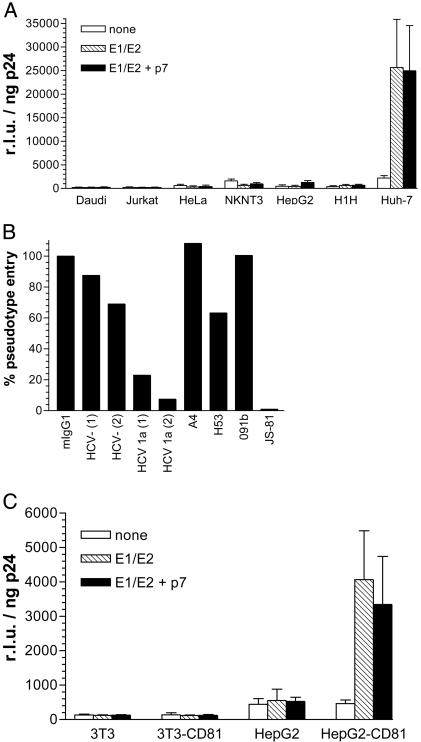
Entry of HCV Pseudoviruses into Human Cell Lines. Entry of HCV pseudoviruses was also tested in several human cell lines that express similarly high levels of CD81, except for HepG2, which are CD81- (data not shown). Entry could not be detected in nonhepatic cell lines such as Daudi (B cell line), Jurkat (T cell line), and HeLa (endothelial cell line). Of the four hepatoma cell lines tested (NKNT3, HepG2, H1H, and Huh-7), only Huh-7 were permissive to E1/E2-mediated infection by luciferase-expressing pseudoviruses, and entry levels were of the same order of magnitude as those observed in primary hepatocytes (Fig. 2a). As with primary cells, no significant differences in viral entry were observed when E1/E2 were coexpressed with p7. HCV pseudovirus entry into Huh-7 cells could be inhibited by >80% with sera (1:100) from two donors infected with HCV subtype 1a. Entry of pseudovirions into Huh-7 cells was not inhibited by anti-E1 mAb A4 or anti-E2 mAbs 091b and H53 (10 μg/ml) (Fig. 2b). We conclude that these mAbs do not recognize functionally relevant domains of E1 and E2. In contrast, entry was completely inhibited by anti-CD81 mAb JS-81 (10 μg/ml), confirming that this molecule is essential for E1/E2-mediated entry (Fig. 2b). Overall, the pattern of inhibition of HCV pseudotype entry with Abs was similar in Huh-7 and primary hepatocytes.
Fig. 2.
HCV pseudovirus entry into human cell lines. (A) Cells (104) indicated along the x axis were infected with NLluc+env- viruses (≈100 ng/ml p24) pseudotyped with no envelope glycoproteins, E1/E2, or E1/E2 coexpressed with p7, and luciferase activities (r.l.u.) were measured 48 h postinfection. Results are means of three independent experiments ± SD. (B) Alternatively, Huh-7 cells (104) were premixed with control murine IgG1 (10 μg/ml), sera (1:100) from two HCV- and two HCV 1a+ donors, or mAbs A4 (anti-E1), H53 (anti-E2), 091b (anti-E2), or JS-81 (anti-CD81) (each at 10 μg/ml), immediately followed by infection with NLluc+env- viruses (≈100 ng/ml p24) pseudotyped with E1/E2. Luciferase activities (r.l.u.) were measured 48 h postinfection and standardized for p24 content. The % pseudovirus entry was calculated by (r.l.u. in the presence of inhibitor)/(r.l.u. in the absence of inhibitor) × 100%. Results are from a representative experiment with an assay error of ±25%. Low levels of inhibition observed with anti-E2 mAb H53 (10 μg/ml) were within the error of the assay. (C) 3T3, HepG2, as well as CD81+ derivatives thereof were infected with NLluc+env- viruses (≈100 ng/ml p24) pseudotyped with no envelope glycoproteins, E1/E2, or E1/E2 coexpressed with p7, and luciferase activities (r.l.u.) were measured 48 h postinfection. Results are means of three independent experiments ± SD.
In light of these observations, CD81- murine 3T3 fibroblasts and human HepG2 hepatoma cells were engineered to express human CD81 and tested for their ability to support HCV pseudovirus entry. 3T3 and HepG2, as well as CD81+ derivatives, expressed comparable levels of luciferase after infection with NLluc+env- pseudotyped with vesicular stomatitis virus G protein (data not shown), demonstrating that the HIV-1 genome was similarly expressed in both cell types. However, no luciferase activity was detected in 3T3 CD81+ cells infected with HCV pseudotypes, indicating that they remain resistant to E1/E2-mediated entry. In contrast, HepG2 CD81+ cells acquired an entry-permissive phenotype, although luciferase activity was about an order of magnitude lower than that observed in Huh-7 cells (Fig. 2c). Using quantitative flow cytometry, the density of CD81 molecules per cell was determined to be ≈2.5 × 105 for Huh-7, 1.4 × 106 for HepG2-CD81, and 4.6 × 105 for 3T3-CD81 (data not shown), demonstrating that efficiency of pseudovirus entry does not directly correlate to CD81 expression levels. Taken together, these experiments indicate that other factors present in human hepatocytes are required for HCV E1/E2-mediated fusion and entry.
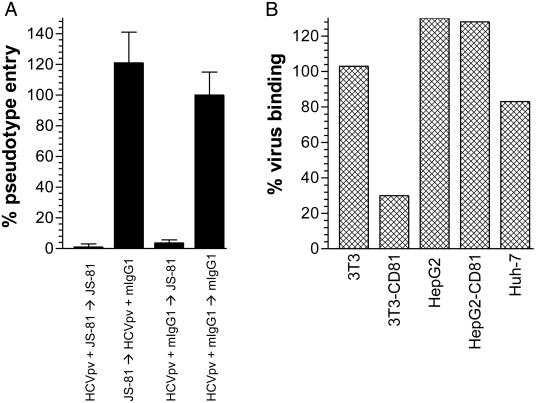
HCV Binding to CD81+ and CD81- Permissive and Nonpermissive Cell Lines. HCV binding and entry experiments were performed to elucidate the role of CD81 in E1/E2-mediated entry. We reasoned that if CD81 was the primary HCV receptor, both virus binding to and entry into target cells would be CD81-dependent. In contrast, if CD81 was an HCV coreceptor, only entry but not binding would be CD81-dependent. To test these hypotheses, we performed inhibition of binding and entry experiments with an anti-CD81 mAb. Simultaneously adding HCV pseudoviruses and anti-CD81 mAb JS-81 (10 μg/ml) to target cells abrogated entry, whereas washing out the anti-CD81 mAb before addition of pseudoviruses allowed entry to occur (Fig. 3a). When HCV pseudoviruses were prebound to target cells at 4°C, a temperature that is nonpermissive for fusion, entry could still be inhibited with the anti-CD81 mAb but not by control murine IgG1, implying that virus binding had occurred through E1/E2 interactions with cell-surface molecules other than CD81 (Fig. 3a).
Fig. 3.
Effect of anti-CD81 mAb on HCV binding and entry into different cells. (A) Huh-7 cells (104) were preincubated at 4°C with NLluc+env- viruses (≈100 ng/ml p24) pseudotyped with E1/E2 (HCVpv) in the presence of anti-CD81 mAb JS-81 (10 μg/ml, first bar) or a control murine IgG1 (mIgG1, 10 μg/ml, third and fourth bars). Cells were washed, and a second incubation at 4°C was performed with a control mIgG1 (10 μg/ml, second and fourth bars) or JS-81 (10 μg/ml, first and third bars). Alternatively, cells were first incubated with JS-81 followed by washing and a second incubation with HCVpv and a mlgG1 (second bar). After a second wash, cells were placed at 37°C, and luciferase activities were measured 48 h later. Results are means of three independent experiments ± SD. (B) Cells (104) indicated along the x axis were preincubated with sera of HCV+ individuals for 2 h at 4°C in the presence of JS-81 or a control murine IgG (10 μg/ml each). Viral RNA remaining bound to cells after washing was quantified. Results are expressed as % virus binding = (RNA copies in the presence of JS-81)/(RNA copies in the absence of JS-81) × 100%. Results are from a representative experiment with an assay error of ±20%.
In a complementary set of experiments, we tested the ability of the anti-CD81 mAb JS-81 to inhibit binding of HCV virions from patient sera to different target cells. Absolute levels of virus binding varied between sera, but was weak to 3T3, moderate to HepG2, and high to Huh-7. We note that HCV virion binding levels seem to correlate to relative pseudovirus entry levels into HepG2 and Huh-7. Binding of HCV virions was observed to both parental and CD81+ 3T3 cells, but binding to the latter was ≈3-fold higher and was inhibited by ≈70% with JS-81(10 μg/ml) (Fig. 3b). In contrast, similarly high levels of virion binding were observed to HepG2 and HepG2-CD81 cells, and binding was not inhibited by JS-81, consistent with the presence of other HCV-attachment molecules on these cells. The highest virus binding was observed to Huh-7 cells and was inhibited by <20% with JS-81. Together, these results indicate that HCV binding to target cells mostly occurs through an interaction of the envelope glycoproteins with cell-surface molecules other than CD81.
Discussion
HCV tropism and mechanism of entry into target cells have been difficult to study due to a lack of key experimental systems. Until recently, it was believed that modifying the transmembrane domains of HCV envelope glycoproteins was necessary to achieve their expression on the cell surface, which is a prerequisite for developing viral entry and fusion assays. Surprisingly, we and others recently discovered that unmodified HCV E1/E2 heterodimers are associated with the plasma membrane and can effectively pseudotype retroviral particles to deliver reporter genes into target cells (12-15). Furthermore, entry of HCV E1/E2-pseudotyped particles is inhibited by certain anti-E2 mAbs, an anti-CD81 mAb as well as HCV+ sera (12, 14). In the current study, HCV pseudovirus entry mediated by E1/E2 from a subtype 1a isolate was inhibited by sera from individuals infected with subtypes 1a or 4, suggesting the existence of shared neutralization epitopes. This experimental system therefore seems to authentically replicate the early steps of the viral life cycle, making it possible to unambiguously identify HCV-permissive cells as well as the cellular receptor(s) that mediate attachment and entry.
Numerous studies indicate that HCV-permissive cells include hepatocytes as well as B and T lymphocytes. Virus-like particles have been visualized in liver biopsies of HCV+ individuals (3-5), and in vitro infection, albeit inefficient, of primary hepatocytes and hepatocyte cell lines has been reported (6-8). The existence of extrahepatic reservoirs of HCV is suggested by the detection of viral RNA in serum and PBMC of infected individuals (9-11). The notion that B and T lymphocytes are infected in vivo is supported by in vitro infection of B and T cell lines (33-35). According to one study, however, replicating forms of HCV RNA are exclusively present in hepatocytes, whereas only non-replicating forms are found in B lymphocytes and none in T lymphocytes (36). Our results indicate that hepatocytes are the sole bona fide HCV target cells because the envelope glycoproteins from a prototypical HCV subtype 1a isolate mediate entry of retroviral pseudotypes only into primary hepatocytes and one hepatoma cell line of four that were tested. All of the primary cells and cell lines that we used, except HepG2 and 3T3, express high levels of CD81. We assume that CD81+ hepatoma cell lines that are resistant to entry have down-regulated expression of the primary HCV receptor. We did not detect HCV pseudovirus entry into resting or phytohemagglutinin-stimulated PBMC, nor into several nonhepatic cell lines of different origins. It is possible, however, that only a minor subpopulation of PBMC is susceptible to HCV infection and is undetectable in our assay.
Several studies have documented an uneven distribution of HCV quasispecies in tissues from patients with end-stage liver disease, including serum, PBMC, and liver (9-11). Quasispecies from different host compartments vary mostly at the level of E2 sequences, suggesting that these organs differentially adsorb and replicate HCV subpopulations (37, 38). Levels of viral diversity depend on the tissue sampled (e.g., blood vs. liver), disease stage, drug treatment, and the strength of the humoral and cellular immune responses (37, 38). The H77 isolate that was used for these studies was derived from the serum of an HCV+ individual, yet it seems to be exclusively hepatotropic. This may be fortuitous or may indicate that no other cell types are infected by HCV. Clearly, additional HCV isolates from different host compartments need to be characterized to determine the full range of HCV tropism.
We and others recently demonstrated that L-SIGN and DC-SIGN bind soluble E2 as well as HCV viral and pseudoviral particles and mediate trans-infection of target cells (refs. 16 and 17; E.G.C., unpublished observations). These capture receptors may be important for targeting HCV to the liver and/or lymph nodes but do not mediate viral entry. CD81, a ubiquitously expressed tetraspanin, was also shown to bind soluble E2 and more recently to directly mediate HCV pseudovirus entry into target cells (12, 21-24). In this report, we show that only CD81- human HepG2 cells are rendered susceptible to HCV pseudovirus entry when they are modified to express CD81; murine 3T3 CD81+ cells remain resistant. Furthermore, HCV pseudovirus entry is inhibited by an anti-CD81 mAb, even when virus is prebound to target cells at a temperature that is nonpermissive for fusion. However, binding of HCV to CD81+ human target cells is not inhibited by the anti-CD81 mAb. Taken together, these observations indicate that other cellular factors act in concert with CD81 to mediate HCV binding and entry into target cells.
Recent advances in generating retroviral reporter viruses pseudotyped with HCV envelope glycoproteins have added great impetus to studies of HCV tropism and entry. Here, we further validate this experimental system by confirming HCV pseudovirus tropism for hepatocytes and neutralization of pseudovirus entry by HCV+ patient sera and an anti-CD81 mAb. Most importantly, we show that HCV enters target cells by a mechanism that involves attachment to a primary receptor followed by interactions with CD81, which serves as an entry coreceptor. Others have reported that HCV pseudoviruses enter target cells by a pH-dependent mechanism, which would constitute a third step in the viral entry process (14). A major challenge now is the identification of the primary HCV receptor(s), perhaps through the application of gene transfer and functional cloning into CD81+ entry-resistant cells. The discovery of this molecule would enable detailed studies of HCV tropism and mechanism of entry as well as the identification and characterization of antiviral agents targeting different stages of HCV entry.
Acknowledgments
We thank Dr. I. Fox and Dr. R. Chowdhurry for kindly providing essential cell lines, Dr. S. Porcelli for providing PBMC, and Dr. J. Dubuisson for providing mAbs A4 and H53. We are also grateful to Drs. R. Klein and P. Deny for supplying HCV+ and HCV- human sera. Primary human hepatocytes were provided by Dr. S. Strom through the National Institutes of Health Liver Tissue Procurement and Distribution System program. This work was supported by National Institutes of Health Grant AI060390 and The Speaker's Fund for Biomedical Research Young Investigators' Award (to T.D.), as well as National Institutes of Health Grant AI051134 (to J.P.G.). This work was also supported in part by National Institute of Allergy and Infectious Diseases Centers for AIDS Research Grant AI051519 (to Albert Einstein College of Medicine).
Abbreviations: HCV, hepatitis C virus; PBMC, peripheral blood mononuclear cell; r.l.u., relative light unit.
References
- 1.Cooper, S., Erickson, A. L., Adams, E. J., Kansopon, J., Weiner, A. J., Chien, D. Y., Houghton, M., Parham, P. & Walker, C. M. (1999) Immunity 10, 439-449. [DOI] [PubMed] [Google Scholar]
- 2.Lechner, F., Wong, D. K., Dunbar, P. R., Chapman, R., Chung, R. T., Dohrenwend, P., Robbins, G., Phillips, R., Klenerman, P. & Walker, B. D. (2000) J. Exp. Med. 191, 1499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Vos, R., Verslype, C., Depla, E., Fevery, J., Van Damme, B., Desmet, V. & Roskams, T. (2002) J. Hepatol. 37, 370-379. [DOI] [PubMed] [Google Scholar]
- 4.Bosman, C., Valli, M. B., Bertolini, L., Serafino, A., Boldrini, R., Marcellini, M. & Carloni, G. (1998) Res. Virol. 149, 311-314. [DOI] [PubMed] [Google Scholar]
- 5.Iacovacci, S., Manzin, A., Barca, S., Sargiacomo, M., Serafino, A., Valli, M. B., Macioce, G., Hassan, H. J., Ponzetto, A., Clementi, M., et al. (1997) Hepatology 26, 1328-1337. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda, M., Sugiyama, K., Mizutani, T., Tanaka, T., Tanaka, K., Sekihara, H., Shimotohno, K. & Kato, N. (1998) Virus Res. 56, 157-167. [DOI] [PubMed] [Google Scholar]
- 7.Fournier, C., Sureau, C., Coste, J., Ducos, J., Pageaux, G., Larrey, D., Domergue, J. & Maurel, P. (1998) J. Gen. Virol. 79, 2367-2374. [DOI] [PubMed] [Google Scholar]
- 8.Castet, V., Fournier, C., Soulier, A., Brillet, R., Coste, J., Larrey, D., Dhumeaux, D., Maurel, P. & Pawlotsky, J. M. (2002) J. Virol. 76, 8189-8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afonso, A. M., Jiang, J., Penin, F., Tareau, C., Samuel, D., Petit, M. A., Bismuth, H., Dussaix, E. & Feray, C. (1999) J. Virol. 73, 9213-9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabot, B., Martell, M., Esteban, J. I., Sauleda, S., Otero, T., Esteban, R., Guardia, J. & Gomez, J. (2000) J. Virol. 74, 805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laskus, T., Radkowski, M., Wang, L. F., Nowicki, M. & Rakela, J. (2000) J. Virol. 74, 1014-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartosch, B., Dubuisson, J. & Cosset, F. L. (2003) J. Exp. Med. 197, 633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumonceaux, J., Cormier, E., Kajumo, F., Donovan, G., Gardner, J., Olson, W. & Dragic, T. (2003) J. Virol. 77, 13418-13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu, M., Zhang, J., Flint, M., Logvinoff, C., Cheng-Mayer, C., Rice, C. M. & McKeating, J. A. (2003) Proc. Natl. Acad. Sci. USA 100, 7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummer, H. E., Maerz, A. & Poumbourios, P. (2003) FEBS Lett. 546, 385-390. [DOI] [PubMed] [Google Scholar]
- 16.Gardner, J., Durso, R. J., Arrigale, R. R., Donovan, G. P., Maddon, P. J., Dragic, T. & Olson, W. C. (2003) Proc. Natl. Acad. Sci. USA 100, 4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pohlmann, S., Zhang, J., Baribaud, F., Chen, Z., Leslie, G. J., Lin, G., Granelli-Piperno, A., Doms, R. W., Rice, C. M. & McKeating, J. A. (2003) J. Virol. 77, 4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flint, M., Quinn, E. R. & Levy, S. (2001) Clin. Liver Dis. 5, 873-893. [DOI] [PubMed] [Google Scholar]
- 19.Scarselli, E., Ansuini, H., Cerino, R., Roccasecca, R. M., Acali, S., Filocamo, G., Traboni, C., Nicosia, A., Cortese, R. & Vitelli, A. (2002) EMBO J. 21, 5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartosch, B., Vitelli, A., Granier, C., Goujon, C., Dubuisson, J., Pascale, S., Scarselli, E., Cortese, R., Nicosia, A. & Cosset, F. L. (2003) J. Biol. Chem. 278, 41624-41630. [DOI] [PubMed] [Google Scholar]
- 21.Pileri, P., Uematsu, Y., Campagnoli, S., Galli, G., Falugi, F., Petracca, R., Weiner, A. J., Houghton, M., Rosa, D., Grandi, G. & Abrignani, S. (1998) Science 282, 938-941. [DOI] [PubMed] [Google Scholar]
- 22.Flint, M., Thomas, J. M., Maidens, C. M., Shotton, C., Levy, S., Barclay, W. S. & McKeating, J. A. (1999) J. Virol. 73, 6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higginbottom, A., Quinn, E. R., Kuo, C. C., Flint, M., Wilson, L. H., Bianchi, E., Nicosia, A., Monk, P. N., McKeating, J. A. & Levy, S. (2000) J. Virol. 74, 3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petracca, R., Falugi, F., Galli, G., Norais, N., Rosa, D., Campagnoli, S., Burgio, V., Di Stasio, E., Giardina, B., Houghton, M., et al. (2000) J. Virol. 74, 4824-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy, S., Todd, S. C. & Maecker, H. T. (1998) Annu. Rev. Immunol. 16, 89-109. [DOI] [PubMed] [Google Scholar]
- 26.Lee, B., Sharron, M., Montaner, L. J., Weissman, D. & Doms, R. W. (1999) Proc. Natl. Acad. Sci. USA 96, 5215-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trkola, A., Pomales, A. P., Yuan, H., Korber, B., Maddon, P. J., Allaway, G. P., Katinger, H., Barbas, C. F., III, Burton, D. R., Ho, D. D. & Moore, J. P. (1995) J. Virol. 69, 6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrini, J. B., Pichard, L., Domergue, J. & Maurel, P. (1997) Chem. Biol. Interact. 107, 31-45. [DOI] [PubMed] [Google Scholar]
- 29.Dubuisson, J., Hsu, H. H., Cheung, R. C., Greenberg, H. B., Russell, D. G. & Rice, C. M. (1994) J. Virol. 68, 6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connor, R. I., Chen, B. K., Choe, S. & Landau, N. R. (1995) Virology 206, 935-944. [DOI] [PubMed] [Google Scholar]
- 31.Dragic, T., Trkola, A., Lin, S. W., Nagashima, K. A., Kajumo, F., Zhao, L., Olson, W. C., Wu, L., Mackay, C. R., Allaway, G. P., et al. (1998) J. Virol. 72, 279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolykhalov, A. A., Agapov, E. V., Blight, K. J., Mihalik, K., Feinstone, S. M. & Rice, C. M. (1997) Science 277, 570-574. [DOI] [PubMed] [Google Scholar]
- 33.Kato, N., Nakazawa, T., Mizutani, T. & Shimotohno, K. (1995) Biochem. Biophys. Res. Commun. 206, 863-869. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu, Y. K., Iwamoto, A., Hijikata, M., Purcell, R. H. & Yoshikura, H. (1992) Proc. Natl. Acad. Sci. USA 89, 5477-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sung, V. M., Shimodaira, S., Doughty, A. L., Picchio, G. R., Can, H., Yen, T. S., Lindsay, K. L., Levine, A. M. & Lai, M. M. (2003) J. Virol. 77, 2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boisvert, J., He, X. S., Cheung, R., Keeffe, E. B., Wright, T. & Greenberg, H. B. (2001) J. Infect. Dis. 184, 827-835. [DOI] [PubMed] [Google Scholar]
- 37.Zein, N. N. (2000) Clin. Microbiol. Rev. 13, 223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stumpf, M. P. & Pybus, O. G. (2002) FEMS Microbiol. Lett. 214, 143-152. [DOI] [PubMed] [Google Scholar]