Abstract
Hyperacute rejection of porcine organs by old world primate recipients is mediated through preformed antibodies against galactosyl-α-1,3-galactose (Galα-1,3-Gal) epitopes expressed on the pig cell surface. Previously, we generated inbred miniature swine with a null allele of the α-1,3-galactosyltransferase locus (GGTA1) by nuclear transfer (NT) with gene-targeted fibroblasts. To expedite the generation of GGTA1 null pigs, we selected spontaneous null mutant cells from fibroblast cultures of heterozygous animals for use in another round of NT. An unexpectedly high rate of spontaneous loss of GGTA1 function was observed, with the vast majority of null cells resulting from loss of the WT allele. Healthy piglets, hemizygous and homozygous for the gene-targeted allele, were produced by NT by using fibroblasts that had undergone deletional and crossover/gene conversion events, respectively. Aside from loss of Galα-1,3-Gal epitopes, there were no obvious phenotypic differences between these null piglets and WT piglets from the same inbred lines. In fact, congenital abnormalities observed in the heterozygous NT animals did not reappear in the serially produced null animals.
Antibodies against galactosyl-α-1,3-galactose (Galα-1,3-Gal) residues on cell surface glycoproteins of pig cells mediate hyperacute rejection of porcine organs in primate model recipients and are the most immediate barrier to successful clinical xenotransplantation (1, 2). High levels of preformed “natural” antibodies against the Galα-1,3-Gal epitope are found in humans and old world primates, following evolutionary loss of the corresponding galactosyltransferase activity (encoded by GGTA1) (3). The presence of these antibodies, along with the high density of Galα-1,3-Gal residues on most pig cells (4), suggests that elimination of GGTA1 function would provide a practical means of overcoming both hyperacute rejection and subsequent acute or chronic tissue damage associated with antibody binding to this epitope.
The lack of GGTA1 function in humans and old world primates, along with the viability of GGTA1 knockout mice produced with embryonic stem cell technology (5, 6), suggested that a knockout strategy might be biologically feasible in pigs. The cloning of sheep (7) and subsequently pigs (8-10) by nuclear transfer with somatic cells has made attempts to knockout the GGTA1 locus in pigs technically feasible.
We have previously reported the generation of GGTA1 heterozygous inbred miniature swine using nuclear transfer with gene-targeted fibroblasts (11). Starting with heterozygous fibroblasts from such animals, we now report the isolation of GGTA1 null cells with spontaneous loss of the WT allele. The rate of loss of heterozygosity (LOH) was several orders of magnitude greater than typically expected, an observation that may be related to the inbred background of the heterozygous animals. LOH resulted in some cases from deletion of the WT allele and in others from either somatic crossing over or gene conversion. Similarly high rates of somatic recombination, subject to modulation by genetic background and chromosomal structure, have been reported in the mouse (12). Generation of healthy piglets with both hemizygous and homozygous GGTA1 null cells demonstrates that such somatic LOH mutations can be introduced into large animal genomes by nuclear transfer, in a manner analogous to that using murine embryonic stem cell chimeras (13).
Methods
GGTA1 Heterozygous Cell Lines. 355-F1. Fetus 355-F1 was generated by nuclear transfer from cultured ear fibroblasts of pig O212-2, a GGTA1 heterozygote in which one allele has been inactivated by homologous recombination with vector pGalGTΔS-Neo (11). Cells were isolated at day 33 of gestation by digestion with collagenase/thermolysin (Blendzyme 3, Roche Diagnostics, Indianapolis, IN) and cultured in Ham's Nutrient Mixture F10 (Invitrogen Life Technologies, Baltimore) containing 20% FBS. PL556. Piglet PL556 was derived by nuclear transfer. The donor cell clone, F501-F4, was produced by targeting of fetal fibroblasts from WT fetus F501 with vector pGalGTΔS-Neo, as described (11). PL556 cells were cultured in high glucose DMEM (Invitrogen Life Technologies, Baltimore) containing 10% FBS and 0.1 mM 2-mercaptoethanol.
Cell culture for all work reported here, beginning with tissue acquisition, was done in the absence of G418.
Nuclear Transfer (NT). For generation of piglets from clonal null cell lines, oocytes from sow ovaries were purchased (BoMed, Madison, WI), and NT was performed as described (11). The surviving embryos, possessing an intact plasma membrane, were selected for transfer into recipients after culture for 18-22 h. Potential domestic recipients were heat checked twice a day. Depending upon the exact time of estrus, 100-180 NT-derived embryos were transferred into recipient oviducts 5-17 h or 20-36 h after the onset of estrus for day 0 and day 1 recipients, respectively.
For NT with nonclonal null cell-enriched populations, oocyte maturation, NT, embryo culture, and embryo transfer into recipient females were performed as described (8), except that female recipients were selected that exhibited first standing estrus within 12 h of cybrid activation.
Baboon Natural Antibody. Anti-Galα-1,3-Gal antibodies from naive baboon plasma (natural antibody, NAb) were affinity-purified by absorption to Galα-1,3-Gal LB-VI matrix columns (14) (Alberta Research Council, Alberta, Canada). The bound NAb was eluted from the column in 0.25% acetic acid, neutralized, and dialyzed against PBS before concentration and sterile filtration.
Null Cell Selection. To enrich for GGTA1 null cells in fibroblast cultures, 355-F1 cells and PL556 cells were cultured in F10 medium containing 20% FBS and 20 μg/ml gentamycin on collagen I-coated dishes at 5% CO2,3%O2, and 37°C. The above cell lines were treated in suspension at 2 × 106 cells/ml in 100 μg/ml affinity-purified baboon NAb in media for 30 min at room temperature with mixing. After washing, cells were then treated with 12.5% baby rabbit complement (Pel-Freez Biologicals) containing DNase I (10 μg/ml) in media for 45 min at room temperature with mixing. Surviving cells were counted and plated in bulk culture and expanded for subsequent treatments. This selection was repeated three times for 355-F1 cells and twice for PL556 cells. Selections were performed every 7-10 days, with the fourth selection of 355-F1 performed 3 days after the third selection. Before each NAb/complement selection, cells were analyzed for the presence of Galα-1,3-Gal epitopes with FITC-conjugated BS-I-B4.
For clonal selection, 355-F1 cells were treated twice in suspension as above with 50 μg/ml NAb and 12.5% complement, with 4 days between treatments. After the second treatment, cells were plated at 5 and 10 cells/well in collagen I coated 96-well plates. In situ treatments with 100-500 μg/ml NAb for 1 h at 37°C and 12.5% complement for 1 h at 37°C were performed every other day for treatments 3-5. Wells containing patches of cells covering >15% of the well were transferred to a 48-well plate and treated the following day in situ with 500 μg/ml NAb and complement. Cells were passaged for molecular analysis, BS-I-B4 analysis, and freezing.
Flow Cytometry Analysis. Galα-1,3-Gal epitope expression was analyzed with FITC-conjugated BS-I-B4 lectin (Sigma). Unfixed cells were stained for 5 min at 37°C in 4 μg/ml lectin, washed, and then resuspended in buffer containing propidium iodide (PI). Fluorescence data were collected on a Becton Dickinson FACScan, and analysis of PI excluding cells was performed by using cellquest flow cytometry software (BD Immunocytometry Systems).
Quantitative Southern Blots. Genomic DNA was digested with AflIII, which generates a 1,280-bp fragment of the WT GGTA1 allele (sites at bp 9 and bp 1,289 of GenBank accession number AF221517) and a 2,330-bp fragment of the pGalGTΔS-neo targeted allele (11). Southern blots were simultaneously probed with 32P-labeled RNA transcripts from the exon 9 portion of this fragment (bp 776-891 of GenBank accession number AF221517) and a portion of the porcine DQ-β locus (bp 901-1015 of GenBank accession number M31497). Phosphor-screen autoradiography was performed on a STORM 820 Optical Scanner, and area quantitation was done with imagequant 5.2 software (Molecular Dynamics).
Microsatellite Analysis. PCR was performed by using WellRED-labeled primers, and the reactions were analyzed on a CEQ2000 sequence analyzer (Beckman Coulter). Markers Sw2518 and Sw1430 map to porcine chromosome 1 at ≈67 cM and 58 cM, respectively; combined radiation hybrid and genetic data place the GGTA1 locus at ≈115-122 cM (www.genome.iastate.edu/pig). Heterozygosity of marker Sw2518 in fetus 355-F1 and Sw1430 in piglet PL556 was confirmed by segregation of alleles within the respective inbred miniature swine lineages.
Complement-Mediated Lysis. Lysis [lactate dehydrogenase (LDH) release] and metabolism {conversion of MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt]} were measured by using the Cytotox 96 NonRadioactive Cytotoxicity and CellTiter 96 Aqueous One Solution Cell Proliferation assays (Promega), respectively. Human serum was prepared by heat inactivation of a pool from 10 untyped individuals. Preparation of affinity-purified baboon NAb was as described above. Baby rabbit complement was obtained from Pel-Freez Biologicals. Normal human dermal fibroblasts (Cambrex Bioscience, Walkersville, MD) served as Galα-1,3-Gal negative cell controls.
Two days after plating in triplicate wells, subconfluent fibroblasts were incubated in medium (Ham's Nutrient Mixture F10, 5% FBS) containing NAb or human serum for 30 min at 37°C. The cells were washed twice with Ham's Nutrient Mixture F10 and then incubated for 60 min at 37°C in the above medium containing 12.5% rabbit complement. Medium from this incubation was assayed for LDH release. Remaining cells were incubated in medium containing 16.5% MTS for 2.5-3 h. After incubation, 25 μl of 10% SDS was added to all wells, and the medium was assayed for MTS conversion. Samples from triplicate “no cell” controls for each treatment condition were used to correct for assay background. For MTS assays, corrected average values are expressed as the percentage of corrected absorbance without NAb or serum for each line. For LDH assays, corrected average values are expressed as the percentage of corrected average absorbance after detergent lysis for each line. LDH release using anti-pig pan tissue mAb 1030h-1-19 (BD Biosciences PharMingen) was used as a positive control for porcine cell lysis, with similar titrations obtained for all three porcine fibroblast lines (data not shown).
Results
Selection of GGTA1 Null Lines and Clones from Fetal and Neonatal Heterozygous Cell Lines. Two sources of heterozygously targeted primary fibroblasts were chosen for selection of GGTA1 null cells (Fig. 1). Fibroblasts from fetus 355-F1 were isolated at 33 days gestation after NT using ear fibroblasts from GGTA1 heterozygous gilt O212-2. O212-2 was itself generated by NT by using gene-targeted fetal fibroblast clone F7-H6 as described (11). Similarly, ear fibroblasts from male GGTA1 heterozygous neonate PL556 were isolated after NT by using gene-targeted fibroblast clone F501-F4.
Fig. 1.
Derivation of GGTA1 null pigs.
Approximately 1.5 × 107 cells from established cultures of both sources were depleted of Galα-1,3-Gal epitope-bearing cells by lysis with affinity-purified baboon antibodies against the epitope in combination with complement. Enrichment for Galα-1,3-Gal-negative cells was monitored by flow cytometry analysis with FITC-labeled BS-I-B4 lectin, which binds specifically to Galα-1,3-Gal epitopes. Initial depletions resulted in recovery of ≈0.1% of the cells, 7-15% of which were Galα-1,3-Gal negative. After 3-4 rounds of selection and expansion, these populations were essentially devoid of BS-I-B4 binding cells (Fig. 2). DNA prepared from the null selected populations was analyzed by PCR that distinguishes WT and targeted GGTA1 alleles. The absence of a readily detectable WT band indicated that the vast majority of these cells had undergone at least partial loss of the WT allele (Fig. 2). This loss of heterozygosity in the GGTA1 null cells is compatible with chromosome loss and reduplication, interstitial deletion, or somatic recombination.
Fig. 2.
Selection of GGTA1 null cells from heterozygously targeted fibroblasts. Fibroblasts from heterozygous fetus 355-F1 (A) and neonatal piglet PL556 (B) were analyzed by flow cytometry for binding to FITC-conjugated Galα-1,3-Gal-specific lectin BS-I-B4. 355-F1 4X and PL556#3R populations (solid lines) were selected four and three times, respectively, by lysis with affinity-purified baboon NAb and complement. Stained cells before selection (broken lines) and unstained selected populations (solid lines) served as positive and negative controls for BS-I-B4 binding. (C) Genomic DNA from the above cell populations (and WT fetus F7) was analyzed by PCR by using a forward primer upstream of the selection cassette of the GGTA1 targeting vector (F527) and a reverse primer (GR2520) downstream of the vector end, as described (11). SacI digestion yields a 2,300-bp band from the targeted GGTA1 allele, a 1,250-bp band from the WT allele, and a 7,900-bp band common to both alleles. The WT band is not detected after NAb/complement selection.
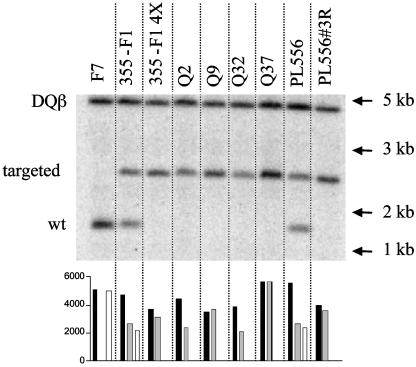
Clonal null lines were isolated from a 355-F1 cell population that had been previously enriched for null cells by NAb/complement depletion. The recovery rate of null clones from the initial heterozygous lines was ≈10-4 in two separate trials. Microsatellite analysis of 28 clones with centromere proximal marker Sw2518 (heterozygous in 355-F1) revealed that all clones remained heterozygous at this locus, indicating that chromosome loss and reduplication was not the mechanism for loss of heterozygosity (LOH) in these clones. DNA samples from four of these clonal lines (Q2, Q9, Q32, and Q37), along with the 355-F1 and PL556 null selected populations, were analyzed by quantitative Southern blotting by using an exon 9 GGTA1 probe present in both the targeted and WT alleles. A probe for the nonlinked porcine SLA DQβ gene served as a diploid copy number control (Fig. 3). In both null selected populations and all four clones, only a targeted length GGTA1 allele was detected. In two null clones (Q2 and Q32) a DQβ/GGTA1 signal ratio of ≈2:1 was obtained, demonstrating deletion of at least a portion of the WT GGTA1 allele present in 355-F1. In comparison, clones Q9 and Q37 had DQB/GGTA1 signal ratios of ≈1:1, indicating that loss of heterozygosity in these clones occurred through either somatic crossing over or gene conversion. Efforts to distinguish between these mechanisms by microsatellite analysis were unsuccessful because no heterozygous markers distal to Sw2185 were identified in fetus 355-F1 from among 24 tested. Sequencing of the junctions between exon 9 and the G418 selection cassette in clones Q9 and Q32 revealed no evidence of nucleotide heterozygosities (data not shown).
Fig. 3.
Quantitative Southern blot analysis of NT donor lines. Genomic DNA from the indicated sources was digested with restriction enzyme AflIII, Southern blotted, and hybridized simultaneously with a 116-bp probe from exon 9 of the GGTA1 locus and a 107-bp probe from the porcine SLA DQβ locus. The GGTA1 probe hybridizes a 1.3-kb WT fragment and a 2.3-kb gene-targeted fragment containing an IRES-neo selection cassette. DNA from WT (F7) and heterozygous (355-F1 and PL556) fibroblasts, before NAb/complement selection, served as controls. 355-F1 4X and PL556#3R samples were prepared from cell populations selected four and three times, respectively, with affinity-purified baboon NAb and complement. Q series samples were from clonal cell lines isolated from 355-F1 fetal fibroblasts. Signal quantitation was performed on a Storm 820 PhosphorImager and graphed as absolute values for the DQβ locus (▪), targeted GGTA1 allele ( ), and WT GGTA1 allele (□).
), and WT GGTA1 allele (□).
Generation of GGTA1 Null Pigs by NT. NT was performed by using the four clonal fetal cell lines characterized by Southern analysis (Q2, Q9, Q32, and Q37), as well as the null cell population selected from neonate PL556. Embryo transfer results are summarized in Table 1.
Table 1. Nuclear transfer with GGTAI null cells.
| Cell line | Transfers | Pregnant | Births |
|---|---|---|---|
| Q2 | 4 | 2 | 1 |
| Q9 | 3 | 2 | 0 |
| Q32 | 6 | 3 | 1 O177-1 and -2 born 11/18/02 |
| Q37 | 5 | 3 | 0 |
| PL556#3R | 30 | 7 | 2 PL742-744 born 1/13/03 |
Transfers with embryos reconstructed using the 355-F1-derived clonal lines Q2 and Q32 each resulted in one pregnancy to term. A single mummy was recovered from the recipient of Q2-derived embryos. The recipient carrying Q32-derived embryos delivered two live born female piglets, O177-1 and O177-2, by means of caesarian section. O177-1 weighed 575 g at birth, was healthy, and continued normal growth thereafter. Littermate O177-2 was undersized (275 g) and died shortly after delivery.
Transfers with embryos reconstructed using the null selected cell population from neonate PL556 also resulted in two pregnancies to term. Two dead, late-stage fetuses were obtained by caesarian section from one surrogate. The other surrogate farrowed three healthy male piglets (PL742, PL743, and PL744) weighing 550, 320, and 450 g, respectively.
No evidence of cataract formation, seen previously in GGTA1 knockout mice (5, 6), or other phenotypic differences between the GGTA1 null pigs and naturally produced WT miniature swine were observed.
Molecular Analysis of GGTA1 Null Pigs. DNA from the five NT piglets described above was analyzed by quantitative genomic Southern blotting and microsatellite analysis. Piglets O177-1 and O177-2 were found to be hemizygous for the targeted GGTA1 allele, consistent with their derivation from the Q32 donor cell clone (Fig. 4). Also as expected, both piglets were heterozygous for marker Sw2518.
Fig. 4.
Quantitative Southern blot analysis of GGTA1 null piglets. DNA from piglets O177-1 and O177-2 (produced by NT with null fibroblast clone Q32) and piglets PL742-744 (produced by NT with the PL556#3R NAb/complement selected fibroblast population) was analyzed as described in the legend to Fig. 3. DNA from heterozygous 355-F1 and PL556 fibroblasts, without NAb/complement selection, served as controls. Shown are DQβ locus (▪), targeted GGTA1 allele ( ), and WT GGTA1 allele (□).
), and WT GGTA1 allele (□).
In contrast to the Q32 derived piglets, all three piglets (PL742-744) derived from the nonclonal PL556 null selected cell line were homozygous for the gene-targeted GGTA1 allele (Fig. 4). A single heterozygous microsatellite marker, Sw1430, was found from among 26 markers tested in piglet PL556. This centromere proximal marker remained heterozygous in all three PL556 derived GGTA1 homozygous piglets. As with homozygous cell clones Q9 and Q37, no nucleotide heterozygosities were found at the junctions of GGTA1 exon 9 and the G418 selection cassette in the three homozygous piglets (data not shown). Consistent with the hemizygous and homozygous targeted genotypes of these piglets, only RNA compatible with transcription from a targeted locus was observed upon Northern blot analysis (Fig. 5).
Fig. 5.
GGTA1 expression in fibroblasts from null piglets and progenitors. A Northern blot of poly(A)+ RNA was hybridized to a 1.4-kb probe containing portions of exons 2-9 of the GGTA1 gene. WT F7 fibroblasts express a 3.6-kb transcript whereas cells from 355-F1 and PL556 heterozygotes express both a 3.6-kb transcript from the WT locus and a 4.7-kb transcript from the targeted locus. Only the 4.7-kb transcript is detected in fibroblasts from the null piglets.
Phenotypic Characterization of Cells from GGTA1 Null Piglets. Fibroblast cultures from ear explants of the four surviving null piglets were stained with FITC-labeled BS-I-B4 lectin and examined by flow cytometry for evidence of cell surface Galα-1,3-Gal epitopes (Fig. 6 A and B). No fluorescence above that obtained with unstained fibroblasts was observed. Similar results were obtained with Galα-1,3-Gal-specific mAb M86 (not shown). Epitope expression was also examined on multilineage white blood cells from O177-1 at 6 weeks of age, with none detectable (Fig. 6C).
Fig. 6.
Flow cytometry analysis of Galα-1,3-Gal epitopes on GGTA1 null piglets and progenitors using BS-I-B4 lectin. Stained heterozygous or WT cells (red) and unstained cells (black) served as positive and negative controls. (A) Stained (blue) and unstained (black) ear fibroblasts from GGTA1 null piglet O177-1 and fetal fibroblasts from heterozygous progenitor 355-F1 (red). (B) Stained ear fibroblasts from null piglets PL742 (orange), PL743 (blue), and PL744 (green); stained (red) and unstained (black) fetal fibroblasts from heterozygous progenitor 355-F1. (C) Stained (blue) and unstained (black) multilineage white blood cells from O177-1 at 6 weeks of age and stained WBC from an age-matched WT control (red).
Susceptibility of fibroblasts from null piglet O177-1 to complement-mediated lysis by purified baboon anti-Galα-1,3-Gal antibodies and heat inactivated human sera was assessed by using enzyme release and metabolic activity assays (Fig. 7). With purified antibodies, the EC50 for WT and GGTA1 heterozygous fibroblasts was <20 μg/ml in both assays whereas concentrations up to 250 μg/ml had no effect on O177-1 cells. Similarly, O177-1 fibroblasts also have far greater resistance to human sera and complement although partial lysis and metabolic inhibition were seen at the highest serum concentration.
Fig. 7.
Complement-mediated lysis of cells after incubation with purified baboon NAb or human sera. Fibroblasts from GGTA1 null piglet O177-1, WT progenitor fetus F7, and heterozygous progenitor fetus 355-F1 were incubated with the indicated concentrations of affinity-purified polyclonal baboon NAb or heat-inactivated pooled human sera before lysis with rabbit complement. Normal human dermal fibroblasts (NHDF) served as a Galα-1,3-Gal negative control. Release of LDH is expressed as percent of total activity after detergent lysis. Residual metabolic activity, measure as MTS conversion, is expressed as percent of conversion without incubation in complement. Data are the average of three trials.
Discussion
Using NT with cells selected for loss of GGTA1 expression from gene-targeted heterozygous cell populations, we have produced healthy null piglets with two distinct LOH genotypes. The stringent selection for loss of function available for this locus permitted efficient selection of GGTA1 null cell clones from heterozygous fetal cell cultures and sufficient enrichment from heterozygous neonatal cell cultures for use directly in NT.
The WT fetal progenitors for the piglets reported here were two inbred miniature swine, with inbreeding coefficients of 0.86 and 0.91 for fetuses F7 and F501, respectively. Mouse cloning experiments have demonstrated a severe decrease in viability of highly inbred embryos generated entirely by NT and, from many strains, an absolute failure to obtain viable mice (15, 16). Inbreeding in the miniature swine lines, at least to this point, has not resulted in a dramatic decrease in viability although some decrease in NT efficiency in comparison with commercial lines used in unrelated studies seems likely. However, the nature and frequency of spontaneous second allele mutations seems to be greatly influenced by the inbred genetic background. The recovery rate of mutant cells we observe resulting from LOH, ≈10-4, is several orders of magnitude greater than that typically expected in mammalian somatic cells. We have obtained similar results after null selection of fibroblasts from inbred GGTA1 heterozygous piglets produced by mating (D.K-S. and D.J.J.R., unpublished observations), indicating that the high LOH rate is unrelated to the NT process itself. Furthermore, our results contrast markedly with those obtained in attempts to isolate GGTA1 null cells on a commercial genetic background, in which mutants were obtained at rates of <10-6. Phelps et al. (17) used Toxin A from Clostridium difficile to select against Galα-1,3-Gal epitope-bearing cells after transfection of heterozygously targeted cells with a second targeting vector. Although no doubly targeted cell clones were obtained, a single clone with a missense mutation in the nontargeted allele was isolated. Sharma et al. (18), using a similar antibody and complement selection with heterozygously targeted cells, isolated 11 resistant cell clones from a starting population totaling 2 × 107 cells. Southern blot analysis of two of these cell clones indicated loss of the nontargeted allele although the mechanism of loss was not further investigated and no pigs were produced by using the antibody resistant lines.
Although not observed in noninbred pigs, the rate of LOH in our inbred pig lines is almost identical to that reported for 2,6-diaminopurine (DAP) selection of spontaneously generated null fibroblasts from heterozygous mice bearing a gene-targeted Aprt allele. Shao et al. (19), using 129/Sv × C3H/HeJ hybrids, observed LOH in 92 of 113 DAP-resistant clones. In all cases, LOH resulted from somatic crossovers that occurred at various points, with a distribution biased toward the region just proximal to the Aprt locus. The propensity for somatic crossing over in this system was subsequently found to be dependent on chromosomal homology because hybrid mice bearing the relevant homologs from distantly related strains yielded much lower frequencies of spontaneous Aprt mutant fibroblasts and none of the mutant cell clones recovered were recombination derived (20). Thus, it seems likely that the GGTA1 homozygous cells used in NT to produce the PL742-44 null piglets also arose through somatic crossing over. However, due to the lack of heterozygous markers in the F501 fetal progenitor, a gene conversion mechanism cannot be formally excluded. It is interesting to note that, if the homozygous donor cells did arise through a somatic crossover, then the pigs would carry a partial uniparental disomy for chromosome 1 distal to the crossover.
In slight contrast to the above Aprt studies, Ponomareva et al., using the same gene-targeted Aprt allele on a C57BL/6 × DBA/2 background, selected spontaneously generated mutant cell clones with similarly high rates of both somatic crossover events and interstitial deletions (21). We have also recovered both hemizygous and homozygous null lines in all clonal null selection experiments performed with GGTA1 heterozygous lines. Whereas neither deletional breakpoint seems to map within the GGTA1 locus in hemizygous piglet O177-1, interstitial deletion mutant cell clones with one or both breakpoints within the GGTA1 locus have been isolated in selections of other heterozygous fetal and neonatal ear fibroblast lines (D.K-S. and D.J.J.R., unpublished observations). Thus, for deletional events at least, LOH in our inbred derived fibroblast lines is a heterogeneous process.
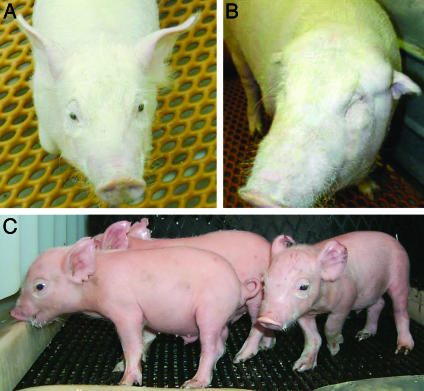
Heterozygous piglets O212-2 and PL556, from which ear fibroblasts were isolated for second allele mutation selection, were produced early in the program and both were developmentally deficient. PL556 was undersized and died shortly after birth from acute respiratory distress. Although in good health and reproductively sound to date, O212-2 was born with one eye, small earflaps, and no patent ear canals (11). A variety of congenital abnormalities have been reported in cloned pigs (11, 22) as well as other species (22, 23), some of which arise through epigenetic errors in reprogramming (24). Unlike the joint and cardiopulmonary defects not uncommonly observed, the deficits in O212-2 must have arisen at a relatively early stage of development. Whereas it might be assumed that these aberrant phenotypes would be magnified only by additional in vitro manipulation and NT, the generation of normal, healthy piglets reported here (Fig. 8) clearly demonstrates that production of normal NT-derived progenitor animals is neither required, nor necessarily advantageous, when performing sequential genetic modifications.
Fig. 8.
GGTA1 null and heterozygous pigs produced by NT. (A) Third-round NT piglet O177-1, produced using GGTA1 null donor cells selected from second-round heterozygous fetus 355-F1 (age 66 days). (B) First-round NT pig O212-2 (11), ear fibroblasts from which served as NT donor cells for fetus 355-F1 (age 3 months). Eye and ear defects in this pig are not observed in O177-1, nor were they apparent at 33 days gestation in fetus 355-F1 or any of its 11 clonal littermates. (C) Second-round NT piglets PL742-744, produced using GGTA1 null donor cells selected from first round heterozygous neonate PL556 (age 9 days).
Serial NT has allowed us to produce α-1,3-galactosyltransferase null piglets in a considerably shorter timeframe than would be required for standard breeding from heterozygotes. Although the inbred miniature swine lines used here were chosen specifically for their advantages for xenotransplantation (25), they also serve to demonstrate the dramatic effect that genetic background can have on rates of spontaneous somatic mutational events in a large animal model. That somatic recombinational and deletional events are not necessarily associated with other deleterious events suggests that appropriate strain selection or construction may be useful in introducing some genetic modifications by means of NT.
Acknowledgments
We thank K. Barnhart, A. Bonk, T. Bruss, M. Duggan, J. Endres, P. Golueke, R. Koppang, G. Lange, K. Mallon, C. Patience, M. Samuel, and H. Schuurman for their contributions to this work. This work was supported in part by grants from the Advanced Technology Program (National Institute of Standards and Technology) and the Small Business Innovative Research Program (National Center for Research Resources, National Institutes of Health).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NAb, natural antibody; LOH, loss of heterozygosity; NT, nuclear transfer; Galα-1,3-Gal, galactosyl-α-1,3-galactose; LDH, lactate dehydrogenase; MTS, [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt].
References
- 1.Cooper, D. K., Koren, E. & Oriol, R. (1993) Lancet 342, 682-683. [DOI] [PubMed] [Google Scholar]
- 2.Lambrigts, D., Sachs, D. H. & Cooper, D. K. (1998) Transplantation 66, 547-561. [DOI] [PubMed] [Google Scholar]
- 3.Galili, U., Clark, M. R., Shohet, S. B., Buehler, J. & Macher, B. A. (1987) Proc. Natl. Acad. Sci. USA 84, 1369-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanemura, M., Maruyama, S. & Galili, U. (2000) Transplantation 69, 187-190. [DOI] [PubMed] [Google Scholar]
- 5.Tearle, R. G., Tange, M. J., Zannettino, Z. L., Katerelos, M., Shinkel, T. A., Van Denderen, B. J., Lonie, A. J., Lyons, I., Nottle, M. B., Cox, T., et al. (1996) Transplantation 61, 13-19. [DOI] [PubMed] [Google Scholar]
- 6.Thall, A. D., Maly, P. & Lowe, J. B. (1995) J. Biol. Chem. 270, 21437-21440. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, K. H., McWhir, J., Ritchie, W. A. & Wilmut, I. (1996) Nature 380, 64-66. [DOI] [PubMed] [Google Scholar]
- 8.Betthauser, J., Forsberg, E., Augenstein, M., Childs, L., Eilertsen, K., Enos, J., Forsythe, T., Golueke, P., Jurgella, G., Koppang, R., et al. (2000) Nat. Biotechnol. 18, 1055-1059. [DOI] [PubMed] [Google Scholar]
- 9.Park, K. W., Cheong, H. T., Lai, L., Im, G. S., Kuhholzer, B., Bonk, A., Samuel, M., Rieke, A., Day, B. N., Murphy, C. N., et al. (2001) Anim. Biotechnol. 12, 173-181. [DOI] [PubMed] [Google Scholar]
- 10.Polejaeva, I. A., Chen, S. H., Vaught, T. D., Page, R. L., Mullins, J., Ball, S., Dai, Y., Boone, J., Walker, S., Ayares, D. L., et al. (2000) Nature 407, 86-90. [DOI] [PubMed] [Google Scholar]
- 11.Lai, L., Kolber-Simonds, D., Park, K. W., Cheong, H. T., Greenstein, J. L., Im, G. S., Samuel, M., Bonk, A., Rieke, A., Day, B. N., et al. (2002) Science 295, 1089-1092. [DOI] [PubMed] [Google Scholar]
- 12.Tischfield, J. A. & Shao, C. (2003) Nat. Genet. 33, 5-6. [DOI] [PubMed] [Google Scholar]
- 13.You, Y., Bergstrom, R., Klemm, M., Lederman, B., Nelson, H., Ticknor, C., Jaenisch, R. & Schimenti, J. (1997) Nat. Genet. 15, 285-288. [DOI] [PubMed] [Google Scholar]
- 14.Sablinski, T., Gianello, P. R., Bailin, M., Bergen, K. S., Emery, D. W., Fishman, J. A., Foley, A., Hatch, T., Hawley, R. J., Kozlowski, T., et al. (1997) Surgery 121, 381-391. [DOI] [PubMed] [Google Scholar]
- 15.Eggan, K., Akutsu, H., Loring, J., Jackson-Grusby, L., Klemm, M., Rideout, W. M., 3rd, Yanagimachi, R. & Jaenisch, R. (2001) Proc. Natl. Acad. Sci. USA 98, 6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakayama, T. & Yanagimachi, R. (2001) Mol. Reprod. Dev. 58, 376-383. [DOI] [PubMed] [Google Scholar]
- 17.Phelps, C. J., Koike, C., Vaught, T. D., Boone, J., Wells, K. D., Chen, S. H., Ball, S., Specht, S. M., Polejaeva, I. A., Monahan, J. A., et al. (2003) Science 299, 411-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma, A., Naziruddin, B., Cui, C., Martin, M. J., Xu, H., Wan, H., Lei, Y., Harrison, C., Yin, J., Okabe, J., et al. (2003) Transplantation 75, 430-436. [DOI] [PubMed] [Google Scholar]
- 19.Shao, C., Stambrook, P. J. & Tischfield, J. A. (2001) Nat. Genet. 28, 169-172. [DOI] [PubMed] [Google Scholar]
- 20.Shao, C., Deng, L., Henegariu, O., Liang, L., Raikwar, N., Sahota, A., Stambrook, P. J. & Tischfield, J. A. (1999) Proc. Natl. Acad. Sci. USA 96, 9230-9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponomareva, O. N., Rose, J. A., Lasarev, M., Rasey, J. & Turker, M. S. (2002) Cancer Res. 62, 1518-1523. [PubMed] [Google Scholar]
- 22.Carter, D. B., Lai, L., Park, K. W., Samuel, M., Lattimer, J. C., Jordan, K. R., Estes, D. M., Besch-Williford, C. & Prather, R. S. (2002) Cloning Stem Cells 4, 131-145. [DOI] [PubMed] [Google Scholar]
- 23.Hill, J. R., Roussel, A. J., Cibelli, J. B., Edwards, J. F., Hooper, N. L., Miller, M. W., Thompson, J. A., Looney, C. R., Westhusin, M. E., Robl, J. M., et al. (1999) Theriogenology 51, 1451-1465. [DOI] [PubMed] [Google Scholar]
- 24.Rideout, W. M., 3rd, Eggan, K. & Jaenisch, R. (2001) Science 293, 1093-1098. [DOI] [PubMed] [Google Scholar]
- 25.Sachs, D. H. (1994) Vet. Immunol. Immunopathol. 43, 185-191. [DOI] [PubMed] [Google Scholar]