Abstract
L3MBTL encodes a member of the Polycomb family of proteins, which, together with Trithorax group proteins, is responsible for the coordinated regulation of patterns of gene activity. Members of the Polycomb family also regulate self renewal of normal and malignant hematopoietic stem cells. L3MBTL lies in a region of chromosome 20, deletion of which is associated with myeloid malignancies and represents a good candidate for a 20q target gene. However, mutations of L3MBTL have not been identified in patients with 20q deletions or in cytogenetically normal patients. Here we demonstrate that monoallelic methylation of two CpG islands correlates with transcriptional silencing of L3MBTL, and that L3MBTL transcription occurs from the paternally derived allele in five individuals from two families. Expression of the paternally derived allele was observed in multiple hematopoietic cell types as well as in bone marrow derived mesenchymal cells. Deletions of 20q associated with myeloid malignancies resulted in loss of either the unmethylated or methylated allele. Our results demonstrate that L3MBTL represents a previously undescribed imprinted locus, a vertebrate Polycomb group gene shown to be regulated by this mechanism, and has implications for the pathogenesis of myeloid malignancies associated with 20q deletions.
Establishing and maintaining cellular identity require the coordinated regulation of patterns of gene activity. Polycomb group (PcG) and Trithorax group genes are central to this process and respectively repress or activate the transcription of multiple loci (1, 2). Two types of PcG complexes can be distinguished in Drosophila and mammals. One class (PcGe) is primarily involved in establishing the silent state and includes the extra sex combs and enhancer of zeste proteins. The second type of complex (PcGm) is mainly required for stable maintenance of gene silencing and is exemplified in Drosophila by the PRC1 complex. Several lines of evidence suggest that these two categories of PcG complexes regulate normal and malignant mammalian hematopoiesis in contrasting ways (2, 3). For example, PcGe components such as ectoderm embryonic development repress the self renewal of hematopoietic progenitors, function as tumor suppressor genes, and contribute to the maintenance of repression at imprinted loci (4, 5), whereas bmi-1, a constituent of PcGm complexes, is essential for self renewal of normal and malignant hematopoietic stem cells (6, 7).
L3MBTL is a member of a growing family of Polycomb-like proteins characterized by at least two conserved mbt domains that are also present in the Drosophila Polycomb group gene Sex comb on midleg (Scm) (8). Other human members include SCMH1 (9), SCML2 (10), SFMBT/RU1 (11), and L3MBTL2 (12). With the exception of L3MBTL2, all these proteins also possess a conserved C-terminal SPM domain, which, in the case of L3MBTL, mediates interaction with translocation Ets leukemia (TEL) (13).
L3MBTL was identified as a candidate tumor suppressor gene by virtue of its homology to the Drosophila tumor suppresser gene, lethal(3) malignant brain tumor (14, 15). Inactivation of the D-l(3)mbt gene results in overgrowth of adult optic neuroblasts and ganglion mother cells of the larval brain (16). D-l(3)mbt is expressed throughout larval development in a number of organ systems, including the hematopoietic system (15). L3MBTL and D-l(3)mbt show extensive similarity especially within the “mbt repeat” domain, a C2HC zinc finger domain, and the C-terminal SPM domain (14).
L3MBTL localizes to the nucleus and has been shown to associate with condensed chromosomes during mitosis in U2OS osteosarcoma cells. Overexpression of L3MBTL in U251MG glioma cells leads to multinucleate cells suggesting that it plays a role in the correct progression of a cell through mitosis (14). Recent data have suggested that L3MBTL functions as an histone deacetylase-independent transcriptional repressor, which can be recruited by direct binding to translocation Ets leukemia (TEL) (13). It is widely expressed in normal adult tissues and cancer cell lines (14) and, of particular note, it is expressed within purified CD34-positive hematopoietic progenitors (17).
L3MBTL maps to the long arm of human chromosome 20, a region that undergoes acquired deletion in a subset of patients with myeloid malignancies. A 2-Mb common deleted region has been defined by using samples from patients with a myeloproliferative disorder, a myelodysplastic syndrome, or acute myeloid leukemia. This region contains 16 genes, of which 6 are expressed within normal CD34+ cells (ref. 17 and A.J.B., J.L., B. J. P. Huntly, E. Delabesse, N.F., A. R. Hunt, P. Deloukas, and A.R.G., unpublished work). Because myeloid malignancies are thought to result from transformation of multipotent progenitors within the CD34+ population, these six genes, which include L3MBTL, are prime candidates for a 20q tumor suppressor gene. We have characterized the intron/exon structure of L3MBTL and demonstrated that it is not mutated in 27 patients with 20q deletions or in 19 cytogenetically normal patients with a myeloproliferative disorder (A.J.B., J.L., B. J. P. Huntly, E. Delabesse, N.F., A. R. Hunt, P. Deloukas, and A.R.G., unpublished work). Here we demonstrate that L3MBTL is imprinted with exclusive expression of the paternally derived allele in hematopoietic cells. This observation has implications for the pathogenesis of myeloid malignancies associated with 20q deletions and is a previously undescribed member of the vertebrate PcG family, shown to undergo imprinting.
Materials and Methods
Samples. Bone marrow and peripheral blood were obtained after informed consent from normal individuals and patients carrying an acquired 20q deletion. Bone marrow, granulocytes, and T cells were isolated as described (18). Peripheral blood platelets were purified by three steps of centrifugation at 150× g for 20 min at room temperature. The purity of granulocytes, T cells, and platelets was >95%. Genomic DNA was prepared by standard methods (19). Total RNA was extracted from cells by using TRI Reagent according to the manufacturer's instructions (Sigma-Aldrich).
Purification of Hematopoietic Stem Cells. Mononuclear cells were obtained from normal bone marrow by centrifugation over Histopaque 1077 (Sigma-Aldrich). CD34+ hematopoietic progenitor cells were isolated by immunomagnetic selection with the Miltenyi MiniMACs CD34 Multisort Isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany). Purity of the CD34+ cells was 88%. After staining of the cells with PE-CD34 (Becton Dickinson) and PC5-CD38 (Beckman Coulter), CD34+/CD38- (representing early stem cells) and CD34+/CD38+ (representing progenitor cells) subpopulations were simultaneously purified by fluorescence-activated cell sorting.
Bone Marrow Mesenchymal Cells. Mononuclear cells were obtained from a bone marrow donor by centrifugation over Histopaque 1077 (Sigma-Aldrich). CD34- cells were collected after isolation of CD34+ cells as described above and were immediately cultured as described (20). After 4 weeks, the adherent bone marrow mesenchymal cells were harvested, and flow cytometry by using FITC-CD45 (marker for all hematopoietic lineages) and PE-CD105 (marker for bone marrow stromal mesenchymal cells) showed them to be 99.5% CD105+/CD45-.
Methylation Analysis. One microgram of genomic DNA was modified with sodium bisulfite by using the CpGenome DNA modification kit according to the manufacturer's instructions (Intergen, Purchase, NY). Universally methylated male control DNA (Intergen) was treated in the same way. The CpG island regions were amplified from modified DNA by two rounds of PCR by using primers designed to recognize the bisulfite-modified sequence. Details of primers and PCR conditions are available upon request.
In all cases, PCR products obtained from the second round of amplification were purified, sequenced, and assessed by single-strand conformation polymorphism (SSCP) as described (21), except gel conditions were 0.6× mutation enhancement detection with 5% glycerol for CpG island 3 and 0.5 × MDE for CpG island 4. In addition, the PCR products for all four CpG islands were sequenced. A number of PCR products were cloned into pGEM-T Easy (Promega) according to the manufacturer's instructions, and clones were sequenced.
Identification of Transcribed Polymorphisms in the L3MBTL Gene. Using oligonucleotide primers spanning the entire transcribed region, PCR amplification was performed on DNA from 150 individuals as described (A.J.B., J.L., B. J. P. Huntly, E. Delabesse, N.F., A. R. Hunt, P. Deloukas, and A.R.G., unpublished work). PCR products were subjected to SSCP and heteroduplex analysis as described (21), and those demonstrating an unusual pattern of bands were purified and sequenced.
RT-PCR for Analyzing Allele-Specific Expression. RT-PCR was performed on total RNA from informative samples. Total RNA was treated with DNase I (Sigma-Aldrich) to remove DNA in a final volume of 10 μl containing 1 unit of DNase I, and the reaction was terminated by adding 1 μl of stop buffer (Sigma-Aldrich). Four hundred picomole of pd(n)6 random hexamers (Amersham Pharmacia) was added to the treated RNA, and the mixture was denatured at 70°C for 10 min followed by cooling to 2°C for 2 min to allow the hexamers to anneal. The RT reaction was performed at 37°C for 90 min followed by 70°C for 10 min in a 20-μl volume containing 1× first-strand buffer (Invitrogen), 10 mM DTT (Invitrogen), 20 units of RNasin (Promega), 1 mM each dNTP, and 200 units of M-MLV Reverse Transcriptase (Invitrogen). One to two microliters of the RT reaction was used for PCR by using the primers indicated in Table 1, and all RT-PCR products were directly sequenced. PCR amplification of both genomic DNA and RNA of individual 1 was performed by using P7 and P8 and was analyzed by digestion with restriction enzyme MscI (New England Biolabs).
Table 1. RT-PCR primers for allelic expression analysis.
| Primers | Primer sequence (5′-3′) | Product size, bp | Location within sequence (GenBank accession no.) | Annealing temperatures, °C | Conditions, mM MgCl2 |
|---|---|---|---|---|---|
| P1 | AAGAAGCCTCGCCATCACG | 112 | 2080-2192 | 56 | 1 |
| P2 | GACATGAAGAGGGACTGGTGC | (U89358) | |||
| P3 | ACCAGTCCCTCTTCATGTCAG | 289 and 407 | 2174-2463 | 59 | 1.5 |
| P4 | AAGATGATCTGAGCACACAAGG | (U89358) | |||
| P5 | AGCGAAGGTTGGGTTTACAA | 580 | 27461-28041 | 59 | 1.5 |
| P6 | GCCAGGGGCCAAAATATAAG | (Z98752) | |||
| P7 | AAATGGAGATGCTGAGGACACTG | 313* | 160466-160976 | 59 | 1.5 |
| P8 | GCCTCTGTCCATTCCAGAAG | (AL031681) |
511-bp PCR product obtained from genomic DNA.
Results
The L3MBTL Gene Undergoes Monoallelic Methylation. The L3MBTL gene lies on the long arm of human chromosome 20, contains 24 exons, and spans 45 kb (Fig. 1A). Two putative promoters have been identified, one upstream of exon 1 and the other upstream of exon 5a (A.J.B., J.L., B. J. P. Huntly, E. Delabesse, N.F., A. R. Hunt, P. Deloukas, and A.R.G., unpublished work). Four CpG islands were predicted by using genebuilder (22): CpG islands 1 and 2 span promoter 1, and CpG islands 3 and 4 are found in the vicinity of exons 4 and 5.
Fig. 1.
Allelic methylation of the L3MBTL gene. (A) Locus of the L3MBTL gene and intron/exon structure of its 5′ and 3′ regions. Coding and noncoding exons are shown in black and gray boxes, respectively. Bent arrows indicate two putative promoters. Patterned horizontal bars represent the four CpG islands. Asterisks indicate the locations of SNPs used in the study, and primers spanning the respective SNPs are indicated (P1-P8). (B) Methylation analysis of the CpG island 3 by using SSCP. Lane 1, universally methylated genomic control DNA; lanes 2-5, normal bone marrow; lanes 6-9, normal granulocytes; lanes 10-13, granulocytes from patients, with an acquired 20q deletion. (C) Bisulfite sequencing of CpG island 3. Sequences relate to samples corresponding to individual lanes from B, as indicated. Underlined thymidines (T) are those converted from cytosines.
To assess the methylation status of these CpG islands, methylation analysis was performed by using bisulfite modification followed by SSCP analysis and sequencing. The results for CpG island 3 are shown in Fig. 1B. The PCR product amplified from fully methylated control DNA gave rise to a single SSCP band (lane 1). By contrast, PCR products amplified from normal bone marrow and peripheral blood granulocytes gave rise to two bands of equal intensity (lanes 2-9), one of which exhibited the same mobility as the methylated control band. Direct sequencing of the PCR products demonstrated that the CpG dinucleotides (but not other cytosines) were partially protected from the effects of bisulfite modification (Fig. 1C). Cloning and sequence analysis of PCR products from six normal individuals confirmed that the CpG dinucleotides were either all methylated or all unmethylated in each individual clone (data not shown).
The methylation status of CpG island 3 was then assessed in four patients with myeloid malignancies associated with an acquired deletion of the long arm of chromosome 20, which removes one copy of the L3MBTL locus. In these four patients, microsatellite PCR had previously demonstrated that, in purified peripheral blood granulocytes or bone marrow cells, >90% of the cells contained the 20q deletion (18). Bisulfite modification followed by SSCP analysis or sequencing of PCR products showed that two patients retained a methylated allele (Fig. 1 B and C, lanes 11 and 13), whereas the other two patients retained an unmethylated allele (Fig. 1 B and C, lanes 10 and 12). These data are consistent with monoallelic methylation of CpG island 3 and loss of either the methylated allele or the unmethylated allele in patients with a 20q deletion.
Analysis of CpG island 4 gave similar results. Two SSCP bands of equal intensity were observed in all normal samples, whereas samples from patients with an acquired 20q deletion retained a single band corresponding to either the methylated or unmethylated allele (data not shown). Moreover, the methylation results for CpG islands 3 and 4 were fully concordant in all four patients. Taken together, these data imply monoallelic methylation of CpG islands 3 and 4, which span promoter 2. By contrast, CpG islands 1 and 2 were completely unmethylated in all normal individuals and in the four patients (data not shown).
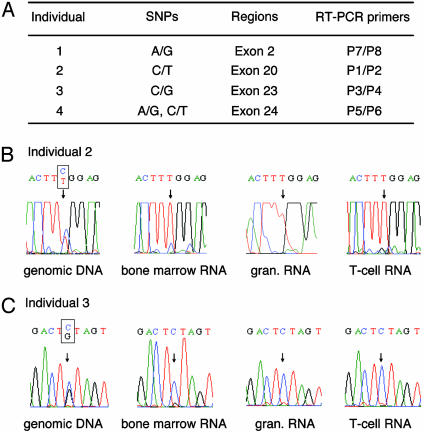
Monoallelic Expression of the L3MBTL Gene in Hematopoietic Cells. To determine whether L3MBTL exhibits monoallelic expression, it was necessary to identify transcribed single-nucleotide polymorphisms (SNPs) within L3MBTL. Samples from 150 individuals were screened for SNPs by SSCP analysis followed by sequencing. Four informative individuals were identified (Fig. 2A); individual 1 with an A/G SNP in exon 2, individual 2 with a C/T SNP in exon 20, individual 3 with a C/G SNP in exon 23, and individual 4 with two SNPs of A/G and C/T in exon 24. RT-PCR was performed by using RNA from peripheral blood granulocytes, peripheral blood T cells, and bone marrow cells or peripheral blood nucleated cells obtained from all four individuals. Sequence analysis of the RT-PCR products demonstrated monoallelic expression in all samples; the results for individual 2 and 3 are shown in Fig. 2 B and C, respectively.
Fig. 2.
Monoallelic expression of the L3MBTL gene. (A) Detail of SNPs identified in four individuals. Individual 1, patient with polycythemia vera and an acquired 20q deletion; individuals 2 and 3, patients with myeloproliferative disorders with normal karotype; individual 4, normal individual. (B) Sequences of genomic DNA and RT-PCR products from individual 2. (C) Sequences of genomic DNA and RT-PCR products from individual 3.
The Paternally Derived Allele of the L3MBTL Is Expressed in Hematopoietic Cells. Two different mechanisms may produce monoallelic expression of mammalian autosomal genes. Imprinting is characterized by monoallelic expression that is determined by the parental origin of each allele. By contrast, the term allelic exclusion is used to describe apparently random monoallelic expression at the single cell level, which appears to reflect stochastic inactivation of one or the other allele.
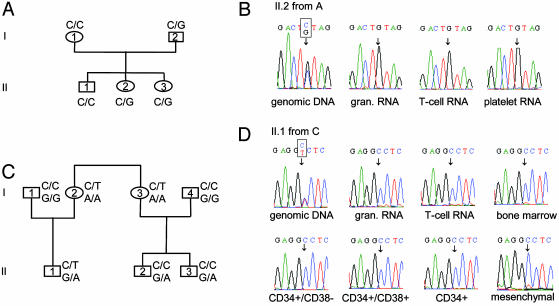
To distinguish between these mechanisms, family members of the four individuals were screened for relevant SNPs, and informative relatives were identified for individuals 3 and 4. The family of individual 3 is shown in Fig. 3A. The father and both daughters were informative for a C/G SNP in exon 23. RNA was purified from peripheral blood granulocytes, peripheral blood T cells, and platelets obtained from one daughter. RT-PCR was performed by using primers P3 and P4 (Fig. 1 A) and the products analyzed by direct sequencing. As shown in Fig. 3B, the paternally derived “G” allele was expressed in all three cell types. The same result was obtained by using RNA extracted from total peripheral blood nucleated cells obtained from the second daughter (data not shown).
Fig. 3.
Paternal expression of the L3MBTL gene. (A) Family tree of individual 3 indicating the exon 23 SNP. (B) Sequences of genomic DNA and RT-PCR products from II.2 in A demonstrating expression of paternal “G” allele in peripheral blood granulocytes, T cells, and platelets. (C) Family tree of individual 4 indicating the exon 24 SNPs. (D) Sequences of genomic DNA and RT-PCR products from II.1 in C demonstrating expression of paternal “C” allele in all cell populations studied.
The family of individual 4 is shown in Fig. 3C. The proband (II.1) carries a C/T SNP as well as an A/G SNP in exon 24. RT-PCR analysis by using primers P5 and P6 demonstrated that peripheral blood granulocytes, T cells, and bone marrow all displayed exclusive expression of the paternally derived “C” allele (Fig. 3D). Total peripheral blood nucleated cells from family members II.2 and II.3 (Fig. 3C) were analyzed for expression of the A/G SNP in exon 24. Again, exclusive expression of the paternally derived “G” allele was demonstrated (data not shown). To investigate whether L3MBTL is monoallelically expressed in the hematopoietic stem cell compartment, CD34+ and CD34+CD38- cells, a population known to be enriched for hematopoietic stem cells, were purified from bone marrow obtained from II.1. There was exclusive expression of the paternally derived “C” allele in both these populations of cells as well as in CD34- and CD34+ CD38+ cells (Fig. 3D and data not shown). To investigate whether L3MBTL is monoallelically expressed in nonhematopoietic cells, the bone marrow of II.1 was cultured to obtain mesenchymal cells (99.5% CD105+ CD45-), and monoallelic expression of the paternal allele was again demonstrated (Fig. 3D).
Taken together, these data demonstrate expression of the paternally derived allele in five individuals from two different families. Moreover, monoallelic expression of L3MBTL was observed in the hematopoietic stem cell compartment, in differentiated cells representing three different hematopoietic lineages as well as in bone marrow mesenchymal cells.
Methylation of CpG Islands 3 and 4 Is Associated with Transcriptional Silencing. At some imprinted loci, the methylated allele corresponds to the inactive allele, whereas for other imprinted genes, the methylated allele corresponds to the active allele. To define the relationship between methylation and transcription, we studied individual 1 (Fig. 2A), a patient with polycythemia vera associated with an acquired 20q deletion, who carries an inherited A/G SNP in exon 2 of L3MBTL. PCR products derived from the “A” but not the “G” allele contain a site for the restriction enzyme MscI. Fig. 4A shows the predicted digestion patterns of PCR products from both genomic DNA and RNA. PCR amplification by using granulocyte and T cell DNA was performed by using primers P7 and P8 and was followed by digestion with MscI. T cell genomic DNA gave rise to two bands corresponding to the “A” and “G” alleles (Fig. 4B, lane 3). By contrast, genomic DNA from peripheral blood granulocytes gave rise to a single band corresponding to the “A” allele (Fig. 4B, lane 2). These results are consistent with the fact that granulocytes but not T cells carry the 20q deletion. Because peripheral blood granulocytes retained the methylated allele in this patient (Fig. 1B, lane 11), these data demonstrate that the “A” allele corresponds to the methylated allele.
Fig. 4.
Methylation of CpG islands 3 and 4 is associated with transcriptional silencing. (A) Diagram of 5′ region of L3MBTL locus showing the MscI site, PCR primers (P7, P8) and PCR products derived from either RNA or genomic DNA. *, A/G SNP in exon 2. (B) PCR amplification of genomic DNA or RNA from individual 1 was followed by MscI digestion. Similar results were obtained in two independent experiments. Lane 1, undigested PCR product of granulocyte genomic DNA; lanes 2 and 3, MscI digested PCR products of genomic DNA from granulocytes and T cells, respectively; lane 4, undigested RT-PCR product from T cells; lane 5, MscI digested RT-PCR product from T cells.
To determine which L3MBTL allele was transcribed, RT-PCR was performed by using RNA from T cells, because they contain both alleles of L3MBTL. Digestion with MscI gave rise to a single predominant band corresponding to the “G” allele (Fig. 4B, lane 5), demonstrating that the “G” allele was transcribed. Direct sequencing of the PCR product confirmed this observation (data not shown). Taken together, our results demonstrate that in individual 1, the L3MBTL allele, which lacks methylation of CpG islands 3 and 4, is transcriptionally active.
Discussion
In this paper, we demonstrate that L3MBTL represents a previously undescribed imprinted locus, a vertebrate PcG gene found to be regulated by this mechanism. L3MBTL was shown to exhibit exclusive expression of the paternally derived allele in multiple hematopoietic progenitors and in several differentiated hematopoietic cell types. Moreover, methylation of two CpG islands correlates with transcriptional inactivation in normal T cells.
The L3MBTL encodes a complex transcriptional unit with both 5′ and 3′ alternative splicing events and two putative promoters (A.J.B., J.L., B. J. P. Huntly, E. Delabesse, N.F., A. R. Hunt, P. Deloukas, and A.R.G., unpublished work). We have identified four 5′ CpG islands, two of which represent differentially methylated regions. CpG islands 3 and 4 flank promoter 2 and are monoallelically methylated, whereas CpG islands 1 and 2 span promoter 1 and are fully unmethylated. These observations raise the possibility that the two promoters might be differentially imprinted as reported at other loci including GNAS-1 (23). However, our results show this is highly unlikely. L3MBTL has two poly(A) signals contained in exons 23 and 24, suggesting that all transcripts, regardless of promoter usage, terminate in one of these two exons. Analysis of individual 3, who carries a SNP in exon 23, demonstrates that all transcripts ending in exon 23 are monoallelically expressed. Similarly, analysis of individual 4, who carries a SNP in exon 24, demonstrates that all transcripts ending in exon 24 are also monoallelically expressed. These data therefore suggest that in hematopoietic cells, all L3MBTL transcripts are monoallelically expressed. Because transcripts derived from both promoters are readily detectable in normal peripheral blood granulocytes and T cells (data not shown), our results indicate that neither promoter 1 nor promoter 2 exhibit biallelic activity.
Differentially methylated regions play varying roles in epigenetic regulation operating at distinct imprinted loci. Usually allelic methylation correlates with transcriptional silencing (24-26), but there are exceptions; for example, IGF2 is methylated on the active paternal copy (27). To correlate allelic methylation with transcription, we took advantage of a rare patient who carried an acquired 20q deletion in peripheral blood granulocytes, as well as being informative for a transcribed SNP in exon 2. Analysis of T cell samples in which both alleles were intact demonstrated that the same allele was methylated and transcriptionally inactive. However, it is important to note that this individual suffered from polycythemia vera, and we cannot exclude the possibility of aberrant methylation in the patient's clonally derived granulocytes.
Analysis of the parental origin of the expressed allele demonstrated that five informative individuals from two families exhibited exclusive expression from the paternally derived allele. Given these results, allelic exclusion seems highly unlikely, especially because all known examples involve stochastic allelic inactivation at the single cell level and do not give rise to inactivation of the same allele in all cells within a polyclonal population (28-31). Instead, our data indicate that L3MBTL undergoes imprinting with inactivation of the maternally derived allele.
Imprinted genes are functionally haploid and as a consequence are frequent targets for inherited or acquired alterations associated with disease (32, 33). The L3MBTL gene is located in a region that is commonly deleted in myeloid malignancies and within a large chromosomal region subject to epigenetic regulation. Two other imprinted loci, NNAT (34) and GNAS-1 (35), are known to be present on the long arm of chromosome 20 but lie, respectively, 6 Mb centromeric and 15 Mb telomeric to the L3MBTL. L3MBTL may therefore represent a previously undescribed imprinted domain, and it will be important to characterize the mechanisms responsible for the epigenetic regulation of the locus.
Is imprinting of L3MBTL relevant to the pathogenesis of myeloid malignancies associated with 20q deletions? In nonhematological malignancies, several imprinted genes, including WT1 (36, 37), p57KIP2 (38-40), and M6P/IGF2R (41-43), have been reported to have tumor suppressive functions. L3MBTL is a good candidate for 20q target gene. It lies in the common deleted region; is expressed in CD34+ cells, which include the hematopoietic progenitors from which myeloid malignancies arise; and is homologous to a Drosophila tumor suppressor protein (A.J.B., J.L., B. J. P. Huntly, E. Delabesse, N.F., A. R. Hunt, P. Deloukas, and A.R.G., unpublished work). Moreover, L3MBTL is a member of the Polycomb family, other members of which regulate self renewal of normal and malignant hematopoietic progenitors and stem cells (6, 7). A systematic survey of all coding exons, noncoding exons, and splice sites has failed to identify any mutations in patients either with or without 20q deletions (A.J.B., J.L., B. J. P. Huntly, E. Delabesse, N.F., A. R. Hunt, P. Deloukas, and A.R.G., unpublished work). However, the demonstration that L3MBTL is monoallelically expressed raises the possibility that deletion of the paternally derived chromosome 20 may contribute to disease pathogenesis.
Interestingly, our results demonstrate that 20q deletions resulted in loss of either the methylated or the unmethylated allele. We were not able to examine the parental origin of the deleted chromosome, because the myeloid malignancies associated with 20q deletions usually occur in elderly patients, and parental samples are usually unavailable. These data are consistent with at least three interpretations. First, despite circumstantial evidence to the contrary, it remains possible that L3MBTL does not contribute to the pathogenesis of myeloid malignancies. Second, all patients with a 20q deletion may retain the inactive allele with aberrant hypomethylation, explaining why the retained allele appears unmethylated. Third, L3MBTL may function as a target gene in some patients with a 20q deletion (i.e., those who have lost the active paternal allele), with different mechanisms operating in other patients who have lost the inactive maternal allele. This latter possibility is reminiscent of two related but distinct inherited syndromes, Prader-Willi Syndrome (PWS) and Angelman Syndrome (AS), which are associated with 3- to 4-Mb deletions of human chromosome 15q affecting respectively the paternally derived or maternally derived chromosome (44-46). The different phenotype of the two syndromes is thought to reflect the fact that the region contains several imprinted genes, some of which display expression of their paternally derived allele, the loss of which gives rise to PWS, and others with the expression of their maternally derived allele, the loss of which results in AS. We speculate that a similar scenario may apply to acquired 20q deletions, with related but distinct subtypes of myeloid malignancies associated with deletions of the maternally or paternally derived chromosome 20. This model predicts the existence of L3MBTL neighboring genes on 20q, which are expressed from the maternally derived chromosome. It will therefore be important to investigate whether neighboring genes undergo imprinting and to define the extent of the imprinted domain that contains L3MBTL.
Acknowledgments
We thank Dr. Brian Huntly, Dr. Kim Champion, Liz Delaney, Phyllis Paterson, and Dr. Peter Campbell for assistance with the sample collection. We are also grateful to Emma Frith, Dr. Mike Scott, Kevin Jestice, Nigel Miller, and Dr. Asif Qasim for assistance with purification of the hematopoietic progenitors and culturing of the bone marrow mesenchymal cells and to Prof. John Todd (Cambridge Institute for Medical Research) for normal DNA samples. This work was supported by the Leukaemia Research Fund.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PcG, Polycomb group; SNP, single-nucleotide polymorphism; SSCP, single-strand conformation polymorphism.
References
- 1.Orlando, V. (2003) Cell 112, 599-606. [DOI] [PubMed] [Google Scholar]
- 2.Lessard, J. & Sauvageau, G. (2003) Exp. Hematol. (Charlottesville, Va.) 31, 567-585. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs, J. J. (2002) Biochim Biophys Acta 1602, 151-161. [DOI] [PubMed] [Google Scholar]
- 4.Lessard, J., Schumacher, A., Thorsteinsdottir, U., van Lohuizen, M., Magnuson, T. & Sauvageau, G. (1999) Genes Dev. 13, 2691-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mager, J., Montgomery, N. D., de Villena, F. P. & Magnuson, T. (2003) Nat. Genet. 33, 502-507. [DOI] [PubMed] [Google Scholar]
- 6.Park, I. K., Kiel, M., Becker, M. W., Pihalja, M., Weissman, I. L., Morrison, S. J. & Clarke, M. F. (2003) Nature 423, 302-305. [DOI] [PubMed] [Google Scholar]
- 7.Lessard, J. & Sauvageau, G. (2003) Nature 423, 255-260. [DOI] [PubMed] [Google Scholar]
- 8.Bornemann, D., Miller, E. & Simon, J. (1996) Development (Cambridge, U.K.) 122, 1621-1630. [DOI] [PubMed] [Google Scholar]
- 9.Berger, J., Kurahashi, H., Takihara, Y., Shimada, K., Brock, H. W. & Randazzo, F. (1999) Gene 237, 185-191. [DOI] [PubMed] [Google Scholar]
- 10.Montini, E., Buchner, G., Spalluto, C., Andolfi, G., Caruso, A., den Dunnen, J. T., Trump, D., Rocchi, M., Ballabio, A. & Franco, B. (1999) Genomics 58, 65-72. [DOI] [PubMed] [Google Scholar]
- 11.Morel, S., Levy, F., Burlet-Schiltz, O., Brasseur, F., Probst-Kepper, M., Peitrequin, A. L., Monsarrat, B., Van Velthoven, R., Cerottini, J. C., Boon, T., et al. (2000) Immunity 12, 107-117. [DOI] [PubMed] [Google Scholar]
- 12.Wismar, J. (2001) FEBS Lett. 507, 119-121. [DOI] [PubMed] [Google Scholar]
- 13.Boccuni, P., MacGrogan, D., Scandura, J. M. & Nimer, S. D. (2003) J. Biol. Chem. 278, 15412-15420. [DOI] [PubMed] [Google Scholar]
- 14.Koga, H., Matsui, S., Hirota, T., Takebayashi, S., Okumura, K. & Saya, H. (1999) Oncogene 18, 3799-3809. [DOI] [PubMed] [Google Scholar]
- 15.Wismar, J., Loffler, T., Habtemichael, N., Vef, O., Geissen, M., Zirwes, R., Altmeyer, W., Sass, H. & Gateff, E. (1995) Mech. Dev. 53, 141-154. [DOI] [PubMed] [Google Scholar]
- 16.Gateff, E., Loffler, T. & Wismar, J. (1993) Mech. Dev. 41, 15-31. [DOI] [PubMed] [Google Scholar]
- 17.Bench, A. J., Nacheva, E. P., Hood, T. L., Holden, J. L., French, L., Swanton, S., Champion, K. M., Li, J., Whittaker, P., Stavrides, G., et al. (2000) Oncogene 19, 3902-3913. [DOI] [PubMed] [Google Scholar]
- 18.Asimakopoulos, F. A., Gilbert, J. G. R., Aldred, M. A., Pearson, T. C. & Green, A. R. (1996) Blood 88, 2690-2698. [PubMed] [Google Scholar]
- 19.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 20.Oreffo, R. O. C., Virdi, A. S. & Triffitt, J. T. (1997) Eur. J. Cell Biol. 74, 251-261. [PubMed] [Google Scholar]
- 21.Gayther, S. A., Batley, S. J., Linger, L., Bannister, A., Thorpe, K., Chin, S. F., Daigo, Y., Russell, P., Wilson, A., Sowter, H. M., et al. (2000) Nat. Genet. 24, 300-303. [DOI] [PubMed] [Google Scholar]
- 22.Milanesi, L., D'Angelo, D. & Rogozin, I. B. (1999) Bioinformatics 15, 612-621. [DOI] [PubMed] [Google Scholar]
- 23.Peters, J., Wroe, S. F., Wells, C. A., Miller, H. J., Bodle, D., Beechey, C. V., Williamson, C. M. & Kelsey, G. (1999) Proc. Natl. Acad. Sci. USA 96, 3830-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferguson-Smith, A. C., Sasaki, H., Cattanach, B. M. & Surani, M. A. (1993) Nature 362, 751-755. [DOI] [PubMed] [Google Scholar]
- 25.Stoger, R., Kubicka, P., Liu, C. G., Kafri, T., Razin, A., Cedar, H. & Barlow, D. P. (1993) Cell 73, 61-71. [DOI] [PubMed] [Google Scholar]
- 26.Riesewijk, A. M Hu, L., Schulz, U., Tariverdian, G., Hoglund, P., Kere, J., Ropers, H. H. & Kalscheuer, V. M. (1997) Genomics 42, 236-244. [DOI] [PubMed] [Google Scholar]
- 27.Reik, W., Brown, K. W., Slatter, R. E., Sartori, P., Elliott, M. & Maher, E. R. (1994) Hum. Mol. Genet. 3, 1297-1301. [DOI] [PubMed] [Google Scholar]
- 28.Chess, A. S., Simon, I., Cedar, H. & Axel, R. (1994) Cell 78, 823-834. [DOI] [PubMed] [Google Scholar]
- 29.Nutt, S. L. & Busslinger, M. (1999) Biol. Chem. 380, 601-611. [DOI] [PubMed] [Google Scholar]
- 30.Sano, Y., Shimada, T., Nakashima, H., Nicholson, R. H., Eliason, J. F., Kocarek, T. A. & Ko, M. S. (2001) Genome Res. 11, 1833-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereira, J. P., Girard, R., Chaby, R., Cumano, A. & Vieira, P. (2003) Nature Immunol. 4, 464-470. [DOI] [PubMed] [Google Scholar]
- 32.Morison, I. M. & Reeve, A. (1998) Hum. Mol. Genet. 7, 1599-1609. [DOI] [PubMed] [Google Scholar]
- 33.Falls, J. Pulford, D. J., Wylie, A. A. & Jirtle, R. L. (1999) Am. J. Pathol. 154, 635-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans, H. K., Wylie, A. A., Murphy, S. K. & Jirtle, R. L. (2001) Genomics 77, 99-104. [DOI] [PubMed] [Google Scholar]
- 35.Hayward, B. E., Kamiya, M., Strain, L, Moran, V., Campbell, R., Hayashizaki, Y. & Bonthron, D. T. (1998) Proc. Natl. Acad. Sci. USA 95, 10038-10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jinno, Y., Yun, K., Nishiwaki, K., Kubota, T., Ogawa, O., Reeve, A. E. & Niikawa, N. (1994) Nat. Genet. 6, 305-309. [DOI] [PubMed] [Google Scholar]
- 37.Rauscher, F. J. (1993) FASEB J. 7, 896-903. [PubMed] [Google Scholar]
- 38.Chung, W. Y., Yuan, L., Feng, L., Hensle, T. & Tycko, B. (1996) Hum. Mol. Genet. 5, 1101-1108. [DOI] [PubMed] [Google Scholar]
- 39.Hatada, I., Inazawa, J., Abe, T., Nakayama, M., Kaneko, Y., Jinno, Y., Niikawa, N., Ohashi, H., Fukushima, Y., Iida, K., et al. (1996) Hum. Mol. Genet. 5, 783-788. [DOI] [PubMed] [Google Scholar]
- 40.Kondo, M., Matsuoka, S., Uchida, K., Osada, H., Nagatake, M., Takagi, K., Harper, J. W., Takahashi, T., Elledge, S. J. & Takahashi, T. (1996) Oncogene 12, 1365-1368. [PubMed] [Google Scholar]
- 41.De Souza, A. T., Hankins, G. R., Washington, M. K., Orton, T. C. & Jirtle, R. L. (1995) Nat. Genet. 11, 447-449. [DOI] [PubMed] [Google Scholar]
- 42.Hankins, G. R., De Souza, A. T., Bentley, R. C., Patel, M. R., Marks, J. R., Iglehart, J. D. & Jirtle, R. L. (1996) Oncogene 12, 2003-2009. [PubMed] [Google Scholar]
- 43.Yamada, T., De Souza, A. T., Finkelstein, S. & Jirtle, R. L. (1997) Proc. Natl. Acad. Sci. USA 94, 10351-10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malcolm, S., Clayton-Smith, J., Nichols, M., Robb, S., Webb, T., Armour, J. A., Jeffreys, A. J. & Pembrey, M. E. (1991) Lancet 337, 694-697. [DOI] [PubMed] [Google Scholar]
- 45.Nicholls, R. D., Knoll, J. H., Butler, M. G., Karam, S. & Lalande, M. (1989) Nature 342, 281-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson, W. P., Bottani, A., Xie, Y. G., Balakrishman, J., Binkert, F., Machler, M., Prader, A. & Schinzel, A. (1991) Am. J. Hum. Genet. 49, 219-234. [PMC free article] [PubMed] [Google Scholar]