Abstract
Pre-erythrocytic malaria vaccines, including those based on whole-parasite approaches, have shown protective efficacy in animal and human studies. However few pre-erythocytic antigens other than the immunodominant circumsporozoite protein (CSP) have been studied in depth with the goal of developing potent subunit malaria vaccines that are suited for use in endemic areas. Here we describe a novel technique to identify pre-erythrocytic malaria antigens that contribute to protection elicited by whole-parasite vaccination in the mouse model. Our approach combines immunization with genetically attenuated parasites and challenge with DNA plasmids encoding for potential protective pre-erythrocytic malaria antigens as luciferase fusions by hydrodynamic tail vein injection. After optimizing the technique, we first showed that immunization with Pyfabb/f−, a P. yoelii genetically attenuated parasite, induces killing of CSP-presenting hepatocytes. Depletion of CD8+ but not CD4+ T cells diminished the killing of CSP-expressing hepatocytes, indicating that killing is CD8+ T cell-dependent. Finally we showed that the use of heterologous prime/boost immunization strategies that use genetically attenuated parasites and DNA vaccines enabled the characterization of a novel pre-erythrocytic antigen, Tmp21, as a contributor to Pyfabb/f− induced protection. This technique will be valuable for identification of potentially protective liver stage antigens and has the potential to contribute to the understanding of immunity elicited by whole parasite vaccination, as well as the development of effective subunit malaria vaccines.
Introduction
Malaria remains a daunting public health challenge in spite of global elimination efforts that have significantly reduced incidence and death caused by this parasitic disease. An estimated 2.5 billion people are at risk of infection, which causes ∼250 million clinical cases and ∼840,000 child deaths per year in sub-Saharan Africa [1]. Since all clinical symptoms derive from the blood stages (BS), malaria vaccines that block parasite development during pre-erythrocytic (PE) stages prevent all human disease symptoms [2]. With up to 100% efficacy in human trials, live attenuated whole-parasite vaccines have been most effective to date, and include sporozoites that have been radiation-, drug-, or genetically-attenuated (reviewed in [3]). All of these can invade hepatocytes but subsequently arrest at different points during the liver stage (LS) or early in the BS of the life cycle of the parasite, while simultaneously inducing immune responses that protect against subsequent challenge with wild type sporozoites (wt spz). For example, by knocking out genes that are essential for LS parasite development, genetically attenuated parasite (GAP) vaccines have been shown to induce sterile and long-lasting protective immunity against challenge with wt spz in mice [4]–[6]. Similarly, immunization through the bite of mosquitos infected with P. falciparum or P. vivax irradiation-attenuated sporozoites (irr-spz) can protect humans from infection after challenge with wt spz [7]–[9]. Importantly, the PfSPZ vaccine was recently reported to protect 80% of volunteers who received 4–5 doses of intravenously administered irr-spz [10], in line with the vaccine efficacy required for eradication as per recent WHO guidelines [11].
In spite of their promise, currently available whole-parasite malaria vaccines require inoculation with as many as 1,000 bites of P. falciparum-infected mosquitoes, or intravenous administration of high doses of purified, cryopreserved irradiated sporozoites. Thus, a rational malaria vaccine development plan should also include subunit vaccines, which circumvent the logistical constraints of generating and administering live attenuated parasites and thus constitute a complementary approach that could be used to boost and maintain protective immunity elicited by live-attenuated parasite vaccinations. However, the most advanced subunit malaria vaccine, RTS,S, which is based on the immunodominant circumsporozoite protein (CSP), only exhibited a 32–50% anti-disease (but not anti-infection) efficacy among African children in Phase IIIb trials [12], [13].
Animal model studies and human clinical trials have demonstrated that CD8+ T cells play an important role in protection induced by live-attenuated sporozoite vaccination [10], [14]–[19]. In particular, we have used in vitro live-cell imaging to demonstrate that cytotoxic CD8+ T cells from mice immunized with PyGAP can directly kill LS parasite-infected hepatocytes [14]. In addition, antigen-specific CD8+ T cells have also been shown to correlate to protection induced by subunit vaccines [20]–[22].
Immune responses elicited by vaccination with whole parasites are biased towards CSP; however, several studies have shown that protection against malaria parasites can be achieved in the absence of CSP [23]–[25]. Despite these observations, few PE antigens other than CSP have been evaluated as vaccine candidates, including LSA1, CelTOS and TRAP [26]. The identification of novel PE antigens is hindered by the difficult culturing of infected primary hepatocytes. A recent attempt to expand the repertoire of PE antigens characterized antigen-specific IFN-γ production by splenic lymphocytes in CSP-tolerant mice immunized with irr spz [27]. The authors concluded that although immunization with several non-CSP antigens generated large numbers of specific CD8+ T cells that produced IFN-γ levels comparable to those elicited by CSP, only CSP was able to protect mice against challenge with wt spz. Another recent study aimed at identifying pre-erythrocytic antigens conducted a systematic profiling of H2b-restricted P. berghei peptides recognized by CD8+ T cells from mice immunized with whole malaria parasites [28]. PbS20 and PbTRAP were identified as targets of CD8+ T cells although only TRAP-specific cytotoxic CD8+ T cells were shown to contribute to protection against sporozoite challenge. These results support the importance of developing new methods to identify PE antigens that contribute to vaccine protection.
We report the optimization of a technical approach that characterizes the ability of antigen-specific CD8+ T cells to eliminate hepatocytes in vivo following immunization with whole parasite vaccines as a means to validate potential vaccine candidates. This method combines the use of Hydrodynamic Tail Vein Injection (HTVI) to deliver naked DNA encoding luciferase-tagged malaria LS antigens directly to the liver [29]–[31] with an in vivo imaging system (IVIS) that allows real-time monitoring of the abundance of the luciferase-tagged antigens in the liver [32]. After validating this method in the P. yoelii murine immunization/challenge model using CSP as a positive control, we used it to confirm that a potential new LS antigen, PyTmp21, which reduces liver stage burden in vivo, contributes to the protection elicited by whole parasite vaccines. This technology will be useful to down-select candidate LS antigens.
Materials and Methods
Animal and ethics statement
Five to ten week-old female BALB/c mice were obtained from Jackson Laboratory. All murine studies and procedures were approved by the institutional Animal Care and Use Committee of the Seattle Biomedical Research Institute (Seattle BioMed) following the NIH guidelines for animal housing and care.
Construction of DNA plasmids
phCMV-PyCSP-Luc was constructed by cloning an amino-terminal Myc-tagged synthetic P. yoelii CSP gene fragment (IDT) containing a CD4+ epitope (aa 57–70), 3 units of the central repeat (aa 139–156), and the carboxy-terminus (aa 280–345) of CSP into the phCMV-Luc vector as a carboxy-terminal fusion with the firefly luciferase reporter gene using restriction enzymes XhoI and HindIII (Figure S1A).
phCMV-PyTmp21-Luc was constructed by cloning an amino-terminal Myc-tagged fragment of transmembrane protein PyTmp21 (Plasmodium yoelii yoelii str. 17XNL PY06414, PlasmoDB, aa 26 to 181) excluding the amino-terminal endoplasmic reticulum targeting signal sequence and the carboxy-terminal transmembrane domain that was PCR amplified from P. yoelii cDNA using the following primers: F PyTmp21: 5′-CGATCTCGAGATGGAACAAAAACTCATCTCAGAAGAGGATCTGATATATTTATCCT TAAAACC-3′ and R PyTmp21 5′-CGTAAAGCTTGCTAATGTATCATTTAA TTTTTCG-3′ (Figure S1A). After confirming the orientation of the inserts by PCR and double digest with XhoI and HindIII restriction enzymes, positive clones were sequenced to ensure accurate amplification and in-frame cloning with the luciferase open reading frame. Plasmid DNA was prepared by using the Qiagen EndoFree Mega Plasmid Kit (Qiagen, Valencia, CA). Full-length PyTmp21 was PCR amplified from non-lethal P. yoelii 17X NL clone 1.1 and the resulting PCR product was cloned into the gWIZ vector (Gentlantis, CA, USA) using SalI and NotI restriction sites (McLab, CA, USA).
Luciferase activity assay
COS-7 cells (obtained from the American Type Culture Collection, ATCC) were cultured in DME medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine and 100 U of penicillin-streptomycin/ml. One day before transfection, 1×106 cells were plated in 10 cm2 plates in growth medium without antibiotics. Twenty-four hours later, a total of 4 µg of DNA was transfected with lipofectamine 2000, following the manufacturer's instructions (Invitrogen). After 48 hours, cells were collected and lysed for luciferase expression analysis. Protein concentration was determined using a Bradford protein assay kit (Bio-Rad). The luciferase activity of cell lysates was measured using the Bright-Glo luciferase assay system (Promega). Luminescence was measured on a CentroXS3 LB 960 luminometer. Measurements were taken in triplicate. Light emission was integrated over a 10-second time period.
Western Blot analyses
Twenty µg of total protein from transfected COS-7 cell lysates containing protease inhibitor (Roche) were resolved by electrophoresis on 12% SDS-PAGE gels (Bio-Rad), and transferred to a nitrocellulose membrane using a wet transfer cell apparatus (Bio-Rad). The membrane was blocked for 2 h in TBS buffer containing 0.05% Tween 20 (TBS-T) and 5% not-fat dry milk and incubated at 4°C overnight in a 1∶1,000 dilution of anti-luciferase antibody (GeneTex) in 5% milk/TBS-T. Next, the membrane was washed in TBS-T, incubated for 2 h with 1∶20,000 dilution of goat anti-mouse IgG-HRP in 5% milk/TBS-T, and detected by enhanced chemiluminescence (ECL plus) according to the manufacturer's instructions.
Whole-parasite immunization strategies
The genetically attenuated parasite P. yoelii fabb/f− (Pyfabb/f−), which carries a targeted deletion of FabB/F, a critical enzyme in fatty acid synthesis and therefore arrests late in the liver phase of the malaria life cycle [19], was isolated from the salivary glands of infected A. stephensi mosquitoes.
For the first group of experiments, BALB/c mice were vaccinated intravenously with two doses of either 50,000 Pyfabb/f− sporozoites or mosquito salivary gland debris two weeks apart. For the heterologous immunization strategies, mice were vaccinated 3 times by intramuscular injection with 20 µg of gWIZ/PyTmp21 recombinant plasmid DNA, followed by boosting with a single dose of Pyfabb/f− sporozoites, or, alternatively with a single dose of 50,000 Pyfabb/f− sporozoites followed by boosting with 20 µg of gWIZ/PyTmp21 recombinant plasmid DNA.
DNA vaccination and protection studies
BALB/c mice were vaccinated by administration of 20 µg of gWIZ recombinant plasmid DNA encoding for PyTmp21, PyCSP or empty vector into the tibialis muscle at weeks 0, 3 and 6, using the Ichor Tri-GridTM delivery system (Ichor Medicals, CA, USA). Mice were challenged with 20,000 wild type P. yoelii sporozoites by i.v. injection 10 days after the third immunization. Inhibition of liver stage parasite development was measured by qRT-PCR at 42 hours post challenge [33]. The ratio of P. yoelii 18S RNA expression was compared to that of the mouse housekeeping gene GAPDH.
Challenge by Hydrodynamic Tail Vein Injection
Ten week old BALB/c naïve mice or mice immunized as described above were injected with phCMV-Luc, phCMV-PyCSP-Luc or phCMV-PyTmp21-Luc recombinant plasmid DNA by HTVI as previously described [29]. Challenge was performed 14 or 30 days after the immunized mice received the last dose of Pyfabb/f− sporozoites. In brief, mice were placed in a restrainer tube and injected rapidly in the tail vein within 5 seconds with approximately 10% volume/body weight of phosphate-buffered saline (PBS, Gibco, UK) containing 25 µg of plasmid DNA, using a 27 1/2 gauge needle.
In vivo imaging of bioluminescence
Four hours, eight hours, 1 day, 2 days, 4 days and 7 days after HTVI, mice were anesthetized with 2% isoflurane and administered 150 mg/kg of luciferase substrate (D-luciferin, Xenogen-Caliper) intraperitoneally. Five minutes later, the mice were positioned in the imaging chamber (IVIS Lumina II) for data collection. Living Image (Xenogen-Caliper) software was used to measure photons emitted by region of interest (ROI), which were analyzed using the ICOR image analysis software. Data was quantified as photon sec/cm2/sr. Values are shown as mean ± SD.
In vivo T cell Depletion
Mock-immunized mice and mice immunized with Pyfabb/f− sporozoites as described above were injected intraperitoneally with 0.5 mg of anti-CD8 monoclonal antibody 2.43 (TIB210; American Type Culture Collection), anti-CD4 monoclonal antibody GK1.5, or an equivalent dose of rat IgG2b isotype control for two consecutive days before being challenged with plasmid DNA by HTVI. The dose and regimen was optimized to deplete >95% of CD8+ T cells or CD4+ T cells (data not shown). Depletion of specific cell types was confirmed by surface staining of PBMC with Pacific Blue-conjugated anti-CD3, PerCP/cy5.5-conjugated anti-CD8 and APC-conjugated anti-CD4 antibodies (Biolegend) by flow cytometric analysis one day before challenge.
Hepatocyte and lymphocyte purification
Twenty hours or seven days after mice were challenged with PyCSP-Luc recombinant plasmid DNA by HTVI, hepatocytes and infiltrating lymphocytes were harvested and purified as previously described [15]. Briefly, mouse livers were perfused with 10 mL of perfusion media (1X HBSS, 5 mM HEPES, and 0.5 mM EDTA) via the portal vein and treated in situ with 0.5 mg/mL collagenase in 10 mL of collagenase buffer (1X HBSS (without Ca/Mg), 0.5 mM CaCl, 0.5 mM MgCl, 5 mM HEPES). Afterwards, livers were collected and homogenized, and hepatocytes were isolated by low speed centrifugation at 500 rpm. The pellet was washed 3 times with R10 media (complete RPMI containing 10% fetal bovine serum (FBS), penicillin-streptomycin and glutamine) and suspended in 10 ml R10. Lymphocytes were isolated from the supernatant by centrifugation on a gradient of 44% Percoll buffer underlaid with 67% Percoll buffer (GE Life Sciences). Percoll gradients were centrifuged at 2,000 rpm, and mononuclear cells at the gradient interface were extracted, washed 3 times with R10, and suspended in complete FACs buffer (1X PBS containing 2% FBS).
Intracellular staining and flow cytometry
For cytokine staining, 1×106 liver lymphocytes were incubated for 6 hours at 37°C in the presence of Brefeldin A (10 mg/ml, Sigma-Aldrich) followed by 1 hour incubation with PE-Cy7-conjugated anti-CD3 antibody (Biolegend), PerCP/cy5.5-conjugated anti-CD8 antibody (Biolegend) and APC-labeled anti-H2Kd/CSP280–288 (SYVPSAEQI) tetramer antibody (obtained from the NIH tetramer core facility). Cells were subsequently washed with FACS buffer (1% BSA, 0.05% sodium azide in PBS), fixed and permeabilized by incubation at 4°C for 20 min in 100 µL BD Cytofix/Cytoperm (BD Biosciences)). After washing in Perm/Wash buffer (BD Biosciences), intracellular staining for IFN-γ was determined using a Pacific Blue-conjugated anti-IFN-γ clone XMG1.2 antibody (eBioscience).
To determine the subset and proportion of liver lymphocytes expressing PyCSP, 1×106 lymphocytes were surface stained with BV421-conjugated anti-CD3 antibody, PerCP/Cy5.5-conjugated anti-B220 antibody, APC-conjugated Tie-2 antibody, APC-Cy7-conjugated anti-F4/80 antibody, PE-Cy7-conjugated CD11b antibody and BV605-conjugated CD11c antibody (BioLegend). Cells were washed and fixed as described above before being incubated with a polyclonal anti-rabbit PyCSP antibody (AbBiotech, Inc.) at a 1∶100 dilution in Perm/Wash buffer for 30 minutes at room temperature. After 3 washes with Perm/Wash buffer cells were incubated with Alexa-Fluor 488-conjugated anti-rabbit antibody for another 30 minutes. Cells were washed in Perm/Wash buffer and suspended in 120 µL of FACs buffer.
To determine the proportion of liver hepatocytes expressing PyCSP, 500 µL of cell suspension prepared as described above was pelleted and resuspended in 500 µL of Fixing Solution (1% Paraformaldehyde in PBS) for 10 minutes at RT. Cells were washed 3 times with FACs buffer before being stained with polyclonal rabbit anti-PyCSP antibodies as described above.
Data were acquired using a BD LSRII instrument and analyzed with FlowJo software (Tree Star, Inc.)
Statistical analysis
All data are presented as mean ± SD. Differences between groups were determined by unpaired, two tailed Mann-Whitney tests. All p-values <0.05 were considered statistically significant.
Results
Plasmid construction and in vitro expression of a P. yoelii CSP luciferase fusion protein
We first established the HTVI/IVIS system in the Pyfabb/f− murine model, using CSP as a positive control. We constructed an amino-terminal Myc-tagged, carboxy-terminal luciferase fusion protein (PyCSP-Luc) by synthesizing a DNA fragment that encodes for a previously identified CD4 CSP epitope, followed by 3 copies of the central antigenic repeat and the carboxy-terminus of CSP, which has been shown to contain overlapping CD8 and CD4 epitopes [34]. This DNA fragment was cloned into the phCMV-Luc vector to generate phCMV-PyCSP-Luc (Figure S1A, left panel). To confirm the expression of the resulting fusion protein, we transfected COS-7 cells with either phCMV-Luc or phCMV-PyCSP-Luc, harvested these cells 48 h post transfection and prepared protein extracts. Both Western blot analysis using a polyclonal firefly luciferase antibody and a luminometry assay showed that PyCSP-Luc was expressed at a similar level to that of luciferase alone (Figure S1B and C).
In vivo expression of PyCSP-Luc
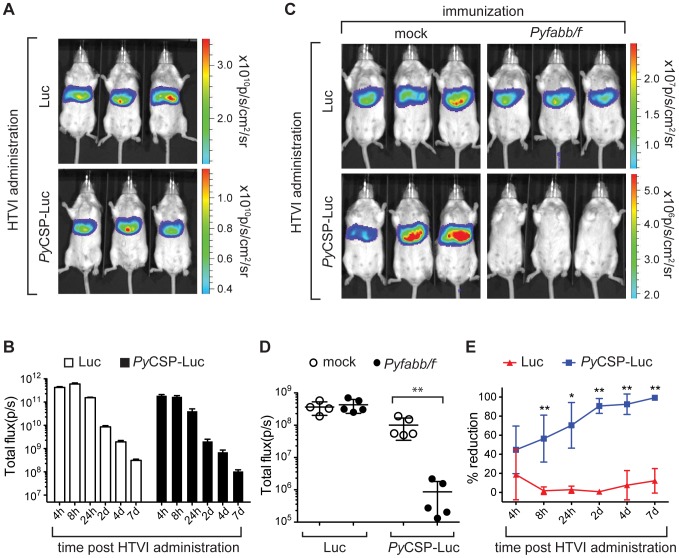
To evaluate the expression of PyCSP-Luc fusion protein in vivo, we injected groups of BALB/c mice (n = 3) by HTVI with 25 µg of either plasmid DNA encoding for luciferase (Luc) or PyCSP-Luc fusion protein each, followed by administration of D-luciferase and whole-body bioluminescence detection by IVIS 4 h, 8 h, 24 h, 2 d, 4 d, and 7 d later. Using this approach, we detected high levels of luciferase activity in the liver of all mice in both groups as early as 4 h post HTVI, with expression peaking 4 to 8 h post HTVI and remaining high 7 d post HTVI (Figure 1A and B). This result shows that the PyCSP-luciferase fusion protein is expressed in a stable and persistent manner in the liver of mice injected with plasmid DNA through HTVI, and that by measuring whole-body bioluminescence in vivo, IVIS allows the real-time monitoring of the abundance of luciferase-fused tagged proteins.
Figure 1. Immunization of mice with Pyfabb/f− reduces PyCSP-Luc in vivo luminescence.
(A–B) Expression of luciferase (Luc) and PyCSP-Luc in the liver of mice immunized by HTVI. (A) Luciferase signal in live naïve BALB/c mice injected by HTVI with 25 µg of either phCMV-Luc plasmid DNA (top panel) or phCMV-PyCSP-Luc (bottom panel) and imaged 8 h later by IVIS, after injection of D-luciferin. The scale indicates radiance expressed as p/s/cm2/sr. (B) Kinetics of luciferase signal in live naïve BALB/c mice (shown as total flux p per second) during the course of the experiment. Data is representative of two individual experiments, with 3 mice each per group. (C–E) Reduction of luciferase signal in the liver of Pyfabb/f−-immunized mice upon HTVI challenge with PyCSP-Luc plasmid DNA (C) Luciferase signal in 3 representative live mice immunized twice with 50,000 Pyfabb/f− salivary gland sporozoites (right panel) or mock-immunized with salivary gland debris (left panel), challenged by HTVI with 25 µg of phCMV-Luc plasmid DNA (top panel) PyCSP-Luc (bottom panel) 30 days after the last immunization, imaged 7 d post challenge as described. Each group contained 4 to 5 mice. (D) Quantification of luciferase signal (shown as total flux p per second) from mice in the four groups described in part C. The data represents 4 to 5 individually analyzed mice in each group and correspond to mean ± SD; significant differences between the mean of the mock vs. Pyfabb/f−-immunized mice for each plasmid calculated using the Mann-Whitney test are indicated (** = p<0.01). (E) Inhibition of luciferase signal over the course of the experiment for mice challenged with plasmid DNA encoding for luciferase alone (Luc, red triangles) or PyCSP-Luc (blue squares), calculated as percentage reduction vs. the mock-immunization control). The data represents 4 to 5 individually analyzed mice in each group and correspond to mean ± SD; significant differences between the mean of Luc vs. PyCSP-Lus calculated using the Mann-Whitney test are indicated for each plasmid (* = p<0.005; ** = p<0.001).
Previous studies using microscopy of liver sections have reported that between 1 and 40% of hepatocytes express proteins encoded by plasmids administered by HTVI, and that the level of expression from non-parenchymal cells is negligible [29], [35]. We confirmed that hepatocytes correspond to the major liver cell type expressing PyCSP-Luc by isolating liver hepatocytes and lymphocytes 20 hours after administration of empty Luc vector or PyCSP-Luc by HTVI (Figure S2). Expression of luciferase in the liver was confirmed by IVIS before livers were perfused and extracted (data not shown). Using flow cytometry, we observed that between 7.5% and 31.5% of the hepatocytes in the animals that received PyCSP-Luc by HTVI expressed PyCSP, as compared to 1.6% of hepatocytes in animals injected with empty Luc vector (Figure S2A). In comparison, expression levels of PyCSP-Luc were similar between both groups of mice for the majority of the lymphocyte cell types tested, including Kupffer cells (F4/80+), dendritic cells (CD11b+/CD11c+), sinusoidal liver endothelial cells (Tie2+) and B cells (B220+). We only observed PyCSP expression in CD3+ T cells in one out of three mice that received PyCSP-Luc by HTVI. In conclusion, our data shows that the PyCSP-Luc plasmid is stably expressed in the liver for at least 7 days after HTVI administration, and is preferentially expressed by hepatocytes.
Immunization of mice with Pyfabb/f− induces PyCSP-specific hepatocyte killing
Next, we determined whether the HTVI/IVIS method could be used as a tool to measure antigen-specific killing of hepatocytes upon whole parasite vaccination. To do this, mice were immunized intravenously (i.v.) twice, 14 days apart, with 50,000 Pyfabb/f− sporozoites dissected from the salivary glands of infected A. stephensi mosquitoes. As a control, mice were mock-immunized with salivary gland debris. Thirty days after the second dose, mice were challenged by HTVI with 25 µg of luciferase (Luc) or PyCSP-luc plasmid DNA, and bioluminescence was measured 4 h, 8 h, 24 h, 2 d, 4 d and 7 d later using IVIS. There was no significant difference in the kinetics of luciferase expression detected in the liver of Pyfabb/f− or mock-immunized mice challenged with plasmid encoding for luciferase (Figure 1C top panel and Figure 1D). However, the luciferase signal was dramatically decreased in Pyfabb/f−-immunized mice challenged with PyCSP-Luc plasmid DNA, as compared to mock-immunized mice (Figure 1C bottom panel and Figure 1D). Specifically, the luciferase signal was reduced by 45% 4 h post challenge, and continued to diminish over the course of the experiment, reaching a maximum reduction of 97% on d7 post challenge (Figure 1E). This result was highly reproducible, as very similar values were obtained in 3 independent experiments (data not shown).
The reduction in luciferase signal observed in the liver of mice upon HTVI challenge with PyCSP-Luc plasmid DNA after immunization with Pyfabb/f− suggests that vaccination with attenuated whole parasites induces specific killing of hepatocytes that express CSP epitopes [14], [17], [23], [36], [37]. Moreover, our data show that the combination of HTVI challenge with the non-invasive IVIS imaging technique constitutes a powerful tool that can be used to measure reductions of luciferase expression that are indicative of the elimination of specific antigen-presenting hepatocytes in vivo.
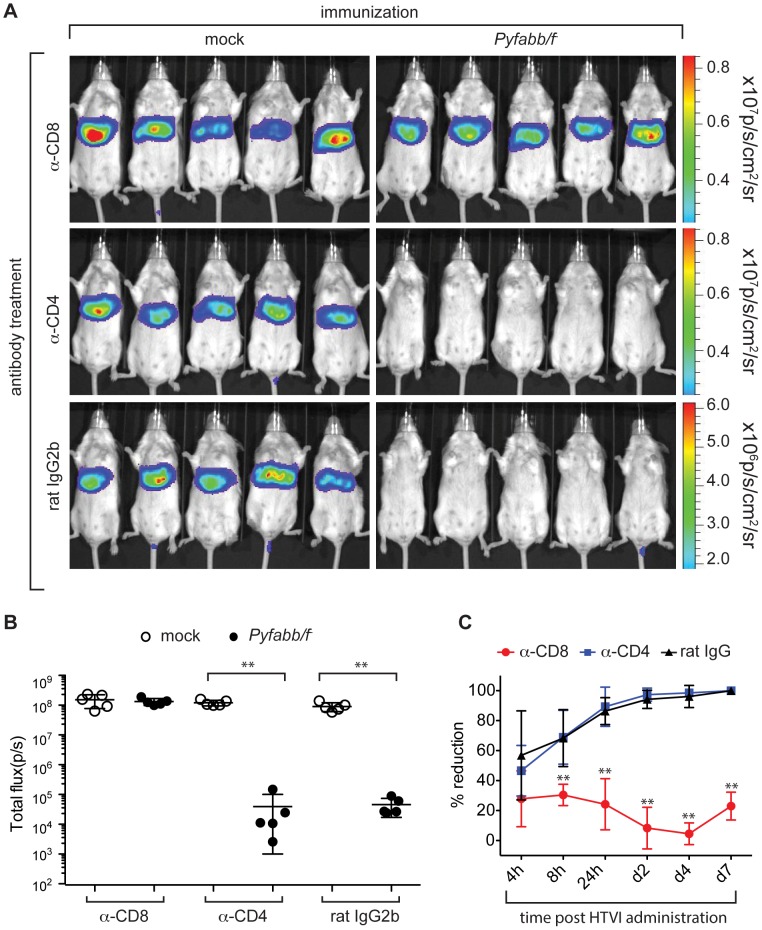
CD8+ T cells are required for the elimination of PyCSP-Luc expressing liver cells in Pyfabb/f− immunized mice
We and others have previously shown that CD8+ T cells are critical for protection from challenge with wt spz after immunization of mice with whole parasite vaccinations [14], [15], [17], [37]. Thus, we investigated the role of CD8+ T cells in eliminating hepatocytes that present malaria specific antigens after immunization with Pyfabb/f−. To do this, we depleted CD4+ or CD8+ T cells in BALB/c mice immunized with Pyfabb/f− sporozoites (and in mock-immunized mice) thirteen days after the last dose of Pyfabb/f− through administration of monoclonal antibodies against CD4+ or CD8+ T cell receptors, or rat IgG2b as a negative control. Successful depletion of both CD4+ and CD8+ T cells was confirmed by analyzing the peripheral blood of individual mice by flow cytometry the day before challenge (data not shown). Twenty-four hours later, mice were challenged by HTVI with 25 µg of plasmid DNA encoding for PyCSP-Luc fusion protein. Depletion of CD8+ T cells (but not depletion of CD4+ cells nor treatment with rat IgG2b antibodies) abrogated the inhibition of luciferase signal induced by Pyfabb/f− immunization (Figure 2A–C). This result suggests that GAP immunization induces CSP-specific CD8+ T cells that are capable of eliminating hepatocytes that present CSP epitopes on their surface. Importantly, it agrees with in vitro data by our group and others showing that elimination of malaria-infected hepatocytes is mainly mediated by CD8+ T cells [14], [38].
Figure 2. Elimination of hepatocytes that express PyCSP-Luc is mediated by CD8+ cells.
(A) Luciferase signal in live mice immunized twice with 50,000 Pyfabb/f− salivary gland sporozoites (right panel) or mock immunized (left panel) with salivary gland debris, and treated 14 days later for two consecutive days with antibodies anti-CD8 (top panel), anti-CD4 (middle panels) or an equivalent amount of control rat IgG2b, before HTVI challenge with 25 µg of phCMV-PyCSP-Luc and imaged 7 d post challenge as described. Each group contained 5 mice. The scale indicates radiance expressed as p/s/cm2/sr. (B) Quantitation of the data shown in part A. Radiance is shown as total flux (p/s). The data represents 5 individually analyzed mice in each group and correspond to mean ± SD; significant differences between the mean of the mock vs. Pyfabb/f−-immunized mice for each treatment calculated using the Mann-Whitney test are indicated (** = p<0.01). (C) Kinetics of inhibition of luciferase expression upon depletion of CD8+ (red circles) or CD4+ (blue squares) T cells, or mock depletion (black triangles), calculated as percentage reduction compared to the mock immunization control for each condition. The data represents 5 individually analyzed mice in each group, and correspond to mean ± SD; significant differences between the CD8 or CD4 T cell depletion vs. the mock depletion are indicated (** = p<0.01).
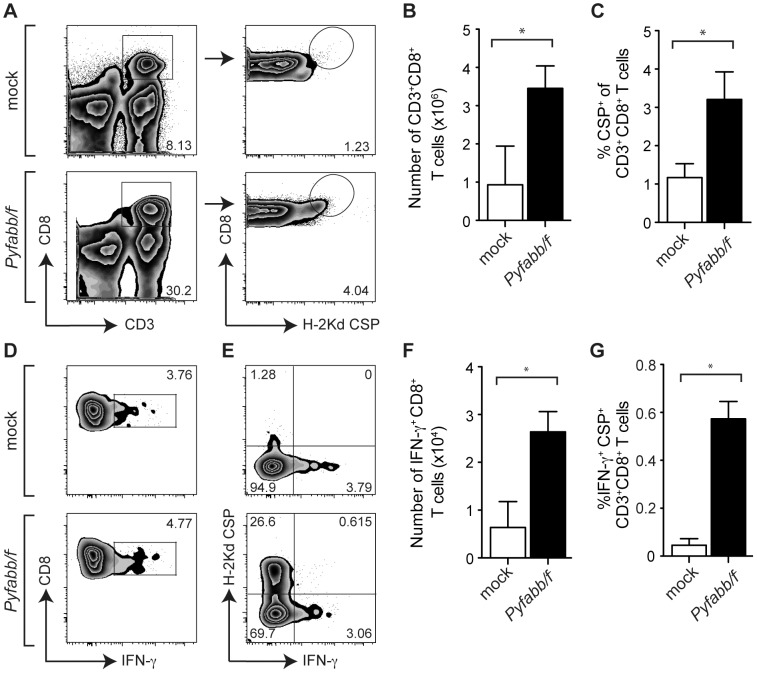
PyCSP-specific CD8+ T cells produce high levels of IFN-γ
Next, we addressed the specificity and mechanism by which hepatocytes presenting specific epitopes are killed by malaria-specific CD8+ T cells after whole parasite vaccination. We isolated infiltrating liver lymphocytes from mice immunized with Pyfabb/f − as well as from mock-immunized mice 7 days after HTVI challenge with plasmid encoding for PyCSP-Luc fusion protein. The number of total and CSP-specific CD8+ T cells was analyzed by flow cytometry using the gating strategy shown in Figure 3A (left panel). We observed a significant increase in the number of CD8+ T cells present in Pyfabb/f −-immunized mice challenged with PyCSP-Luc plasmid DNA vs. mock-immunized mice (Figure 3B). To determine whether these cells were specific for PyCSP, we used a CSP H-2Kd tetramer, as previously described [15] (Figure 3A, right panel), and established that CSP-specific CD8+ T cells were significantly increased in Pyfabb/f −-immunized mice challenged with PyCSP-Luc plasmid DNA as compared to mock-immunized mice (Figure 3C).
Figure 3. PyCSP-specific CD8+ T cells produce high levels of IFN-γ.
(A) Characterization of infiltrating liver lymphocytes isolated from mice immunized with Pyfabb/f − sporozoites (bottom panel) or mock-immunized (top panel) 7 days after HTVI challenge with plasmid DNA encoding for PyCSP-Luc. Representative dot plots gated on live lymphocytes showing CD8+ versus CD3+ (left panel) and CD8+ versus H-2Kd CSP tetramer (left panel) expression (B) Absolute number of CD8+ T cells in mock-immunized mice and in mice immunized with Pyfabb/f − sporozoites. (C) Percentage of CSP-specific CD8+ T cells in mock-immunized mice and in mice immunized with Pyfabb/f − sporozoites. (D) Characterization of IFN-γ producing liver CD8+ T cells in mice immunized with Pyfabb/f − sporozoites (bottom panel) or mock-immunized (top panel) as determined by intracellular cytokine staining. Representative dot plots gated on live CD3+ lymphocytes showing CD8+ versus IFN-γ expression. (E) Representative dot plot gated on live CD3+ CD8+ T cells showing H-2Kd-CSP versus IFN-γ expression. (F) Absolute number of IFN-γ+ CD8+ T cells in in mock-immunized mice and in immunized with Pyfabb/f − sporozoites. (G) Percentage of CSP-specific CD8+ T cells producing IFN-γ in mock-immunized mice and in mice immunized with Pyfabb/f − sporozoites. (B–C and F–G) The data represents 3–5 individually analyzed mice in each group, and correspond to mean ± SD; significant differences between the mean of the mock vs. Pyfabb/f −-immunized mice calculated using the Mann-Whitney test are indicated (* = p<0.05).
IFN-γ is a critical component in CD8+ T cell-mediated protection induced by attenuated whole parasites [39]–[41]. We used intracellular flow cytometry analysis to determine what proportion of CD8+ T cells produced IFN-γ in response to GAP immunization. We saw that IFN-γ-producing CD8+ T cells were significantly increased in Pyfabb/f −-immunized mice as compared to mock-immunized mice after challenge with plasmid DNA encoding for PyCSP-Luc (Figure 3D and F). Finally, we analyzed whether these cells were specific for CSP by using the H-2Kd tetramer described above. As shown in Figure 3E and G, the number of PyCSP+ IFN-γ+ CD8+ T cells was also significantly increased in Pyfabb/f −-immunized mice challenged with plasmid DNA encoding for PyCSP-Luc as compared to mock-immunized mice. Taken together, our results suggest that a CSP-specific recall response involving IFN-γ secretion is generated during challenge of Pyfabb/f −-immunized mice with PyCSP-Luc plasmid DNA, and that this response correlates with the elimination of hepatocytes that present CSP epitopes, as measured by the reduction of bioluminescence.
Identification of PyTmp21 as a potential protective antigen that reduces liver stage parasite burden
We previously identified PVA022 (PVX_082595) as a potential PE antigen recognized by PBMCs obtained from Duffy receptor negative donors who display naturally acquired immunity to P. vivax [42], in agreement with the observation that its P. falciparum ortholog (PF3D7_1333300) is expressed by sporozoites. To determine if this antigen can induce a protective response in vivo, we generated a DNA vaccine by cloning the P. yoellii ortholog of PVA022 (PyTmp21, or PY06414) into the gWIZ vector. Empty gWIZ plasmid and gWIZ-PyCSP (encoding for full-length P. yoelii CSP) were used as negative and positive controls, respectively. BABL/c mice (5 per group) were vaccinated three times at 3-week intervals by intramuscular (i.m.) administration of 20 µg of each of these plasmids and challenged with 20,000 P. yoelii 17XNL sporozoites ten days after the last immunization. The inhibition of liver stage parasite development was measured by qRT-PCR 24 h later. The parasite burden was calculated as the ratio of P. yoelii 18S RNA to that of mouse housekeeping gene GAPDH. Vaccination with PyCSP reduced liver stage parasite load by 98.4%, as compared to the negative control (Figure 4). Similarly, vaccination with PyTmp21 reduced parasite load by 71.8%. This result shows that immunization with a PyTmp21 DNA vaccine triggers an immune response that is sufficient to reduce liver stage burden.
Figure 4. DNA vaccination of mice with PyTmp21 reduces liver stage parasite burden.
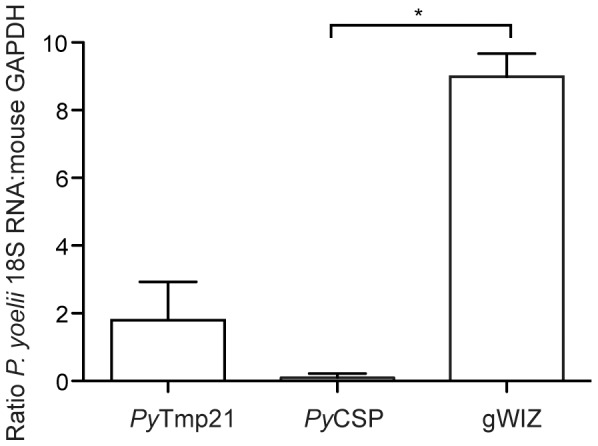
Shown is the average ratio of P. yoelii 18S RNA to GAPDH mouse housekeeping gene mRNA for 3–5 individually analyzed mice immunized with gWIZ, PyCSP or PyTmp21 plasmid DNA. Statistical significance between the PyCSP or PyTmp21 as compared to gWIZ plasmid was calculated using the Mann-Whitney test (* = p<0.05).
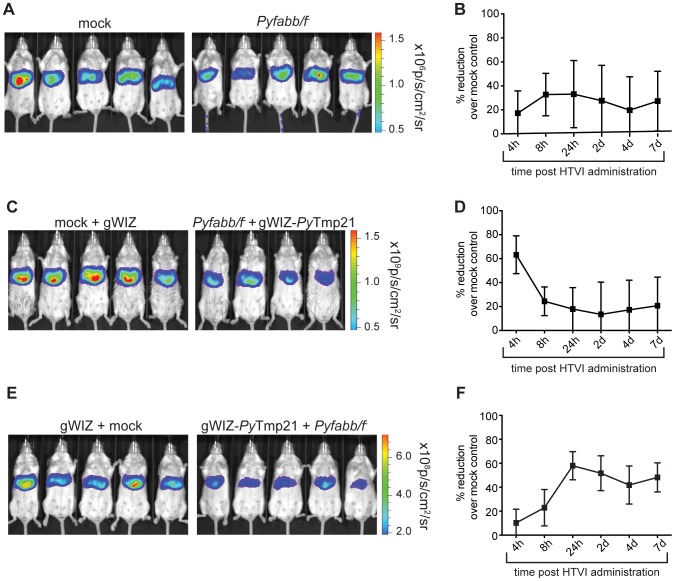
PyTmp21 contributes to the protective immune response elicited by Pyfabb/f − immunization
We next determined whether the HTVI/IVIS technique optimized with PyCSP can be used to validate other PE antigens as contributing to protection elicited by whole parasite vaccines by evaluating the ability of PyTmp21 to induce hepatocyte killing in vivo. To do this, we cloned the central region of PyTmp21 (excluding the amino-terminal signal peptide and the carboxy-terminal trans-membrane domain) into the pHCMV-Luc vector as an amino-terminal Myc tagged, carboxy-terminal luciferase fusion protein (Figure S1A, right panel). We then tested the in vitro expression and luciferase signal of the fusion protein, as described above for PyCSP-Luc (Figure S1B–C). Although the in vitro expression level of PyTmp21-Luc protein was much lower than that of PyCSP-Luc as measured by Western blot analysis (Figure S1B), the luciferase signal of the cell lysate was only one order of magnitude lower than that of PyCSP-Luc (Figure S1C). This could be due to differences in the stability of the fusion protein, as suggested by the strong band corresponding to luciferase in the PyTmp21-luc lane of the Western blot (Figure S1B).
We then immunized mice with two doses of 50,000 Pyfabb/f− sporozoites each, followed by HTVI challenge with 25 µg of plasmid DNA encoding for PyTmp21-Luc thirty days after the last dose. As a control, we mock-immunized mice with mosquito salivary gland debris. In contrast to the result previously obtained for PyCSP using this strategy, in this case we failed to observe a reduction in the luciferase signal upon challenge (Figure 5A and B). Therefore, in spite of our preliminary data showing that immunization with PyTmp21 as a DNA vaccine can protect mice against malaria infection, HTVI challenge with PyTmp21 after whole parasite vaccination did not result in hepatocyte destruction. This observation could be explained by a recent report suggesting that exposure to whole parasite vaccines results in a bias of the immune response towards the immunodominant CSP, and away from less abundant antigens expressed during PE stages [43].
Figure 5. PyTmp21 contributes to the protective immune response elicited by GAP vaccines.
(A) Quantification of luciferase signal (shown as total flux p per second) in mice immunized twice with 50,000 Pyfabb/f− salivary gland sporozoites (right panel) or mock-immunized with salivary gland debris (left panel), challenged by HTVI with 25 µg of phCMV-PyTmp21-Luc 30 days after the last immunization. Each group contained 5 mice. (B) Inhibition of luciferase signal over the course of the experiment calculated as percentage reduction vs. the mock-immunization control. The data represents 5 individually analyzed mice in each group and correspond to mean ± SD. (C) Quantification of luciferase signal (shown as total flux p per second) in mice primed with 50,000 Pyfabb/f− salivary gland sporozoites and boosted with gWIZ-PyTmp21 (right panel), or mock-immunized and injected with gWIZ (left panel), challenged as described in part A. Each group contained 4 to 5 mice. (D) Inhibition of luciferase signal over the course of the experiment calculated as percentage reduction vs. the mock-immunization control. The data represents 5 individually analyzed mice in each group and correspond to mean ± SD. (E) Quantification of luciferase signal (shown as total flux p per second) in mice primed with gWIZ-PyTmp21 and boosted with 50,000 Pyfabb/f− salivary gland sporozoites (right panel), or injected with gWIZ and mock-immunized (left panel), challenged as described in part A. Each group contained 5 mice. (F) Inhibition of luciferase signal over the course of the experiment calculated as percentage reduction vs. the mock-immunization control. The data represents 5 individually analyzed mice in each group and correspond to mean ± SD.
To overcome this obstacle, we designed a heterologous prime/boost strategy consisting of an initial vaccination with a single dose of 50,000 Pyfabb/f− sporozoites, followed by boosting with plasmid DNA encoding for PyTmp21 (gWIZ-PyTmp21). Using this method, we observed a 60% reduction in luciferase signal 4 h after HTVI challenge of Pyfabb/f−-primed, gWIZ-PyTmp21 boosted mice with plasmid DNA encoding for PyTmp21-Luc, as compared to mock-primed, gWIZ-boosted mice (Figure 5C and D). This initial high level of inhibition of luciferase signal diminished to ∼20% at the 8 h time point, and remained at this level until 7 d post challenge (Figure 5D). This result suggests that priming with Pyfabb/f− elicits PyTmp21-specific T cells that can be boosted by the subsequent administration of plasmid DNA encoding for this antigen, resulting in the killing of hepatocytes that display the antigen upon HTVI challenge.
Finally, we also tested the effect of priming with plasmid DNA encoding for PyTmp21, followed by boosting with a single dose of Pyfabb/f− parasites. Interestingly, in contrast to the fast response observed when priming with attenuated whole parasites followed by boosting with plasmid DNA, in this case we observed a delayed reduction in the luciferase signal (Figure 5E and F). In other words, we saw an initial 10% reduction 4 h after HTVI challenge of gWIZ-PyTmp21-primed, Pyfabb/f−-boosted mice, as compared to gWIZ-primed, mock-boosted mice, which then steadily increased to 60% at 24 h and remained constant at approximately 50% until 7 d post challenge (Figure 5F). Taken together, our data suggest that although immunization with whole parasites was not sufficient to generate a significant T-cell based response to PyTmp21, likely because of the bias of the immune response towards immunodominant CSP, the use of heterologous immunization strategies that combine whole parasite and DNA vaccines generates a detectable immune response against PyTmp21, validating it as a PE antigen that contributes to the protective effect of whole parasite vaccines.
Discussion
Ideal anti-infection malaria vaccines should target the silent pre-erythrocytic stages, blocking sporozoite invasion and/or subsequent development in the hepatocyte, hence preventing malaria infection and disease [2]. In fact, the most effective malaria vaccines tested to date are all based on attenuated parasites [8], [10], [44]. The use of immunization strategies that result in late LS or early BS arrest results in the development of immunity against a broad spectrum of antigens expressed by LS-infected hepatocytes and as consequence, in enhanced protection against sporozoite challenge [19], [44].
Human and mouse studies have shown that immunization with whole parasites elicits both humoral and T-cell responses, and that the bulk of this response is directed towards the immunodominant sporozoite surface protein CSP [27], [43], [45]. Thus, subsequent boosts with whole parasites will repeatedly enhance the response to CSP, leading to significant reductions of hepatocyte invasion by spz and LS parasite load in the liver. As an unintended consequence, however, this immunization strategy prevents the development of robust T cell responses against non-CSP antigens expressed by LS parasites [43], perhaps explaining why extremely high doses of irr-spz are required in order to reach the peak of anti-CSP immunity that allows efficient inhibition of hepatocyte invasion by spz after challenge with wt P. falciparum spz [10]. Although CSP-specific CD8+ T cell responses were induced by P. yoelii GAP in this study, CSP has been proven to be dispensable in C57BL/6 mice following vaccination with P. berghei sporozoites attenuated through irradiation [23], and was only marginally involved in protection of BALB/c mice immunized with P. berghei under chloroquine coverage [25]. These results highlight the importance of identifying pre-erythrocytic antigens other than CSP that contribute to the protection elicited by attenuated whole parasite vaccines, which will be important in designing effective immunization strategies that result in a broad immune response that can control LS parasite infections that might have escaped CSP-dependent responses.
To enable the identification of bona fide LS antigens that contribute to the protection induced by whole parasite immunization, we took advantage of the P. yoelii FabB/F gene knock-out GAP (Pyfabb/f−) that arrests late in the LS parasite development [46]. Immunization with Pyfabb/f− induces sterile long-term protection and a broad CD8+ T cell response [19], therefore allowing the induction of immunity against LS antigens. Thus, this parasite is ideal model for identifying LS antigens that contribute to Pyfabb/f− -induced CD8+ responses. Moreover, to measure the ability of antigen-specific CD8+ T cells to eliminate hepatocytes in vivo after immunization of mice with Pyfabb/f−, we utilized a highly efficient, non-viral method for delivery of DNA into the liver, known as HTVI. This technique consists of the rapid injection of a large volume of plasmid DNA into the tail vein, resulting in uptake of the DNA into the cytoplasm of liver cells [35], and has been widely used in recent years to deliver DNA and RNA for gene function, gene therapy and for establishment of disease animal models (reviewed in [47]). Although HTVI is a valid approach to deliver antigens directly into the liver, it has been recently shown that it does not constitute and effective vaccination strategy because it leads to defective CD8+ responses [48]. In contrast, this method has been used to show that immunization of mice with adenovirus vectors that encode for the well-established P. yoelii PE antigens PyCSP or PyCelTOS, followed by HTVI challenge with the same proteins as luciferase fusions results in significant reductions in the luciferase signal in the liver [49].
In this study, we first used CSP as a model protein to optimize the HTVI challenge of Pyfabb/f−-immunized mice. We observed that HTVI successfully delivered plasmid DNA into hepatocytes, resulting in the expression of luciferase-tagged PyCSP (Figure 1 and Figure S2). Importantly, mice immunized with two doses of Pyfabb/f− sporozoites and challenged with plasmid DNA encoding for luciferase-tagged PyCSP displayed a reduced luciferase signal (Figure 1), suggesting that immunization with whole parasites can induce a CSP-specific immune response that is able to eliminate hepatocytes that present CSP on their surface. Furthermore, depletion of CD8+, but not of CD4+ T cells, abrogated the suppression of the luciferase signal (Figure 2), demonstrating that hepatocyte killing is CD8+ dependent. We also showed an increase in the level of total and IFN-γ producing CSP-specific CD8+ T cells in the liver of mice immunized with Pyfabb/f− and challenged with PyCSP-Luc (Figure 3). These results agree with previous data from our group and others suggesting that the elimination of malaria-infected hepatocytes is mediated by CD8+ T cells [14], [16], [28].
Next, as a proof of concept of the ability of this method to identify novel LS antigens that associate with the protective immunity induced by whole parasite vaccination, we used it to test PyTmp21. The P. vivax orthologue of PyTmp21 was originally identified as a potential novel PE antigen recognized by PBMCs of Duffy receptor negative donors with naturally acquired malaria immunity to P. vivax [42], who do not support blood stage P. vivax infection as a result of the mutation. We showed that PyTmp21 elicits functional immunity that significantly reduces LS parasite burden after challenge with wt P. yoelii spz (Figure 4). However, we were initially unable to confirm PyTmp21 as an antigen that contributes to the protection elicited by Pyfabb/f− sporozoites using the immunization strategy that worked for PyCSP (Figure 5A and B). This result agrees with recent data suggesting that repeated exposure to whole parasite vaccines results in a bias of the immune response towards the immunodominant CSP, and away from less abundant antigens expressed during PE stages [43].
To overcome this obstacle, we explored different heterologous priming/boost immunization strategies. First, we vaccinated mice with a single dose of Pyfabb/f− sporozoites followed by boosting with plasmid DNA encoding for PyTmp21. In contrast to the initial negative result obtained for PyTmp21 upon repeated immunization with Pyfabb/f−, this strategy resulted in reduction of the luciferase signal upon HTVI challenge with PyTmp21 as a luciferase fusion (Figure 5C and D). Vaccination of mice with plasmid DNA encoding for PyTmp21 followed by boosting with a single dose of Pyfabb/f− also resulted in a reduction of luciferase signal upon challenge with PyTmp21-Luc plasmid DNA (Figure 5E and F). These data suggest that PyTmp21 is expressed by Pyfabb/f− parasites, and that it contributes to protection induced by whole parasite vaccination. Interestingly, the kinetics of these two immunization strategies were strikingly different: whereas priming with Pyfabb/f− followed by boosting with plasmid DNA resulted in 60% reduction of luciferase signal 4 h post challenge, which then declined to about 20% for the duration of the experimental time course, priming with DNA followed by boosting with Pyfabb/f− yielded an initially low response that only peaked at 60% 24 h post challenge and remained fairly constant thereafter (Figure 5D and F). It is possible that these different responses could be due to the different location of memory T cells elicited by DNA and whole parasite vaccinations [50]. Priming the immune system with a DNA vaccine administered through the i.m. route results in a T cell response that is localized to peripheral sites, so that upon boosting with Pyfabb/f− these cells have to migrate to the liver in order to respond to the HTVI challenge, resulting in a delayed response. In contrast, priming with Pyfabb/f− results in a liver infection that induces tissue resident T cells, so that boosting with DNA leads to a rapid expansion of these cells and a quick induction of cytotoxic effector cells that can destroy hepatocytes presenting specific antigens.
Based on our observation that elimination of PyCSP-Luc signal is abrogated in the absence of CD8+ cells, in addition to previous data from our group and others that demonstrate the essential role of CD8+ T cells in controlling pre-erythrocytic malaria infection [14], [16], [17], [28], we hypothesize that PyTmp21-expressing hepatocytes are most likely eliminated by mechanisms that depend on CD8+ T cells. Future studies will be aimed at determining the specific mechanism involved in the reduction of luciferase signal observed upon challenge with PyTmp21-Luc of mice immunized with heterologous strategies that combine whole parasites and DNA vaccines.
In conclusion, using CSP as a model, we were able to confirm that the HTVI/IVIS method enables the detection of hepatocytes that are killed as a consequence of presenting specific parasite antigens, and that this killing depends on CD8+ T cells. Furthermore, the data presented herein show that the use of a heterologous immunization strategy coupled with the HTVI/IVIS method constitutes a powerful tool to validate pre-erythrocytic antigens that contribute to the protection elicited by whole parasite vaccines. In particular, we confirmed that PyTmp21, which we previously identified as a novel pre-erythrocytic antigen, contributes to the protective immunity elicited by whole parasite vaccinations. Ultimately, the method described herein can be used to validate new malaria vaccine candidates and increase our understanding of how whole parasite immunization protects against malaria, thus paving the way for intelligent vaccine design.
Supporting Information
Cloning and expression of PyCSP and PyTmp21 luciferase-fusion proteins. (A) Representation of the structure of PyCSP-Luc and PyTmp21-Luc. The diagram shows the amino-terminal Myc tag, the carboxy-terminal luciferase fusion protein, and the regions and elements of the P. yoelii proteins included in the constructs. (B–C) Assessment of expression of the fusion proteins ex vivo. (B) Cell lysates purified from COS-7 cells transfected with phCMV-PyCSP-Luc, phCMV-PyTmp21-Luc, phCMV-Luc or not transfected were separated by protein gel electrophoresis and transferred to a nitrocellulose membrane. Luciferase fusion proteins were identified by probing the membrane with a polyclonal anti-luciferase antibody by Western blot. (C) Cell lysates prepared as described in part B were assayed for luciferase activity 48 hours after transfection, using a luminometer.
(TIF)
Expression of PyCSP by hepatocytes and liver lymphocytes. Representative dot plots showing the percentage of PyCSP positive cells gated on hepatocytes (A), Kupffer cells (B); dendritic cells (C), Sinusodial Liver Endothelial Cells (D), T cells (E) and B cells (F), obtained from mice injected with phCMV-Luc (left panel, n = 2) or phCMV-PyCSP-Luc (middle panel, n = 3). The graphs on the right show the data for all mice. The horizontal bar indicates the mean.
(TIF)
Acknowledgments
The anti-H2Kd/CSP tetramer antibody was obtained from the NIH Tetramer Core Facility. We are grateful to William W. Betz, Mark F. Kennedy, Heather Kain and Jen C.C. Hume of the Seattle Biomedical Research Institute Insectary Facility for mosquito and sporozoite production, and Tim Dawe and the Vivarium personnel for animal care.
Funding Statement
This work was supported by the National Institutes of Health grant R01AI076498 to R.W., Bill and Melinda Gates Foundation grant OPP1016829 to R.W., National Natural Science Foundation of China #30972570 to F.H., and by Seattle BioMed. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO (2013) World Malaria Report 2013. Geneva: World Health Organization. [Google Scholar]
- 2. Vaughan AM, Wang R, Kappe SH (2010) Genetically engineered, attenuated whole-cell vaccine approaches for malaria. Hum Vaccin 6: 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Epstein JE, Richie TL (2013) The whole parasite, pre-erythrocytic stage approach to malaria vaccine development: a review. Curr Opin Infect Dis 26: 420–428. [DOI] [PubMed] [Google Scholar]
- 4. VanBuskirk KM, O'Neill MT, De La Vega P, Maier AG, Krzych U, et al. (2009) Preerythrocytic, live-attenuated Plasmodium falciparum vaccine candidates by design. Proc Natl Acad Sci U S A 106: 13004–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mueller AK, Labaied M, Kappe SH, Matuschewski K (2005) Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature 433: 164–167. [DOI] [PubMed] [Google Scholar]
- 6. van Dijk MR, Douradinha B, Franke-Fayard B, Heussler V, van Dooren MW, et al. (2005) Genetically attenuated, P36p-deficient malarial sporozoites induce protective immunity and apoptosis of infected liver cells. Proc Natl Acad Sci U S A 102: 12194–12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clyde DF (1975) Immunization of man against falciparum and vivax malaria by use of attenuated sporozoites. Am J Trop Med Hyg 24: 397–401. [DOI] [PubMed] [Google Scholar]
- 8. Hoffman SL, Goh LM, Luke TC, Schneider I, Le TP, et al. (2002) Protection of humans against malaria by immunization with radiation-attenuated Plasmodium falciparum sporozoites. J Infect Dis 185: 1155–1164. [DOI] [PubMed] [Google Scholar]
- 9. Nussenzweig RS, Vanderberg J, Most H, Orton C (1967) Protective immunity produced by the injection of x-irradiated sporozoites of Plasmodium berghei . Nature 216: 160–162. [DOI] [PubMed] [Google Scholar]
- 10. Seder RA, Chang LJ, Enama ME, Zephir KL, Sarwar UN, et al. (2013) Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science 341: 1359–1365. [DOI] [PubMed] [Google Scholar]
- 11.Malaria Vaccine Funders Group (2013) Malaria Vaccine Technology Roadmap.
- 12. Kester KE, Cummings JF, Ofori-Anyinam O, Ockenhouse CF, Krzych U, et al. (2009) Randomized, double-blind, phase 2a trial of falciparum malaria vaccines RTS,S/AS01B and RTS,S/AS02A in malaria-naive adults: safety, efficacy, and immunologic associates of protection. J Infect Dis 200: 337–346. [DOI] [PubMed] [Google Scholar]
- 13. Kester KE, Cummings JF, Ockenhouse CF, Nielsen R, Hall BT, et al. (2008) Phase 2a trial of 0, 1, and 3 month and 0, 7, and 28 day immunization schedules of malaria vaccine RTS,S/AS02 in malaria-naive adults at the Walter Reed Army Institute of Research. Vaccine 26: 2191–2202. [DOI] [PubMed] [Google Scholar]
- 14. Trimnell A, Takagi A, Gupta M, Richie TL, Kappe SH, et al. (2009) Genetically attenuated parasite vaccines induce contact-dependent CD8+ T cell killing of Plasmodium yoelii liver stage-infected hepatocytes. J Immunol 183: 5870–5878. [DOI] [PubMed] [Google Scholar]
- 15. Cooney LA, Gupta M, Thomas S, Mikolajczak S, Choi KY, et al. (2013) Short-lived effector CD8 T cells induced by genetically attenuated malaria parasite vaccination express CD11c. Infect Immun 81: 4171–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodrigues MM, Cordey AS, Arreaza G, Corradin G, Romero P, et al. (1991) CD8+ cytolytic T cell clones derived against the Plasmodium yoelii circumsporozoite protein protect against malaria. Int Immunol 3: 579–585. [DOI] [PubMed] [Google Scholar]
- 17. Cockburn IA, Amino R, Kelemen RK, Kuo SC, Tse SW, et al. (2013) In vivo imaging of CD8+ T cell-mediated elimination of malaria liver stages. Proc Natl Acad Sci U S A 110: 9090–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roestenberg M, McCall M, Hopman J, Wiersma J, Luty AJ, et al. (2009) Protection against a malaria challenge by sporozoite inoculation. N Engl J Med 361: 468–477. [DOI] [PubMed] [Google Scholar]
- 19. Butler NS, Schmidt NW, Vaughan AM, Aly AS, Kappe SH, et al. (2011) Superior antimalarial immunity after vaccination with late liver stage-arresting genetically attenuated parasites. Cell Host Microbe 9: 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barbosa A, Naniche D, Aponte JJ, Manaca MN, Mandomando I, et al. (2009) Plasmodium falciparum-specific cellular immune responses after immunization with the RTS,S/AS02D candidate malaria vaccine in infants living in an area of high endemicity in Mozambique. Infect Immun 77: 4502–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ewer KJ, O'Hara GA, Duncan CJ, Collins KA, Sheehy SH, et al. (2013) Protective CD8+ T-cell immunity to human malaria induced by chimpanzee adenovirus-MVA immunisation. Nat Commun 4: 2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang R, Doolan DL, Charoenvit Y, Hedstrom RC, Gardner MJ, et al. (1998) Simultaneous induction of multiple antigen-specific cytotoxic T lymphocytes in nonhuman primates by immunization with a mixture of four Plasmodium falciparum DNA plasmids. Infect Immun 66: 4193–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gruner AC, Mauduit M, Tewari R, Romero JF, Depinay N, et al. (2007) Sterile protection against malaria is independent of immune responses to the circumsporozoite protein. PLoS One 2: e1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mauduit M, Gruner AC, Tewari R, Depinay N, Kayibanda M, et al. (2009) A role for immune responses against non-CS components in the cross-species protection induced by immunization with irradiated malaria sporozoites. PLoS One 4: e7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mauduit M, Tewari R, Depinay N, Kayibanda M, Lallemand E, et al. (2010) Minimal role for the circumsporozoite protein in the induction of sterile immunity by vaccination with live rodent malaria sporozoites. Infect Immun 78: 2182–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duffy PE, Sahu T, Akue A, Milman N, Anderson C (2012) Pre-erythrocytic malaria vaccines: identifying the targets. Expert Rev Vaccines 11: 1261–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mishra S, Rai U, Shiratsuchi T, Li X, Vanloubbeeck Y, et al. (2011) Identification of non-CSP antigens bearing CD8 epitopes in mice immunized with irradiated sporozoites. Vaccine 29: 7335–7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hafalla JC, Bauza K, Friesen J, Gonzalez-Aseguinolaza G, Hill AV, et al. (2013) Identification of targets of CD8(+) T cell responses to malaria liver stages by genome-wide epitope profiling. PLoS Pathog 9: e1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu F, Song Y, Liu D (1999) Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther 6: 1258–1266. [DOI] [PubMed] [Google Scholar]
- 30. Sebestyen MG, Budker VG, Budker T, Subbotin VM, Zhang G, et al. (2006) Mechanism of plasmid delivery by hydrodynamic tail vein injection. I. Hepatocyte uptake of various molecules. J Gene Med 8: 852–873. [DOI] [PubMed] [Google Scholar]
- 31. Bell JB, Podetz-Pedersen KM, Aronovich EL, Belur LR, McIvor RS, et al. (2007) Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat Protoc 2: 3153–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller JL, Murray S, Vaughan AM, Harupa A, Sack B, et al. (2013) Quantitative bioluminescent imaging of pre-erythrocytic malaria parasite infection using luciferase-expressing Plasmodium yoelii . PLoS One 8: e60820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bruna-Romero O, Hafalla JC, Gonzalez-Aseguinolaza G, Sano G, Tsuji M, et al. (2001) Detection of malaria liver-stages in mice infected through the bite of a single Anopheles mosquito using a highly sensitive real-time PCR. Int J Parasitol 31: 1499–1502. [DOI] [PubMed] [Google Scholar]
- 34. Franke ED, Corradin G, Hoffman SL (1997) Induction of protective CTL responses against the Plasmodium yoelii circumsporozoite protein by immunization with peptides. J Immunol 159: 3424–3433. [PubMed] [Google Scholar]
- 35. Zhang G, Budker V, Wolff JA (1999) High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther 10: 1735–1737. [DOI] [PubMed] [Google Scholar]
- 36. Butler NS, Schmidt NW, Harty JT (2010) Differential effector pathways regulate memory CD8 T cell immunity against Plasmodium berghei versus P. yoelii sporozoites. J Immunol 184: 2528–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cockburn IA, Tse SW, Zavala F (2014) CD8+ T cells eliminate liver-stage Plasmodium berghei parasites without detectable bystander effect. Infect Immun 82: 1460–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cabrera M, Pewe LL, Harty JT, Frevert U (2013) In vivo CD8+ T cell dynamics in the liver of Plasmodium yoelii immunized and infected mice. PLoS One 8: e70842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jobe O, Lumsden J, Mueller AK, Williams J, Silva-Rivera H, et al. (2007) Genetically attenuated Plasmodium berghei liver stages induce sterile protracted protection that is mediated by major histocompatibility complex Class I-dependent interferon-gamma-producing CD8+ T cells. J Infect Dis 196: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krzych U, Schwenk J (2005) The dissection of CD8 T cells during liver-stage infection. Curr Top Microbiol Immunol 297: 1–24. [DOI] [PubMed] [Google Scholar]
- 41. Nganou-Makamdop K, van Gemert GJ, Arens T, Hermsen CC, Sauerwein RW (2012) Long term protection after immunization with P. berghei sporozoites correlates with sustained IFNgamma responses of hepatic CD8+ memory T cells. PLoS One 7: e36508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang R, Arevalo-Herrera M, Gardner MJ, Bonelo A, Carlton JM, et al. (2005) Immune responses to Plasmodium vivax pre-erythrocytic stage antigens in naturally exposed Duffy-negative humans: a potential model for identification of liver-stage antigens. Eur J Immunol 35: 1859–1868. [DOI] [PubMed] [Google Scholar]
- 43. Murphy SC, Kas A, Stone BC, Bevan MJ (2013) A T-cell response to a liver-stage Plasmodium antigen is not boosted by repeated sporozoite immunizations. Proc Natl Acad Sci U S A 110: 6055–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bijker EM, Bastiaens GJ, Teirlinck AC, van Gemert GJ, Graumans W, et al. (2013) Protection against malaria after immunization by chloroquine prophylaxis and sporozoites is mediated by preerythrocytic immunity. Proc Natl Acad Sci U S A 110: 7862–7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumar KA, Sano G, Boscardin S, Nussenzweig RS, Nussenzweig MC, et al. (2006) The circumsporozoite protein is an immunodominant protective antigen in irradiated sporozoites. Nature 444: 937–940. [DOI] [PubMed] [Google Scholar]
- 46. Vaughan AM, O'Neill MT, Tarun AS, Camargo N, Phuong TM, et al. (2009) Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol 11: 506–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herweijer H, Wolff JA (2007) Gene therapy progress and prospects: hydrodynamic gene delivery. Gene Ther 14: 99–107. [DOI] [PubMed] [Google Scholar]
- 48. Obeng-Adjei N, Hutnick NA, Yan J, Chu JS, Myles DJ, et al. (2013) DNA vaccine cocktail expressing genotype A and C HBV surface and consensus core antigens generates robust cytotoxic and antibody responses in mice and Rhesus macaques. Cancer Gene Ther 20: 652–662. [DOI] [PubMed] [Google Scholar]
- 49. Rai UH, Huang J, Mishra S, Li X, Shiratsuchi T, et al. (2012) A new method to determine antigen-specific CD8+ T cell activity in vivo by hydrodynamic injection. Biomolecules 2: 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Woodland DL, Kohlmeier JE (2009) Migration, maintenance and recall of memory T cells in peripheral tissues. Nat Rev Immunol 9: 153–161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cloning and expression of PyCSP and PyTmp21 luciferase-fusion proteins. (A) Representation of the structure of PyCSP-Luc and PyTmp21-Luc. The diagram shows the amino-terminal Myc tag, the carboxy-terminal luciferase fusion protein, and the regions and elements of the P. yoelii proteins included in the constructs. (B–C) Assessment of expression of the fusion proteins ex vivo. (B) Cell lysates purified from COS-7 cells transfected with phCMV-PyCSP-Luc, phCMV-PyTmp21-Luc, phCMV-Luc or not transfected were separated by protein gel electrophoresis and transferred to a nitrocellulose membrane. Luciferase fusion proteins were identified by probing the membrane with a polyclonal anti-luciferase antibody by Western blot. (C) Cell lysates prepared as described in part B were assayed for luciferase activity 48 hours after transfection, using a luminometer.
(TIF)
Expression of PyCSP by hepatocytes and liver lymphocytes. Representative dot plots showing the percentage of PyCSP positive cells gated on hepatocytes (A), Kupffer cells (B); dendritic cells (C), Sinusodial Liver Endothelial Cells (D), T cells (E) and B cells (F), obtained from mice injected with phCMV-Luc (left panel, n = 2) or phCMV-PyCSP-Luc (middle panel, n = 3). The graphs on the right show the data for all mice. The horizontal bar indicates the mean.
(TIF)