Abstract
Acid rain (AR) is a serious environmental issue inducing harmful impacts on plant growth and development. It has been reported that Liquidambar formosana, considered as an AR-sensitive tree species, was largely injured by AR, compared with Schima superba, an AR-tolerant tree species. To clarify the different responses of these two species to AR, a comparative proteomic analysis was conducted in this study. More than 1000 protein spots were reproducibly detected on two-dimensional electrophoresis gels. Among them, 74 protein spots from L. formosana gels and 34 protein spots from S. superba gels showed significant changes in their abundances under AR stress. In both L. formosana and S. superba, the majority proteins with more than 2 fold changes were involved in photosynthesis and energy production, followed by material metabolism, stress and defense, transcription, post-translational and modification, and signal transduction. In contrast with L. formosana, no hormone response-related protein was found in S. superba. Moreover, the changes of proteins involved in photosynthesis, starch synthesis, and translation were distinctly different between L. formosana and S. superba. Protein expression analysis of three proteins (ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, ascorbate peroxidase and glutathione-S-transferase) by Western blot was well correlated with the results of proteomics. In conclusion, our study provides new insights into AR stress responses in woody plants and clarifies the differences in strategies to cope with AR between L. formosana and S. superba.
Introduction
Acid rain (AR) emerged as a serious environmental issue as a consequence of the increasing industrial activities throughout the world [1]. Forty percent of the territory in China is seriously affected by AR since the late 1970s, especially in southern China [2]. The harmful impacts of AR on plants are observed in a wide array of biological processes. AR decreases seed germination [3], strips the protective wax from leaves [4], induces visible injury symptoms [1], disturbs plant nitrogen metabolism [5]. AR also decreases chlorophyll content and photosynthetic efficiency [3], [6], increases reactive oxygen species (ROS) production [7], accelerates the leaching of nutrients from plant foliage [8], which further inhibits tree radial growth, vertical growth and total tree biomass [4], [9].
Liquidambar formosana and Schima superba are both dominant broad-leaf tree species and are distributed over large surface areas in the forest of southern China [10]. Some field observations and laboratory experiments reported that, when compared with S. superba, L. formosana was largely injured by AR during the past decade, which had negative impacts on forest ecosystem [9]. Our previous study also found that AR more easily affected some physiological parameters in L. formosana seedlings than S. superba’s seedlings, e.g., seed germination, seedling growth, photosynthesis, antioxidant system, etc. [3], [11]. Thus, L. formosana is considered as an AR-sensitive species, while S. superba is an AR-tolerant species. Although the differential responses of L. formosana and S. superba to AR have been analyzed at the morphological and physiological level, a comprehensive elucidation of the molecular mechanisms underlying the different strategies to cope with AR between two tree species is still needed.
Proteomics is a powerful tool for providing new insights into complete proteomes at the organ, tissue and cell levels [12]. A number of proteomic analyses help us understand the molecular mechanisms of plants in responses to various environmental stresses including salinity [13], cold [14], heavy metal [15], etc. In our previous work, a wide array of proteins related to AR-resistance has been identified by comparative proteomic analysis in a model plant, Arabidopsis thaliana [5], [16]. However, little information is available in proteome analysis for tree species subjected to AR stress, and a comparison between AR-sensitive and AR-resistant broad-leaf tree species has not been fully conducted at the proteome level.
In the present study, we initiated a comparative proteomic study to systematically investigate the changes in protein profile in two broad-leaf tree species, L. formosana and S. superba, that are different sensitive to AR tolerance, when submitted to simulated AR (pH 3.0) treatment for one month. Based on two-dimensional electrophoresis (2-DE) and mass spectrometry (MS) analysis, a comprehensive inventory of proteins regulated by AR was established in the two tree species. The overall objectives of this study are (1) to provide valuable insights into AR stress responses in woody plants; (2) to clarify the differences in strategies to cope with AR between L. formosana and S. superba.
Materials and Methods
Plant materials and treatments
Seeds of L. formosana and S. superba were purchased from Tree Seed Centre of Shuyang County in Jiangsu Province, China. The seeds have been mixed together when they were collected from independent individuals and families. Seeds were surface-sterilized with 0.5% hypochlorite for 30 min, then washed thoroughly with distilled water. Then the seeds were germinated in a soil/vermiculite (1∶1) mixture in an environmentally controlled growth chamber. For each species, three weeks old healthy seedlings with similar size were randomly transplanted into individual pots, each with a dimension of 24 cm (open top) × 13 cm (height) × 15 cm (flat bottom), and filled with soil/vermiculite (1∶1) mixture. Fifteen seedlings were planted in one pot. All seedlings were cultivated in the same controlled growth chamber with a daily temperature regime of 28/25°C (day/night), relative humidity of 60–70% and a 12-h photoperiod at 210 µmol m−2 s−1 photosynthetically active radiation (PAR). Three months later, the seedlings were divided into control group (CK) and simulated AR treatment group (AR). Each group had at least three replicates. The control group and simulated AR treatment group were sprayed once per day with the control (pH 5.6) solution and AR (pH 3.0) solution, respectively. The ion compositions of the control solution was adapted from Fan and Wang [4], AR solution was made from control solution and the pH was adjusted by adding a mixture of H2SO4 and HNO3 in the ratio of 5∶1. The final concentration of H2SO4 and HNO3 were 0.45 and 0.09 mM, respectively, which represents the average ion composition of rainfall in southern China [4]. After one month of treatment, a portion of fresh leaves was used for measuring some physiological parameters such as necrosis percentage, chlorophyll content, net photosynthetic rate, H2O2 content and so on. The remaining leaves were stored at −80°C for proteomic and Western blot analysis.
Necrosis percentage and chlorophyll content measurements
At least 30 fully expanded leaves of each species were randomly selected from control and AR treatment groups and photographed using a digital camera. The necrosis percentage was calculated as described previously [16]. Chlorophyll was extracted from approximately 0.1 g fresh leaf slices directly into 10 ml ice-cold acetone (80%, v/v). The chlorophyll content (mg g−1 fresh weight (FW)) was determined as described previously [17].
Net photosynthetic rate and chlorophyll fluorescence measurements
Three seedlings per species were randomly chosen from different pots and at least two fully emerged leaves per plant were selected for net photosynthetic rate (Pn) and chlorophyll fluorescence measurements. Pn was performed with a portable photosynthesis system (LI-6400, Li-Cor Inc., Lincoln, Nebraska, USA), as described previously [18]. According to the method of Liu et al [16], leaf chlorophyll fluorescence was measured using a pluse-amplitude-modulation fluorometer (PAM-2100, Heinz Walz, Effeltrich, Germany).
Measurement of proline, malondialdehyde (MDA) and ROS production
Proline content was measured according to the method of Jiang et al [19]. The level of lipid peroxidation was measured by estimating MDA content using thiobarbituric acid (TBA) reaction [20]. Superoxide radical (O2 •-) and H2O2 content was measured following the method of Chen et al [11].
Protein extraction
Protein extraction was performed using phenol-based protocol described by Liu et al [5], with slight modifications. Briefly, frozen leaves (1.0 g) were ground with a mortar and pestle with liquid nitrogen, the ground powder was homogenized in pre-cooled extraction buffer (20 mM Tris-HCl, pH 7.5, 250 mM sucrose, 10 mM ethylene diamine tetraacetic acid (EDTA), 1 mM phenylmethyl-sulfonyl fluoride, 1 mM 1,4-dithiothreitol (DTT) and 1% Triton X-100) on ice. Then an equal volume of ice-cold Tris-HCl (pH 7.5) saturated phenol was added and the mixture was centrifuged (15,000 g, 4°C) for 15 min. The phenol phase was collected and proteins were precipitated with ammonium acetate in methanol for 10 h at −20°C. After centrifugation, the supernatant was discarded and the pellet was washed for three times using cold acetone containing 10 mM DTT. The washed protein pellets were dissolved in a lysis buffer (8 M urea, 2 M thiourea, 4% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propane sulfonate (CHAPS), 1% (w/v) DTT and 0.5% (w/v) IPG buffer pH 4–7) at room temperature. The total protein concentration of the lysates was determined using a Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA, USA).
Two-dimensional electrophoresis, image and data analysis
Two-dimensional electrophoresis was conducted according to the methods of Bai et al [13] and Hu et al [21]. Isoelectric focusing (IEF) was done using an Ettan IPGphor system (GE Healthcare) PROTEAN electrophoresis system and immobilized IPG dry gel strips with a linear pH range (18 cm long, pH 4–7 linear) (GE Healthcare Amersham Bioscience, Little Chalfont, UK). Protein samples (800 µg) were loaded during the rehydration step at room temperature for 12 h. IEF was performed at 300 voltage (V), 500 V and 1,000 V for 1 h, a gradient to 8,000 V over 4 h, and kept at 8,000 V for a total of 80,000 volt-hours (Vh) at 20°C. Subsequently, focused strips were equilibrated in equilibration buffer as described by Yang et al [12]. For the second dimension, proteins were separated on 15% SDS polyacrylamide gels. Proteins spots were detected by staining the gels with Coomassie Brilliant Blue R-250. The 2-DE gels were scanned with a scanner (Uniscan M3600, China) and the gel images were analyzed with PDQuest software (Version 8.01, Bio-Rad, Hercules, CA), on the basis of their relative volume. Only those protein spots with significant (more than 2-fold change) and reproducible changes in three replicates were selected for next MS analysis.
In-gel digestion, protein identification and classification analysis
The protein spots, which were differentially displayed under AR treatment, were excised from the preparative 2-D gels and digested by trypsin. After digestion, the peptide solution was collected and peptide mass fingerprint (PMF) was acquired using matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS) analysis (ReFlexTM III, Bruker, Bremen, Germany) as described previously [16]. The PMF spectra were used in online searches combined with the Mascot program search engine (http://www.matrixscience.com) and National Center for Biotechnology Information (NCBI) protein database (http://www.ncbi.nlm.nih.gov) (NCBInr, 17751536 entries, downloaded on April 17, 2012). PMF search parameters were set up as described previously [22]. Proteins with a MOWSE score >73 were considered as positive identifications. The identified proteins were searched with against the UniProt (http://www.uniprot.org) and/or NCBI protein database (http://www.ncbi.nlm.nih.gov) for updated annotation and homologous proteins identification. Afterwards, the successfully identified proteins were further classified using Functional Catalogue software (http://mips.gsf.de/projects/funcat).
Western blot analysis
Western blot analysis was performed as described previously [11]. Total proteins (40 µg) extracted from L. formosana and S. superba leaves were separated by 12% w/v standard sodium dodecyl sulfate polyacrylamide gel electrophoresis and then electrophoretically blotted to polyvinylidene difluoride membrane for 50 min. The membranes were blocked over-night with Western Blocking Buffer (TIANGEN, China). Protein blots were probed with primary antibodies of Rubisco large subunit (RuBISCO LSU) (AS03037-200, Agrisera, Sweden), ascorbate peroxidase (APX) (AS08368, Agrisera, Sweden) and glutathione-S-transferase (GST) (AS09479, Agrisera, Sweden), at dilution of 1∶5000, 1∶2000 and 1∶1000, respectively, for 4 h at room temperature with agitation. Next, the membranes were washed in phosphate buffered saline with Tween-20 solution (PBST) solution (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% Tween 20, v/v) three times and incubated with anti-rabbit IgG horseradish peroxidase (HRP) conjugated to alkaline phosphatase (Abcam, U.K., 1∶5000 dilution) for 1 h at room temperature to detect primary antibodies. β-actin (1∶5000, Santa Cruz, California, USA) was used as an internal control. After several washes with PBST solution, membranes were incubated in an enhanced chemiluminescence (ECL) substrate detection solution (TIANGEN, China) according to the manufacturer’s instructions. Images of protein blots were obtained using a CCD imager (FluorSMax Bio-Rad, Hercules, CA, USA). The optical density values of the protein signals were quantified using the Quantity One software (Bio-Rad, Hercules, CA, USA).
Statistical analysis
For physiological measurements, at lease four independent repetitions were used. Values in figures and tables were expressed as means ± se. The statistical significance of the data was analyzed using a univariate analysis of variance (One-way ANOVA, Duncan’s multiple range test, p<0.05) with the SPSS 20.0 package (SPSS, Chicago, Illionis USA). For proteomic experiment, protein samples for 2-DE gel image analysis were extracted from three independent seedlings grown in three different pots in the same growth chamber. Thus, for each species, three independent biological replicates were performed in 2-DE gel image analysis. Statistic analysis for protein spot on 2-DE gels was performed using Student’s t-test (p<0.05) provided by PDQuest software as mentioned earlier.
Results
Morphological and physiological responses of L. formosana and S. superba to AR
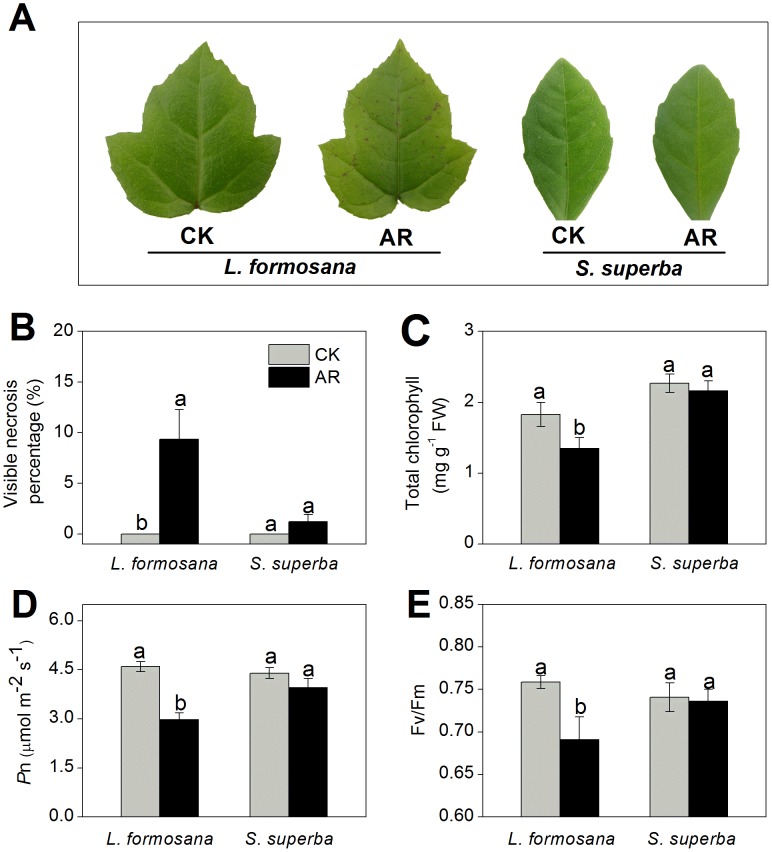
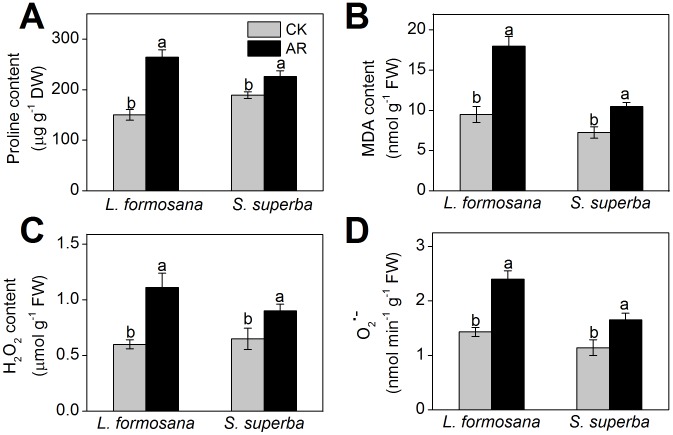
As shown in Fig. 1A and B, remarkable yellowing symptom and significant necrosis emerge in L. formosana leaves after one-month exposure to AR. There was an important decrease in total chlorophyll content in AR-treated L. formosana leaves, however, no statistically significant change was found in S. superba (Fig. 1C). Pn and Fv/Fm in AR-treated L. formosana seedlings were remarkably inhibited, whereas AR slightly decreased Pn and Fv/Fm in S. superba (Fig. 1D and E). After AR treatment, proline content in L. formosana and S. superba increased by 76.0% and 19.7%, respectively, compared with the control (Fig. 2A). MDA contents in AR-treated L. formosana and S. superba increased by 89.4% and 44.8%, respectively (Fig. 2B). As shown in Fig. 2C and D, the levels of H2O2 and O2 •- in both L. formosana and S. superba were significantly stimulated by AR. In particular, compared with the control, H2O2 and O2 •- content increased by 83.3% and 67.8% in L. formosana, and by 38.4% and 44.7% in S. superba, respectively (Fig. 2C and D).
Figure 1. Effects of one-month AR on morphology and photosynthesis of L. formosana and S. superba.
The pH of AR solution was adjusted to 3.0 by adding a mixture of H2SO4 and HNO3 in the ratio of 5∶1. The final concentration of H2SO4 and HNO3 were 0.45 and 0.09 mM, respectively. (A) Leaf injury phenotype. (B) Leaf necrosis percentage. (C) Total chlorophyll content. (D) Net photosynthetic rate (Pn). (E) Quantum efficiency of open PSII centers in a dark-adapted state (Fv/Fm). Columns labeled with different letters indicate significant differences at p<0.05.
Figure 2. Changes in proline (A), MDA (B), H2O2 (C) and O2 •- (D) content after AR treatment.
Columns labeled with different letters indicate significant differences at p<0.05.
Identification of AR-responsive proteins
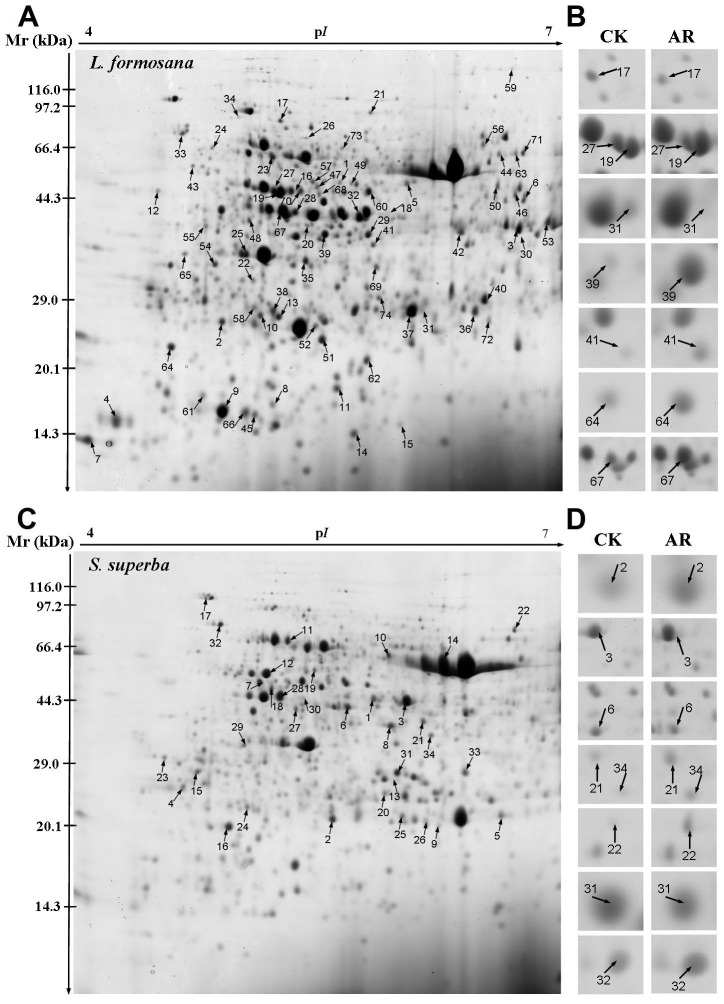
To investigate the differentially expressed proteins in L. formosana and S. superba exposed to AR treatment, comparative proteomic analysis was performed on Coomassie-stained 2-DE maps shown in Fig. 3. Over 1000 protein spots reproducibly separated and matched between control and AR gels, 74 protein spots in L. formosana, and only 34 protein spots in S. superba had at least a 2-fold greater abundance in either AR or control (Fig. 3). The identified proteins in L. formosana and S. superba by MALDI-TOF MS analysis are presented in Tables 1 and 2, respectively. Because the complete annotated sequences of L. formosana and S. superba genomes are not yet available, all identified proteins were functionally classified by UniProt and NCBI databases according to their homology with other proteins. Functional annotations in databases existed for the majority of the protein spots, while 12 proteins (spots L63–L74) in L. formosana and 6 proteins (spots S29–S34) in S. superba were annotated as predicted or unknown proteins (Tables 1 and 2).
Figure 3. 2D gel analysis of proteins extracted from L. formosana and S. superba leaves.
The numbers assigned to the proteins spots correspond to those listed in Tables 1 and 2. (A) Representative 2-DE gels of L. formosana in which 74 spots showing at least 2-fold changes (p<0.05) under AR were identified by MALDI-TOF MS. (B) Close-up views of differentially expressed protein spots in L. formosana (highlighted by arrows). (C) Representative 2-DE gels of S. superba in which 34 spots showing at least 2-fold changes (p<0.05) under AR were identified by MALDI-TOF MS. (D) Close-up views of differentially expressed protein spots in S. superba (highlighted by arrows).
Table 1. Identification of AR-responsive proteins in L. formosana.
| Spota | NCBI accessionb | Protein identityc | Thero. Da/pI d | Exper. Da/pIe | SCf | MP/TPg | Scoreh | Ci | Organism matched |
| Material metabolism | |||||||||
| L1 | gi|297835714 | glucose-1-phosphate adenylyltransferase (GPAT) | 57.66/6.54 | 46.40/5.48 | 15% | 6/7 | 81 | D | Arabidopsis lyrata subsp |
| L2 | gi|226500818 | shikimate kinase family protein | 30.34/5.87 | 27.41/4.74 | 36% | 7/8 | 110 | U | Zea mays |
| L3 | gi|162458456 | zeta-carotene desaturase | 63.49/8.62 | 39.22/6.95 | 10% | 5/5 | 74 | U | Zea mays |
| L4 | gi|356504466 | haloalkane dehalogenase-like | 45.01/8.43 | 15.10/4.12 | 21% | 8/10 | 99 | U | Glycine max |
| L6 | gi|95117792 | glutamate dehydrogenase (GDH) | 44.82/6.38 | 44.21/7.00 | 23% | 8/8 | 112 | U | Vitis vinifera |
| L7 | gi|15081239 | glycine-rich protein 17 (GRP17) | 53.32/10.31 | 14.09/4.02 | 21% | 5/5 | 87 | U | Arabidopsis thaliana |
| L8 | gi|15218536 | stearoyl-acyl-carrier protein desaturase-like protein (SACPDLP) | 44.36/6.10 | 17.36/5.09 | 14% | 6/6 | 97 | U | Arabidopsis thaliana |
| L9 | gi|38426301 | 6-phosphogluconate dehydrogenase | 51.78/6.58 | 16.60/4.74 | 12% | 5/5 | 78 | U | Oryza sativa |
| L11 | gi|255567778 | cysteine synthase (CS) | 43.38/7.60 | 20.47/5.49 | 21% | 9/10 | 114 | D | Ricinus communis |
| L12 | gi|3341511 | cinnamoyl-CoA reductase | 40.63/5.73 | 47.97/4.40 | 16% | 5/5 | 81 | U | Saccharum officinarum |
| L39 | gi|309951612 | flavanone 3-hydroxylase (F3H) | 41.24/5.39 | 39.35/5.41 | 28% | 7/9 | 103 | U | Litchi chinensis |
| L46 | gi|114795072 | chalcone synthase (CHS) | 42.83/6.05 | 45.37/6.91 | 22% | 8/10 | 84 | U | Pyrus communis |
| Photosynthesis and energy production | |||||||||
| L18 | gi|1022805 | phosphoglycerate kinase (PGK) | 41.99/4.93 | 36.02/5.79 | 24% | 8/11 | 108 | D | Arabidopsis thaliana |
| L19 | gi|355329944 | actin | 40.37/5.67 | 41.03/5.13 | 44% | 15/28 | 144 | U | Malus domestica |
| L20 | gi|225423755 | photosystem II stability/assembly factor HCF136 | 44.47/6.92 | 34.91/5.31 | 36% | 12/27 | 109 | D | Vitis vinifera |
| L21 | gi|357438645 | chlorophyllide a oxygenase (CAO) | 25.99/8.95 | 90.68/5.69 | 30% | 4/4 | 76 | D | Medicago truncatula |
| L22 | gi|183217735 | ATP synthase CF1 alpha subunit | 55.62/5.20 | 32.35/4.95 | 24% | 10/11 | 138 | U | Guizotia abyssinica |
| L23 | gi|114421 | ATP synthase subunit beta | 59.93/5.95 | 59.97/5.09 | 25% | 12/25 | 88 | U | Nicotiana plumbaginifolia |
| L24 | gi|225428086 | V-type proton ATPase subunit B | 54.37/5.04 | 66.71/4.68 | 32% | 16/40 | 107 | U | Vitis vinifera |
| L25 | gi|147945622 | oxygen-evolving enhancer protein (OEE) | 34.72/6.08 | 36.12/4.91 | 24% | 7/8 | 109 | U | Leymus chinensis |
| L26 | gi|5758863 | ATP synthase beta subunit | 53.56/5.16 | 73.58/5.31 | 42% | 17/24 | 177 | U | Colchicum autumnale |
| L27 | gi|158726716 | ribulose 1,5-bisphosphate carboxylase/oxygenase activase | 48.81/6.10 | 46.81/5.10 | 26% | 10/13 | 115 | D | Flaveria bidentis |
| L28 | gi|15222551 | phosphoribulokinase (PPK) | 44.72/5.71 | 43.22/5.22 | 24% | 9/18 | 87 | D | Arabidopsis thaliana |
| L29 | gi|79322651 | fructose-bisphosphate aldolase | 41.95/5.94 | 39.20/5.66 | 15% | 7/7 | 96 | D | Arabidopsis thaliana |
| L30 | gi|2108252 | P-glycoprotein-2 (PGP2) | 135.75/8.97 | 38.31/6.98 | 6% | 7/7 | 77 | D | Arabidopsis thaliana |
| L31 | gi|162946539 | ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit | 20.80/8.23 | 28.36/6.01 | 32% | 5/5 | 97 | D | Solanum tuberosum |
| L32 | gi|1022805 | phosphoglycerate kinase (PGK) | 41.99/4.93 | 42.14/5.61 | 24% | 8/17 | 87 | D | Arabidopsis thaliana |
| L33 | gi|356539332 | RuBisCO large subunit-binding protein subunit alpha-like isoform 1 | 61.73/5.23 | 79.85/4.53 | 19% | 9/11 | 111 | D | Glycine max |
| L34 | gi|146188415 | ribulose-1,5-biphosphate carboxylase/oxygenase (Rubisco) | 46.17/6.44 | 87.79/4.87 | 26% | 11/21 | 104 | D | Podalyria canescens |
| L61 | gi|297816654 | metal ion binding protein | 26.66/9.55 | 16.60/4.74 | 25% | 4/4 | 73 | U | Arabidopsis lyrata subsp |
| Stress and defense | |||||||||
| L35 | gi|42568255 | TIR-NBS-LRR class disease resistance protein | 121.85/6.36 | 35.08/5.30 | 7% | 7/7 | 81 | U | Arabidopsis thaliana |
| L36 | gi|241989446 | NBS-LRR class disease resistance protein | 19.52/5.43 | 24.37/6.42 | 29% | 4/4 | 75 | U | Oryza sativa |
| L37 | gi|224111296 | cc-nbs-lrr resistance protein | 149.57/5.73 | 24.48/5.95 | 13% | 12/16 | 106 | U | Populus trichocarpa |
| L38 | gi|289157416 | 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase | 51.64/5.63 | 28.42/5.08 | 16% | 5/5 | 77 | U | Artemisia annua |
| L40 | gi|14210363 | ascorbate peroxidase (APX) | 27.50/5.13 | 29.55/6.55 | 22% | 4/4 | 73 | U | Zantedeschia aethiopica |
| L41 | gi|110289462 | glutathione S-transferase (GST) | 18.04/10.13 | 37.47/5.70 | 36% | 4/4 | 74 | U | Oryza sativa |
| L43 | gi|18404004 | TSK-associating protein 1 | 84.20/4.58 | 56.88/4.55 | 10% | 6/7 | 74 | U | Arabidopsis thaliana |
| L44 | gi|17530547 | class III peroxidase ATP32 | 35.02/6.88 | 58.60/6.71 | 30% | 6/6 | 107 | U | Arabidopsis thaliana |
| L45 | gi|356559803 | stromal 70 kDa heat shock-related protein | 73.88/5.20 | 16.24/4.95 | 19% | 12/23 | 100 | U | Glycine max |
| L47 | gi|357490825 | NBS-LRR resistance protein | 136.09/6.05 | 48.01/5.37 | 8% | 9/9 | 102 | U | Medicago truncatula |
| Signal transduction | |||||||||
| L48 | gi|333441302 | phytochrome C | 42.23/6.08 | 41.46/4.94 | 21% | 6/7 | 86 | U | Digoniopterys microphylla |
| L49 | gi|371940268 | truncate phytochrome A2 protein | 110.22/6.65 | 48.59/5.56 | 9% | 9/9 | 103 | U | Glycine max |
| L50 | gi|18405351 | abscisic acid receptor PYL6 | 24.06/6.17 | 46.80/6.69 | 22% | 4/4 | 74 | U | Arabidopsis thaliana |
| L51 | gi|359475476 | serine carboxypeptidase-like 18 | 58.18/6.16 | 25.30/5.39 | 18% | 7/7 | 108 | U | Vitis vinifera |
| L62 | gi|77553062 | cyclic nucleotide-gated ion channel 14 | 72.47/8.15 | 23.34/5.66 | 10% | 7/7 | 91 | D | Oryza sativa |
| Transcription | |||||||||
| L52 | gi|187369233 | topoisomerase I | 102.34/9.50 | 22.01/5.39 | 13% | 11/16 | 88 | U | Catharanthus roseus |
| L53 | gi|308802618 | DNA-damage-inducible protein F | 48.73/4.50 | 34.58/6.99 | 16% | 6/8 | 80 | U | Ostreococcus tauri |
| L54 | gi|20196900 | putative RNA helicase A | 124.80/6.54 | 34.89/4.69 | 8% | 8/10 | 74 | U | Arabidopsis thaliana |
| L55 | gi|126022792 | RNA polymerase beta subunit | 155.17/9.36 | 41.14/4.63 | 10% | 10/12 | 104 | U | Spinacia oleracea |
| L56 | gi|308808201 | minichromosome maintenance protein 10 isoform 1-like | 64.87/9.42 | 65.31/6.50 | 17% | 9/11 | 95 | U | Ostreococcus tauri |
| L57 | gi|323690255 | maturase K | 45.41/9.80 | 48.01/5.37 | 16% | 5/5 | 80 | U | Pitraea cuneato-ovata |
| L58 | gi|183529139 | maturase K | 60.34/9.28 | 28.91/4.96 | 12% | 7/8 | 82 | U | Jacqueshuberia brevipes |
| L59 | gi|21629786 | maturase K | 46.79/9.62 | 134.26/6.81 | 21% | 7/8 | 100 | U | Hyalochlamys globifera |
| L60 | gi|255567202 | putative transcription elongation factor s-II | 38.76/9.57 | 46.80/5.67 | 14% | 5/5 | 77 | D | Ricinus communis |
| Post-translational modification | |||||||||
| L13 | gi|224089629 | f-box family protein | 45.92/6.43 | 27.54/5.11 | 16% | 5/5 | 82 | U | Populus trichocarpa |
| L14 | gi|308802882 | ubiquitin-protein ligase/hyperplastic discs protein | 92.22/5.72 | 14.48/5.58 | 8% | 7/7 | 81 | U | Ostreococcus tauri |
| L15 | gi|304322967 | translational elongation factor Tu (EF-Tu) | 45.64/6.16 | 14.88/5.89 | 20% | 6/6 | 94 | D | Floydiella terrestris |
| L16 | gi|225429488 | eukaryotic initiation factor 4A-11 | 47.07/5.38 | 47.10/5.26 | 17% | 10/10 | 118 | D | Vitis vinifera |
| L17 | gi|30684767 | cell division protease ftsH-2 | 74.28/6.00 | 83.96/5.15 | 15% | 9/9 | 123 | D | Arabidopsis thaliana |
| Hormone response | |||||||||
| L5 | gi|3024127 | S-adenosylmethionine synthase (SAM synthase) | 43.43/5.51 | 48.33/5.91 | 31% | 10/29 | 82 | U | Catharanthus roseus |
| L10 | gi|33342178 | ABA inducible protein | 17.52/5.95 | 27.92/5.00 | 24% | 5/5 | 84 | U | Triticum aestivum |
| L42 | gi|350535769 | ethylene-responsive transcriptional coactivator | 16.08/10.03 | 39.11/6.27 | 36% | 6/6 | 112 | U | Solanum lycopersicum |
| Others | |||||||||
| L63 | gi|168047657 | predicted protein | 113.56/6.45 | 58.41/6.85 | 12% | 10/13 | 83 | U | Physcomitrella patens subsp |
| L64 | gi|302772723 | hypothetical protein | 84.02/9.40 | 24.97/4.47 | 10% | 6/6 | 79 | U | Selaginella moellendorffii |
| L65 | gi|302823293 | hypothetical protein ELMODRAFT-449095 | 84.19/9.43 | 36.83/4.51 | 15% | 8/11 | 84 | U | Selaginella moellendorffii |
| L66 | gi|167998464 | predicted protein | 35.11/9.51 | 16.16/4.89 | 22% | 7/8 | 101 | U | Physcomitrella patens subsp |
| L67 | gi|297797753 | predicted protein | 41.46/6.47 | 42.91/5.14 | 11% | 5/5 | 78 | U | Arabidopsis lyrata subsp |
| L68 | gi|147834872 | hypothetical protein VITISV-040309 | 46.87/6.06 | 46.55/5.38 | 32% | 17/33 | 116 | U | Vitis vinifera |
| L69 | gi|168004878 | predicted protein | 46.87/5.63 | 34.19/5.72 | 27% | 9/22 | 92 | U | Physcomitrella patens subsp |
| L70 | gi|168000362 | predicted protein | 120.48/9.19 | 47.05/5.29 | 12% | 10/13 | 86 | U | Physcomitrella patens subsp |
| L71 | gi|116779860 | unknown | 23.62/7.77 | 59.88/6.98 | 29% | 6/7 | 99 | U | Picea sitchensis |
| L72 | gi|218201086 | hypothetical protein OsI-29089 | 83.26/6.03 | 27.24/6.61 | 16% | 8/10 | 86 | D | Oryza sativa |
| L73 | gi|49389230 | hypothetical protein | 38.17/6.67 | 65.34/5.50 | 21% | 7/7 | 99 | D | Oryza sativa |
| L74 | gi|297721931 | Os03g0229600 | 11.76/9.77 | 29.86/5.71 | 41% | 5/5 | 107 | D | Oryza sativa |
The seedlings were treated with AR (pH 3.0) for one month. The pH of AR solution was adjusted with a mixture of H2SO4 and HNO3 in the ratio of 5∶1. The final concentration of H2SO4 and HNO3 were 0.45 and 0.09 mM, respectively.
Spot No. is the unique differentially expressed protein spot number. L, protein spot in L. formosana gel. b Database accession numbers according to NCBInr. c Description of the proteins identified by MALDI-TOF MS. d Theoretical mass (kDa) and pI of identified proteins. e Experimental mass (kDa) and pI of identified proteins. f Amino acid sequence coverage for the identified proteins. g Number of the matched peptides and the total searched peptides. hMascot searched score against the database NCBInr. Protein score is −10*Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 73 are significant (p<0.05). i Spot abundance change. D decreased abundance of proteins, U increased abundance of protein.
Table 2. Identification of AR-responsive proteins in S. superba.
| Spota | NCBI accessionb | Protein identityc | Thero. Da/pI d | Exper. Da/pI e | SCf | MP/TPg | Scoreh | Ci | Organism matched |
| Material metabolism | |||||||||
| S1 | gi|62321345 | glutamate-ammonia ligase | 23.45/5.70 | 44.39/5.55 | 20% | 6/8 | 93 | U | Arabidopsis thaliana |
| S2 | gi|205277664 | granule-bound starch synthase I (GBSS) | 15.08/6.41 | 20.84/5.29 | 34% | 4/4 | 78 | U | Thinopyrum intermedium |
| S3 | gi|60101355 | glutamine synthetase (GS) | 31.14/5.75 | 43.58/5.76 | 23% | 6/8 | 91 | U | Vigna radiata |
| S19 | gi|303280145 | glycosyltransferase family 7 protein | 48.84/9.23 | 53.35/5.17 | 16% | 5/5 | 79 | U | Micromonas pusilla |
| Photosynthesis and energy production | |||||||||
| S6 | gi|255544584 | phosphoglycerate kinase (PGK) | 50.11/8.74 | 42.11/5.39 | 36% | 13/25 | 143 | D | Ricinus communis |
| S7 | gi|37721507 | photosystem II subunit H | 2.53/11.72 | 49.18/4.85 | 62% | 3/4 | 75 | D | Ixiolirion tataricum |
| S8 | gi|5708095 | ATP synthase gamma chain | 33.48/6.12 | 37.32/5.66 | 35% | 9/20 | 99 | D | Arabidopsis thaliana |
| S9 | gi|6688696 | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 52.03/6.22 | 20.08/5.97 | 20% | 10/18 | 95 | D | Morkillia mexicana |
| S11 | gi|290490212 | ATP synthase CF1 alpha subunit protein | 55.58/5.04 | 70.29/5.01 | 32% | 17/27 | 173 | U | Staphylea colchica |
| S12 | gi|13430334 | rubisco activase | 37.25/6.70 | 51.86/4.85 | 33% | 10/18 | 99 | D | Zantedeschia aethiopica |
| S13 | gi|308320553 | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 30.68/6.24 | 26.45/5.67 | 22% | 5/6 | 78 | D | Madia sp |
| S14 | gi|170664996 | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 52.85/6.00 | 53.02/6.05 | 23% | 11/16 | 125 | U | Lycoseris crocata |
| S10 | gi|81301612 | protein Ycf2 | 268.41/8.58 | 61.08/5.64 | 9% | 16/20 | 99 | U | Nicotiana tomentosiformis |
| S20 | gi|303283276 | beta carbonic anhydrase | 26.07/6.21 | 24.62/5.62 | 27% | 5/5 | 92 | D | Micromonas pusilla |
| Stress and defense | |||||||||
| S16 | gi|3328221 | thioredoxin peroxidase (TPx) | 28.40/6.34 | 19.81/4.56 | 37% | 5/8 | 86 | U | Secale cereale |
| S17 | gi|2654208 | heat shock 70 | 76.27/5.19 | 105.00/4.39 | 21% | 12/22 | 116 | U | Spinacia oleracea |
| S18 | gi|116323 | endochitinase 3 | 37.17/8.72 | 47.33/4.89 | 30% | 5/6 | 84 | U | Nicotiana tabacum |
| S21 | gi|384247250 | clavaminate synthase-like protein | 41.35/5.60 | 38.59/5.86 | 17% | 5/5 | 81 | U | Coccomyxa subellipsoidea |
| Signal transduction | |||||||||
| S15 | gi|350536755 | 14-3-3 protein 4 | 29.44/4.66 | 28.05/4.37 | 43% | 6/9 | 99 | U | Solanum lycopersicum |
| S22 | gi|115393868 | phytocyanin-like arabinogalactan-protein (PLA) | 18.87/9.17 | 77.80/7.03 | 34% | 4/4 | 77 | U | Gossypium hirsutum |
| S27 | gi|110532561 | calmodulin (CaM) | 17.10/4.06 | 41.79/5.06 | 39% | 4/4 | 81 | U | Aegiceras corniculatum |
| S28 | gi|224131906 | calcium dependent protein kinase 6 (CDPK) | 62.93/5.37 | 45.18/4.96 | 13% | 7/8 | 93 | U | Populus trichocarpa |
| Transcription | |||||||||
| S23 | gi|108861639 | transposase | 15.02/9.08 | 30.68/4.32 | 47% | 5/7 | 82 | U | Oligostachyum sulcatum |
| S24 | gi|379041605 | maturase K | 40.48/10.01 | 22.60/4.71 | 18% | 8/11 | 80 | D | Bromus commutatus |
| S25 | gi|255660958 | pentatricopeptide repeat-containing protein | 39.59/5.95 | 21.25/5.73 | 21% | 8/10 | 98 | U | Verbena macdougalii |
| S26 | gi|11993344 | marpoflo protein | 27.93/9.19 | 20.25/5.88 | 19% | 5/5 | 74 | D | Marchantia polymorpha |
| Post-translational modification | |||||||||
| S4 | gi|225441985 | proteasome subunit alpha type-5 isoform 1 | 26.13/4.65 | 25.55/4.35 | 31% | 6/8 | 84 | U | Vitis vinifera |
| S5 | gi|356545337 | mitochondrial import receptor subunit TOM6 homolog isoform 1 | 6.27/9.36 | 21.22/6.80 | 62% | 4/5 | 79 | U | Glycine max |
| Others | |||||||||
| S29 | gi|224064392 | predicted protein | 37.25/5.62 | 33.24/4.68 | 37% | 7/10 | 102 | U | Populus trichocarpa |
| S30 | gi|388496926 | unknown | 39.81/8.46 | 44.78/5.11 | 24% | 8/10 | 120 | D | Lotus japonicus |
| S31 | gi|242052501 | hypothetical protein SORBIDRAFT_03g010120 | 48.42/9.53 | 27.93/5.71 | 22% | 9/12 | 106 | D | Sorghum bicolor |
| S32 | gi|168071263 | predicted protein | 23.61/9.95 | 81.80/4.52 | 26% | 4/4 | 80 | U | Physcomitrella patens subsp |
| S33 | gi|77554095 | hypothetical protein LOC_Os12g13240 | 23.16/10.25 | 27.91/6.30 | 34% | 6/9 | 92 | U | Oryza sativa |
| S34 | gi|145347277 | predicted protein | 107.30/5.81 | 35.29/5.92 | 9% | 7/7 | 78 | U | Ostreococcus lucimarinus |
The seedlings were treated with AR (pH 3.0) for one month. The pH of AR solution was adjusted with a mixture of H2SO4 and HNO3 in the ratio of 5∶1. The final concentration of H2SO4 and HNO3 were 0.45 and 0.09 mM, respectively.
Spot No. is the unique differentially expressed protein spot number. S, protein spot in S. superba gel. b Database accession numbers according to NCBInr. c Description of the proteins identified by MALDI-TOF MS. d Theoretical mass (kDa) and pI of identified proteins. e Experimental mass (kDa) and pI of identified proteins. f Amino acid sequence coverage for the identified proteins. g Number of the matched peptides and the total searched peptides. hMascot searched score against the database NCBInr. Protein score is −10*Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 73 are significant (p<0.05). i Spot abundance change. D decreased abundance of proteins, U increased abundance of proteins.
Of the 74 protein spots identified in L. formosana, the abundances of 53 proteins were increased and those of 21 proteins were decreased in response to AR (Table 1). In S. superba, 21 proteins were increased in their abundances and 11 proteins were decreased under AR stress (Table 2). Among these affected proteins, phosphoglycerate kinase (PGK, spot L32, L8, S6) and ATP synthase CF1 alpha subunit (spot L22, S11), were decreased in their abundances in both L. formosana and S. superba (Tables 1 and 2). Remarkably, after AR treatment, abundance of maturase K was increased in L. formosana, but decreased in S. superba (Tables 1 and 2). Further analysis on the results revealed that some proteins were represented by more than one spot. These proteins included phosphoglycerate kinase (PGK, spot L32, L8), ATP synthase beta subunits (spot L22, L26) and maturase K (spot L57–59) in L. formosana and ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit (spot S9, S13, S14) in S. superba. The multiple spots might represent isoforms or different post-translation modification of individual proteins [13].
Functional classification of AR-responsive proteins
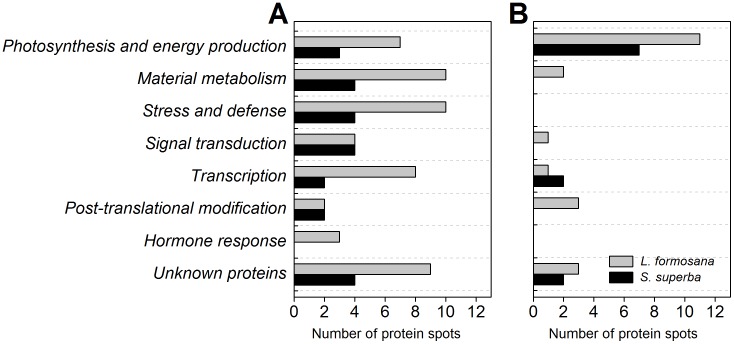
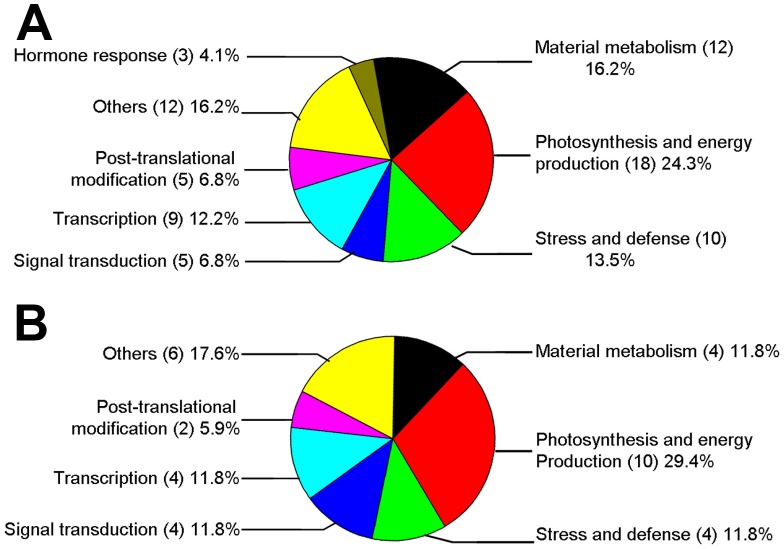
In order to obtain annotation of AR-responsive protein, all identified proteins were further classified according to their biological function and cellular component categories in UniProt (http://www.uniprot.org) and/or NCBI protein database (http://www.ncbi.nlm.nih.gov). AR-responsive proteins were found to be involved in a wide range of biological processes. After AR treatment, with the exception of photosynthesis and energy production related proteins, the abundance of most proteins were decreased in both L. formosana and S. superba (Fig. 4). As shown in Fig. 5, a higher percentage of proteins were involved in photosynthesis and energy production, which accounted for 24.3% and 29.4% of the total proteins in L. formosana and S. superba, respectively. The following groups were proteins involved in material metabolism, stress and defense, transcription, post-translational and modification, and signal transduction. In opposition to L. formosana, no protein related to hormone response was found in S. superba.
Figure 4. Number of protein spots significantly changed in AR-treated L. formosana and S. superba.
(A) Protein spots increased in their abundances. (B) Protein spots decreased in their abundances.
Figure 5. Functional classification of AR-responsive proteins in L. formosana (A) and S. superba (B).
The proportion of identities in each functional group was the sum of this identity accounting for all protein quantities.
Protein expression analysis by Western blot
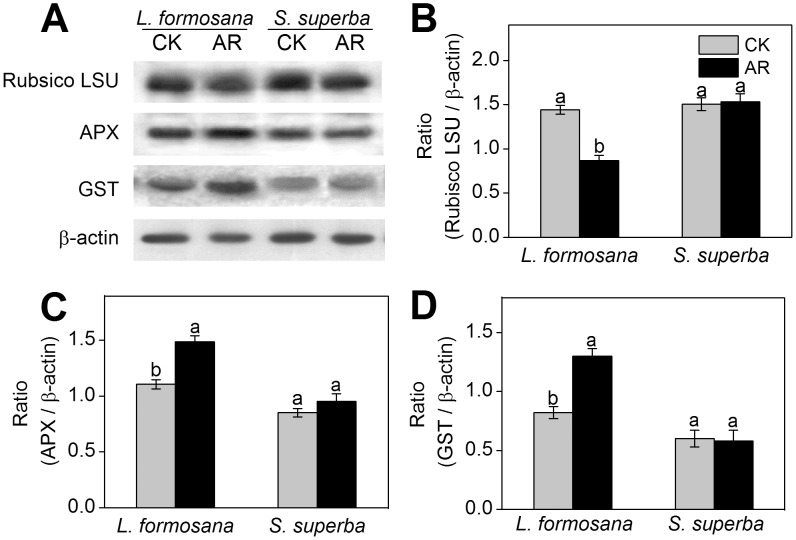
Our above proteomic results revealed that the abundance of Rubisco was decreased (spot L31, L34), while APX (spot L40) and GST (spot L41) were increased in L. formosana under AR treatment (Table 1). As shown in Fig. 6A and B, compared with the control, the protein expression of Rubisco large subunit analyzed by Western blot was significantly decreased in AR-treated L. formosana. In contrast, the protein expression level of APX and GST increased 1.3-fold and 1.6-fold, respectively, compared to control, in L. formosana (Fig. 6C and D). No significant change in the expression of Rubisco large subunit, APX and GST was observed in S. superba (Fig. 6).
Figure 6. Western blot analysis showing the expression of three protein spots.
(A) Expression of rubulose-1,5-bisphoshate carboxylase large subunit (RuBisco LSU), ascorbate peroxidase (APX) and glutathione-S-transferase (GST) in L. formosana and S. superba seedlings after AR treatment. Relative expression level of RuBisco LSU (B), APX (C) and GST (D) were analyzed with the Quantity One software. β-actin was used as the internal control. Means with different letters indicate significantly difference (p<0.05) with regard to AR treatments.
Discussion
AR has negative effects on plant growth and development [1], [6]. Neves et al [23] found that simulated AR (pH 3.1) caused chlorosis and necrosis in leaves and led to chlorophyll loss and photosynthetic depression in Eugenia uniflora. Moreover, chlorophyll content and photosynthesis in L. formosana were also remarkably suppressed by AR treatment in this study (Fig. 1). Compared with S. superba, reductions on chlorophyll content and photosynthetic ability by AR were more obvious in L. formosana (Fig. 1), which is consistent with the results of previous studies [9], [11]. Besides chlorophyll content and photosynthesis, proline content, MDA content and ROS (e.g. H2O2 and O2 •-) production are commonly used as biochemical markers to monitor the damage level in plants under environmental stress [24]. In this study, AR increased proline content, MDA content and ROS production in both L. formosana and S. superba (Fig. 2), which were consistent with the results obtained by Chen et al [11]. However, the increase in these physiological changes caused by AR was less pronounced in S. superba than those in L. formosana (Fig. 1 and 2), suggesting that S. superba, a tolerant species, had less cell damage than L. formosana, a sensitive species.
To further reveal the different strategies to cope with AR between the two species, 74 protein spots in L. formosana and 34 protein spots in S. superba caused by AR were identified by proteomic analysis in this study. Interestingly, similar results were also reported in previous studies, which reported more changes in protein abundance in sensitive species, Arabidopsis thaliana, than in tolerant species, Thellungiella halophila, under salt stress [25]. Since L. formosana had much higher changes in its protein profile, our results proved that this species is more sensitive to AR than S. superba.
Photosynthesis and energy production-related proteins
Photosynthesis is an essential metabolic process of plants and is vulnerable to environmental stress. It is well known that AR can remarkably reduce photosynthesis [3], [23]. In this study, two light reaction-related proteins, including chlorophyllide a oxygenase (CAO, spot L21) and photosystem II (PSII) stability/assembly factor HCF136 (spot L20), were identified in L. formosana. CAO, that converts chlorophyll a to chlorophyll b, regulates the stabilization of light-harvesting chlorophyll a/b proteins [26]. Villeth et al [27] reported that pathogen infection could decrease the accumulation of PSII stability/assembly factor HCF136, which is important for the accurate assembly of PSII. In this study, AR remarkably decreased the abundance of CAO and PSII stability/assembly factor HCF136 in L. formosana, but no light reaction-related proteins was depressed by AR in S. superba (Table 1 and 2). These results suggest that photosynthesis apparatus of L. formosana is more sensitive to AR stress than S. superba.
It has been reported that the expression of Calvin cycle enzymes were down-regulated in Arabidopsis under salinity stress [25]. Our previous study also reported that the reduction in photosynthesis was linked to Calvin cycle enzymes in AR-treated Arabidopsis [16]. Consistent with previous results, our proteomic data from this study confirmed that the abundances of Calvin cycle-related proteins including phosphoribulokinase (PPK, spot L28), Rubisco (spot L31, L34) and Rubisco activase (spot L27) were significantly decreased in L. formosana (Table 1). Rubisco, the CO2 fixing enzyme in Calvin cycle, is the primary limiting factor of net photosynthesis under environmental stress [16]. Rubisco activase promotes and maintains the catalytic activity of Rubisco [28]. The decreased expression of Rubisco and Rubisco activase in AR-treated L. formosana may disturb Calvin cycle activity, leading to the reduction in photosynthetic CO2 assimilation and thus the inhibition in plant growth. Moreover, the results of Western blot analysis showed that the decrease in protein expression of Rubisco large subunit was more obvious in L. formosana than in S. superba under AR stress (Fig. 6A and B). These results may explain why AR-induced damage to photosynthesis was more serious in S. superba than in L. formosana (Fig. 1D and E). Compared with L. formosana, less damage of AR to photosynthesis-related proteins probably result from higher tolerance to AR in S. superba.
Besides photosynthesis-related proteins, AR also affected the abundances of energy production-related proteins in L. formosana and S. superba. It is well known that increased ATP production is required in response to abiotic stress in plants [29]. For example, the abundance of ATP synthase was considerably increased in salt-stressed rice [30] and osmotic-stressed wheat [31]. Thus, the increased abundances of ATP synthase subunits in L. formosana (spot L22, L23, L26) and S. superba (spot S11) in our study demonstrate the prime role of ATP synthase in the adaptation of two tree species to AR stress.
Material metabolism-related proteins
In L. formosana, AR affected a series of protein abundances involved in several metabolism processes, including nitrogen metabolism (spot L2, L6, L11), starch and sugar metabolism (spot L1, L9), lipid metabolism (spot L7, L8), secondary metabolism (spot L39, L46), vitamin metabolism (spot L3) and lignin biosynthesis (spot L12) (Table 1). However, only two nitrogen metabolism-related proteins (spot S1, S3) and one starch biosynthesis-related protein (spot S2) were remarkably induced by AR in S. superba (Table 2). It is clear that AR disturbed more metabolism processes in L. formosana than in S. superba.
AR stress can change free amino acid levels and disturbs N metabolism in plants [19]. Cysteine synthase (CS, spot L11), which is responsible for the terminal step of cysteine biosynthesis, is a critical enzyme involved in environmental stress response in plants [12]. The abundance of CS was decreased in L. formosana (Table 1), indicating that metabolic processes related to cysteine biosynthesis might be strongly depressed by AR. In addition, two glutamine metabolism-related proteins, such as glutamate dehydrogenase (GDH, spot L6) and glutamine synthetase (GS, spot S3), were identified in L. formosana and S. superba, respectively. Abiotic stresses, such as salinity, drought and metal toxicity, can up-regulate GDH gene expression and enhance GDH activity in plants [32]. GS catalyzes ATP-dependent incorporation of ammonium into glutamate and other reduced N compounds [33]. It has been found that GS accumulation was also stimulated by salt and drought, and this helped improve the tolerance of plants to stresses [25]. In agreement with previous findings, our study found that the abundance of GDH in L. formosana, as well as that of GS in S. superba, were increased (Table 1), suggesting that GDH and GS play critical roles in N metabolic acclimation of plants when exposed to AR.
Abiotic stress can also affect starch synthesis [34]. In this study, two starch biosynthesis-related proteins including glucose-1-phosphate adenylyltransferase (GPAT, spot L1) in L. formosana and granule-bound starch synthase (GBSS, spot S2) in S. superba were identified (Tables 1 and 2). Interestingly, the responses of GPAT and GBSS to AR were different in two broad-leaf species. The abundance of GPAT was decreased in L. formosana. However, GBSS, the only enzyme implicated in amylose synthesis [35], was increased in AR-treated S. superba. Basically, this result was consistent with the observation where GBSS activity and starch content in rice was found to increase under cold stress [36]. It is well known that starch is required to synthesize sucrose which serves as a carbon and energy source for plant growth and stress response [35]. Thus we believe that the increased abundance of GBSS may contribute to higher AR-tolerance in S. superba through enhancing starch synthesis and energy production. It also should be noted that AR stress changed the abundances of lipid metabolism-related proteins including glycine-rich protein 17 (GRP17, spot L7) and stearoyl-acyl-carrier protein desaturase-like protein (SACPDLP, spot L8), as well as secondary metabolism-related proteins including flavanone 3-hydroxylase (F3H, spot L39) and chalcone synthase (CHS, L46) in L. formosana. The abundance of SACPDLP was increased, which may contribute to lipid synthesis and membrane integrity in AR-treated L. formosana [25]. F3H and CHS are two key enzymes that catalyze the biosynthesis of flavonoids and chalcones, both of which play critical roles in enhancing secondary metabolism under environmental stress [37]. The increased abundances of these proteins imply that secondary metabolism pathway may be activated to cope with AR stress in AR-sensitive species, L. formosana.
Stress defense-related proteins
The majority of environmental stresses generate a secondary oxidative stress in plants [13]. Oxidative stress occurs when there is a serious imbalance between ROS production and antioxidant defense [24]. Overproduction of ROS induced by a number of adverse environmental factors can attack proteins, lipids, and nucleic acids [38]. Under long-term heavy metal or AR stress, significant accumulation of ROS was observed in previous studies [7], [19], [39]. For instance, Kovacik et al [38] observed the increased ROS in four Tillandsia species under 2 µM Cd2+ treatment over 30 days. In this study, H2O2 and O2 •- content, and thus oxidative stress, induced by AR was remarkably higher in L. formosana than in S. superba (Fig. 2C and D).
To avoid oxidative damage, plants developed an antioxidant system consisting of antioxidative enzymes as well as non-enzymatic antioxidants [38]. In L. formosana, three antioxidant-related proteins, including APX (spot L40), GST (spot L41) and class III peroxidase ATP32 (spot L44), were identified by proteomic analysis. APX plays an important role in scavenging H2O2 from cells [34]. GST is also an important enzyme that counteracts cellular damage induced by oxidative stress [12]. Enhanced activity of APX and GST has also regularly been detected in plants after exposure to salt, cold, heavy metal, and heat stresses [40]. Our recently published work found that both APX and GST were increased in their abundance in an AR-sensitive conifer tree species, Piuns massoniana [41]. Similarity, APX (spot L40) and GST (spot L41) with higher abundances and expression levels (Table 1 and Fig. 6) were also found in AR-treated L. formosana in this study, implying that antioxidant defense system was provoked by AR in L. formosana. However, the expression levels of APX and GST were not changed in S. superba seedlings under AR treatment (Fig. 6). A likely reason is that there may be other pathways that can remove excessive ROS in S. superba. Interestingly, thioredoxin peroxidase (TPx, spot S16), which appears to be a key enzyme in H2O2 detoxification [42], was found to have an increased abundance in AR-treated S. superba. The increased TPx may function in resisting AR-induced oxidative damage by reducing ROS production in S. superba. In addition, endochitinase, a glycosyl hydrolase that catalyzes chitin degradation, plays an essential role in forming the fine cell-wall matrix that enhances the physical barrier against abiotic stresses [43]. Tapia et al [44] reported that endochitinase could be induced by heat, drought, and salinity. In this study, the increased abundance of endochitinase (spot S18) in S. superba (Table 2) may improve physical interactions at the plasma membrane-cell wall interface to cope with AR stress.
Signal transduction-related proteins
Signal transduction plays a crucial role in triggering a cascade of defense events [12]. Phytochrome is a chromoprotein that regulates the expressions of a large number of light-responsive genes and controls plant growth and development [45]. Recently, phytochrome has been found to modulate both biotic and abiotic stresses, such as salinity, drought, cold or herbivory [46]. Cross-talk between phytochrome-mediated light signals and some stress signaling pathways has been reported in diverse plants [47]. Thus it is possible that phytochrome is involved in the modulation of AR stress. A recent study found that increased abundance of phytochrome was needed to resist cold stress in cucumber [48]. Likewise, AR also increased the abundance of both phytochrome C (spot L48) and truncate phytochrome A2 protein (spot L49) in L. formosana but not in S. superba in this study (Table 1), which suggest a role of phytochrome signaling in response to AR in this sensitive species. In addition to the phytochrome signaling pathway, antioxidant enzymes have been found to be modulated by phytochromes under stress conditions [46]. In this study, the increased expression of antioxidant enzymes (APX and GST) was observed in L. formosana under AR treatment (Fig. 6). We suggest that the enhancement of phytochromes induced by AR modulates the antioxidant system in L. formosana seedlings. Further research need to widen our understanding of the role of phytochrome in AR-stressed plants.
Calcium (Ca) also plays a crucial role in regulating plant defense responses to various environmental stimuli [49]. Recently, Kovacik et al [50] found that oxidative stress evoked by hexavalent chromium were evidently suppressed by Ca in Matricaria chamomilla using microscopic visualization method. Our previous study also reported that Ca addition dramatically alleviated the negative effects of AR on seed germination, seedling growth and photosynthesis [3]. Free Ca ion within cell is a second messenger for conveying internal and external signals by Ca sensors that subsequently regulate diverse cellular processes in plants [17]. Calmodulin (CaM) and calcium-dependent protein kinase (CDPK), two major types of Ca sensor, play important roles in Ca signaling and further response to diverse stresses in plants [51]. Saijo et al [52] found that over-expression of OsCDPK gene enhanced tolerance to cold, salt and drought in transgenic rice. Moreover, 14-3-3 protein can also regulate proteins involved in stress response and activate CDPK signal transduction pathway in plants [13]. In S. superba, the abundances of CaM (spot S27), CDPK (spot S28) and 14-3-3 protein (spot S15) were increased after AR treatment, while these proteins were not identified in L. formosana. In agreement with the proteomic analysis results, the enhanced gene expression of CaM and CDPK by real-time quantitative PCR was more obvious in S. superba than in L. formosana under AR treatment (Fig. S1). These results indicate that AR activated different signaling transduction pathways in two tree species and that Ca sensor-dependent signaling pathway might play a critical role in enhancing AR tolerance in S. superba.
AR affected transcription, protein synthesis and modification
Transcription machinery plays an important role in abiotic stress adaptation [25]. Nine proteins (spots L52–L60) and four proteins (spots S23–S26) that involve in gene transcription were identified in response to AR stress in L. formosana and S. superba, respectively (Tables 1 and 2). Four of these proteins were maturase K (spots L57–59, S24). In plants, maturase K catalyzes intron RNA binding during reverse transcription and spicing and directly affects gene expression at transcriptional level [53]. The changes in protein expression of maturase K are very complex in plants under abotic stress, depending on plant species and the type of stress [54]. Pandey et al [54] reported that maturase K was induced when plant suffered from high ROS pressure. On the contrary, maturase K protein was down-regulated in salt-treated maize [55]. In this study, the abundance of maturase K (spots L57–59) was increased in L. formosana, but was decreased in S. superba (Tables 1 and 2). Based on our physiological data, AR induced higher ROS (H2O2 and O2 •-) production in L. formosana than that in S. superba (Fig. 2), which might be one reason to explain the increased abundance of maturase K in L. formosana. However, the decreased abundance of maturase K in S. superba indicates that its gene transcription had been affected by AR, thought no significant visible damage symptom emerged in S. superba leaves.
Translational elongation factor Tu (EF-Tu) plays important roles in response to abiotic stresses including high and low temperatures, salinity, and water deficit [56]. Pandey et al [54] reported that the expression of EF-Tu was down-regulated in chickpea with the increased treatment period of water deficit, which was consistent with the decrease in EF-Tu abundance (spot L15) after AR stress in L. formosana observed in our study. This result indicates that AR has induced a lower protein synthesis in L. formosana. However, EF-Tu was not identified in S. superba, instead, proteasome subunit alpha type-5 isoform 1 (spot S4) and mitochondrial import receptor subunit TOM6 homolog isoform 1 (spot S5) were found in AR-treated leaves (Table 2). Proteasome degrades the proteins with translational errors and proteins damaged by stress which can aggregate and become toxic to the cell [57]. Proteasome has been implicated in regulating numerous plant signaling and metabolic pathways under stress condition [57]. Our pervious study found that the expression of 20 S proteasome subunit was up-regulated in Arabidopsis after 32 h of AR treatment [16]. In this study, the abundance of proteasome subunit alpha type-5 isoform 1 was also increased in AR-treated S. superba, indicating that the control of protein degradation by the proteasome is likely to play an important role in enhancing AR-tolerance in S. superba. In addition, mitochondrial import receptor subunit TOM complex mediates the translocation and uptake of nuclear-encoded mitochondrial preproteins from the cytosol [58]. Increased abundance of mitochondrial import receptor subunit TOM6 by AR may accelerate the import of mitochondrial preproteins and enhance cellular metabolism and energy production in mitochondria, finally contributing to improve resistant to AR in S. superba.
Hormone response-related proteins
Plant hormones are not only important in plant growth and development, but also closely related to environmental stresses response [59]. It is known that increased ethylene biosynthesis is a general response of plants to stress conditions [60]. S-adenosyl-L-methionine (SAM), a major methyl donor in plants, is used as a substrate for ethylene biosynthesis [60]. SAM synthetase catalyzes SAM biosynthesis from L-methionine and ATP [61]. It has been found that both activity and expression of SAM synthetase were increased in tomato and rice under salt stress [62]. Accordingly, our present results indicate that AR increased the abundance of SAM synthetase (spot L5) in L. formosana (Table 1), which may further contribute to ethylene biosynthesis. Another ethylene-related protein, ethylene-responsive transcriptional coactivator (spot L42), which was also increased by AR stress in L. formosana, can positively control the expression of ethylene-responsive genes in plants [63]. In addition, ABA inducible protein (spot L10), which is involved in ABA stimulus and abiotic stresses response, was increased in L. formosana when exposed to AR. It appeared that ethylene biosynthesis and ABA signaling pathways might be activated in AR-treated L. formosana. It is interesting to note that, in S. superba, no hormone response-related proteins was identified, partly due to the high resistance to AR stress in S. superba.
Conclusion
Using the approach of proteomic analysis, this study investigated the differential responses to AR in two broad-leaf tree species, L. formosana and S. superba, an AR-sensitive species and an AR-tolerant species, respectively. After AR treatment, more proteins were significantly changed in their abundances in L. formosana than in S. superba. It should be noted that hormone response-related protein was only found in L. formosana. After AR treatment, signaling pathways, energy production and antioxidant system were activated in both L. formosana and S. superba. Due to higher AR-tolerance in S. superba, AR induced less damage to photosynthesis-related proteins in this species. Moreover, the proteins related to starch synthesis and translation were depressed in AR-treated L. formosana, but enhanced in S. superba, implying that these proteins may greatly contribute to enhance AR-tolerance in S. superba. The identification of novel AR-responsive proteins in this study provides not only new insights into AR stress responses, but also a good starting point for further exploration of the differential AR adaptation strategies between sensitive and tolerant tree species.
Supporting Information
Effects of AR on CaM and CDPK gene expressions in L. formosana and S. superba. The relative gene expression level was calculated based on AR/CK.
(TIF)
Details of identified proteins and peptides list of each protein in AR-treated L. formosana.
(DOC)
Details of identified proteins and peptides list of each protein in AR-treated S. superba.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was financially supported by the Natural Science Foundation of China (NSFC) (31300505, 30930076, 31260057, 30770192, 30670317), China Postdoctoral Science Foundation (2012M521278), the Foundation of the Chinese Ministry of Education (20070384033, 209084), the Program for New Century Excellent Talents in Xiamen University (NCETXMU X07115), and the Scholarship Award for Excellent Doctoral Student granted by the Chinese Ministry of Education. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Larssen T, Lydersen E, Tang D, He Y, Gao J, et al. (2006) Acid rain in China. Environ Sci Technol 40: 418–425. [PubMed] [Google Scholar]
- 2. Dai Z, Liu X, Wu J, Xu J (2012) Impacts of simulated acid rain on recalcitrance of two different soils. Environ Sci Pollut Res 20: 4216–4224. [DOI] [PubMed] [Google Scholar]
- 3. Liu TW, Wu FH, Wang WH, Chen J, Li ZJ, et al. (2011) Effects of calcium on seed germination, seedling growth and photosynthesis of six forest tree species under simulated acid rain. Tree Physiol 31: 402–413. [DOI] [PubMed] [Google Scholar]
- 4. Fan HB, Wang YH (2000) Effects of simulated acid rain on germination, foliar damage, chlorophyll contents and seedling growth of five hardwood species growing in China. Forest Ecol Manag 126: 321–329. [Google Scholar]
- 5. Liu TW, Jiang XW, Shi WL, Chen J, Pei ZM, et al. (2011) Comparative proteomic analysis of differentially expressed proteins in beta-aminobutyric acid enhanced Arabidopsis thaliana tolerance to simulated acid rain. Proteomics 11: 2079–2094. [DOI] [PubMed] [Google Scholar]
- 6. Sun Z, Wang L, Chen M, Liang C, Zhou Q, et al. (2012) Interactive effects of cadmium and acid rain on photosynthetic light reaction in soybean seedlings. Ecotox Environ Safe 79: 62–68. [DOI] [PubMed] [Google Scholar]
- 7. Kovacik J, Klejdus B, Backor M, Stork F, Hedbavny J (2011) Physiological responses of root-less epiphytic plants to acid rain. Ecotoxicology 20: 348–357. [DOI] [PubMed] [Google Scholar]
- 8. DeHayes DH, Schaberg PG, Hawley GJ, Strimbeck GR (1999) Acid rain impacts on calcium nutrition and forest health. BioScience 49: 789–800. [Google Scholar]
- 9. Feng ZW, Miao H, Zhang FZ, Huang YZ (2002) Effects of acid deposition on terrestrial ecosystems and their rehabilitation strategies in China. J Environ Sci-China 14: 227–233. [PubMed] [Google Scholar]
- 10. Liu J, Zhou G, Yang C, Ou Z, Peng C (2006) Responses of chlorophyll fluorescence and xanthophyll cycle in leaves of Schima superba Gardn. & Champ. and Pinus massoniana Lamb. to simulated acid rain at Dinghushan Biosphere Reserve, China. Acta Physiol Plant 29: 33–38. [Google Scholar]
- 11. Chen J, Wang WH, Liu TW, Wu FH, Zheng HL (2013) Photosynthetic and antioxidant responses of Liquidambar formosana and Schima superba seedlings to sulfuric-rich and nitric-rich simulated acid rain. Plant Physiol Biol 64: 41–51. [DOI] [PubMed] [Google Scholar]
- 12. Yang Q, Wang Y, Zhang J, Shi W, Qian C, et al. (2007) Identification of aluminum-responsive proteins in rice roots by a proteomic approach: cysteine synthase as a key player in Al response. Proteomics 7: 737–749. [DOI] [PubMed] [Google Scholar]
- 13. Bai X, Yang L, Yang Y, Ahmad P, Hu X (2011) Deciphering the protective role of nitric oxide against salt stress at the physiological and proteomic levels in maize. J Proteome Res 10: 4349–4364. [DOI] [PubMed] [Google Scholar]
- 14. Sehrawat A, Gupta R, Deswal R (2013) Nitric oxide-cold stress signalling crosstalk-evolution of a novel regulatory mechanism. Proteomics 13: 1816–1835. [DOI] [PubMed] [Google Scholar]
- 15. Hossain Z, Komatsu S (2012) Contribution of proteomic studies towards understanding plant heavy metal stress response. Front Plant Sci 3: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu TW, Fu B, Niu L, Chen J, Wang WH, et al. (2011) Comparative proteomic analysis of proteins in response to simulated acid rain in Arabidopsis. J Proteome Res 10: 2579–2589. [DOI] [PubMed] [Google Scholar]
- 17. Huang SS, Chen J, Dong XJ, Patton J, Pei ZM, et al. (2011) Calcium and calcium receptor CAS promote Arabidopsis thaliana de-etiolation. Physiol Plant 144: 73–82. [DOI] [PubMed] [Google Scholar]
- 18. Chen J, Wu FH, Liu TW, Chen L, Xiao Q, et al. (2012) Emissions of nitric oxide from 79 plant species in response to simulated nitrogen deposition. Environ Pollut 160: 192–200. [DOI] [PubMed] [Google Scholar]
- 19. Jiang H, Korpelainen H, Li C (2013) Populus yunnanensis males adopt more efficient protective strategies than females to cope with excess zinc and acid rain. Chemosphere 91: 1213–1220. [DOI] [PubMed] [Google Scholar]
- 20. Montillet JL, Chamnongpol S, Rusterucci C, Dat J, van de Cotte B, et al. (2005) Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol 138: 1516–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu WJ, Chen J, Liu TW, Liu X, Chen J, et al. (2013) Comparative proteomic analysis on wild type and nitric oxide-overproducing mutant (nox1) of Arabidopsis thaliana. Nitric Oxide 36C: 19–30. [DOI] [PubMed] [Google Scholar]
- 22.Hu WJ, Chen J, Liu TW, Wu Q, Wang WH, et al. (2014) Proteome and calcium-related gene expression in Pinus massoniana needles in response to acid rain under different calcium levels. Plant Soil doi: 10.1007/s11104-014-2086-9.
- 23. Neves NR, Oliva MA, da Cruz Centeno D, Costa AC, Ribas RF, et al. (2009) Photosynthesis and oxidative stress in the restinga plant species Eugenia uniflora L. exposed to simulated acid rain and iron ore dust deposition: potential use in environmental risk assessment. Sci Total Environ 407: 3740–3745. [DOI] [PubMed] [Google Scholar]
- 24. Wyrwicka A, Skłodowska M (2006) Influence of repeated acid rain treatment on antioxidative enzyme activities and on lipid peroxidation in cucumber leaves. Environ Exp Bot 56: 198–204. [Google Scholar]
- 25. Pang Q, Chen S, Dai S, Chen Y, Wang Y, et al. (2010) Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J Proteome Res 9: 2584–2599. [DOI] [PubMed] [Google Scholar]
- 26. Yamasato A, Nagata N, Tanaka R, Tanaka A (2005) The N-terminal domain of chlorophyllide a oxygenase confers protein instability in response to chlorophyll B accumulation in Arabidopsis. Plant Cell 17: 1585–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Villeth GR, Reis FB Jr, Tonietto A, Huergo L, de Souza EM, et al. (2009) Comparative proteome analysis of Xanthomonas campestris pv. campestris in the interaction with the susceptible and the resistant cultivars of Brassica oleracea. FEMS Microbiol Lett 298: 260–266. [DOI] [PubMed] [Google Scholar]
- 28. Portis ARJ (2003) Rubisco activase – Rubisco’s catalytic chaperone. Photosynth Res 75: 11–27. [DOI] [PubMed] [Google Scholar]
- 29. Jiang Y, Yang B, Harris NS, Deyholos MK (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 58: 3591–3607. [DOI] [PubMed] [Google Scholar]
- 30. Kim DW, Rakwal R, Agrawal GK, Jung YH, Shibato J, et al. (2005) A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophoresis 26: 4521–4539. [DOI] [PubMed] [Google Scholar]
- 31. Flagella Z, Trono D, Pompa M, Di Fonzo N, Pastore D (2006) Seawater stress applied at germination affects mitochondrial function in durum wheat (Triticum durum) early seedlings. Funct Plant Biol 33: 357–366. [DOI] [PubMed] [Google Scholar]
- 32. Beato VM, Teresa Navarro-Gochicoa M, Rexach J, Begona Herrera-Rodriguez M, Camacho-Cristobal JJ, et al. (2011) Expression of root glutamate dehydrogenase genes in tobacco plants subjected to boron deprivation. Plant Physiol Biol 49: 1350–1354. [DOI] [PubMed] [Google Scholar]
- 33. Liu TW, Niu L, Fu B, Chen J, Wu FH, et al. (2013) A transcriptomic study reveals differentially expressed genes and pathways respond to simulated acid rain in Arabidopsis thaliana. Genome 56: 49–60. [DOI] [PubMed] [Google Scholar]
- 34. Wang L, Liang W, Xing J, Tan F, Chen Y, et al. (2013) Dynamics of chloroplast proteome in salt-stressed mangrove Kandelia candel (L.) Druce. J Proteome Res 12: 5124–5136. [DOI] [PubMed] [Google Scholar]
- 35. Cheng J, Khan MA, Qiu WM, Li J, Zhou H, et al. (2012) Diversification of genes encoding granule-bound starch synthase in monocots and dicots is marked by multiple genome-wide duplication events. PLoS ONE 7: e30088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang SJ, Liu LF, Chen CK, Chen LW (2006) Regulations of granule-bound starch synthase I gene expression in rice leaves by temperature and drought stress. Biol Plant 50: 537–541. [Google Scholar]
- 37. Cheng H, Wang J, Chu S, Yan HL, Yu D (2013) Diversifying selection on flavanone 3-hydroxylase and isoflavone synthase genes in cultivated soybean and its wild progenitors. PLoS ONE 8: e54154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399. [DOI] [PubMed] [Google Scholar]
- 39. Kovacik J, Babula P, Klejdus B, Hedbavny J (2014) Comparison of oxidative stress in four Tillandsia species exposed to cadmium. Plant Physiol Biochem 80C: 33–40. [DOI] [PubMed] [Google Scholar]
- 40. Kovacik J, Klejdus B, Stork F, Hedbavny J (2012) Physiological responses of Tillandsia albida (Bromeliaceae) to long-term foliar metal application. J Hazard Mater 239–240: 175–182. [DOI] [PubMed] [Google Scholar]
- 41. Hu WJ, Chen J, Liu TW, Simon M, Wang WH, et al. (2014) Comparative proteomic analysis of differential responses of Pinus massoniana and Taxus wallichiana var. mairei to simulated acid rain. Int J Mol Sci 15: 4333–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vieira Dos Santos C, Rey P (2006) Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci 11: 329–334. [DOI] [PubMed] [Google Scholar]
- 43. Kwon Y, Kim SH, Jung MS, Kim MS, Oh JE, et al. (2007) Arabidopsis hot2 encodes an endochitinase-like protein that is essential for tolerance to heat, salt and drought stresses. Plant J 49: 184–193. [DOI] [PubMed] [Google Scholar]
- 44. Tapia G, Morales-Quintana L, Inostroza L, Acuna H (2011) Molecular characterisation of Ltchi7, a gene encoding a Class III endochitinase induced by drought stress in Lotus spp. Plant Biol 13: 69–77. [DOI] [PubMed] [Google Scholar]
- 45. Foo E, Ross JJ, Davies NW, Reid JB, Weller JL (2006) A role for ethylene in the phytochrome-mediated control of vegetative development. Plant J 46: 911–921. [DOI] [PubMed] [Google Scholar]
- 46. Carvalho RF, Campos ML, Azevedo RA (2011) The role of phytochrome in stress tolerance. J Integr Plant Biol 53: 920–929. [DOI] [PubMed] [Google Scholar]
- 47. Liu J, Zhang F, Zhou J, Chen F, Wang B, et al. (2012) Phytochrome B control of total leaf area and stomatal density affects drought tolerance in rice. Plant Mol Biol 78: 289–300. [DOI] [PubMed] [Google Scholar]
- 48. Sysoeva MI, Markovskaya EF, Sherudilo EG (2013) Role of phytochrome B in the development of cold tolerance in cucumber plants under light and in darkness. Russ J Plant Physiol 60: 383–387. [Google Scholar]
- 49. Tang RH, Han SC, Zheng HL, Cook CW, Choi CS, et al. (2007) Coupling diurnal cytosolic Ca2+ oscillations to the CAS-IP3 pathway in Arabidopsis. Science 315: 1423–1426. [DOI] [PubMed] [Google Scholar]
- 50. Kovacik J, Babula P, Hedbavny J, Klejdus B (2014) Hexavalent chromium damages chamomile plants by alteration of antioxidants and its uptake is prevented by calcium. J Hazard Mater 273C: 110–117. [DOI] [PubMed] [Google Scholar]
- 51. Ho SL, Huang LF, Lu CA, He SL, Wang CC, et al. (2013) Sugar starvation- and GA-inducible calcium-dependent protein kinase 1 feedback regulates GA biosynthesis and activates a 14-3-3 protein to confer drought tolerance in rice seedlings. Plant Mol Biol 81: 347–361. [DOI] [PubMed] [Google Scholar]
- 52. Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23: 319–327. [DOI] [PubMed] [Google Scholar]
- 53. Ji XL, Gai YP, Zheng CC, Mu ZM (2009) Comparative proteomic analysis provides new insights into mulberry dwarf responses in mulberry (Morus alba L.). Proteomics 9: 5328–5339. [DOI] [PubMed] [Google Scholar]
- 54. Pandey A, Chakraborty S, Datta A, Chakraborty N (2008) Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L.). Mol Cel Proteomics 7: 88–107. [DOI] [PubMed] [Google Scholar]
- 55. Zorb C, Schmitt S, Muhling KH (2010) Proteomic changes in maize roots after short-term adjustment to saline growth conditions. Proteomics 10: 4441–4449. [DOI] [PubMed] [Google Scholar]
- 56. Fu J, Ristic Z (2010) Analysis of transgenic wheat (Triticum aestivum L.) harboring a maize (Zea mays L.) gene for plastid EF-Tu: segregation pattern, expression and effects of the transgene. Plant Mol Biol 73: 339–347. [DOI] [PubMed] [Google Scholar]
- 57. Kurepa J, Smalle JA (2008) Structure, function and regulation of plant proteasomes. Biochimie 90: 324–335. [DOI] [PubMed] [Google Scholar]
- 58. Rapaport D (2002) Biogenesis of the mitochondrial TOM complex. Trends Biochem Sci 27: 191–197. [DOI] [PubMed] [Google Scholar]
- 59. Symons GM, Smith JJ, Nomura T, Davies NW, Yokota T, et al. (2008) The hormonal regulation of de-etiolation. Planta 227: 1115–1125. [DOI] [PubMed] [Google Scholar]
- 60. Wang KL, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14 Suppl: S131–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ravanel S, Gakiere B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95: 7805–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sanchez-Aguayo I, Rodriguez-Galan JM, Garcia R, Torreblanca J, Pardo JM (2004) Salt stress enhances xylem development and expression of S-adenosyl-L-methionine synthase in lignifying tissues of tomato plants. Planta 220: 278–285. [DOI] [PubMed] [Google Scholar]
- 63. Firon N, Pressman E, Meir S, Khoury R, Altahan L (2012) Ethylene is involved in maintaining tomato (Solanum lycopersicum) pollen quality under heat-stress conditions. AoB Plants 2012: pls024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of AR on CaM and CDPK gene expressions in L. formosana and S. superba. The relative gene expression level was calculated based on AR/CK.
(TIF)
Details of identified proteins and peptides list of each protein in AR-treated L. formosana.
(DOC)
Details of identified proteins and peptides list of each protein in AR-treated S. superba.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.