Abstract
Recently, several systems designed to trigger RNA interference by using small hairpin RNA driven by polymerase III promoters have been described. Here, we report a lentiviral-mediated small interfering RNA delivery system that can be induced by CRE recombinase. The system consists of a lentiviral vector carrying a mouse U6 promoter that is separated from a small hairpin RNA by a random DNA stuffer sequence flanked by modified loxP sites. The silencing cassette is not expressed until activated by addition of CRE recombinase delivered by a lentiviral vector. We have used this system to show specific down-regulation of GFP and two endogenous genes (the tumor suppressor p53 and the NF-κB transcription factor subunit p65) in vitro. Furthermore, down-regulation of both p53 and p65 resulted in the expected effect on downstream genes and cellular phenotype. We foresee multiple applications of this system both in vitro and in vivo to down-regulate specific targets in a tissue-specific and localized manner.
In the postgenomic era, one of the pressing biological questions is how to decipher the function of myriad unknown genes. Gene knockout by homologous recombination has become the mainstay of understanding gene function. In mouse, perhaps one of the most successful mammalian model systems, gene knockout technology has been a powerful tool of dissecting gene function, but it is laborious and expensive. This technology has been less used for in vitro studies because all of the alleles need to be knocked out, so researchers have taken advantage of antisense and ribozyme technology, which are not always reliable or robust. RNA interference (RNAi) has emerged as a pathway that offers great hope and promise to study the functions of a vast number of genes (1-14). In invertebrates, long double-stranded RNA molecules are processed by the endonuclease Dicer into 21- to 23-nucleotide small interfering RNAs (siRNAs), which are then incorporated into an RNA-induced silencing complex, a multicomponent nuclease complex that selects and degrades mRNAs that are homologous to the initially delivered double-stranded RNA (15, 16). However, in mammalian systems, introduction of long double-stranded RNA (>50 bp) results in systemic, nonspecific inhibition of translation due to activation of the double-stranded RNA-dependent protein kinase R (PKR) response. This problem can be solved by the use of synthetic siRNA (<30 bp) that can be delivered either exogenously (17) or expressed endogenously from RNA polymerase pol III promoters, resulting in a powerful tool for achieving specific down-regulation of target mRNAs (11, 12, 18, 19). Currently, in mammalian systems, the method of choice to generate a whole-genome knockdown screening by using RNAi is to systematically generate synthetic or pol III transcribed siRNA hairpins for every known target gene. Algorithms have been developed to predict effective siRNA sequences for efficient screening of large numbers of genes (20).
We have shown that the lentiviral vector system can express integrated siRNA efficiently in a wide variety of cell lines, tissues, and even preimplantation embryos (14). More interestingly, we and others (7) have reported that the resulting progeny expressing siRNA had reduced expression of a specific gene. We now want to extend the use of lentiviral siRNA transgenesis further by generating regulatable siRNA vectors. We chose to use a loxP-CRE system because it has extensively been used to generate conditional knockout mice. Furthermore, many investigators have generated cell or tissue-specific CRE mice. Here, we report the design and construction of lentiviral siRNA vectors capable of suppressing GFP, p53, and p65 expression only upon expression of CRE recombinase.
Methods
Primer Design. The following primers (5′ to 3′) were used throughout this study. F1, CCCAAGCTTATCCGACGCCGCCATCTCTAGGC; F2, GATCGATATCCGACGCCGCCATCTCTAGG; F3, GCTTGTTTGCAAGCTGACCCTGAAGTTCTTCAAGAGAGAACTTCAGGGTCATGCTTGCTTTTTGTCGAC; F4, AGCTTGGATCCAGCTCTAGAATAACTTCGTATAGTATAAATTATACGAAGTTATA; F5, GCATTAGGATCCTTTTTGCGTTAACCGTCACGAGCATCAT; F6, CTAGAATAACTTCGTATAGTATAAATTATACGAAGTTATAAGCTTGTTTgtacatgtgtaatagctccTTCAAGAGAggagctattacacatgtacTTTTTGTC GAC; F7, CTAGAATAACTTCGTATAGTATAAATTATACGAAGTTATAAGCTTGTTTGgagtttcagcagctcctgaacTTCAAGAGAgttcaggagctgctgaaactcTTTTTGTCGAC; F8, CGGCTGTTGGGCACTGA; R1, GGGATCCGAAGACCACAAACAGGCTTTTCTCCAAGGGATATTTAT; R2, GGTCGACAAAAAgcaagctgaccctgaagttcTCTCTTGAAgaacttcagggtcagcttgcAAACAGGCTTTTCTCCAAGGGATA; R3, AACTACTTTACAGTTAGGGTGAGTTCTCGAGTGTGCTGTTTTTTAAAATAATAATTTAGT; R4, GCTTGCAAACAAGCTTATAACTTCGTATAATTTATACTATACGAAGTTATACTAACTTTACAGTTAGGGTGAG; R5, CATGGTCGACAAAAAGCAAGCTGACCCTGAAGTTCTCTCTTGAAGAACTTCAGGGTCAGCTTGCAAACA; R6, AGCTTATAACTTCGTATATTTATACTATACGAAGTTATTCTAGAGCTGGATCCA; R7, TACAGCGAGGTGTTCCATTTGTCAAGATCTATTACG; R8, CATGGTCGACAAAAAgtacatgtgtaatagctccTCTCTTGAAggagctattacacatgtacAAACAAGCTTATAACTTCGTATAATTTATACTATACGAAGTTATT; R9, CATGGTCGACAAAAAgagtttcagcagctgctgaacTCTCTTGAAgttcaggagctgctgaaactcAAACAAGCTTATAACTTCGTATAATTTATACTATACGAAGTTATT; and R10, GGAAGGTCCGCTGGATTGA.
Note that R2 contains the siRNA hairpin against GFP originally published by Brummelkamp et al. (12); R8 contains the siRNA hairpin against mouse p53 originally published by Dirac and Bernards (21). The pol III termination signal is underlined, the sense and antisense strands of the GFP hairpin are in lowercase, and the loop is italicized. TATA box sequences are shown in bold, with loxP spacer mutations in bold italics.
The following siRNA targets were used in this study: GFP, GCA AGCTGACCCTGA AGT; p65, GAGT T TCAGCAGCTCCTGAAC; and p53, GTACATGTGTAATAGCTCC.
Construct Design. Cloning of LV-shGFP-WT. The mouse U6 (mU6) promoter (GenBank accession no. X06980) was amplified by PCR from mouse genomic DNA using primers F1and R1 and cloned into pGEM-T Easy (PGTE) (Promega), creating PGTE+mU6-WT. This clone was used as template for generation of an mU6-shGFP-WT PCR product with a 5′ end ClaI site and a 3′ end SalI site by PCR with primers F2 and R2, which was cloned first into PGTE (creating PGTE+mU6-shGFP-WT) and then into the fourth-generation lentivector (LV#5, described in ref. 22) digested with ClaI-SalI (Fig. 1A).
Fig. 1.
Diagram of lentiviral constructs (integrated proviral form) used in this study. (A) LV-shGFP-WT: lentiviral vector carrying a wild-type silencing cassette. The mU6 promoter consists of three elements: distal sequence element, PSE, and TATA box. The circled numbers (1, 2, and 3) indicate sites where unsuccessful attempts were made to introduce loxP sites. (B) LV-shRNA-OFF: the introduction of a DNA stuffer sequence (LacZ) flanked by modified loxP sites (loxP*). loxP* is a loxP site containing two mutations in the 8-bp spacer region (GCATACAT to GTATAAA), resulting in a modified loxP site (loxP*) with a spacer that resembles a U6 TATA box. The presence of a stuffer fragment puts the promoter in an OFF configuration. The promoter will be silent until CRE recombinase excises the fragment, leaving one loxP* site (LV-shRNA-ON) (C) and switching the silencing cassette from OFF to ON. The shRNA is thus expressed, leading to siRNA synthesis and target shutdown. (D) LV-shRNA+GFP: lentiviral construct allowing the delivery of both a silencing cassette and a GFP marker. PPT, polypyrimidine tract; WPRE, woodchuck hepatitis virus posttranscriptional response element; all viral elements are shown on a white background; DSE, distal sequence element; TATA, mU6 TATA box; loxP*, modified loxP site with spacer resembling mU6 TATA box; all mU6 promoter elements are shown on a violet background, except for T5 (pol III transcription termination signal), which is shown on a red background. The stuffer fragment (randomly chosen 1-kb region of LacZ) is shown on a yellow background and is present only in LV-shRNA-OFF. The GFP marker in LV-shRNA is shown on a green background. shRNA hairpins are shown in black.
Cloning of LV-shGFP-ON. The sequence of the mU6 promoter was modified to introduce (i) a XhoI site between -69 and -72, (ii) a modified loxP site (loxP*) (ATAACTTCGTATAGTATAAAT- TATACGAAGTTAT) between -11 and -44 (with the spacer sequence mutated from GCATACAT to GTATAAAT and engineered to conserve the original TATA box position, -24 to -30), and (iii)a HindIII site between -5 and -10. This was done by nested PCR, using PGTE+mU6-shGFP-WT as template with F2 as forward primer and reverse primers R3 and R4. The resulting PCR product was cloned into PGTE (creating PGTE+mU6-ON). This cloning step eliminated the short hairpin GFP (shGFP) hairpin, which was reintroduced by annealing primers F3 and R5 and cloning the resulting product into PGTE+mU6-ON digested with HindIII and NcoI, creating PGTE+mU6-shGFP-ON. The ClaI-SalI mU6-shGFP-ON fragment was cloned into LV#5 cut with ClaI-SalI (Fig. 1C).
Cloning of LV-shRNA-OFF. PGTE+mU6-shGFP-ON was modified further by introducing a sequence containing a second loxP* site preceded by BamHI and XbaI sites. This was achieved by annealing primers F4 and R6, followed by cloning of the resulting reannealed product into a unique HindIII site in PGTE+mU6-shGFP-ON. A clone in the correct orientation (with the BamHI and XbaI sites between the two loxP* sites, PGTE+mU6-loxP*loxP*shGFP) was then modified by introducing a stuffer fragment (S) consisting of 1 kb of LacZ fragment generated by PCR from pcDNA 3.1 His-myc LacZ (Invitrogen) using primers F5 and R7 (F5 contains a pol III termination signal near the 5′ end) by cloning into the BamHI and XbaI sites, generating PGTE+mU6-shGFP-OFF (Fig. 1B).
Cloning of LV-shp53-OFF or LV-shp65-OFF. PGTE+mU6-shGFP-OFF was cut with XbaI-NcoI, and the XbaI-NcoI insert was replaced with annealed primers F6 and R8 (p53) or F7 and R9 (p65), generating PGTE+mU6-shp53-OFF and PGTE+mU6-shp65-OFF, respectively. A sequence for shp65 was not available in the literature; thus, we cloned six shp65 candidates into PE/U6 (Invitrogen) and tested their silencing capability by cotransfection of each candidate with a p65-GFP fusion expression vector in 293T cells (data not shown). The ClaI-SalI mU6-shRNA-OFF fragments were cloned into LV#5 cut with ClaI-SalI, as described above.
Cloning of LV-shRNA-WT+GFP. LV#5 was modified by introducing a Gateway destination cassette consisting of attR1-CmR-ccdB-attR2 into a unique HpaI site in LV#5, thus adapting it to Gateway cloning technology (Invitrogen) (Fig. 1D). Small hairpin RNA (shRNA) hairpins for p53 and p65 were cloned into PE/U6 entry vector (Invitrogen) and transferred by an LR clonase reaction into LV-shRNA-WT+GFP according to manufacturer's protocol.
Cell Culture and Reagents. GFP-expressing 293T cells were cultured in DMEM plus 10% FBS (HyClone). Spontaneously immortalized murine embryo fibroblasts were cultured in DMEM supplemented with 10% FCS (HyClone)/100 μg/ml penicillin/100 μg/ml streptomycin/250 ng/ml of amphotericin B (GIBCO). Doxorubicin was purchased from Sigma and dissolved in Milli-Q water at a concentration of 10 mg/ml as stock. Tumor necrosis factor α was obtained from Calbiochem and stored in PBS containing 0.1% BSA. The antibodies against p65 (C-20), IκBα (C-21), IκBβ (C-20), p50 (NLS), p21 (C-19), and p53 (FL-393)-G were all obtained from Santa Cruz Biotechnology. Anti-β actin antibody (Sigma) was used to detect levels of actin, which served as loading controls in all of the experiments. The levels of mdm2 were determined by using the Ab-2 from Oncogene Research Products.
Viral Production and Transduction. Viral Production was done as described (22, 23). Briefly, 293T cells were transfected by the CaCl2 precipitation method. Supernatants were collected 48 and 72 h after transfection, filtered, and concentrated by two successive ultracentrifugations. Viral preparation titers were determined by p24 ELISA (Alliance; NEN) and by TaqMan real-time PCR determination of transduced proviral genomes using primers F8 and R10 for woodchuck hepatitis virus posttranscriptional response element. On average, the vector preparations were 1.5 E4 particles per nanogram of p24 (see below). In vitro transduction was done by plating cells on 24-well plates pretreated with 0.002% poly(L-lysine) in PBS. After overnight culture in DMEM plus 10% FBS, viral preparation aliquots were diluted to achieve the estimated range of multiplicity of infection (moi) required to test our vectors, and infection was carried out in a 200-μl total volume. After 24 h, cells were washed with PBS, and fresh medium was added. Cells were cultured and passaged for 5 days (p53 and p65) or 9 days (GFP).
Assays. GFP levels were determined by fluorescence-activated cell sorting (FACS) (Becton Dickinson).
Western blots. Typically, cells were cultured in six-well dishes. To detect p65, IκBα, IκBβ, and p50 proteins, whole-cell extracts of confluent cells were prepared by lysis in radioimmunoprecipitation assay (RIPA) buffer, and these lysates were resolved on Bis-Tris SDS gels (4-12%) in Mops buffer (Invitrogen). To analyze the levels of p53 and its target genes, cells were either left untreated or treated with 0.4 μg/ml doxorubicin for 12-14 h when indicated, washed with PBS, and harvested in 2× SDS loading buffer. After electrophoresis on 10% Tris glycine gels, proteins were transferred on poly(vinylidene difluoride) membrane (Immobilon P, Millipore). All membranes were blocked in PBS without Mg2+ and Ca2+ containing 0.2% Tween 20 and 5% nonfat milk and probed with the indicated antibodies in PBS without Mg2+ and Ca2+ containing 0.2% Tween 20 and 1% milk (24).
Cell-survival assays. Cells typically were plated in 12-well dishes. Confluent murine embryo fibroblasts infected with LV-CRE (control), LV-shp65, and LV-shp65-OFF were either left untreated or were treated with fresh 100 ng/ml tumor necrosis factor α for 3 consecutive days. Surviving cells were estimated by staining with crystal violet and reading the stain leached in methanol at 595 nm by using a spectrophotometer. The untreated samples in each cell population were considered as 100% survival and were used to calculate the percentage of survival. TaqMan PCR. Primer sets used for quantitative TaqMan PCR to detect proviral DNA were F8 and R10 for woodchuck hepatitis virus posttranscriptional response element. Reactions contained TaqMan universal mastermix (Perkin-Elmer), 300 nM each primer, 100 nM probe, and a 100-ng template. Reactions were analyzed by using the ABI Prism 7700 sequence detection system (Applied Biosystems) using the following PCR cycles: 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C. To analyze the proviral DNA content of the samples, a standard curve ranging from 10 to 1 × 106 copies was measured in every experiment. All samples and standards were analyzed in triplicate.
Results and Discussion
We have designed a system that consists of a lentiviral vector carrying mU6 that is separated from a shRNA by a random DNA stuffer sequence flanked by modified loxP sites. The expression cassette is silent until activated by addition of CRE recombinase delivered by a lentiviral vector.
The general structure of a lentiviral vector carrying a silencing cassette consisting of mU6 driving an shRNA hairpin targeting GFP is shown in Fig. 1A. To engineer a vector that would only be active after expression of CRE recombinase, we sought to design an inactive cassette in which the mU6 promoter is separated from the shRNA hairpin by a stuffer fragment flanked by loxP sites. The stuffer fragment should contain a pol III termination signal (25) near its 5′ end to ensure the promoter is silent. Delivery of CRE recombinase would result in excision of the intervening sequence, thus allowing the promoter to drive expression of the shRNA hairpin and down-regulation of the target. Whereas the general strategy is straightforward, the fact that CRE recombinase-mediated excision of a loxP flanked sequence leaves a loxP site at the excision site posed the problem of finding a site in the silencing cassette where the leftover loxP site would not interfere with efficient expression and processing of the shRNA hairpin. LoxP sites are 34-bp long and consist of two inverted repeats of 13 bp separated by a spacer fragment of 8 bp (26). The mU6 promoter consists of the distal sequence element, the proximal sequence element (PSE), and a TATA box (TATA) (27-29). Three potential sites in an mU6-shGFP silencing cassette, namely (i) between the distal sequence element and the PSE, (ii) between the transcriptional start site and the shRNA hairpin, and (iii) within the shRNA hairpin loop, were tested to determine the effect of a loxP site in the mU6 promoter on silencing of GFP (Fig. 1A). All three positions were found to be unsuitable (data not shown), but interestingly, a recent report (see ref. 35) obtained CRE-inducible RNA interference by positioning an intervening sequence within the shRNA loop, suggesting that the small differences in DNA stuffer design may have considerable effects on the efficiency of inducibility or down-regulation of the target mRNA. Furthermore, the PSE, TATA box, and transcriptional start site are separated by 17 and 24 bp, respectively; the arrangement of these elements is compact, and expression of the promoter is extremely sensitive to alterations in the spacing between them (29). Thus, positioning the 34-bp loxP site either between the PSE and the TATA box or between the TATA box and the transcriptional start site was not deemed suitable. We therefore took advantage of the similarity of the loxP 8-bp spacer sequence (GCATACAT) and the mU6 TATA box (TATAAA), noting that mutating the first and second C bases of the spacer to T and A, respectively, created a mutant loxP site (referred to as loxP*), whose 8-bp spacer region resembles a mU6 TATA box. A published report examined the effect of a large number of single and double-base substitutions in the spacer sequence of loxP and found that mutations in this region of the loxP site had varied effects on the efficiency of CRE-mediated recombination. Interestingly, this particular double mutant (loxP*) resulted in a loxP site capable of recombination, albeit at a frequency of ≈35% of wild-type loxP levels (30). We surmised that any loss of recombination efficiency could be compensated for by increasing viral moi, and therefore, we designed the two following lentiviral constructs driving a shRNA against GFP: (i) a silencing cassette in the ON configuration, i.e., an mU6 promoter containing a loxP* site between the PSE and the transcriptional start site (LV-shGFP-ON, Fig. 1C), and (ii) a silencing cassette in the OFF configuration, i.e., an mU6 promoter containing an intervening sequence flanked by loxP* sites between the PSE and the transcriptional start site (LV-shGFP-OFF, Fig. 1B). We arbitrarily chose a 1-kb fragment of the LacZ ORF to use as a stuffer sequence and introduced a pol III termination signal (TTTTT) near the 5′ end. These lentiviral vectors were used to infect a 293T cell line stably expressing two copies of GFP. Quantification of GFP expression was carried out by FACS analysis 9 days after infection. LV-CRE alone (moi = 70) had no effect on GFP expression levels (Fig. 2Aa). As expected, although the LV-shGFP-OFF (moi = 100) was incapable of shutting down GFP expression, the LV-shGFP-ON (moi = 100) was as efficient as the LV-shGFP-WT (moi = 100) in reducing GFP levels to ≈5% of the levels found in uninfected cells (Fig. 2Ab-d). To test how efficiently coexpression of CRE recombinase could switch the silencing cassette from OFF to ON, we coinfected the GFP-expressing 293T cell line with LV-shGFP-OFF (moi = 100) and LV-CRE (moi = 70), a CRE recombinase expressing self-deleting lentiviral vector described in ref. 31. Use of this self-deleting vector allowed us to express CRE transiently, avoiding potential toxicity problems (31, 32). Cells infected with LV-CRE (moi = 70) expressed CRE recombinase within 24 h, with a peak of expression 48 h after infection; CRE recombinase levels decreased thereafter and were almost undetectable beyond 96 h after infection (data not shown). Infection of LV-CRE at higher mois resulted in visible toxicity to the cells (data not shown). Remarkably, coinfection of LV-shiGFP-OFF (moi = 100) and LV-CRE (moi = 70) resulted in down-regulation of GFP to levels similar to those seen in cells infected with the LV-shGFP-ON alone (moi = 100), indicating that expression of CRE recombinase efficiently excised the stuffer fragment and resulted in activation of the GFP silencing cassette (Fig. 2Ae). To quantitate the silencing power of the system, we titrated down levels of LV-shGFP-OFF (moi = 60, 20, or 10) while maintaining the same levels of infection for LV-CRE (moi = 70). This resulted in increasingly lower levels of silencing of the target; under the conditions of this assay, a minimum LV-shGFP-OFF moi of 60 was required to obtain satisfactory silencing of GFP. Conversely, lowering the moi of LV-CRE below 70 (moi = 35, 7, and 3) also resulted in increasingly lower levels of silencing of GFP expression (Fig. 2B, bars f-k). Representative results of composite FACS analysis are summarized in Fig. 2B. We conclude that the levels of expression of presumably any given target gene for which an effective shRNA is available can be controlled by careful titration of LV-shRNA-OFF and LV-CRE. It is thus possible that expression of CRE under the control of tissue-specific promoters can regulate the expression of a given target gene to any desired level in a cell type-specific manner.
Fig. 2.
(A) 293T cells expressing two copies of GFP were infected with different lentiviral constructs, with mois as shown in B. Vector titers were determined by p24 ELISA and by TaqMan real-time PCR determination of transduced proviral genomes. On average, the vector preparations were 1.5 E4 particles per nanogram of p24. Cells were cultured for 9 days, and GFP expression levels were quantitated by FACS analysis. GFP expression levels were unaffected by infection with LV-CRE alone (Aa). Infection with LV-shGFP-OFF in the absence of LV-CRE resulted in no change in GFP levels (Ab), whereas LV-shGFP-WT and LV-shGFP-ON were equally effective in down-regulating the target (A c and d). Coinfection of LV-CRE (moi = 70) and LV-shGFP-OFF (moi = 100) resulted in efficient down-regulation of GFP expression (Ae). (B) FACS analysis composite. Coinfection of LV-CRE (moi = 70) with decreasing mois of LV-siGFP-OFF (moi = 60, 20, or 10) resulted in decreased silencing of GFP (data not shown). Similarly, decreasing of LV-CRE (moi = 70, 35, 7, and 3) while keeping LV-shGFP-OFF constant (moi = 100) diminished silencing (data not shown).
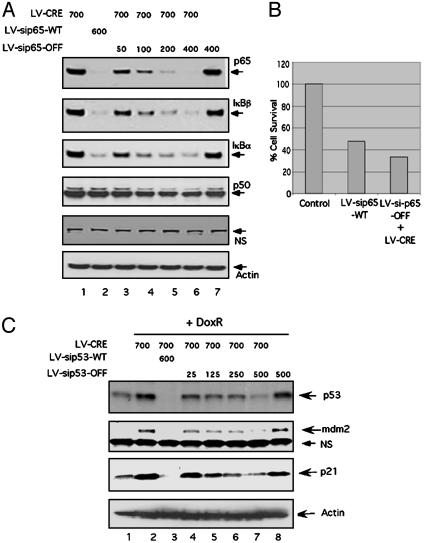
We next tested the use of these vectors in targeting endogenous genes. We chose to analyze the tumor suppressor p53 and the p65 subunit of the NFκB transcription factor, both of which have well characterized cellular phenotypes when down-regulated. Decreased expression of p53 should compromise its ability to induce the up-regulation of downstream targets such as mdm2 and p21 in response to genotoxic stress (34). Similarly, lowering the levels of p65 should cause down-regulation of IκBα and IκBβ proteins (34). IκBα is a transcriptional target of p65; IκBβ protein levels are stabilized by p65. Lentiviral vectors containing shRNAs for both the p53 and p65 genes were generated in both the LV-shRNA-WT+GFP (Fig. 1D) and LV-shRNA-OFF configurations (Fig. 1B). Mouse embryonic fibroblasts were transduced with combinations of LV-shRNAWT+GFP, LV-shRNA-OFF, and LV-CRE, as shown in Fig. 3 A and B. Coinfection of a fixed amount of LV-CRE (moi = 700) with increasing mois of LV-shp65-OFF (moi = 50, 100, 200, and 400) led to a concomitant decrease of p65 (Fig. 3A, lanes 3-6). This decrease of p65 led to reduced levels of both IκBβ and IκBα and also sensitized the cells to tumor necrosis factor α-induced apoptosis (Fig. 3B). Notably, the highest moi of LV-shp65-OFF (moi = 400) used in the absence of LV-CRE had no effect on p65 activity (Fig. 3A, compare lanes 6 and 7). LV-shp65+GFP was used as a positive control in these experiments (Fig. 3A, lane 2). Similarly, coinfection of LV-CRE with increasing mois of LV-shp53-OFF resulted in increased down-regulation of basal and doxorubicin-induced p53 and its target genes p21 and mdm2 (Fig. 3C, lanes 4-7). The LV-shp53+GFP used as a positive control could efficiently reduce the level of doxorubicin-induced p53 stabilization, whereas an equally high moi of LV-shp53-OFF in the absence of LV-CRE had no effect on any of the three genes tested (Fig. 3C, lane 8).
Fig. 3.
Regulated knockdown of endogenous genes. (A) Wild-type murine embryo fibroblasts (MEFs) were infected with mois of 50 (lane 3), 100 (lane 4), 200 (lane 5), and 400 (lanes 6 and 7) of LV-shp65-OFF, with (lanes 3-6) or without (lane 7) coinfection with LV-CRE at an moi of 700. Cells were cultured for 5 days, and lysates were analyzed for the indicated markers by Western blot analysis. LV-shp65-OFF used in combination with LV-CRE (but not when used alone) resulted in highly decreased levels of p65 and downstream targets. Actin was used as a loading control, and p50 and a nonspecific band (NS) are included to show specificity. (B) Tumor necrosis factor α-induced apoptosis. MEFs infected with LV-shp65-WT +GFP (moi = 600) were used as a positive control (lane 2), and LV-shp65-OFF (moi = 400) in combination with LV-CRE (moi = 700) showed increased sensitivity to apoptosis after being assayed as described in Methods.(C) Wild-type MEFs were infected with mois of 25 (lane 4), 125 (lane 5), 250 (lane 6), and 500 (lanes 7 and 8) of LV-shp53-OFF, with (lanes 4-7) or without (lane 8) coinfection with LV-CRE at moi 700. Cells were cultured for 5 days and either left untreated (lane 1) or treated with 0.4 μg/ml doxorubicin (lanes 2-8) and assayed by Western blotting. Cells infected with LV-shp53-WT+GFP (moi = 600, lane 3) were used as a positive control. Increased mois of LV-shp53-OFF (moi = 500) in combination with LV-CRE (moi = 700) resulted in increasingly stronger down-regulation of p53 and downstream targets (lanes 4-7). Actin was used as a loading control, and a nonspecific band is included to show specificity.
In summary, we have developed a lentiviral vector carrying an siRNA-silencing cassette that is inactive until turned on by CRE recombinase. Our results suggest that levels of expression of any target gene can be manipulated by varying the mois of infection with LV-CRE and L-shRNA-OFF, optimizing infection levels depending on the gene target, the level of expression desired, and ease of infection of the particular cell line involved. Two reports describing CRE-inducible siRNA systems have recently been published (35, 36). Our system couples RNA interference-mediated silencing with the ability of lentiviral vectors to transduce both dividing and nondividing cells, and used in combination with lentiviral transgenesis techniques, it will provide a rapid and powerful approach to the study of gene function by generating localized, temporal, and tissue-specific knockdown of specific targets. Use of CRE under the control of tissue-specific promoters will allow precise dissection of signaling pathways between distinct populations of cells in vitro and in vivo. In particular, genes with embryonic lethal phenotypes can now be tested for function by down-regulating the gene target in any cell type, location, and developmental time frame, provided adequate cell type-specific promoters are available.
Acknowledgments
We thank Kenneth Frimpong and Peter Welch of Invitrogen for technical support. We also thank O. Singer, N. Tonnu, and N. Hoong for technical support and critically reviewing the manuscript. G.T. is supported by an Institute of Molecular Medicine training grant (University of California, San Diego). V.T. is supported by a career development fellowship from the Leukemia and Lymphoma Society. I.M.V. is an American Cancer Society Professor of Molecular Biology and is supported in part by grants from the National Institutes of Health, the Larry L. Hillblom Foundation, the Lebensfeld Foundation, the Wayne and Gladys Valley Foundation, and the H. N. and Frances C. Berger Foundation.
Abbreviations: mU6, mouse U6; shRNA, small hairpin RNA; siRNA, small interfering RNA; pol, polymerase; PSE, proximal sequence element; moi, multiplicity of infection; PGTE, PGEM-T Easy.
References
- 1.Wilson, J. A. & Richardson, C. D. (2003) Curr. Opin. Mol. Ther. 5, 389-396. [PubMed] [Google Scholar]
- 2.Zhao, L. J., Jian, H. & Zhu, H. (2003) Gene 316, 137-141. [DOI] [PubMed] [Google Scholar]
- 3.Tomar, R. S., Matta, H. & Chaudhary, P. M. (2003) Oncogene 22, 5712-5715. [DOI] [PubMed] [Google Scholar]
- 4.Sorensen, D. R., Leirdal, M. & Sioud, M. (2003) J. Mol. Biol. 327, 761-766. [DOI] [PubMed] [Google Scholar]
- 5.Somasundaram, K. (2003) Cancer Biol. Ther. 2, 211-212. [DOI] [PubMed] [Google Scholar]
- 6.Scherr, M., Battmer, K., Ganser, A. & Eder, M. (2003) Cell Cycle 2, 251-257. [PubMed] [Google Scholar]
- 7.Rubinson, D. A., Dillon, C. P., Kwiatkowski, A. V., Sievers, C., Yang, L., Kopinja, J., Rooney, D. L., Ihrig, M. M., McManus, M. T., Gertler, F. B., et al. (2003) Nat. Genet. 33, 401-406. [DOI] [PubMed] [Google Scholar]
- 8.Matta, H., Hozayev, B., Tomar, R., Chugh, P. & Chaudhary, P. M. (2003) Cancer Biol. Ther. 2, 206-210. [DOI] [PubMed] [Google Scholar]
- 9.Katahira, T. & Nakamura, H. (2003) Dev. Growth Differ. 45, 361-367. [DOI] [PubMed] [Google Scholar]
- 10.An, D. S., Xie, Y., Mao, S. H., Morizono, K., Kung, S. K. & Chen, I. S. (2003) Hum. Gene Ther. 14, 1207-1212. [DOI] [PubMed] [Google Scholar]
- 11.Miyagishi, M. & Taira, K. (2002) Nucleic Acids Res. 2, Suppl., 113-114. [DOI] [PubMed] [Google Scholar]
- 12.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- 13.Barton, G. M. & Medzhitov, R. (2002) Proc. Natl. Acad. Sci. USA 99, 14943-14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiscornia, G., Singer, O., Ikawa, M. & Verma, I. M. (2003) Proc. Natl. Acad. Sci. USA 100, 1844-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fjose, A., Ellingsen, S., Wargelius, A. & Seo, H. C. (2001) Biotechnol. Annu. Rev. 7, 31-57. [DOI] [PubMed] [Google Scholar]
- 16.Hannon, G. J. (2002) Nature 418, 244-251. [DOI] [PubMed] [Google Scholar]
- 17.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 18.Paul, C. P., Good, P. D., Winer, I. & Engelke, D. R. (2002) Nat. Biotechnol. 20, 505-508. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira, D. M. & Goodell, M. A. (2003) Genesis 36, 203-208. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W. S. & Khvorova, A. (2004) Nat. Biotechnol. 22, 326-330. [DOI] [PubMed] [Google Scholar]
- 21.Dirac, A. M. & Bernards, R. (2003) J. Biol. Chem. 278, 11731-11734. [DOI] [PubMed] [Google Scholar]
- 22.Follenzi, A., Ailles, L. E., Bakovic, S., Geuna, M. & Naldini, L. (2000) Nat. Genet. 25, 217-222. [DOI] [PubMed] [Google Scholar]
- 23.Dull, T., Zufferey, R., Kelly, M., Mandel, R. J., Nguyen, M., Trono, D. & Naldini, L. (1998) J. Virol. 72, 8463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tergaonkar, V., Pando, M., Vafa, O., Wahl, G. & Verma, I. (2002) Cancer Cell 1, 493-503. [DOI] [PubMed] [Google Scholar]
- 25.Allison, D. S. & Hall, B. D. (1985) EMBO J. 4, 2657-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn, R. & Torres, R. M. (2002) Methods Mol. Biol. 180, 175-204. [DOI] [PubMed] [Google Scholar]
- 27.Bark, C., Weller, P., Zabielski, J., Janson, L. & Pettersson, U. (1987) Nature 328, 356-359. [DOI] [PubMed] [Google Scholar]
- 28.Das, G., Henning, D., Wright, D. & Reddy, R. (1988) EMBO J. 7, 503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goomer, R. S. & Kunkel, G. R. (1992) Nucleic Acids Res. 20, 4903-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, G. & Saito, I. (1998) Gene 216, 55-65. [DOI] [PubMed] [Google Scholar]
- 31.Pfeifer, A., Brandon, E. P., Kootstra, N., Gage, F. H. & Verma, I. M. (2001) Proc. Natl. Acad. Sci. USA 98, 11450-11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt, E. E., Taylor, D. S., Prigge, J. R., Barnett, S. & Capecchi, M. R. (2000) Proc. Natl. Acad. Sci. USA 97, 13702-13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine, A. J. (1997) Cell 88, 323-331. [DOI] [PubMed] [Google Scholar]
- 34.Beg, A. A., Sha, W. C., Bronson, R. T., Ghosh, S. & Baltimore, D. (1995) Nature 376, 167-170. [DOI] [PubMed] [Google Scholar]
- 35.Fritsch, L., Martinez, L. A., Sekhri, R., Naguibneva, I., Gerard, M., Vandromme, M., Schaeffer, L. & Harel-Bellan, A. (2004) EMBO Rep. 5, 178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasim, V., Miyagishi, M. & Taira, K. (2003) Nucleic Acids Res. 3, Suppl., 255-256. [DOI] [PubMed] [Google Scholar]