Abstract
DNA mutagenesis is generally considered harmful. Yet activated B cells normally mutate the Ig loci. Because this somatic hypermutation is potentially dangerous, it has been hypothesized that mutations do not occur throughout the genome but instead are actively targeted to the Ig loci. Here we challenge this longstanding and widely accepted hypothesis. We demonstrate that hypermutation requires no Ig gene sequences. Instead, activation-induced cytidine deaminase and other trans-acting hypermutation factors may function as general mutators.
Upon antigenic stimulation of B lymphocytes, the variable (V) regions of Ig loci mutate at a rate orders of magnitude higher than the normal spontaneous rate. This somatic hypermutation requires activation-induced cytidine deaminase (AID) (1), an enzyme expressed exclusively in activated B cells of the germinal center (2). The enzyme converts deoxycytidine to deoxyuridine (3, 4). Except for a short motif, WRC (W = A or T, R = A or G) on the nontemplate DNA strand, purified AID has no sequence preference in vitro (4). In line with biochemical experiments, transgenic expression of AID in fibroblasts (5), hybridomas (6), and even bacteria (7, 8) causes hypermutation.
It has been reasoned that hypermutation must be targeted to the Ig loci, because if mutations occurred at locations outside the V region, this would be dangerous and eventually lead to tumorigenesis. Thus Ig hypermutation has been regarded as the prime example of site-directed mutagenesis (9-13). Surprisingly, the V region itself is not needed for hypermutation (14). Instead, nearby enhancers and other sequences from the Ig intron have been proposed to direct hypermutation to the V regions (15-19).
Recently, three genes that do not encode the V region, BCL-6 (20), B29, and mb1 (21), were reported to hypermutate in normal B lymphocytes. To explain these results within the framework of site-directed mutagenesis, it was proposed that these genes have unidentified “Ig-like” (20) or common cis-acting regulatory sequences (21). Furthermore, the T cell receptor in AID-transgenic mice (22) and a GFP plasmid reporter in AID-transgenic fibroblasts (5) were shown to hypermutate. Although these observations seem to conflict with the notion of site-directed mutagenesis, it has been suggested that transgenic overexpression of AID or lack of B cell factors leads to nonphysiological, nontargeted hypermutation (13).
The paradigm of site-directed mutagenesis is deeply rooted. Despite some observations to the contrary, the interpretation of results has rarely sought to dispute this paradigm. Yet the putative factors that recruit and deliver AID to the V regions remain undiscovered. Given the nondiscriminating enzymatic activity of AID, we hypothesized that hypermutation is not an actively targeted process. Here we challenge the paradigm of site-directed mutagenesis by demonstrating that AID-mediated hypermutation requires no Ig gene elements.
Materials and Methods
Reporter Constructs. Constructs were based on the Moloney murine leukemia virus retroviral vector contained in p102.21 (kindly provided by J. B. Lorens, Rigel Pharma, South San Francisco, CA). They contain the internal ribosome entry site and puromycin resistance gene of pIRESpuro3 (Clontech). The mouse Ig μ intron sequence consisted of the 1-kb XbaI fragment from the major intron. The positive control vectors pGFP-Ipuro and pGFP-Emu-Ipuro contained enhanced GFP (EGFP) from pEGFP-N1 (Clontech). To create the reporter constructs pGFP*-Ipuro and pGFP*-Emu-Ipuro, mutations were introduced at two bases (C321G and A322C) of the EGFP gene to introduce a stop codon (TAG) within an RGYW sequence (Y107*, K108Q).
Cell Lines and Culture. The 70Z/3-YFP-AID (YFP, yellow fluorescent protein) and 70Z/3-YFP cell lines were created by infecting mouse 70Z/3 cells with VSV-G pseudotyped lentiviral vectors, engineered from pHR′GFP-W-SIN18 (23) to deliver either enhanced YFP (EYFP) (Clontech) and mouse AID cDNA or EYFP only. Cultures were sorted for stable fluorescent cells by fluorescence-activated cell sorting (FACS). The 70Z/3, 70Z/3-YFP-AID, 70Z/3-YFP, and 18-81 cell lines were infected with reporter constructs packaged by PhoenixEco cells (ATCC SD 3444). Infected cells were selected by addition of puromycin (70Z/3 2.5 μg/ml, 18-81 5 μg/ml). With both lentiviral and Moloney murine leukemia virus gene delivery, the multiplicity of infection was sufficiently low that nearly all infected cells contained one copy per cell. Cells infected by the reporter constructs were analyzed by flow cytometry and considered GFP-positive if they exhibited a fluorescence intensity at least 50 times that of untransduced 18-81 cells. Cells were sorted for either positive or negative fluorescence by FACS, and 18-81 clones were expanded from GFP-positive single cells. All cultures were grown in RPMI media 1640 at 37°C with 5% CO2.
Identification of Mutations in the Reporter Gene by Sequencing. DNA from the 18-81 clones was isolated, and the GFPstop reporter gene was PCR-amplified by using Pfu polymerase (Stratagene). The PCR products were incubated with Taq polymerase to add deoxyadenosine overhangs and then cloned into pCR2.1-TOPO (Invitrogen). The plasmids were then amplified in Escherichia coli and sequenced.
Identification of Reporter Integration Sites. Integration sites of the reporter vectors were PCR-amplified by using Taq polymerase (24). The first amplification step of the PCR used a biotinylated primer homologous to the vector and a nonspecific primer, degenerate at the 3′ end and constant at the 5′ end. Biotinylated PCR product was recovered by use of streptavidin-coupled magnetic beads (Dynal, Great Neck, NY). The genomic DNA flanking the 3′ LTR of the provirus was then amplified from the biotinylated PCR product. This secondary PCR used a nested LTR-specific primer and a primer homologous to the constant section of the nonspecific primer of the first PCR products were cloned into pCR2.1-TOPO, amplified in E. coli, and then sequenced.
Results and Discussion
Detection of AID-Mediated Mutations. Perhaps hypermutation outside the V region is the norm. We tested this idea by disseminating a reporter construct containing no Ig elements throughout the genome of hypermutating cells (Fig. 1A). The reporter gene consisted of an inactive GFP that fluoresces on reversion of a premature stop codon, TAG. Any single point mutation at the codon activates the reporter, except the transition from G to A, which only creates the stop codon TAA. If only sequences near the V region hypermutate, then the reporter gene would be activated only when it is integrated near or within the Ig loci. If there were other locations in the genome that support hypermutation, then the reporter also would be activated when it is integrated in these locations outside the Ig loci. Because previous data from our lab and others (15-19) indicated that mutation of a V region is markedly increased by Ig intronic enhancers, we also created a construct that contained the μ intronic enhancer adjacent to the reporter gene. If the intronic enhancer indeed directs hypermutation factors to a target, then the frequency of mutation in this construct would be high regardless of genomic location.
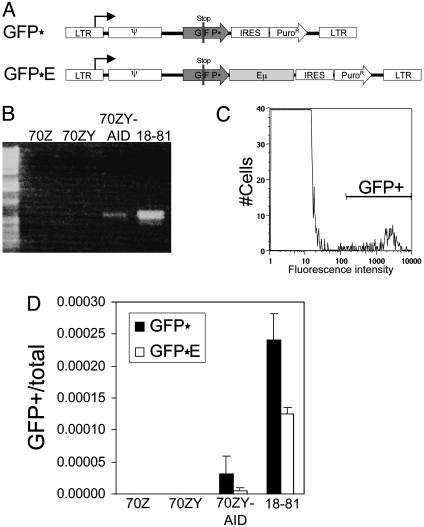
Fig. 1.
Reporter constructs, with or without Ig elements, are activated by AID-mediated hypermutation. (A) Reporters contained either no Ig elements, GFP*, or the Ig enhancer of the large intron, GFP*E. Vector components: ψ, retroviral packaging signal; GFP*, GFP with premature stop codon; IRES, internal ribosome entry site; PuroR, puromycin resistance gene; Eμ, μ enhancer. (B) RT-PCR amplifying AID cDNA in cell lines 70Z/3 (denoted 70Z), 70Z/3-YFP (denoted 70ZY), 70Z/3-YFP-AID (denoted 70ZY-AID), and 18-81. (C) Flow cytometry of GFP expression. Cells 50 times more fluorescent than background (bar labeled GFP+) were counted as GFP-positive. (D) Fraction of GFP-positive cells 8 days after removal of GFP-positive cells (i.e., preexisting mutants) by fluorescence-activated cell sorting. Cells were grown under constant puromycin selection.
To adequately survey the genome, our reporter gene was delivered to cells via retroviral infection. Retroviral delivery was well suited for our experiment, because it is highly efficient, and integration occurs at essentially random locations in the genome. Based on the starting cell number and infection rate, we estimate that the reporter was integrated only once in a given cell with up to 2 × 105 integration sites per culture. The two constructs were transduced into four cell lines. One line was 70Z/3, a pre-B cell line that does not express AID and does not hypermutate. Into this cell line, we introduced either YFP alone (70Z/3-YFP) or YFP together with AID (70Z/3-YFP-AID) (Fig. 1B), thereby creating two cell lines that ought to differ by their ability to hypermutate. The fourth cell line was 18-81, which expresses AID endogenously (Fig. 1B). Because the light chain genes are not rearranged in the majority of its cells, 18-81 may be classified as a pre-B cell, but its ability to both hypermutate and switch its endogenous heavy chain genes mimics the phenotype of an activated B cell. Both the mechanism for switching (25) and hypermutation (26, 27) have been verified in 18-81.
Both constructs hypermutated in an AID-dependent fashion (Fig. 1 C and D). We conclude this because neither construct yielded any GFP-positive cells in the absence of AID. Somewhat surprisingly, the reporter could hypermutate without any Ig sequences. In the 70Z/3-YFP-AID and 18-81 lines, hypermutation actually appeared lower in the presence of the Ig enhancer.
Mutation Spectrum. We also investigated whether the spectrum of mutations reflects those previously observed in AID-expressing cells. To this end, GFP-positive 18-81 cells were isolated by fluorescence-activated cell sorting, and clones were expanded from single cells. Clones were harvested 32 days after infection; GFP genes were PCR-amplified, ligated into plasmids, and sequenced. Indeed, both constructs contained mutations, e.g., C to T and G to A, that likely result from cytidine deamination, as well as mutations, e.g., G to T and C to A, that might also require DNA repair enzymes (28, 29) or polymerases (30) as modifiers. Furthermore, mutations were likely to be within RGYW/WRCY motifs. These biases indicate that our reporter constructs were hypermutated in an AID-mediated fashion (Table 1).
Table 1. Analysis of mutations in 18-81 clones after 32 days.
| Distribution of mutations by type, %
|
Association with RGYW/WRCY, %
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reporter construct | No. of mutations | [mutations]/[bp × day] × 105 | A → C | A → G | A → T | C → A | C → G | C → T | G → A | G → C | G → T | T → A | T → C | T → G | Mutations | Total |
| GFP* | 17 | 1.2 | 0 | 0 | 6 | 24 | 0 | 35 | 18 | 18 | 0 | 0 | 0 | 0 | 47 | 27 |
| GFP*E | 18 | 1.5 | 0 | 0 | 0 | 11 | 11 | 39 | 39 | 0 | 0 | 0 | 0 | 0 | 56 | 27 |
| Total | 35 | 1.3 | 0 | 0 | 3 | 17 | 6 | 37 | 29 | 9 | 0 | 0 | 0 | 0 | 51 | 27 |
| Ref. 5 | 241 | 45 | 0 | 0 | 0.8 | 7 | 5 | 37 | 31 | 11 | 8 | 0 | 0 | 0.4 | 75 | 26 |
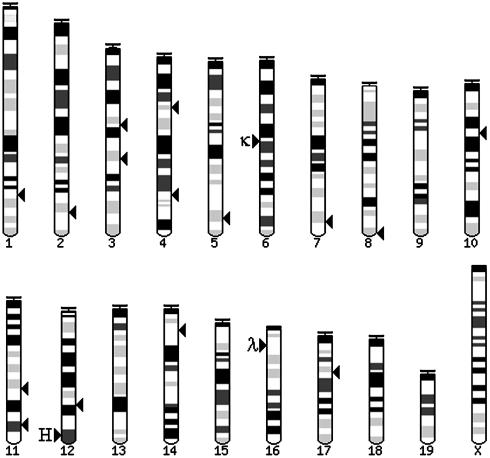
Identification of Reporter Integration Sites. Our findings seem to indicate that the cells do not actively target hypermutation to Ig loci. But even though retroviral integration is generally random throughout the genome, we needed to confirm this for our constructs. It was possible that reporter constructs, even ones containing no Ig sequences, integrated near Ig genes in the genome. Consequently, hypermutation normally occurring at the Ig locus might have also resulted in mutations in nearby reporters. Therefore, we identified the integration sites of 15 GFP-positive clones and found that no reporters were integrated near Ig loci (Fig. 2). Thus we conclude that in the 18-81 cell line, no cis-acting Ig sequences are needed for hypermutation. We further conclude that a large number of genomic sites must be amenable to hypermutation. These sites may or may not harbor non-Ig sequences that affect hypermutation. We also note that because we have used an exogenous reporter construct, we cannot exclude that hypermutation was affected by elements in the construct, such as the retroviral LTRs, IRES, or puromycin resistance gene.
Fig. 2.
Genomic integration sites of hypermutated reporter constructs in 18-81 clones. Arrows to the right of the chromosomes represent integration sites. Location of Ig loci, arrows to the left; H, heavy chain; κ, κ light chain; λ, λ light chain. The exact genomic locations are available upon request.
Dynamics of Mutation Accumulation. Because mutations can both activate and inactivate a reporter gene, the number of active reporter genes at one time point does not necessarily reflect the actual mutation rate. Thus hypermutation in 18-81 cells was monitored continuously (Fig. 3A). In these experiments, we removed puromycin from cultures after 3 days. This helped ensure that mutation and subsequent inactivation of the puromycin selection gene did not affect the number of GFP-positive revertants. The results of this experiment were in line with the single time-point experiment: hypermutation occurred without any Ig sequences, and the presence of the Ig enhancer resulted in fewer GFP-positive revertants.
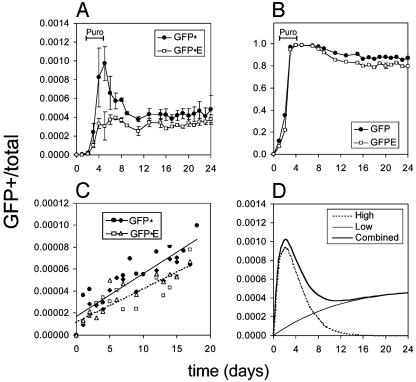
Fig. 3.
The 18-81 cell line mutated both reporter constructs, GFP* and GFP*E, over a long period. y axis, GFP-positive cells as fraction of all cells; x axis, days. Puromycin, Puro, added to cultures where noted. (A) Activation and inactivation of the reporter constructs. (B) Fraction of GFP-positive cells after transduction with GFP control vectors (no premature stop codon) containing either no Ig elements (denoted GFP) or the Ig enhancer of the large intron (denoted GFPE). (C) Cultures sorted for GFP-negative cells after 17 days (differing symbols, circle or triangle, open or closed, represent a single experiment). (D) Model curve fitting experimental results, reflecting the sum total of different reporter integration sites, which may support a high or low mutation rate.
In the time-course experiment, the fraction of GFP-positive mutants increased sharply over the first 3-5 days. At the steepest section of these curves (Fig. 3A), the reversion rates at the reporter stop codon ranged from 2 × 10-4 to 6 × 10-4 per day. This is comparable to hypermutation at the V region of 18-81, which reverts a stop codon at 3.9 × 10-4 per day (27). After the initial increase, we observed a decrease in the number of mutants. Because GFP without the premature stop codon did not markedly lose fluorescence over time (Fig. 3B), the decrease in GFP-expressing cells is likely due not only to general gene silencing but also to mutations that inactivate the reporter gene. In support of this notion, sequences from GFP-negative cells, isolated from cultures initially expanded from a mutation-activated, GFP-positive single cell, showed an increase in missense and nonsense mutations (data not shown). The decrease in GFP-positive cells appeared to be followed by a gradual increase. To ascertain whether the second increase could be due to mutation, we sorted for GFP-negative cells after day 17. Because the sorted culture generated new GFP-positive mutants (Fig. 3C), this indicated that mutations were still accumulating.
If a population of cells contained the reporter construct at sites that support a high rate of hypermutation, then one might project that the number of GFP-positive mutants increases (reporter activation), then decreases (inactivation). If a population had the reporter integrated at sites favoring a low mutation rate, this might result in a gradual accumulation of GFP-positive cells. In our experiments, we surveyed thousands of genomic locations simultaneously. Assuming that cells differ only by the genomic location of the reporter, we infer that the initial phase of mutations represents locations in the genome that support a higher mutation rate, and similarly the second phase represents locations that support a lower mutation rate (Fig. 3D). Thus we propose that many, but not all, locations throughout the genome support hypermutation. Because hypermutation requires transcription (4, 31, 32), only transcribed genes in these locations would be affected. If only such a fraction of genes hypermutates, this might explain why several genes have been reported not to hypermutate (33).
Other Mutator Enzymes. Because our finding of genome-wide AID-mediated mutagenesis goes against the current paradigm, we thought of other artifacts. Recently, it was discovered that another cytidine deaminase, APOBEC-3G, mutates and inactivates retroviruses before integration into the host genome (34, 35). It is also possible that reverse transcriptase, an error prone polymerase, introduces mutations before the reporter constructs are integrated into the genome. So, could the mutations we report result from APOBEC-3G-like activity or reverse transcriptase? We believe that we can rule out such mutations. Whereas APOBEC-3G and reverse transcriptase can mutate only before integration, during first-strand synthesis of the provirus, we observed accumulation of mutations well after integration of our reporter constructs. When we stably transduced cultures and sorted for GFP-negative cells, GFP-positive mutants were detected later. In cultures continuously monitored, the fraction of GFP-positive cells continuously increased for several days after infection. Furthermore, although APOBEC3-G induces only G to A mutations in the reading DNA strand, we observed not only G to A mutations but also many others, such as C to T and G to C. In the case of reverse transcriptase-mediated mutations, it is clearly not possible for reverse transcriptase to contribute to mutations after transduction, because both our retroviral vectors and our cell lines did not express the reverse transcriptase gene.
Conclusion
Before the discovery of Ig gene rearrangement, Brenner and Milstein (12) laid out the paradigm of site-directed hypermutation to explain the generation of Ig V region diversity. Although it was later discovered that the V region does hypermutate (36-38), few studies showed that it is the only region that hypermutates. To prove that hypermutation is V region specific, one would need to sequence the complete genome of single cells. Instead, the paradigm was mostly built on sequencing data that showed few (39) or no (37, 40) mutations in the Ig constant region. Although it is obvious that mutations are less abundant in the constant region, this fact does not require that specific factors target the V region. The constant region may be less mutated merely because it is further from the promoter (31, 39). Certainly, in addition to positive selection for V region mutations in vivo, there will be selection against cells with mutated constant regions. Some Ig sequences might even protect the constant region from mutation. Furthermore, the results presented in this paper do not necessarily contradict earlier findings that Ig intron and enhancer sequences affect V region hypermutation. Although it is certainly possible that these sequences affect the V region, this does not preclude hypermutation of other genes when such sequences are absent. It has recently been reported that chromatin structure (41) and 6-bp nucleotide motifs (42) can enhance hypermutation, and such Ig-nonspecific factors could influence mutagenesis throughout the genome. It is conceivable that any DNA sequence that sufficiently exposes single-stranded DNA (3, 4, 8, 32) is accessible to AID and other hypermutation factors.
In the past, hypermutation outside the V region has been cast as an exception to the rule. Based on the results of this study, we believe that AID-mediated mutations outside the V region should be expected, not excepted. Although the physiological level of mutation of the V region still may differ from that of other genes, we believe that this difference is not nearly as dramatic as previously thought. Although the bias has been that nontargeted mutations would be dangerous, perhaps the brief temporal expression of AID in activated B cells is sufficient to minimize deleterious effects. Clearly, further study with different cell lines, mice, different reporter constructs, and extensive DNA sequencing is now required to test the hypothesis that Ig hypermutation is the work of a general mutator.
Acknowledgments
We are grateful to D. Trono (University of Geneva, Geneva) and J. Lorens (Rigel Pharma, South San Francisco, CA) for providing retroviral vectors. We thank Desirée Yang (University of California, San Francisco), M. Klasen (University of California, San Francisco), B. Wang (Picobella, Burlingame, CA), and T. Wechsler (University of California, San Francisco) for contributions. This study was supported by Grant AG20684 from the National Institutes of Health. C.L.W. was supported by the Leukemia Research Foundation and the National Institutes of Health Immunology Training Grant (NIH/NIAID T32 AI07334).
Abbreviations: AID, activation-induced cytidine deaminase; V, variable; YFP, yellow fluorescent protein.
References
- 1.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102, 553-563. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu, M., Sankaranand, V. S., Anant, S., Sugai, M., Kinoshita, K., Davidson, N. O. & Honjo, T. (1999) J. Biol. Chem. 274, 18470-18476. [DOI] [PubMed] [Google Scholar]
- 3.Bransteitter, R., Pham, P., Scharff, M. D. & Goodman, M. F. (2003) Proc. Natl. Acad. Sci. USA 100, 4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pham, P., Bransteitter, R., Petruska, J. & Goodman, M. F. (2003) Nature 424, 103-107. [DOI] [PubMed] [Google Scholar]
- 5.Yoshikawa, K., Okazaki, I. M., Eto, T., Kinoshita, K., Muramatsu, M., Nagaoka, H. & Honjo, T. (2002) Science 296, 2033-2036. [DOI] [PubMed] [Google Scholar]
- 6.Martin, A., Bardwell, P. D., Woo, C. J., Fan, M., Shulman, M. J. & Scharff, M. D. (2002) Nature 415, 802-806. [DOI] [PubMed] [Google Scholar]
- 7.Petersen-Mahrt, S. K., Harris, R. S. & Neuberger, M. S. (2002) Nature 418, 99-103. [DOI] [PubMed] [Google Scholar]
- 8.Ramiro, A. R., Stavropoulos, P., Jankovic, M. & Nussenzweig, M. C. (2003) Nat. Immunol. 4, 452-456. [DOI] [PubMed] [Google Scholar]
- 9.Neuberger, M. S., Harris, R. S., Di Noia, J. & Petersen-Mahrt, S. K. (2003) Trends Biochem. Sci. 28, 305-312. [DOI] [PubMed] [Google Scholar]
- 10.Fugmann, S. D. & Schatz, D. G. (2002) Science 295, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 11.Green, N. S., Lin, M. M. & Scharff, M. D. (1998) BioEssays 20, 227-234. [DOI] [PubMed] [Google Scholar]
- 12.Brenner, S. & Milstein, C. (1966) Nature 211, 242-243.5965537 [Google Scholar]
- 13.Reynaud, C. A., Aoufouchi, S., Faili, A. & Weill, J. C. (2003) Nat. Immunol. 4, 631-638. [DOI] [PubMed] [Google Scholar]
- 14.Yelamos, J., Klix, N., Goyenechea, B., Lozano, F., Chui, Y. L., Gonzalez Fernandez, A., Pannell, R., Neuberger, M. S. & Milstein, C. (1995) Nature 376, 225-229. [DOI] [PubMed] [Google Scholar]
- 15.Betz, A. G., Milstein, C., Gonzalez-Fernandez, A., Pannell, R., Larson, T. & Neuberger, M. S. (1994) Cell 77, 239-248. [DOI] [PubMed] [Google Scholar]
- 16.Goyenechea, B., Klix, N., Yelamos, J., Williams, G. T., Riddell, A., Neuberger, M. S. & Milstein, C. (1997) EMBO J. 16, 3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klix, N., Jolly, C. J., Davies, S. L., Bruggemann, M., Williams, G. T. & Neuberger, M. S. (1998) Eur. J. Immunol. 28, 317-326. [DOI] [PubMed] [Google Scholar]
- 18.Bachl, J. & Wabl, M. (1996) Immunogenetics 45, 59-64. [DOI] [PubMed] [Google Scholar]
- 19.Bachl, J. & Wabl, M. (1996) Proc. Natl. Acad. Sci. USA 93, 851-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen, H. M., Peters, A., Baron, B., Zhu, X. & Storb, U. (1998) Science 280, 1750-1752. [DOI] [PubMed] [Google Scholar]
- 21.Gordon, M. S., Kanegai, C. M., Doerr, J. R. & Wall, R. (2003) Proc. Natl. Acad. Sci. USA 100, 4126-4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okazaki, I. M., Hiai, H., Kakazu, N., Yamada, S., Muramatsu, M., Kinoshita, K. & Honjo, T. (2003) J. Exp. Med. 197, 1173-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zufferey, R., Dull, T., Mandel, R. J., Bukovsky, A., Quiroz, D., Naldini, L. & Trono, D. (1998) J. Virol. 72, 9873-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorensen, A. B., Duch, M., Jorgensen, P. & Pedersen, F. S. (1993) J. Virol. 67, 7118-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jack, H. M., McDowell, M., Steinberg, C. M. & Wabl, M. (1988) Proc. Natl. Acad. Sci. USA 85, 1581-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer, J., Jack, H. M., Ellis, N. & Wabl, M. (1986) Proc. Natl. Acad. Sci. USA 83, 6950-6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wabl, M., Burrows, P. D., von Gabain, A. & Steinberg, C. (1985) Proc. Natl. Acad. Sci. USA 82, 479-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cascalho, M., Wong, J., Steinberg, C. & Wabl, M. (1998) Science 279, 1207-1210. [DOI] [PubMed] [Google Scholar]
- 29.Di Noia, J. & Neuberger, M. S. (2002) Nature 419, 43-48. [DOI] [PubMed] [Google Scholar]
- 30.Faili, A., Aoufouchi, S., Flatter, E., Gueranger, Q., Reynaud, C. A. & Weill, J. C. (2002) Nature 419, 944-947. [DOI] [PubMed] [Google Scholar]
- 31.Peters, A. & Storb, U. (1996) Immunity 4, 57-65. [DOI] [PubMed] [Google Scholar]
- 32.Chaudhuri, J., Tian, M., Khuong, C., Chua, K., Pinaud, E. & Alt, F. W. (2003) Nature 422, 726-730. [DOI] [PubMed] [Google Scholar]
- 33.Storb, U., Peters, A., Klotz, E., Kim, N., Shen, H. M., Hackett, J., Rogerson, B. & Martin, T. E. (1998) Immunol. Rev. 162, 153-160. [DOI] [PubMed] [Google Scholar]
- 34.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L. & Trono, D. (2003) Nature 424, 99-103. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C. & Gao, L. (2003) Nature 424, 94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weigert, M. G., Cesari, I. M., Yonkovich, S. J. & Cohn, M. (1970) Nature 228, 1045-1047. [DOI] [PubMed] [Google Scholar]
- 37.Gearhart, P. J. & Bogenhagen, D. F. (1983) Proc. Natl. Acad. Sci. USA 80, 3439-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gearhart, P. J., Johnson, N. D., Douglas, R. & Hood, L. (1981) Nature 291, 29-34. [DOI] [PubMed] [Google Scholar]
- 39.Motoyama, N., Okada, H. & Azuma, T. (1991) Proc. Natl. Acad. Sci. USA 88, 7933-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, S., Davis, M., Sinn, E., Patten, P. & Hood, L. (1981) Cell 27, 573-581. [DOI] [PubMed] [Google Scholar]
- 41.Woo, C. J., Martin, A. & Scharff, M. D. (2003) Immunity 19, 479-489. [DOI] [PubMed] [Google Scholar]
- 42.Michael, N., Shen, H. M., Longerich, S., Kim, N., Longacre, A. & Storb, U. (2003) Immunity 19, 235-242. [DOI] [PubMed] [Google Scholar]