Abstract
Arginine methylation is a common posttranslational modification (PTM) that alters roughly 0.5% of all arginine residues in the cells. There are three types of arginine methylation: monomethylarginine (MMA), asymmetric dimethylarginine (ADMA), and symmetric dimethylarginine (SDMA). These three PTMs are enriched on RNA-binding proteins and on histones, and also impact signal transduction cascades. To date, over thirty arginine methylation sites have been catalogued on the different core histones. These modifications alter protein structure, impact interactions with DNA, and also generate docking sites for effector molecules. The primary “readers” of methylarginine marks are Tudor domain-containing proteins. The complete family of thirty-six Tudor domain-containing proteins has yet to be fully characterized, but at least ten bind methyllysine motifs and eight bind methylarginine motifs. In this review, we will highlight the biological roles of the Tudor domains that interact with arginine methylated motifs, and also address other types of interactions that are regulated by these particular PTMs.
1. Introduction
Arginine has the longest side chain of the 20 naturally occurring amino acids, and the end of the side chain bears a positive charge – properties that make it a good anchor for potential protein-protein interactions. Its guanidine group contains five potential hydrogen bond donors that can be used to stabilize interactions with DNA, RNA and proteins [1]. The methylation of arginine changes its shape, does not alter the charge, but removes potential hydrogen bond donors, which would potentially inhibit certain interactions [2]. Importantly, the methylation of arginine residues can also increase their affinity to aromatic rings in cation-pi interactions, thus promoting other interactions [3]. So, protein arginine methylation can both positively and negatively regulate protein-protein interactions, examples of which will be highlighted here.
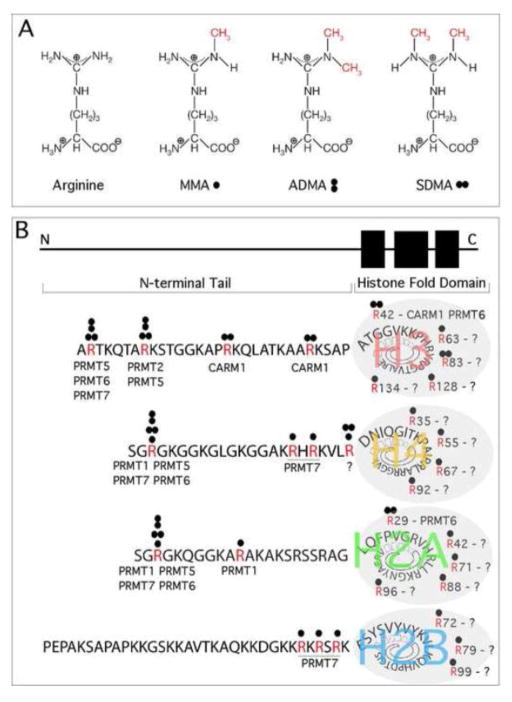
Three distinct types of methylated arginine residues occur in mammalian cells (Figure 1A). The most abundant type is omega-NG,NG-dimethylarginine [4]. In this case, two methyl groups are placed on one of the terminal nitrogen atoms of the guanidino group, and this derivative is commonly referred to as asymmetric dimethylarginine (ADMA). Two other derivatives occur at levels less than 50% that of ADMA. These include the symmetric dimethylated derivative, where one methyl group is placed on each of the terminal guanidino nitrogens (omega-NG,N′G-dimethylarginine; commonly referred to as SDMA) and the monomethylated derivative with a single methyl group on the terminal nitrogen atom (omega-NG-monomethylarginine; commonly referred to as MMA). The three types of arginine methylation are catalyzed by a family of nine AdoMet-dependent enzymes called the protein arginine methyltransferases (PRMTs). Arginine demethylation activity has been reported for the JmjC-domain-containing protein JMJD6 [5, 6].
Figure 1. Types and sites of arginine methylation on histones.
(A) Arginine residues in the tails of histones can be monomethylarginines (MMA - ●), asymmetric dimethylarginines (ADMA -
 ), or symmetric dimethylarginines (SDMA -
), or symmetric dimethylarginines (SDMA -
 ). Methyl groups are marked in red. (B) Positioning of the unstructured histone tail relative to the structure C-terminal core region. The reported sites of histone H3, H4, H2A, and H2B arginine methylation are shown. The references that first reported these methylated sites are listed in Table 1. A number of arginine methylated sites have been identified by mass spectrometric methods, but it has yet to be established which PRMTs modify them, and these sites are thus assigned question marks.
). Methyl groups are marked in red. (B) Positioning of the unstructured histone tail relative to the structure C-terminal core region. The reported sites of histone H3, H4, H2A, and H2B arginine methylation are shown. The references that first reported these methylated sites are listed in Table 1. A number of arginine methylated sites have been identified by mass spectrometric methods, but it has yet to be established which PRMTs modify them, and these sites are thus assigned question marks.
The PRMTs are classified according to the type of methylation they are able to catalyze. Type I, II and III are able to generate a MMA. Type I enzymes (PRMT1, PRMT2, PRMT3, PRMT4/CARM1, PRMT6, and PRMT8) perform a second methylation step to generate the ADMA mark, and the Type II enzyme (PRMT5) generates the SDMA mark. The Type III enzyme (PRMT7) only generates a MMA mark. Most MMA marks are presumed to serve as precursors for the subsequent methylation by Types I and II PRMTs, but certain proteins exist in a heavily monomethylated state [7]. Sequence analysis of all PRMTs shows a highly conserved catalytic core region, containing the signature methyltransferase motifs I, post-I, II and III, which are characteristic of the super-family of seven-beta strand methyltransferases. They also harbor additional “double E” (two glutamate residues) and “THW” (threonine-histidine-tryptophan) sequence motifs, which are particular to the PRMT subfamily of methyltransferases [1]. The catalytic core is highly conserved at the structural level, as revealed by the crystal structure of PRMT1, PRMT3, PRMT4 and PRMT5 [8–14].
It should be noted that the metabolic cost of arginine methylation is high, requiring the use of 12 ATP molecules per methylation event [15]. The fact that such an “expensive” PTM is abundant and has not been lost to evolutionary pressure underscores the biological importance of the methylated motifs that have survived.
2. Sites of Arginine Methylation on Histones
Arginine methylation is an abundant posttranslational modification (PTM), with about 0.5% of arginine residues methylated in mammalian tissues [4, 16], and roughly 2% of arginine residues methylated in rat liver nuclei [17]. The large majority of this type of protein methylation occurs on non-histone proteins and most of these substrates are methylated on Glycine/Arginine-Rich (GAR) motifs. Many of these substrates have recently been catalogued by mass spectrometric analysis [18–20]. Importantly, a number of sites on histone tails are methylated [21], and there is emerging evidence for the existence of arginine-methylated sites within the histone core region [22] (Figure 1B). We have complied a list of the arginine methylation sites that are found on histone, along with the reference that reported each particular PTM (Table 1). We must emphasize that many of these sites are not very well characterized, and often their existence has not been confirmed by alternative approaches, like methyl-specific antibodies or in vitro methylation assays. Furthermore, the enzymes that methylate many of these sites have yet to be elucidated. Finally, modulation of the levels of one type of arginine modification can alter the levels of the other two methylarginine types [7]. This is important, because manipulation of one type of arginine methylation reaction by PRMT knockout, small molecule inhibition or overexpression, may impact the occurrence of other types of arginine methylation.
Table 1.
Arginine methylation sites on histone tails and cores
| SITE & TYPE | ENZYME | REF | SITE & TYPE | ENZYME | REF |
|---|---|---|---|---|---|
| Histone H3 | Histone H4 | ||||
| R2me2a | PRMT6 | [86] [87] [88] | R3me2a | PRMT1 | [120] |
| R2me2s | PRMT5 PRMT7 | [94] | R3me2a | PRMT6 | [87] |
| R8me2a | PRMT2 | [121] | R3me2s | PRMT5 | [97] |
| R8me2s | PRMT5 | [97] | R3me2s | PRMT7 | [122] |
| R17me2a | CARM1 | [123] | R17me1/me2 | PRMT7 | [124] [125] |
| R26me2a | CARM1 | [123] | R19me1/me2 | PRMT7 | [124] [125] |
| R42me2a | CARM1 PRMT6 | [22] | R23me1/me2 | Unknown | [124] |
| R63me1 | Unknown | [126] | R35me1 | Unknown | [127] [126] |
| R83me1/me2 | Unknown | [124] | R55me1 | Unknown | [127] [126] |
| R128me1 | Unknown | [126] | R67me1 | Unknown | [127] [126] |
| R134me1 | Unknown | [124] | R92me1 | Unknown | [128] |
| H2A | H2B | ||||
| R3me2a | PRMT6 | [87] | R29me1 | PRMT7 | [125] |
| R3me2s | PRMT5 | [129] | R31me1 | PRMT7 | [125] |
| R3me2s | PRMT7 | [122] | R33me1 | PRMT7 | [125] |
| R11me1 | PRMT1 | [130] | R72me1 | Unknown | [127] |
| R29me2a | PRMT6 | [130] | R79me1 | Unknown | [126] |
| R42me1 | Unknown | [126] | R99me1 | Unknown | [128] |
| R71me1 | Unknown | [124] | |||
| R88me1 | Unknown | [126] | |||
| R96me1 | Unknown | [127] | |||
3. Tudor Domains
The seminal discovery, made by Tony Pawson over twenty years ago, that SRC homology 2 (SH2) domains bind to short protein motifs that are tyrosine phosphorylated [23], led to the realization that different modular domains bind distinct types of PTMs [24]. For example, lysine methylated motifs are bounds by at least eight different domain types – Chromo, PHD, MBT, Tudor, PWWP, Ank, BAH and WD40 domains. Currently, the only protein domain family known to bind methylated arginine motifs is the Tudor family (although individual PHD and WD40 domains also harbor this ability). Tudor domains were identified simultaneously by two research groups, which both realized that the Drosophila melanogaster Tudor protein contains previously unrecognized repeating domains, which were found in a number of other proteins in many different species [25, 26]. Interestingly, the fly Tudor gene was discovered in a genetic screen for maternally expressed genes that result in lethality or sterility of the progeny [27]. The Tudor gene was named after the English Tudor dynasty because of the fertility issues that plagued Henry VIII, who was desperate for a male heir to continue the Tudor line, and whose many wives had repeated stillbirths and miscarriages. Tudor domains are roughly 60 amino acids in size and fold into four antiparallel β-strands. The Tudor domain is the founding member of the ‘Royal Family’ of domains, which also includes Chromo, MBT and PWWP domains [28]. All ‘Royal Family’ domains with methyl-binding properties have an aromatic cage to facilitate the methyl-dependent protein-protein interaction.
4. Tudor Domains Bind Methyllysine and Methylarginine Motifs
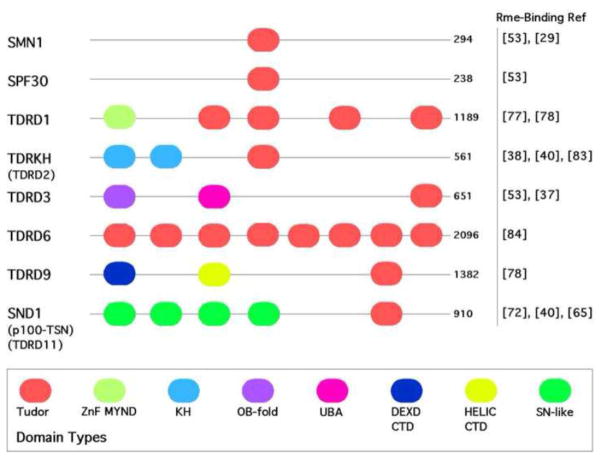
The pioneering work on Tudor domain biochemistry and structure involved studies using the human survival motor neuron (SMN) protein [29, 30], which is mutated in spinal muscular atrophy syndrome [31]. SMN harbors a single Tudor domain and was one of the first proteins identified to interact with a methylated motif [29, 32], along with the chromo domain-containing protein, HP1 [33, 34]. It soon became clear that Tudor domains not only bind methylarginine motifs, but also methyl-lysine marks [35–37]. In humans, there are at least thirty-six proteins that harbor Tudor domains, but there are over 60 Tudor domains in humans, because many proteins have multiple copies of this domain. Recently, the structures of a number of different Tudor domains, in complex with their methylarginine motif ligands, have been solved [38–43]. Based on their primary amino acid sequence, it is not possible to predict the binding specificity of Tudor domains. However, structural studies suggest that the aromatic cage of the methylarginine binders is narrower than that of methyllysine binders. This favors the docking of the planar methyl-guanidinium group of arginine [41]. In the next section, we will discuss only those Tudor domain-containing proteins that have been clearly shown to participate in methylarginine driven protein-protein interactions (Figure 2). These include tudor domain-containing proteins that are implicated in the regulation of splicing (SMN and SPF30), in the regulation of gene expression (TDRD3 and SND1), and in a gonad-specific small RNA silencing pathway (TDRD1, 6 & 9, and TDRKH).
Figure 2. Domain architecture of methylarginine-binding Tudor domain-containing proteins.
Alternative names are given in brackets. The proteins are not drawn to scale. The size of the human Tudor domain-containing protein in given in amino acid number at the end of each stick diagram. The references are given to the original manuscripts that present the methylarginine-binding properties of each particular tudor domain-containing protein. ZnF MYND, MYND-type zinc-finger domain; KH, K homology domain; OB-fold, oligonucleotide/oligosaccharide-binding fold; UBA, ubiquitin-associated domain; DEXD, DEAD-like helicase domain; HELIC, helicase superfamily; SN-like, staphylococcal nuclease-like domain.
5. The Tudor Domains That Bind Methylarginine Motifs
5.1. SMN
Spinal muscular atrophy (SMA) is an autosomal recessive disease resulting from the loss of SMN1 gene function. SMA is among the leading genetic causes of infant death, with a prevalence of ~1 in 6000 live births [44]. SMN regulates the assembly of RNA-protein complexes called small nuclear ribonucleoproteins (snRNPs), and binds the spliceosomal core proteins SmD1, SmD3 and SmB/B′ through its Tudor domain. This binding is driven by symmetric methylation of arginine residues in the C-terminal regions of these splicing factors [29, 32]. SMN is proposed to assemble RNPs along motor neuron axon, but why motor neurons are ultra-sensitive to SMN changes in comparison to other cell types is unknown. Point mutations within the SMN Tudor domain have been identified in SMA patients, establishing a clear link between the methyl-binding requirements of SMN and SMA pathogenesis [45]. The SMN Tudor domain can bind both ADMA and SDMA motifs, and interacts with a number of PRMT5 and CARM1 substrates [46, 47]. These findings are supported by structural studies, which confirm that the Tudor domain of SMN binds both SDMA and ADMA motifs, with a slightly lower affinity for ADMA motifs [41, 43]. It was recently reported that the SMN Tudor domain also has the ability to bind methyllysine marks, in particular the DOT1L-generated H3K27me1 and me2 motifs [48]. If confirmed, this would be the first example of a Tudor domain that can bind both methylarginine and methyllysine motifs.
5.2. SPF30
SPF30, also referred to as SMNrp or SMNDC1, is the Tudor domain most closely related to SMN, displaying almost 50% identity [49]. It was identified in a proteomic screen of the human spliceosome [50], and subsequent functional analysis revealed that it is an essential splicing factor required for spliceosome maturation [51, 52]. Peptide pull-down experiments revealed that the SPF30 Tudor domain binds symmetrically dimethylated arginine motifs [53]. Structural studies have shown that, like the Tudor domain of SMN, SPF30 binds both SDMA and ADMA motifs, with generally a slightly lower affinity for ADMA [41, 43]. Also, when the Tudor domains of SMN, SPF30 and TDRD3 were tested against a panel of methylated peptides, SPF30 displayed the weakest methylarginine binding of the three domains [41].
5.3. TDRD3
The methylarginine binding properties of the TDRD3 Tudor domain were first discovered using a peptide pull-down experiment [53]. A protein microarray approach reaffirmed the ability of Tudor domain of TDRD3 to read methylarginine modified peptides [37]. TDRD3 harbors a functional UBA domain, between its Tudor domain and OB fold [54]. Thus, it could also be directed to specific genomic loci or subcellular sites where particular ubiquitin and arginine methylation signals are enriched.
PRMT1 and CARM1 are the primary transcriptional coactivators in the PRMT enzyme family, and they deposit the H4R3me2a and H3R17me2a marks respectively [55]. Both of these marks are recognized by the Tudor domain of TDRD3 [56]. The Tudor domain of TDRD3 also “reads” a methylarginine mark (R1810) imbedded in the C-terminal domain (CTD) of RNA Polymerase II (RNAP II) [57]. ChIP-seq analysis has revealed that TDRD3 is enriched at the promoters of highly-transcribed genes. TDRD3 has no enzymatic activity of its own, but it is tightly complexed with DNA topoisomerase IIIβ (TOP3B), and TDRD3’s coactivator activity is bestowed on it, in part, by this interaction. TOP3B is a member of the 1A subfamily of DNA topoisomerases, and as such, targets underwound or negatively supercoiled DNA [58]. This subfamily of topoisomerases has been implicated in the resolution of R-loops [59], which are nucleic acid structures formed by a RNA/DNA hybrid and the displacement of the DNA strand. Thus, the TDRD3/TOP3B relaxes negative supercoiled DNA, reduces transcription-generated R-loops, and promotes transcription [60].
Recently, two research groups identified the TDRD3/TOP3B heterodimer as a binding partner for fragile X mental retardation protein (FMRP) [61, 62], thus expanding on the finding that FMRP and its homologs (FXR1 & 2) are complexed with TDRD3 [54]. FMRP is a RNA-binding protein that associates with polyribosomes, and can function as a regulator of translation [63]. Indeed, the TDRD3/TOP3B/FMRP complex is associated with the mRNA pool that is actively undergoing translation [62]. Importantly, TOP3B possesses not only DNA- but also RNA-directed topoisomerase activity [61, 62]. Although, it is not yet clear how this RNA-directed activity is implicated in FMRP function at the ribosomes, it has been proposed to reduce mRNA topological stress, thereby promoting translation [61]. Thus, the TDRD3/TOP3B heterodimer takes on two forms, one nuclear and the other cytoplasmic, and regulates both transcription and translation.
The Tudor domain of TDRD3 preferentially recognizes ADMA marks over SDMA and MMA marks. This was first shown using peptide pull-down approaches with histone H3 tail [56] and the CTD of RNAP II [57], and more recently by Isothermal Titration Calorimetry (ITC) [41]. The crystal structure of the TDRD3 Tudor domain with a methylarginine mimic has facilitated the modeling of methyl-dependent interaction with this particular aromatic cage [41]. An elegant structural analysis of TDRD3 bound to the CTD R1810me2a mark shows that a tyrosine residue in the Tudor domain is critical for the ADMA specificity displayed by this domain. Indeed, when this tyrosine (Y566) is altered to a tryptophan residue, the mutant TDRD3 Tudor domain now binds both the ADMA and SDMA mark [42].
5.4. SND1
SND1, also referred to as TSN-p100 or TDRD11, harbors a single Tudor domain, as well as four tandem repeats of staphylococcal nuclease (SN)-like domains. It was first identified in a protein complex with the Epstein-Bar virus nuclear antigen 2 (EBNA2) [64]. In this context, SND1 functions as a transcriptional coactivator. It has since been found to interact with a number of transcription factors, including STAT6 and E2F1 [65–67]. At least with regards to the E2F1-SND1 interaction, the binding is arginine methylation dependent [65]. SND1 is also able to interact with RNA polymerase II [66], where it associates with the heavily arginine methylated protein SAM68 and regulates alternative splicing [68].
SND1 likely functions as a transcriptional coactivator by also facilitating the acetylation of histone. Studies have shown that the SN domain of SND1 associates with the histone acetyltransferase (HAT), CBP, and recruits it to STAT6 response elements, resulting in an increased histone H4 acetylation [67]. Interestingly, the H4K5ac mark promotes the symmetric methylation of H4R3 [69], and an in vitro study has shown that the extended Tudor domain of SND1 is able to bind methylated H4R3 [40]. Thus, the Tudor domain of SND1 may bind arginine methylation sites on histone tails, then recruit HAT activity, which will acetylate the tails and prime them for arginine methylation by PRMTs. This, in turn, will provide more SND1 docking sites, and promote spreading of the open chromatin state to facilitate transcription.
Apart from transcriptional regulation, SND1 also 1) interacts with the RISC complex and plays a role in miRNA processing [70, 71]; 2) interacts with methylated Sm proteins and facilitates pre-mRNA splicing [72, 73]; and 3) associates with PIWIL1/Miwi in germ cells in a methylarginine-dependent manner [40]. SND1 is thus implicated in many RNA processing steps. It is overexpressed in a number of different cancers, and is a candidate gene for autism susceptibility [68, 74–76].
5.5. TDRD1
Piwi proteins interact with a class of small noncoding RNAs, piwi-interacting RNAs (piRNAs), in germ cells. The piRNA pathway is responsible for repressing transposable elements to prevent genomic instability in germ cells. The mouse Piwi proteins (Mili, Miwi & Miwi2) are all heavily arginine methylated, and can interact with a number of Tudor domain-containing proteins [77, 78]. In the case of TDRD1, its four Tudor domains interact with the N-terminal end of Mili. A SDMA peptide from this region can pull-down TDRD1 from cell lysates [77], and an R/K mutant of Mili is unable to interact with TDRD1 [78]. Furthermore, small molecule inhibitors of arginine methylation block the TDRD1-Mili interaction [78]. Although TDRD1 expression is usually restricted to germline stem cells, it is often overexpressed in human prostate cancer [79, 80]. This inappropriate expression of TDRD1 may impact epigenetic signatures in prostate cancer because of its ability to “read” methylarginine marks. Indeed, it is becoming apparent that the PIWI proteins and PIWI-interacting RNAs have emerging functions in somatic tissues [81].
5.6. TDRKH
TDRKH, also referred to as TDRD2, is the only Tudor domain-containing protein to also harbor KH domains, which bind RNA [82]. Initial RNA-based expression studies showed that TDRKH is present in a number of different tissues (high in brain and heart) [82], and Western analysis identified expression in the testis and brain [38]. The mouse knockout model of TDRKH revealed that it is essential for spermatogenesis and piRNA biogenesis in the germline [83]. However, because of its broad expression pattern, which is supported by data in The Human Protein Atlas, it may have additional non-germ cell functions. Mutational analysis mapped the Tudor domain of TDRKH as the module that interacts with the arginine-rich N-terminal region of Miwi, which represses transposition, regulates translation, and guides epigenetic programming in the germline [38]. PRMT5 knock-down experiments show that the TDRKH-Miwi interaction is arginine methylation dependent [83]. Importantly, for optimal binding to a symmetrically methylarginine peptide a region larger than the Tudor domain is required, which encompasses the Tudor domain and the C-terminal tail of TDRKH [39].
5.7. TDRD6
TDRD6 is composed solely of Tudor domains, and harbors eight of them. It also interacts with Miwi; and SDMA peptides, but not ADMA peptides, are able to pull-down TDRD6 from mouse testis extracts [84]. It is not clear which of the eight Tudor domains harbors the methylarginine-binding properties. Mice null for TDRD6 display defective spermatogenesis [85].
5.8. TDRD9
TDRD9, like TDRD1, is unable to interact with a R/K mutant of Mili [78]. This is supportive of a methyl-dependent TDRD9-Mili interaction, but not direct evidence of such. Although other Tudor domain-containing proteins (TDRD4 & TDRD7) have been shown to interact with Mili, using a co-immunoprecipitation approach, these interactions have yet to be shown to be methyl-dependent [78].
6. Cross Talk and Regulatory Circuits on the Tips of Histone Tails
6.1. The H3R2 Site as a Node of Transcriptional Regulation – PHD and WD40 Domains
The H3K4 site is a major epigenetic mark, which when tri-methylated defines active promoters. H3K4 methylation and effector molecule recognition can be impacted by methylation of H3R2. H3R2me2a is a major mark deposited by PRMT6 [86–88]. Methylation of the H3R2 site essentially prevents the MLL1 complex from methylating H3K4 [87]. However, PRMT6 can strongly methylate H3K4me1 and H3K4me2 peptides, and weakly methylate a H3K4me3 peptide, so dually modified H3R2me2aK4me3 histone tails likely exist [88].
Many effectors that bind H3K4me3 are blocked from docking if H3R2 is also asymmetrically methylated [88]. Thus, PRMT6 functions as a transcriptional repressor by blocking the recruitment of transcriptional activators to the methylated H3K4 mark. ChIP analysis at 185 human promoters supports this hypothesis, demonstrating a counter-correlation between H3K4me3 and H3R2me2a levels [86]. Further ChIP analysis has revealed PRMT6 activity at the promoters of the HoxA2 gene [87] and the TSP-1 gene [89], which correlate with transcriptional repression. Although most H3K4me3 effector molecules are sensitive to H3R2 methylation [88, 90, 91], some are not impacted, either positively or negatively, by this modification [92].
The opposite is true when H3R2 is symmetrically methylated in conjunction with the H3K4me3 mark; certain effector recognition is enhanced. There are two examples of this: 1) the RAG2 PHD domain preferentially binds the H3R2me2sK4me3 dual mark with a 20-fold increase in binding affinity over the H3K4me3 mark alone [93]; and 2) the WDR5 WD40 domain preferentially binds the H3R2me2s mark at least 10-fold better than the unmodified H3 peptide, although it is still unclear how the H3K4me3 mark will impact this interaction [94].
The RAG2 PHD domain was identified as a “reader” of the H3K4me3 mark [95], and structural analysis of this interaction revealed that an additional binding pocket could accommodate a methylated H3R2 residue [96]. At this point, it was unclear if this dual mark could actually exist. To address this issue, antibodies were developed that specifically recognize the dual H3R2me2sK4me3 mark. Strikingly, ChIP-seq analysis reveals that this dual modification tightly correlates with H3K4me3 [93]. Thus, the dual H3R2me2sK4me3 mark is not only present at high levels at recombinationally active antigen receptor loci in pro-B cells (which recruit RAG2), but also found to mark active promoters. PRMT knockdown experiments in mammalian cells have not been able to identify the enzyme responsible for depositing the H3R2me2s mark [93]. Although, in vitro methylation assays suggest that PRMT5 & 7 may be able to deposit this mark [94]. This dual mark is evolutionarily conserved, and in yeast, prior methylation of the H3K4 site is required for H3R2me2s deposition [93].
In an independent study, the Guccione group developed antibodies that recognize the H3R2me2s mark, and ChIP-seq experiments using these antibodies also revealed significant overlap of this mark with H3K4me3 at active promoters [94]. They found that WDR5 is recruited to the H3R2me2s mark and structural analysis reveals that the symmetric methylation of the R2 residue facilitates the formation of tighter hydrophobic interactions with a phenylalanine residue (F219) present in the WD40 domain of WDR5.
It has been proposed that apart from providing high-affinity docking sites for certain domains (the WDR5 WD40 domain and the RAG2 PHD domain), the H3R2me2sK4me3 dual mark may prevent the demethylation of H3K4me3, thus fixing this activation mark in place for long periods [93].
6.2. The H4R3me2s Mark as a Potential Gateway to DNA Methylation – the ADD Domain
PRMT5 symmetrically methylates both H3R8 and the H4R3 motif [97], but the specificity of the enzyme is directed towards the H4R3 motif by one of its regulatory binding proteins, COPR5 [98]. PRMT5 is generally regarded as a transcriptional repressor. Thus, an effector of the H4R3me2s mark should possess repressor functions that can link PRMT5 to its clearly defined role as an attenuator of transcription. One such link was discovered with the identification of the PHD domain of the de novo DNA methyltransferase DNMT3A as a binder of the H4R3me2s mark [99]. A repressive mechanism can now be envisioned where PRMT5 is recruited to a promoter, generating a patch of H4R3me2s that in turn will facilitate the binding of DNMT3A, thereby stimulating DNA methylation and transcriptional lock-down. This sequence of events has been challenged by Otani et al., who confirmed previous reports that the PHD domain (also called the ADD domain) of DNMT3A binds H3K4me0 [100], but were not able to reproduce the interaction between this PHD and a H4R3me2s peptide [101]. A recent ChIP-seq analysis of H4R3me2s peaks found that they are enriched at CpG-rich promoters [102]. However, this promoter marking is independent of transcriptional activity, and furthermore, PRMT5-depletion resulted in loss of H4R3me2s, but not DNA methylation or other repressive marks. Thus, the link between PRMT5 and DNA methylation remains elusive.
6.3 Trans-tail Cross Talk between H4R3me2s and H3K3me3 Marks – PHD Domains
Mixed-lineage leukemia 4 (MLL4) is a transcriptional coactivator that functions by depositing the H3K4me3 mark. Three of the seven PHD domains (PHD4-6) of MLL4 are prevented from binding the N-terminal tail of histone H4 by symmetrical methylation at the R3 position [103]. Peptides carrying the H4R3me2a mark (and unmethylated peptides) interact robustly with these three PHD domains. This raised the possibility that MLL4 interacts with the histone H4 N-terminal tail when it does not contain a repressive mark, and methylated the histone H3 N-terminal tail on the same nucleosome, or on an adjacent nucleosome. This indeed seems be the case, because MLL4-mediated methylation of nucleosomal H3 tails requires the binding of PHD4-6 to H4 tails. These findings nicely link the known transcriptional repressive role of the H4R3me2s mark with reduced MLL4 chromatin recruitment and decreased promoter-specific levels of H3K4me3.
7. Other Domains and Complexes that Bind Methylarginine Marks
7.1. The PAF1 Complex
The transcriptional coactivator, CARM1, deposits the H3R17me2a mark. Peptides bearing this mark were used to pull-down interacting proteins from HeLa nuclear extract [104]. The methyl-dependent interacting proteins were identified by mass spectrometry, and they turned out to be members of the transcription elongation-associated PAF1 complex. Consistent with this finding, loss of CARM1 results in a reduction of the PAF complex at estrogen-response elements. Unfortunately, it is not clear which component of the PAF complex directly interacts with the H3R17me2a mark, or if a protein domain is involved in this interaction. Interestingly, using a similar peptide pull-down approach, the Wong group found that H3R17me2a mark inhibits the binding of corepressors and protects chromatin from deacetylation [105].
7.2. BRCT Domains
The histone acetyltransferase, p300, is methylated by CARM1 at a number of sites, including R754 in the KIX region [106]. Pull-down experiments using R754 MMA and ADMA peptides revealed that the BRCT domains of BRCA1 interact with the ADMA form of this peptide. BRCT domains are well characterized as phospho-binding modules [107], and in this particular case a few of these domains may have evolved the ability to also interact with a methyl-motifs. This interaction between p300R754me2a and the BRCA1 BRCT domain seems fairly unique, because an expanded screen with a number of methylarginine motifs did not identify additional BRCT domains with methylarginine-binding properties.
7.3. Long Non-Coding RNA (lncRNA) Regulated Protein Interaction
Nuclear lncRNAs have been shown to function as transcriptional regulators that can bind to chromatin-associated proteins [108]. The Chromo domain of the Polycomb 2 protein (Pc2/Cbx4) binds the H3K9me3 mark [109]. Recently, Pc2 was found to bind the lncRNAs TUG1 and MALAT1/NEAT2 [110]. Using a modified histone peptide array, the Rosenfeld group was able to show that the recombinant Pc2 Chromo domain bound H3K9me3, as expected. However, when the Pc2 Chromo domain was used to probe the arrays in the presence of TUG1, then it switched specificity to the H4R3me2s mark. This is a very exciting observation that has yet to be followed up on, and it implies that a Chromo domain can switch from a methyllysine to methylarginine binder in the presence of a lncRNA.
7.4. Small Nuclear RNA (snRNA) as a Direct Effector for Methylarginine Marks
7SK is an abundant snRNA that binds and regulates P-TEFb, a factor that regulates the elongation phase of transcription. Again, the Rosenfeld group found, using a modified histone peptide array, that 7SK snRNA binds lysine acetylated and arginine methylated histone tail peptides [6]. In vitro RNA pull-down assays show that 7SK snRNA interacts with H4R3me2a/s marks but not H3R2me2a/s marks. Thus, 7SK snRNA can bind preferentially to some arginine methylated motifs, but also strongly to the acetylated histone H4 tail.
7.5. PELP1
Proline glutamic acid and leucine-rich protein 1 (PELP1) is as estrogen receptor coactivator, which recruits LSD1 to demethylate the H3K9me2 mark [111]. PELP1 also interacts with CARM1 [112]. Interestingly, PELP1 has the ability to bind both H3K4me2 and H3K9me2 peptides, although it does not harbor a classic methyl-reading domain. The interaction with these methyl-peptides was mapped to a glutamic acid-rich region of PELP1. Histone peptide array experiments revealed that PELP1 also interacts with methylarginine peptides that are asymmetrically dimethylated [112]. Thus, PELP1 seems to be a methyl-dependent histone tail binder, but it is unable to distinguish between dimethylated arginine and lysine motifs.
8. Small Molecule Tools to Investigate Methyl-dependent Protein-Protein interactions
8.1. Chemical Compounds That Mimic an Aromatic Cage
Structural analysis has made it abundantly clear that most the domains that “read” methylarginine marks, do so by using a “cage” in which four faces consist of aromatic residues. An exciting new avenue of research is the synthesis of artificial aromatic cages that function as receptors and recognize methylarginine marks. The Waters group has synthesized these small chemical probes that mimic the aromatic cages observed in protein structure [113]. One of these synthetic receptors (A2D) exhibits a binding affinity of 5 μM for H3R8me2a, with a greater than 7-fold selectivity over H3R8me2s and the unmodified peptide, thus displaying tighter binding than most Tudor domains. Once tagged, these compounds could be used as a probe that could detect and enrich for PRMT substrates.
8.2. Chemical Compounds That Bind an Aromatic Cage
With the first well-characterized inhibitors of Bromo domains (JQ1 and I-BET) already in clinical trials [114], the targeting of protein–protein interactions triggered by PTMs has been validated as tractable, proving that other families of “reader” proteins may be amenable to selective small-molecule intervention as well. Indeed, small molecules that bind MBT and Tudor domains have recently been identified by the Frye group [115–117]. One of the lead compounds, UNC1215, can bind the MBT domains of L3MBTL3 and L3MBTL1, and also the Tudor domains of 53BP1 and PHF20. All four of the domains bound by UNC1215 are methyllysine binding aromatic cages, but it is conceivable that analogs of this compound could bind Tudor domains that interact with methylarginine motifs. Thus, inhibitors of methylarginine-dependent protein-protein interactions may one day be developed into epigenetic drugs.
8.3. Chemical Conjugation of Methylarginine Mimics on Recombinant Proteins
Methyllysine mimics have been successfully engineered into recombinant proteins for a number of years, and these methyllysine analogs (MLAs) are functionally similar to their natural counterparts [118]. Recently, a similar approach was developed by the Fujimori group to install methylarginine analogs into specific sites in a recombinant protein [119]. When introduced into the H4R3 site, these methylarginine analogs are specifically recognized, just like their natural counterparts, by methyl-specific antibodies and the TDRD3 effector molecule. This approach allows for the production of large quantities of recombinant protein with a specified site and type of arginine methylation, and will greatly facilitate the functional analysis of arginine methylation.
9. Perspectives
It is probable that we have identified all the arginine methyltransferases within the “classic” PRMT family. It is possible that convergent evolution has generated other classes of arginine methyltransferases, as it has for lysine methyltransferases and demethylases. Also, the search for additional “readers” of arginine methylated motifs will be important to help us understand the mechanisms of action of this PTM. Because of the abundance of this modification, it is likely that there will be “readers” other than Tudor domains. Mass spectrometric analysis has become extremely sensitive, and as a result a large number of arginine methylation sites have been identified on histones and their tails. It is unclear which are these methylation sites play significant biological functions. Comprehensive ChIP-seq analysis of all the arginine methylated histone code marks has yet to be performed, due to the lack of high-quality antibodies, and this information will obviously be of great value in the future.
Highlights.
0.5% of all arginine residues in the cell are methylated.
There are three types of arginine methylation: mono, asymmetric & symmetric.
The primary “readers” of methylarginine marks are Tudor domain-containing proteins.
Acknowledgments
Mark T. Bedford is supported by CPRIT funding (RP110471) and a NIH grant (DK062248). Sitaram Gayatri is an Epigenetic Scholar and is supported by the Center for Cancer Epigenetics (CCE) at MD Anderson Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275:16030–16036. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- 3.Hughes RM, Waters ML. Arginine methylation in a beta-hairpin peptide: implications for Arg-pi interactions, DeltaCp(o), and the cold denatured state. J Am Chem Soc. 2006;128:12735–12742. doi: 10.1021/ja061656g. [DOI] [PubMed] [Google Scholar]
- 4.Paik WK, Kim S. Natural occurrence of various methylated amino acid derivatives. John Wiley & sons; New York: 1980. [Google Scholar]
- 5.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 6.Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. Brd4 and JMJD6-Associated Anti-Pause Enhancers in Regulation of Transcriptional Pause Release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhar S, Vemulapalli V, Patananan AN, Huang GL, Di Lorenzo A, Richard S, Comb MJ, Guo A, Clarke SG, Bedford MT. Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci Rep. 2013;3:1311. doi: 10.1038/srep01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, Han B, Jungheim LN, Qian Y, Rauch C, Russell M, Sauder JM, Wasserman SR, Weichert K, Willard FS, Zhang A, Emtage S. Crystal structure of the human PRMT5:MEP50 complex. Proc Natl Acad Sci U S A. 2012;109:17960–17965. doi: 10.1073/pnas.1209814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho MC, Wilczek C, Bonanno JB, Xing L, Seznec J, Matsui T, Carter LG, Onikubo T, Kumar PR, Chan MK, Brenowitz M, Cheng RH, Reimer U, Almo SC, Shechter D. Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PLoS One. 2013;8:e57008. doi: 10.1371/journal.pone.0057008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun L, Wang M, Lv Z, Yang N, Liu Y, Bao S, Gong W, Xu RM. Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc Natl Acad Sci U S A. 2011;108:20538–20543. doi: 10.1073/pnas.1106946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J. Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. Embo J. 2007;26:4391–4401. doi: 10.1038/sj.emboj.7601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. Embo J. 2007;26:4402–4412. doi: 10.1038/sj.emboj.7601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Cheng X. Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of Its Binding to Substrate Peptides. Structure (Camb) 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Zhou L, Cheng X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. Embo J. 2000;19:3509–3519. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 16.Matsuoka M. Epsilon-N-methylated lysine and guanidine-N-methylated arginine of proteins. 3. Presence and distribution in nature and mammals. Seikagaku. 1972;44:364–370. [PubMed] [Google Scholar]
- 17.Boffa LC, Karn J, Vidali G, Allfrey VG. Distribution of NG, NG,-dimethylarginine in nuclear protein fractions. Biochem Biophys Res Commun. 1977;74:969–976. doi: 10.1016/0006-291x(77)91613-8. [DOI] [PubMed] [Google Scholar]
- 18.Bremang M, Cuomo A, Agresta AM, Stugiewicz M, Spadotto V, Bonaldi T. Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Mol Biosyst. 2013;9:2231–2247. doi: 10.1039/c3mb00009e. [DOI] [PubMed] [Google Scholar]
- 19.Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, Yang V, Aguiar M, Kornhauser J, Jia X, Ren J, Beausoleil SA, Silva JC, Vemulapalli V, Bedford MT, Comb MJ. Immunoaffinity Enrichment and Mass Spectrometry Analysis of Protein Methylation. Mol Cell Proteomics. 2013 doi: 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlmann T, Geoghegan VL, Thomas B, Ridlova G, Trudgian DC, Acuto O. A method for large-scale identification of protein arginine methylation. Mol Cell Proteomics. 2012;11:1489–1499. doi: 10.1074/mcp.M112.020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585:2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casadio F, Lu X, Pollock SB, Leroy G, Garcia BA, Muir TW, Roeder RG, Allis CD. H3R42me2a is a histone modification with positive transcriptional effects. Proc Natl Acad Sci U S A. 2013;110:14894–14899. doi: 10.1073/pnas.1312925110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson D, Koch CA, Grey L, Ellis C, Moran MF, Pawson T. Binding of SH2 domains of phospholipase C gamma 1, GAP, and Src to activated growth factor receptors. Science. 1990;250:979–982. doi: 10.1126/science.2173144. [DOI] [PubMed] [Google Scholar]
- 24.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 25.Ponting CP. Tudor domains in proteins that interact with RNA. Trends Biochem Sci. 1997;22:51–52. doi: 10.1016/s0968-0004(96)30049-2. [DOI] [PubMed] [Google Scholar]
- 26.Callebaut I, Mornon JP. The human EBNA-2 coactivator p100: multidomain organization and relationship to the staphylococcal nuclease fold and to the tudor protein involved in Drosophila melanogaster development. Biochem J. 1997;321(Pt 1):125–132. doi: 10.1042/bj3210125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boswell RE, Mahowald AP. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 28.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 29.Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- 30.Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 31.Kolb SJ, Kissel JT. Spinal muscular atrophy: a timely review. Arch Neurol. 2011;68:979–984. doi: 10.1001/archneurol.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. Rna. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 34.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 35.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 37.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, Tenaglia E, Xu C, Gish G, Min J, Pawson T. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci U S A. 2009;106:20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 2010;24:1876–1881. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu K, Chen C, Guo Y, Lam R, Bian C, Xu C, Zhao DY, Jin J, MacKenzie F, Pawson T, Min J. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc Natl Acad Sci U S A. 2010;107:18398–18403. doi: 10.1073/pnas.1013106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu K, Guo Y, Liu H, Bian C, Lam R, Liu Y, Mackenzie F, Rojas LA, Reinberg D, Bedford MT, Xu RM, Min J. Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PLoS One. 2012;7:e30375. doi: 10.1371/journal.pone.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sikorsky T, Hobor F, Krizanova E, Pasulka J, Kubicek K, Stefl R. Recognition of asymmetrically dimethylated arginine by TDRD3. Nucleic Acids Res. 2012;40:11748–11755. doi: 10.1093/nar/gks929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM, Sattler M. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol. 2011;18:1414–1420. doi: 10.1038/nsmb.2185. [DOI] [PubMed] [Google Scholar]
- 44.Brzustowicz LM, Lehner T, Castilla LH, Penchaszadeh GK, Wilhelmsen KC, Daniels R, Davies KE, Leppert M, Ziter F, Wood D, et al. Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2-13.3. Nature. 1990;344:540–541. doi: 10.1038/344540a0. [DOI] [PubMed] [Google Scholar]
- 45.Cusco I, Barcelo MJ, del Rio E, Baiget M, Tizzano EF. Detection of novel mutations in the SMN Tudor domain in type I SMA patients. Neurology. 2004;63:146–149. doi: 10.1212/01.wnl.0000132634.48815.13. [DOI] [PubMed] [Google Scholar]
- 46.Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 47.Tadesse H, Deschenes-Furry J, Boisvenue S, Cote J. KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum Mol Genet. 2008;17:506–524. doi: 10.1093/hmg/ddm327. [DOI] [PubMed] [Google Scholar]
- 48.Sabra M, Texier P, El Maalouf J, Lomonte P. The Tudor protein survival motor neuron (SMN) is a chromatin-binding protein that interacts with methylated lysine 79 of histone H3. J Cell Sci. 2013;126:3664–3677. doi: 10.1242/jcs.126003. [DOI] [PubMed] [Google Scholar]
- 49.Talbot K, Miguel-Aliaga I, Mohaghegh P, Ponting CP, Davies KE. Characterization of a gene encoding survival motor neuron (SMN)-related protein, a constituent of the spliceosome complex. Hum Mol Genet. 1998;7:2149–2156. doi: 10.1093/hmg/7.13.2149. [DOI] [PubMed] [Google Scholar]
- 50.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 51.Rappsilber J, Ajuh P, Lamond AI, Mann M. SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome. J Biol Chem. 2001;276:31142–31150. doi: 10.1074/jbc.M103620200. [DOI] [PubMed] [Google Scholar]
- 52.Meister G, Hannus S, Plottner O, Baars T, Hartmann E, Fakan S, Laggerbauer B, Fischer U. SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J. 2001;20:2304–2314. doi: 10.1093/emboj/20.9.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 54.Linder B, Plottner O, Kroiss M, Hartmann E, Laggerbauer B, Meister G, Keidel E, Fischer U. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum Mol Genet. 2008;17:3236–3246. doi: 10.1093/hmg/ddn219. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 56.Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, Bedford MT. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40:1016–1023. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sims RJ, 3rd, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ, 3rd, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 59.Wilson-Sali T, Hsieh TS. Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIbeta. Proc Natl Acad Sci U S A. 2002;99:7974–7979. doi: 10.1073/pnas.122007999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y, MKM, Hensley S, Lu Y, Chedin F, Bedford MT. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu D, Shen W, Guo R, Xue Y, Peng W, Sima J, Yang J, Sharov A, Srikantan S, Fox D, 3rd, Qian Y, Martindale JL, Piao Y, Machamer J, Joshi SR, Mohanty S, Shaw AC, Lloyd TE, Brown GW, Ko MS, Gorospe M, Zou S, Wang W. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat Neurosci. 2013 doi: 10.1038/nn.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stoll G, Pietilainen OP, Linder B, Suvisaari J, Brosi C, Hennah W, Leppa V, Torniainen M, Ripatti S, Ala-Mello S, Plottner O, Rehnstrom K, Tuulio-Henriksson A, Varilo T, Tallila J, Kristiansson K, Isohanni M, Kaprio J, Eriksson JG, Raitakari OT, Lehtimaki T, Jarvelin MR, Salomaa V, Hurles M, Stefansson H, Peltonen L, Sullivan PF, Paunio T, Lonnqvist J, Daly MJ, Fischer U, Freimer NB, Palotie A. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat Neurosci. 2013 doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 64.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng S, Moehlenbrink J, Lu YC, Zalmas LP, Sagum CA, Carr S, McGouran JF, Alexander L, Fedorov O, Munro S, Kessler B, Bedford MT, Yu Q, La Thangue NB. Arginine Methylation-Dependent Reader-Writer Interplay Governs Growth Control by E2F-1. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang J, Aittomaki S, Pesu M, Carter K, Saarinen J, Kalkkinen N, Kieff E, Silvennoinen O. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 2002;21:4950–4958. doi: 10.1093/emboj/cdf463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valineva T, Yang J, Palovuori R, Silvennoinen O. The transcriptional co-activator protein p100 recruits histone acetyltransferase activity to STAT6 and mediates interaction between the CREB-binding protein and STAT6. J Biol Chem. 2005;280:14989–14996. doi: 10.1074/jbc.M410465200. [DOI] [PubMed] [Google Scholar]
- 68.Cappellari M, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Saarikettu J, Silvennoinen O, Sette C. The transcriptional co-activator SND1 is a novel regulator of alternative splicing in prostate cancer cells. Oncogene. 2013 doi: 10.1038/onc.2013.360. [DOI] [PubMed] [Google Scholar]
- 69.Feng Y, Wang J, Asher S, Hoang L, Guardiani C, Ivanov I, Zheng YG. Histone H4 acetylation differentially modulates arginine methylation by an in Cis mechanism. J Biol Chem. 2011;286:20323–20334. doi: 10.1074/jbc.M110.207258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li CL, Yang WZ, Chen YP, Yuan HS. Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res. 2008;36:3579–3589. doi: 10.1093/nar/gkn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- 72.Gao X, Zhao X, Zhu Y, He J, Shao J, Su C, Zhang Y, Zhang W, Saarikettu J, Silvennoinen O, Yao Z, Yang J. Tudor staphylococcal nuclease (Tudor-SN) participates in small ribonucleoprotein (snRNP) assembly via interacting with symmetrically dimethylated Sm proteins. J Biol Chem. 2012;287:18130–18141. doi: 10.1074/jbc.M111.311852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J, Valineva T, Hong J, Bu T, Yao Z, Jensen ON, Frilander MJ, Silvennoinen O. Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Res. 2007;35:4485–4494. doi: 10.1093/nar/gkm470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holt R, Barnby G, Maestrini E, Bacchelli E, Brocklebank D, Sousa I, Mulder EJ, Kantojarvi K, Jarvela I, Klauck SM, Poustka F, Bailey AJ, Monaco AP. Linkage and candidate gene studies of autism spectrum disorders in European populations. Eur J Hum Genet. 2010;18:1013–1019. doi: 10.1038/ejhg.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blanco MA, Aleckovic M, Hua Y, Li T, Wei Y, Xu Z, Cristea IM, Kang Y. Identification of staphylococcal nuclease domain-containing 1 (SND1) as a Metadherin-interacting protein with metastasis-promoting functions. J Biol Chem. 2011;286:19982–19992. doi: 10.1074/jbc.M111.240077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsuchiya N, Ochiai M, Nakashima K, Ubagai T, Sugimura T, Nakagama H. SND1, a component of RNA-induced silencing complex, is up-regulated in human colon cancers and implicated in early stage colon carcinogenesis. Cancer Res. 2007;67:9568–9576. doi: 10.1158/0008-5472.CAN-06-2707. [DOI] [PubMed] [Google Scholar]
- 77.Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 78.Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kacprzyk LA, Laible M, Andrasiuk T, Brase JC, Borno ST, Falth M, Kuner R, Lehrach H, Schweiger MR, Sultmann H. ERG induces epigenetic activation of Tudor domain-containing protein 1 (TDRD1) in ERG rearrangement-positive prostate cancer. PLoS One. 2013;8:e59976. doi: 10.1371/journal.pone.0059976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boormans JL, Korsten H, Ziel-van der Made AJ, van Leenders GJ, de Vos CV, Jenster G, Trapman J. Identification of TDRD1 as a direct target gene of ERG in primary prostate cancer. Int J Cancer. 2013;133:335–345. doi: 10.1002/ijc.28025. [DOI] [PubMed] [Google Scholar]
- 81.Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353–359. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lamb FS, Barna TJ, Goud C, Marenholz I, Mischke D, Schutte BC. Complex RNA processing of TDRKH, a novel gene encoding the putative RNA-binding tudor and KH domains. Gene. 2000;246:209–218. doi: 10.1016/s0378-1119(00)00087-1. [DOI] [PubMed] [Google Scholar]
- 83.Saxe JP, Chen M, Zhao H, Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32:1869–1885. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA. 2010;16:70–78. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol. 2009;19:630–639. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 87.Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. Arginine methylation of the histone h3 tail impedes effector binding. J Biol Chem. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 89.Michaud-Levesque J, Richard S. Thrombospondin-1 is a transcriptional repression target of PRMT6. J Biol Chem. 2009;284:21338–21346. doi: 10.1074/jbc.M109.005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qiu Y, Liu L, Zhao C, Han C, Li F, Zhang J, Wang Y, Li G, Mei Y, Wu M, Wu J, Shi Y. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 2012;26:1376–1391. doi: 10.1101/gad.188359.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, Shi YG, Xu Y, Patel DJ, Shi Y. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fiedler M, Sanchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Muller J, Evans P, Bienz M. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell. 2008;30:507–518. doi: 10.1016/j.molcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yuan CC, Matthews AG, Jin Y, Chen CF, Chapman BA, Ohsumi TK, Glass KC, Kutateladze TG, Borowsky ML, Struhl K, Oettinger MA. Histone H3R2 symmetric dimethylation and histone H3K4 trimethylation are tightly correlated in eukaryotic genomes. Cell Rep. 2012;1:83–90. doi: 10.1016/j.celrep.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Migliori V, Muller J, Phalke S, Low D, Bezzi M, Mok WC, Sahu SK, Gunaratne J, Capasso P, Bassi C, Cecatiello V, De Marco A, Blackstock W, Kuznetsov V, Amati B, Mapelli M, Guccione E. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19:136–144. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 95.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007 doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lacroix M, Messaoudi SE, Rodier G, Le Cam A, Sardet C, Fabbrizio E. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 2008;9:452–458. doi: 10.1038/embor.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Girardot M, Hirasawa R, Kacem S, Fritsch L, Pontis J, Kota SK, Filipponi D, Fabbrizio E, Sardet C, Lohmann F, Kadam S, Ait-Si-Ali S, Feil R. PRMT5-mediated histone H4 arginine-3 symmetrical dimethylation marks chromatin at G + C-rich regions of the mouse genome. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dhar SS, Lee SH, Kan PY, Voigt P, Ma L, Shi X, Reinberg D, Lee MG. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes & development. 2012;26:2749–2762. doi: 10.1101/gad.203356.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu J, Xu W. Histone H3R17me2a mark recruits human RNA polymerase-associated factor 1 complex to activate transcription. Proc Natl Acad Sci U S A. 2012;109:5675–5680. doi: 10.1073/pnas.1114905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu J, Cui N, Wang R, Li J, Wong J. A role for CARM1-mediated histone H3 arginine methylation in protecting histone acetylation by releasing corepressors from chromatin. PLoS One. 2012;7:e34692. doi: 10.1371/journal.pone.0034692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee YH, Bedford MT, Stallcup MR. Regulated recruitment of tumor suppressor BRCA1 to the p21 gene by coactivator methylation. Genes Dev. 2011;25:176–188. doi: 10.1101/gad.1975811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 108.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nair SS, Nair BC, Cortez V, Chakravarty D, Metzger E, Schule R, Brann DW, Tekmal RR, Vadlamudi RK. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-alpha target gene activation by regulating lysine demethylase 1 specificity. EMBO reports. 2010;11:438–444. doi: 10.1038/embor.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mann M, Cortez V, Vadlamudi R. PELP1 oncogenic functions involve CARM1 regulation. Carcinogenesis. 2013;34:1468–1475. doi: 10.1093/carcin/bgt091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.James LI, Beaver JE, Rice NW, Waters ML. A synthetic receptor for asymmetric dimethyl arginine. J Am Chem Soc. 2013;135:6450–6455. doi: 10.1021/ja307907p. [DOI] [PubMed] [Google Scholar]
- 114.Garnier JM, Sharp PP, Burns CJ. BET bromodomain inhibitors: a patent review. Expert opinion on therapeutic patents. 2014;24:185–199. doi: 10.1517/13543776.2014.859244. [DOI] [PubMed] [Google Scholar]
- 115.James LI, Barsyte-Lovejoy D, Zhong N, Krichevsky L, Korboukh VK, Herold JM, MacNevin CJ, Norris JL, Sagum CA, Tempel W, Marcon E, Guo H, Gao C, Huang XP, Duan S, Emili A, Greenblatt JF, Kireev DB, Jin J, Janzen WP, Brown PJ, Bedford MT, Arrowsmith CH, Frye SV. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nature chemical biology. 2013;9:184–191. doi: 10.1038/nchembio.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.James LI, Korboukh VK, Krichevsky L, Baughman BM, Herold JM, Norris JL, Jin J, Kireev DB, Janzen WP, Arrowsmith CH, Frye SV. Small-molecule ligands of methyl-lysine binding proteins: optimization of selectivity for L3MBTL3. Journal of medicinal chemistry. 2013;56:7358–7371. doi: 10.1021/jm400919p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kireev D, Wigle TJ, Norris-Drouin J, Herold JM, Janzen WP, Frye SV. Identification of non-peptide malignant brain tumor (MBT) repeat antagonists by virtual screening of commercially available compounds. Journal of medicinal chemistry. 2010;53:7625–7631. doi: 10.1021/jm1007374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Le DD, Cortesi AT, Myers SA, Burlingame AL, Fujimori DG. Site-specific and regiospecific installation of methylarginine analogues into recombinant histones and insights into effector protein binding. Journal of the American Chemical Society. 2013;135:2879–2882. doi: 10.1021/ja3108214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 121.Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS. beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev Cell. 2010;19:220–231. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Karkhanis V, Wang L, Tae S, Hu YJ, Imbalzano AN, Sif S. Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase delta catalytic subunit gene, POLD1. The Journal of biological chemistry. 2012;287:29801–29814. doi: 10.1074/jbc.M112.378281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- 124.Tweedie-Cullen RY, Brunner AM, Grossmann J, Mohanna S, Sichau D, Nanni P, Panse C, Mansuy IM. Identification of combinatorial patterns of post-translational modifications on individual histones in the mouse brain. PLoS One. 2012;7:e36980. doi: 10.1371/journal.pone.0036980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Feng Y, Maity R, Whitelegge JP, Hadjikyriacou A, Zurita-Lopez C, Al-Hadid Q, Thompson P, Richard S, Bedford MT, Masson J-Y, Clarke SG. Mammalian Protein Arginine Methyltransferase 7 (PRMT7) Specifically Targets Histone H2B at R-X-R sites in the N-terminal Basic Region. JBC. 2013 doi: 10.1074/jbc.M113.525345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Montellier E, Boussouar F, Rousseaux S, Zhang K, Buchou T, Fenaille F, Shiota H, Debernardi A, Hery P, Curtet S, Jamshidikia M, Barral S, Holota H, Bergon A, Lopez F, Guardiola P, Pernet K, Imbert J, Petosa C, Tan M, Zhao Y, Gerard M, Khochbin S. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes & development. 2013;27:1680–1692. doi: 10.1101/gad.220095.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang L, Eugeni EE, Parthun MR, Freitas MA. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma. 2003;112:77–86. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
- 129.Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 130.Waldmann T, Izzo A, Kamieniarz K, Richter F, Vogler C, Sarg B, Lindner H, Young NL, Mittler G, Garcia BA, Schneider R. Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin. 2011;4:11. doi: 10.1186/1756-8935-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]