Abstract
Machine learning (ML)-based segmentation methods are a common technique in the medical image processing field. In spite of numerous research groups that have investigated ML-based segmentation frameworks, there remains unanswered aspects of performance variability for the choice of two key components: ML-algorithm and intensity normalization. This investigation reveals that the choice of those elements plays a major part in determining segmentation accuracy and generalizability. The approach we have used in this study aims to evaluate relative benefits of the two elements within a subcortical MRI segmentation framework. Experiments were conducted to contrast eight machine-learning algorithm configurations and 11 normalization strategies for our brain MR segmentation framework. For the intensity normalization, a stable atlas-based mapped prior (STAMP) was utilized to take better account of contrast along boundaries of structures. Comparing eight machine learning algorithms on down-sampled segmentation MR data, it was obvious that a significant improvement was obtained using ensemble-based ML algorithms (i.e., random forest) or ANN algorithms. Further investigation between these two algorithms also revealed that the random forest results provided exceptionally good agreement with manual delineations by experts. Additional experiments showed that the effect of STAMP-based intensity normalization also improved the robustness of segmentation for multicenter data sets. The constructed framework obtained good multicenter reliability and was successfully applied on a large multicenter MR data set (n > 3000). Less than 10% of automated segmentations were recommended for minimal expert intervention. These results demonstrate the feasibility of using the ML-based segmentation tools for processing large amount of multicenter MR images. We demonstrated dramatically different result profiles in segmentation accuracy according to the choice of ML algorithm and intensity normalization chosen.
Keywords: Segmentation, Machine-Learning, Random Forest, Multi-site Study
1. Introduction
The precise delineation of subcortical structures from brain structural magnetic resonance images (MRIs) can advance our understanding of the disease process for many neurological and psychological diseases. The rich set of soft tissue information from an MRI [1] allows for sensitive quantitative studies that can detect subtle morphological changes associated with neurological disease progression. It is known that morphological trajectories of neuroanatomy in normal aging differs from those affected by disorders such as schizophrenia [2, 3], Alzheimer’s disease [4, 5], autism [6], Huntington’s disease [7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17], and others [18]. The desire to accelerate our understanding of these disease progressions has recently lead to several studies that collect extremely large data sets from multiple collection sites [10, 19, 20].
Although manual delineation remains a common practice for smaller studies, analysis of large longitudinal MRI data sets is impractical due to its high labor costs and low intra- and inter-rater consistency. A robust automated segmentation tool can overcome these hurdles and provide efficient and reliable segmentations in very large longitudinal multicenter data collections. Several approaches have been investigated to provide an automated segmentation procedure [21, 22, 17, 23, 24, 25, 26, 27, 28, 29, 30]. Among the various available methodologies, a popular approach is machine learning (ML)-based segmentation. Different ML algorithms have appeared in segmentation frameworks: support vector machine (SVM) [31, 32, 33, 34, 35], AdaBoost [35], k-nearest neighbor (k-NN) [36, 37], and artificial neural network (ANN) [38, 39] to name a few.
Two fundamental elements for providing consistent segmentation for multi-site data are considered in this paper: 1) choice of ML algorithms and 2) intensity normalization strategies. This study systematically compares and contrasts several of these elements to explore which characteristics are most important for providing consistent segmentation results across sites.
A few pair-wise studies have contrasted ML algorithms with each other for brain MRI segmentation: k-NN and semi-supervised fuzzy c-means [40], SVM and AdaBoost [41], FMRIB automated segmentation tool (FAST), statistical parametric mapping (SPM5), and two different k-NNs [42]. These studies have focused on tissue or lesion segmentation, the following two studies focus specifically on volumetric segmentation: SVM and ANN showing compatible results [38], and preferred AdaBoost and Ada-SVM to manual and FreeSurfer in [35].
We explore the performance of various intensity normalization strategies with regards to the performance of ML-algorithms for large-scale MRI processing. A better understanding of the ML and intensity normalization provides insights into performing robust and consistent segmentations for MRI data sets collected across different sites and scanning hardware.
To understand the contributions of each algorithmic component choice in providing generalizable robust segmentation results, we have contrasted eight ML algorithms and 11 normalization strategies. We found that the choice of both ML algorithm and normalization can vastly affect the accuracy and generalizability of volumetric segmentation results. This paper aims to provide a better understanding of the ML algorithm characteristics and normalization choices that affect the overall segmentation quality, and may guide future designs. Additionally, we conducted a multicenter reliability study to determine an estimated sample size needed to detect 5% to 10% volume changes from data collected in multicenter study setting. This empirical and theoretical comparison of eight ML algorithms and 11 normalization strategies results in a robust segmentation framework offering reliable quantitative measurements from large-scale multicenter data. Finally, we apply the best performing combination of choices from the testing framework to a 32 site 3000 scan session MR data set.
2. Material and Method
We aim to investigate the performance of several ML algorithms and normalization approaches for brain MRI subcortical segmentation. First we describe the experimental data sets to be used and then describe the segmentation framework used for comparisons. Brief overviews of ML algorithms and normalization strategies used in this comparative study are presented.
2.1. Data
Three groups of experimental data are utilized in this report: (1) a complete large-scale multicenter data set, (2) a representative sub-sample of the large-scale multicenter data, and (3) a traveling human phantom (THP) data set where the same individuals were scanned at eight different centers. For all three in-vivo data sets, the framework generates feature vectors as described in [38, 39], where each feature vector F being given as:
| (1) |
where ρs, ϕs, and θs constitute the symmetrical spherical coordinate information, Gi,img is a uniform neighborhood sampling of image intensity along the gradient-descent direction of a deformed prior at the given image location i as described in [38, 39] for img ∈ {ℐT1, ℐT2, ℐSG} where SG is the sum of gradient magnitude images, which are T1- and T2-weighted MRI scans. The target segmentations were six subcortical structures (caudate, putamen, globus, accumben, thalamus, hippocampus) for both left and right hemispheres of the brain.
Large-Scale Multicenter MRI Data. Two large MRI data sets (n = 3010) from PREDICT-HD1 and TRACK-HD2 were obtained. Both studies are multicenter longitudinal studies and were employed as our primary target application.
Sub-Sample Data. From the PREDICT-HD data set, 32 subjects are randomly chosen to construct a smaller sample to test the automated segmentation model at the early stage of development.
Traveling Human Phantom Data. The THP data consists of a set of subjects scanned repeatedly at different sites over a short time period. Eight sites participated in this multicenter image collection and represented both Siemens and Philips vendors with various software and hardware configurations common to multi-site studies. Five healthy control subjects were recruited and they were imaged at the eight sites within a 30-day period [43]. Collected data includes T1- and T2-weighted multi-modal MR images acquired using three-dimensional (3D) T1-weighted (MP-RAGE) and T2 (SPACE) sequences at each center. The THP data is used to assess multicenter reliability at a later stages of development.
2.2. Overview of Segmentation Framework
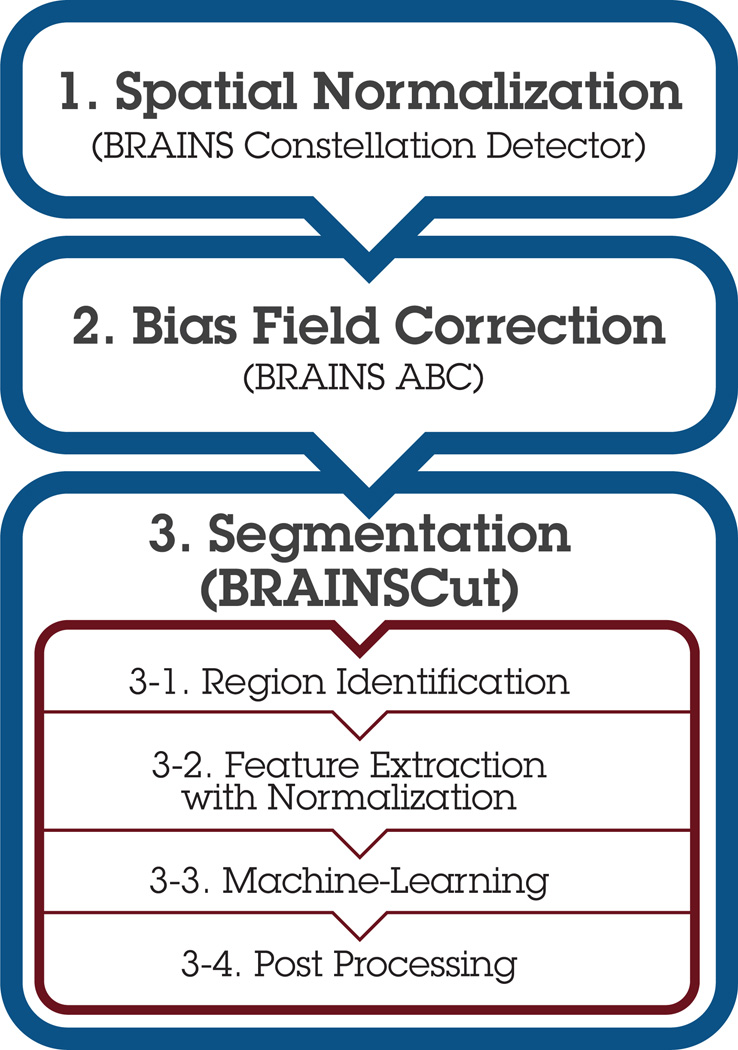
The segmentation framework (Fig. 1) is an adaption from our previous work [38, 39] with the following enhancements: A) landmark-based initial space normalization [44], B) high-deformable registration from the Advanced Normalization Toolkit (ANTS) [45], and C) bias-correction [46]. Each of these enhancements promotes more robust subsequent operations by reducing variations in initial patient placement and varying field of view settings, inter-subject morphometric differences, and different scanner intensity profiles inherent in multi-site data collection.
Figure 1. Segmentation Framework Overview.
Followed by the preprocessing steps: 1. initial space normalization [44], 2. bias field correction [46], and 3. BRAINSCut segmentation [38, 39], which effectively processes MR input in steps 3-1~3-4. This paper aims to determine robust ML and STAMP-based normalization techniques (gray boxes) for robust multicenter scalable data processing.
In our investigation, all performance is reported based on a 10-fold cross-validation against expert manual traces. For the screening study, cross-validation is conducted so that individual voxels are included or excluded. For the full-scale image processing, we designed a custom cross-validation scheme, where an entire subject’s voxel data are included/excluded. For that reason, 32 subjects are roughly subdivided into 10 subsets and cross-validation is conducted to estimate more accurate segmentation performance. This subject-based cross-validation provides more meaningful performance measures by allowing direct volumetric comparison.
The framework is implemented using the on Insight Toolkit [47] (ITK) libraries and conforms to the coding style, testing, and software license guidelines specified by the National Alliance for Medical Image Computing (NAMIC). The implementation is publicly available at github via the BRAINSTools package (git@github.com:BRAINSia/BRAINSTools.git/).
2.3. ML Algorithms
Twelve ML variations from eight algorithms3 are contrasted to construct a reliable and efficient segmentation tool for the multicenter large-scale MR data. We describe and discuss the eight ML algorithms used in this study in terms of theoretical and empirical advantages and disadvantages for large-scale in-vivo MR image processing.
Majority Classifier
A majority classifier simply classifies all the population instances x ∈ 𝕏 as the majority of the training data. If there is no obvious majority, it can be chosen arbitrarily. A majority classifier often serves as a moderate lower bound baseline from which other classifiers can be contrasted against.
Naïve Bayes
Naïve Bayes infers output by estimating posterior probabilities based on Bayes’ Theorem:
| (2) |
Bayes’ Theorem replaces the often-difficult calculation of a ‘posterior’ to the easier computation of two ‘priors’ and one conditional probability. The strength of Naïve Bayes in practice is to isolate noise and irrelevant features F because such data are averaged out when estimating conditional probabilities from the data [48]. As we see from equation (2), however, joint probability calculation assumes statistical independence between all predictors F.
| (3) |
This is often not the case as common input features (e.g., fa and fb, fi ∈ F) often contain partially redundant information. The independent assumption is often not valid in practice and is considered to be violated in the construction of our data because of the redundancy in the feature set chosen to compensate for noise inherent in MRIs.
k-Nearest Neighbor (k-NN)
A k-nearest neighbor (k-NN) classifier predicts a new object using a majority vote of its neighbors. The neighborhood size k determines the number of neighbors participating the majority vote. If k = 1, then the prediction is simply taken from the single closest neighbor, whereas in the case of k = n, where n is a total number of data, the output is induced by taking votes from all data n. The distance to a neighbor from a new object is estimated in feature space; a commonly used distance metric is Euclidean distance [48]. Even though a small k, a fewer neighborhood, generally lowers the training error, it could result in over-fitting of k-NN model to the training data. k-NN usually exhibits good performance without a training phase, but is vulnerable to a bad choice of predictors [48] without proper feature selection strategies.
Support Vector Machine (SVM)
SVM operates by treating each s ∈ 𝕊 as a point in a multi-dimensional hyperspace and then computes the hyperplanes that optimally separate the feature space into regions that are used to assign labels. The distance between hyperplanes is called the “margin,” and the larger the margin the more generalizable the model is. A tradeoff, however, exists between the margin and the error ea: the larger the resulting inter-plane distance (i.e., margin), the better SVM generalizes to data outside the training data set. However, the training error ea also increases because there is a greater chance of data residing in between the separating hyperplanes as the margin grows larger.
Artificial Neural Network (ANN)
ANN is a mathematical model simulating the structural and functional aspects of the human brain [20]. For non-linear problems, a multi-layer perceptron (MLP) can be constructed by connecting multiple artificial neurons, perceptrons, in a layered structure. It is common practice to have one hidden layer 𝕃 = 1 between the input and the output layer, and thus the number of hidden nodes ℍ is only determining factor of model complexity. It has been shown that that an ANN with one hidden layer and with the sigmoid activation function is a universal approximator [49, 50]. There are a few design issues in ANN learning as mentioned in [48], but the most problematic one is based on the fact that ANN is a universal approximator, that can theoretically learn any pattern, but may not be generalizable beyond the training set [48]. Despite these concerns, our previous studies [38, 39] produced very robust results for brain MR subcortical segmentation for single-site data.
Ensemble Methods
Three Ensemble-type methods, Bagging, AdaBoost, and Random forest, are employed in this paper. An ensemble of classifiers is a set of classifiers whose individual decisions are combined in some way (typically by weighted or unweighted voting) to classify new examples [51]. It is based on the intuitive concept that opinions gathered from multiple experts (classifiers) are better than one [52].
Bootstrap Aggregating (Bagging)
Bagging works through voting from base classifiers of choice. Once the base classifier is chosen by an investigator, each base classifier Ci is constructed on the subset Si ⊂ 𝕊, randomly resampled with replacement and bootstrapped from 𝕊. Bagging is known to be most beneficial with methods of high variance in their estimation, such as ANN or a tree structure classifier. A general choice of base classifier is decision stump [53], a tree structure classifier with one root that is immediately connected to the terminal node.
Adaptive Boosting (AdaBoost)
AdaBoost [54] works very similar to Bagging but is distinguished from it in that it builds each classifier in serial. On the serial construction of base classifiers, apparent error ea at the current classifier Ci is used to weigh the data point for the later classifier Ci+1. For the training data that misclassified at run i with Ci, their importance is adaptively increased so that successive classifier Ci+1 can be improved. Theoretically, AdaBoost is particularly sensitive if the training sample includes many misclassified points, (e.g., imperfect manual traces), since latter classifiers become more specialized for the misclassification example.
Random Forest
Random forest combines decisions from multiple tree structure classifiers. One of the appealing properties of random forest is its generalizability. The upper bound for a generalization error of random forests converges when the number of trees 𝕋 is sufficiently large [48]:
| (4) |
where ρ̄ is average correlation among the trees and t is a quantity that measures the strength of the tree classifiers. As the trees become more correlated or the strength of the ensemble decreases, the generalization error bound tends to increase [48]. The random forest model is generally configured with a maximum depth of tree 𝔻, a number of trees 𝕋, and a number of features for split 𝔽.
2.4. Intensity Normalization
For the rigorous evaluation and modeling of a segmentation framework, 11 normalization strategies are compared with segmentation data from six subcortical structures. Stable Atlas-based Mapped Prior (STAMP)-driven robust statistics are developed for robust intensity normalization. STAMP aims to maximize information consistency across scans that originate from different sites and hardware configurations while enhancing image contrast for better separation of neuroanatomy. We describe our idea of normalization with regard to STAMP-driven robust statistics (Sec. 2.4.1) and review several robust statistics and normalization methods investigated in this study (Sec. 2.4.2).
2.4.1. STAMP-driven Robust Statistics
STAMP-driven robust statistics are region-specific statistics that are employed to enhance structural details of an MR image ℐ for a focused region ℛ ⊂ ℐ. For each label l ∈ 𝕃, the focused region is identified by the subject-specific prior pl that is located (‘stamped’) for a subject scan by deforming the template spatial prior with a high-deformable registration 𝒯 into subject space pl(𝒯(x)). The method takes into account all the locally computed, STAMP-driven, statistics according to the spatially bounded region ℛl
| (5) |
In general, normalization techniques in MRI processing utilize statistics computed globally to deal with intrascan intensity variations. This global normalization method, however, is often less sensitive to specific regions of interest. In this study, we hypothesized that normalization methods with STAMP-driven statistics would enhance the robustness of the segmentation framework in multi-site longitudinal data processing where large intensity variations are expected for each subcortical structure region.
2.4.2. Normalization with STAMP-driven Robust Statistics
Seven normalizations with parameter variations are investigated and evaluated in terms of the segmentation accuracy of intraclass correlation (ICCs) to manual segmentations. For each normalization function 𝒩, the STAMP-driven robust statistics aim to provide accurate data description in the presence of gross error. In statistics, robustness can be quantified by the breakdown point [55] describing the largest fraction of arbitrary gross errors tolerated before the statistic ‘breaks down’ and becomes completely unreliable. Detailed discussions of robust statistics of data are well documented in [56, 55, 57]. We have evaluated 11 normalization variations involving six different transform functions and two independent parameters where applicable. The normalization functions with parameter set that we tested are summarized in Table 1.
Table 1.
Seven STAMP-driven normalization functions 𝒩 : ℝ → ℝ given by 𝒩(x) = x′ for robust normalization based on STAMP-driven statistics. α = 0.1 and α = 0.05 are used where applicable.
| Transform f | Notation 𝒩f(·) | 𝒩 : ℝ → ℝ, 𝒩(x) = x′, | where ⋯ | |
|---|---|---|---|---|
| Linear | Min/Max | 𝒩l(m,M) | m =minimum and M =maximum. | |
| Linear | Z-Score | 𝒩l(z) | ||
| Linear | IQR | 𝒩l(IQR) | qj = jth quantile and IQR = q3/4 − q1/4 | |
| Sigmoid | α | 𝒩sg(α) | r = (qα − q(1−α))/2 | |
| Double Sigmoid | α | 𝒩sg2(α) | ||
| Trimming | α | 𝒩trm(α) | x̄t(α) and st(α) are trimmed mean and standard deviation at α level. | |
| Winsorizing | α | 𝒩wz(α) | x̄w(α) and sw(α) are winsorized mean and standard deviation at α level. |
3. Results
First, the ML selection experimental results are reported in two sections: 1) two sets of comparative studies of ML algorithms in Section 3.1 and 2) a STAMP-based normalization comparative study in Section 3.2. The relative superiority of the random forest algorithm compared to the competing ML algorithms are shown using two comparative studies. Next, the benefits of the STAMP-driven normalization approach are presented in Section 3.3. Finally, the best performing ML and normalization techniques were incorporated into a final product, BRAINSCut, and evaluated in terms of both reliability and validity. The multicenter reliability result is presented in Section 3.4, while the sample-size estimation and the validity investigation of the application on a large-scale multicenter data are summarized in Section 3.5.
3.1. Screening Study with WEKA
We contrast 12 variations of eight unique ML algorithms on the down-sampled from the data described in Section 2.1. We use the a publicly available Waikato Environment for Knowledge Analysis (WEKA) [58] ML software package, for a consistent and efficient exploration of the 12 ML approaches. The quality of the 12 ML algorithm variations is thoroughly compared using five metrics: Sensitivity (Recall), Specificity, Precision, F-Measure, and area under the curve (AUC).
The screening study demonstrated that four methods – Bagging, k-NN, ANN, and random forest (Table 2) were the top performers with respect to the specified metrics. Results from caudate segmentation4 are presented in Table 2. Initial WEKA testing used default parameters, then we extended the parameter experiments for k-NN, ANN, Bagging, and random forest.5 Note that three of the four best performing methods belong to the Ensemble type classifier.
Table 2.
Screening study results contrasting performance of 12 machine learning approaches on the identical MR segmentation data for caudate in WEKA. Five measurements are reported for left (L), right (R), background (Bg) of caudate, and the average of all three regions (Avg): sensitivity, specificity, precision, F-measure, and area under the curve (AUC).
| Location | Sensitivity | Specificity | Precision | F-Measure | AUC | Sensitivity | Specificity | Precision | F-Measure | AUC |
|---|---|---|---|---|---|---|---|---|---|---|
| Majority Classifier | Bagging | |||||||||
| L | 0 | 0 | 0 | 0 | 0.500 | 0.876 | 0.020 | 0.881 | 0.879 | * 0.991 |
| R | 0 | 0 | 0 | 0 | 0.500 | 0.880 | 0.021 | 0.887 | 0.883 | * 0.991 |
| Bg | 1 | 1 | 0.702 | 0.825 | 0.500 | * 0.951 | 0.122 | * 0.948 | * 0.950 | * 0.979 |
| Avg | 0.702 | 0.702 | 0.492 | 0.578 | 0.500 | * 0.929 | 0.092 | * 0.929 | * 0.929 | * 0.982 |
| Naïve Bayes | kNN (k=1) | |||||||||
| L | 0.298 | 0.1 | 0.334 | 0.315 | 0.821 | 0.882 | 0.027 | 0.846 | 0.864 | * 0.930 |
| R | 0.717 | 0.272 | 0.325 | 0.447 | 0.807 | 0.894 | 0.028 | 0.852 | 0.872 | * 0.933 |
| Bg | 0.720 | 0.086 | * 0.951 | 0.819 | * 0.901 | * 0.933 | 0.112 | * 0.951 | * 0.942 | * 0.911 |
| Avg | 0.659 | 0.117 | 0.766 | 0.689 | 0.875 | * 0.919 | 0.087 | * 0.921 | * 0.920 | * 0.917 |
| SVM | kNN (k=10) | |||||||||
| L | 0.776 | 0.044 | 0.749 | 0.762 | 0.866 | * 0.925 | 0.030 | 0.836 | 0.878 | * 0.990 |
| R | 0.777 | 0.051 | 0.735 | 0.755 | 0.863 | * 0.902 | 0.023 | 0.877 | 0.889 | * 0.991 |
| Bg | 0.885 | 0.224 | * 0.903 | 0.894 | 0.831 | * 0.935 | 0.087 | * 0.962 | * 0.948 | * 0.980 |
| Avg | 0.853 | 0.171 | 0.855 | 0.854 | 0.841 | * 0.928 | 0.069 | * 0.931 | * 0.929 | * 0.983 |
| AdaBoost | kNN (k=20) | |||||||||
| L | 0 | 0 | 0 | 0 | 0.203 | * 0.902 | 0.027 | 0.848 | 0.874 | * 0.991 |
| R | 0 | 0 | 0 | 0 | 0.791 | * 0.919 | 0.031 | 0.845 | 0.881 | * 0.992 |
| Bg | 1 | 1 | 0.702 | 0.825 | 0.507 | * 0.930 | 0.089 | * 0.961 | * 0.945 | * 0.980 |
| Avg | 0.702 | 0.702 | 0.492 | 0.578 | 0.507 | * 0.924 | 0.071 | * 0.927 | * 0.925 | * 0.983 |
| ANN (HN=20) | Random forest (NT = 10) | |||||||||
| L | 0.880 | 0.024 | 0.860 | 0.870 | * 0.987 | 0.898 | 0.023 | 0.866 | 0.882 | * 0.987 |
| R | * 0.901 | 0.030 | 0.848 | 0.873 | * 0.985 | 0.864 | 0.016 | * 0.906 | 0.884 | * 0.988 |
| Bg | * 0.935 | 0.109 | * 0.953 | * 0.944 | * 0.969 | * 0.952 | 0.119 | * 0.949 | * 0.950 | * 0.976 |
| Avg | * 0.922 | 0.085 | * 0.923 | * 0.922 | * 0.975 | * 0.930 | 0.090 | * 0.931 | * 0.930 | * 0.979 |
| ANN (NH=60) | Random forest (NT = 25) | |||||||||
| L | 0.891 | 0.020 | 0.885 | 0.888 | * 0.989 | 0.884 | 0.018 | 0.894 | 0.889 | * 0.991 |
| R | * 0.904 | 0.022 | 0.881 | 0.893 | * 0.988 | 0.892 | 0.019 | 0.897 | 0.895 | * 0.992 |
| Bg | * 0.949 | 0.102 | * 0.956 | * 0.953 | * 0.976 | * 0.956 | 0.112 | * 0.953 | * 0.954 | * 0.982 |
| Avg | * 0.934 | 0.078 | * 0.934 | * 0.934 | * 0.980 | * 0.936 | 0.084 | * 0.936 | * 0.936 | * 0.985 |
The metrics with preferable performance (> 0.9) are highlighted in bold with an * mark: Bagging, variation of ANN (HN = 20 and 60), kNN (k = 1, 10, and 20), and random forest (NT = 10 and 25).
Exploration continued by focusing on two of the top performing methods, ANN and the random forest, in terms of full-scale data comparison within the segmentation framework. ANN and random forest were chosen for several reasons. First, both ANN and random forest displayed excellent performance compared to other competitor algorithms for a wide range of parameter variations (Table 2). Second, ANN has already successfully been applied to clinical subcortical volumetric studies [39, 15, 26, 29, 59]. Third, the excellent generalizability [60] of the random forest algorithm is very attractive for addressing the inherent challenges of multicenter large-scale data processing.
Despite their similar performance in the initial screening study, we did not investigate bagging and k-NN. k-NN is excluded primarily because of its known susceptibility to noise, or data variation, that we felt would limit its generalizability to multicenter data. Bagging was discarded due to its conceptual similarities to random forest, and that random forest is a specialized version of bagging that we felt was emerging as a preferred technique in the field [61].
3.2. Full-scale ANN and Random Forest Comparative Study with OpenCV
A subject-based cross-validation is conducted for the full-scale sample data from Section 2.1. The choice of a single hidden layer ANN explored is based on previous work [39] with the number of hidden nodes tested for ℍ = [20, 60]. For the random forest algorithm, we fixed the number of features per tree and tested for three different number of trees 𝕋 = [10, 25, 100]. The number of trees for full testing of random forest is decided using an exhaustive parameter space investigation.6
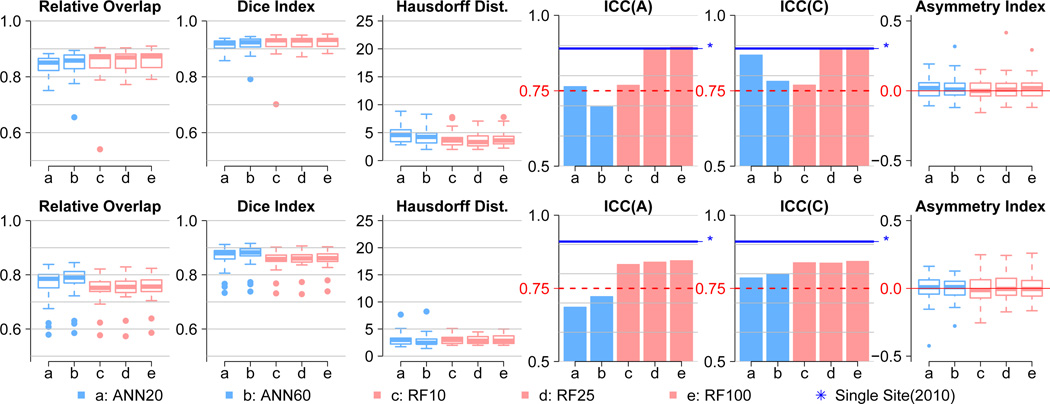
We present here four regions of interest (ROIs) in left hemisphere, five performance metrics comparing the automated results to the manual segmentations in Table 2. The performance metrics include two intraclass correlations (ICC), relative overlap (RO), dice similarity coefficient (DSC, or DI for dice index), Hausdorff distance (HD),
where | · | denotes volume of the region for automated (A) and manual (M) segmentation. Definitions for intraclass correlation of agreement (ICC(A)) and consistency (ICC(C)) follows ICC(2, 1) and ICC(3, 1) as described in [62]. In addition to those five metrics, an asymmetry index between left and right hemispheres is also reported to highlight performance consistency between the hemispheres.
We focused on the ICC metrics because they are more sensitive to segmentation accuracy differences between the various ML algorithm configurations tested than other metrics (RO, DSC, and HD). The two ICC values showed excellent subcortical segmentation accuracy with the random forest algorithm with tree depths of 25 or greater.
3.3. Normalization Selection for STAMP-based Approach
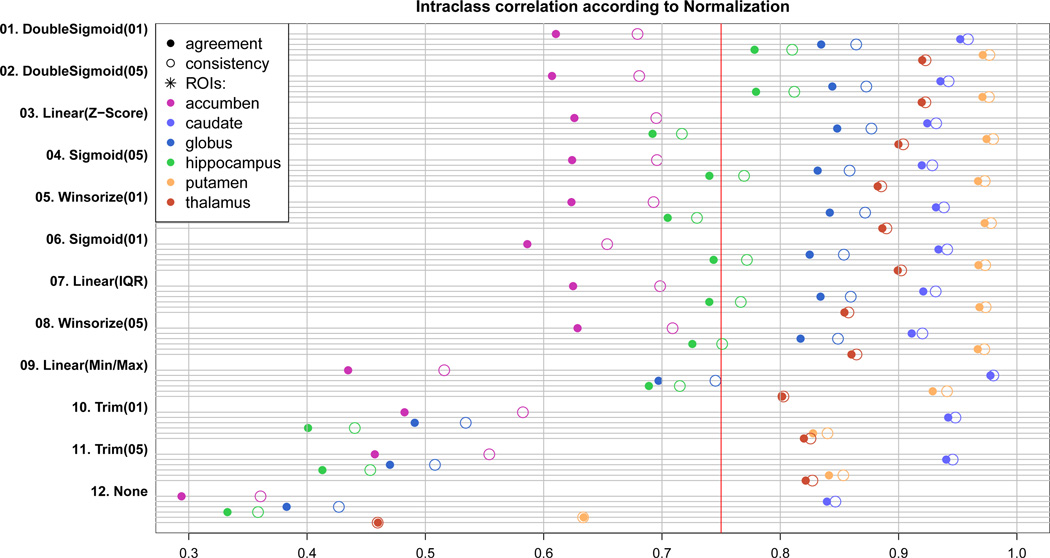
All 11 intensity normalization schemes tested substantially increased the subcortical segmentation accuracy of BRAINSCut in terms of ICCs (Fig. 3). Even though 𝒩trm(α) and 𝒩l(m,M) seem to have under-performed compared to others (01~09) from Fig. 3), note that all 𝒩f(·)s showed statistically significant improvement in segmentation accuracy. The application to the in-vivo multicenter data, however, revealed that improper choice of the STAMP-driven normalization approach may result in failure as shown in Figure 3.
Figure 3.
ICC(A) (solid circle) and ICC(C) (empty circle) is plotted for 11 normalization strategies as well as raw data without normalization (None). Higher ICC means better correspondence of BRAINSCut to manual traces, and thus the greater segmentation accuracy. All six structures are tested and plotted with different colors. The red line is a ICC’s lower bound suggested by Shrout [62]. Experiments are ranked by its average performance over six structures from the top to bottom. One should note that all 11 𝒩f(·)s improved segmentation accuracy of BRAINSCut with statistical significance.
3.4. Multicenter Reliability Results
Multicenter reliability is assessed through the THP results for six subcortical structures. The volume difference across eight sites was formally tested by the analysis of variance (ANOVA) as shown in Table 4. We tested if measured means across subjects are different among eight sites:
H0 : Volume means of five subjects are same across eight different sites.
To employ ANOVA, we first confirmed that our data meets a required assumption of homoscedasticity, and then tested the measure differences across the sites. Our statistical test demonstrate that the BRAINSCut tool does not have significant measurement differences across the eight sites for measuring the subcortical volumes (Table 4).
Table 4.
Statistical test showed no significant measurement differences between sites for six subcortical regions by using BRAINSCut. Statistical test results for the following hypothesis test: ‘H0: Six subcortical volume means are all the same between eight centers’. To test the hypothesis with ANOVA, we first tested homoscedasticity, a homogeneity of variance between groups, for left and right (s = l, r) ROIs with Fligner-Killeen test as their p-values are given Prvar,s. All the tests were not significant, meaning that variances can be regarded as homogeneity. Next, we tested the H0 with ANOVA there were no significant differences at the significant level of α = 0.1 in subcortical measures between sites as shown in p-values Prvar,s > α in all cases.
| ROI | Prvar,l | Prvar,r | Prsite,l | Prsite,r |
|---|---|---|---|---|
| accumben | 0.83 | 0.12 | 0.32 | 0.15 |
| caudate | 0.82 | 0.8 | 0.88 | 0.73 |
| globus | 0.41 | 0.46 | 0.7 | 0.5 |
| thalamus | 0.89 | 0.63 | 0.3 | 0.67 |
| putamen | 0.93 | 0.92 | 0.99 | 0.67 |
| hippocampus | 0.83 | 0.8 | 0.99 | 1 |
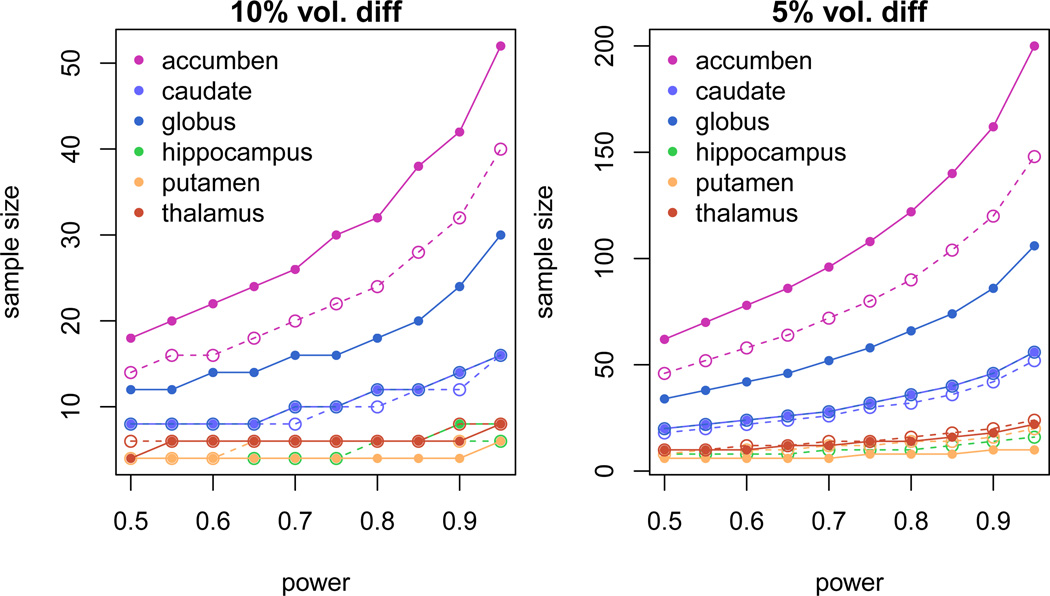
Sample size is also determined based on the mean volume and standard deviation of subjects across sites reported in Table Amp.B2 in the last column. This measurement is important for designing efficient clinical and research trials. The required sample sizes to detect 5% and 10% mean volume difference between two groups are shown in Figure 5. We varied the range of power from 0.5 to 1.0 on each computation. The two-sample t-test formula is used to calculate required sample sizes along the power levels assuming balanced but unpaired with equal variance design.7 To detect 5% and 10% mean volume difference with a power level of 0.8, the appropriate respective sample sizes would be 120 and 30 for nucleus accumben.
Table Amp.B2.
Traveling Human Phantom Data Mean and Standard deviation (sd) measured from MRI by using our proposed approaches. For five subjects, measured volumes of six subcortical structures are shown as well as CVs of each subjects.
| ROI | TPH01 (n=6) mean (sd) |
TPH02 (n=8) mean (sd) |
TPH03 (n=6) mean (sd) |
TPH04 (n=6) mean (sd) |
TPH05 (n=7) mean (sd) |
Mean | |
|---|---|---|---|---|---|---|---|
| accumben | L | 238.8 (18.5) 8% | 302.8 (38.8) 13% | 349.8 (21) 6% | 342.3 (18.6) 5% | 282.9 (50.9) 18% | 303.32 (29.56) 10% |
| R | 294 (13.6) 5% | 309.4 (35.2) 11% | 342.3 (15.5) 5% | 322.8 (19.8) 6% | 300.3 (47) 16% | 313.76 (26.22) 8% | |
| caudate | L | 3282.5 (99.7) 3% | 3019.4 (150.9) 5% | 3741.2 (355.2) 9% | 3642.7 (120.3) 3% | 2553.3 (94.6) 4% | 3247.82 (164.14) 5% |
| R | 3414.3 (175.8) 5% | 3034.8 (124.8) 4% | 3946.5 (147.2) 4% | 3602.5 (243.9) 7% | 2645.9 (115.9) 4% | 3328.8 (161.52) 5% | |
| globus | L | 1323.2 (112.5) 9% | 1350.3 (76.2) 6% | 1285.8 (112.9) 9% | 1500.5 (89.8) 6% | 1327.1 (87.6) 7% | 1357.38 (95.8) 7% |
| R | 1250.2 (58.9) 5% | 1187.1 (58.6) 5% | 1289 (65.1) 5% | 1480 (75.4) 5% | 1299 (72.1) 6% | 1301.06 (66.02) 5% | |
| hippocampus | L | 1834.5 (56.9) 3% | 1800.4 (61.8) 3% | 1421.3 (55.3) 4% | 1749.8 (55.2) 3% | 1858.9 (34.2) 2% | 1732.98 (52.68) 3% |
| R | 1794.8 (46.9) 3% | 1728 (46.6) 3% | 1390.7 (37.9) 3% | 1739 (36.3) 2% | 1959.3 (46.7) 2% | 1722.36 (42.88) 3% | |
| putamen | L | 4501.8 (102.3) 2% | 4741.1 (63.2) 1% | 4665.8 (101.3) 2% | 4960.8 (64.6) 1% | 5167.3 (120.8) 2% | 4807.36 (90.44) 2% |
| R | 4406.8 (135.6) 3% | 4530.5 (190) 4% | 4240.7 (108.5) 3% | 4505.3 (116.6) 3% | 4902.3 (96.8) 2% | 4517.12 (129.5) 3% | |
| thalamus | L | 7415.3 (135.4) 2% | 7498.8 (301.1) 4% | 7719.2 (203.9) 3% | 7815.5 (319) 4% | 6867.1 (165.5) 2% | 7463.18 (224.98) 3% |
| R | 7235.2 (269.7) 4% | 7330.1 (300.6) 4% | 7527.2 (118.5) 2% | 7769.8 (312) 4% | 6904.7 (184.7) 3% | 7353.4 (237.1) 3% | |
Figure 5.
The estimated required sample size to detect 5% (left) and 10% (right) volume changes for the corresponding subcortical structures: 1) Nucleus accumben (accumben), 2) Caudate, 3) Globus Pallidum (globus), 4) Hippocampus, 5) Putamen, and 6) Thalamus. Solid line and dashed line represents structures located in left and right hemisphere, respectively.
3.5. Large-scale Multicenter MRI Application Results
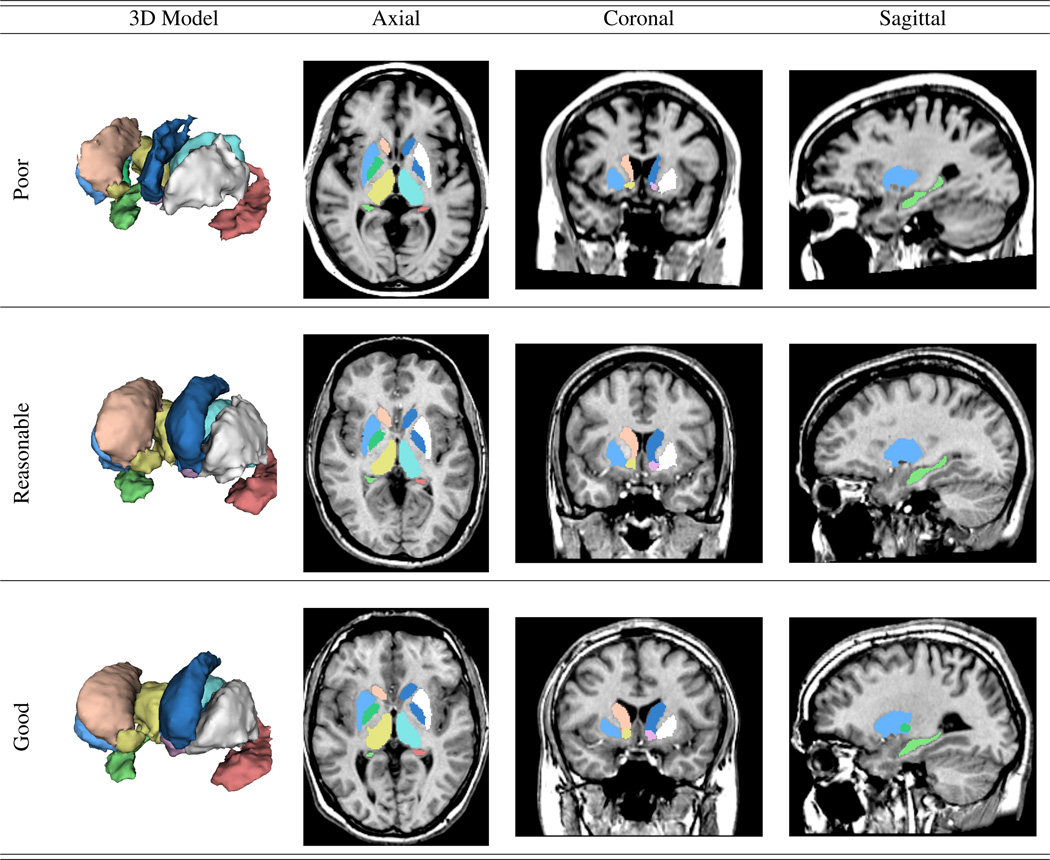
We also evaluated the ability of BRAINSCut to process a wide range of data by applying two large-scale multicenter data, PREDICT-HD (32 sites) and TRACK-HD (4 sites), to derive six subcortical structures. The software’s robustness and segmentation quality were quantified with a success ratio through manual visual inspection. The proportion of scans that completed without error was very high for both the PREDICT-HD and TRACK-HD data. The quality of derived subcortical structures was visually rated according to the provided guideline8 and the results, in terms of three-level grading, are shown in Table 3. The segmentation quality results rated at a poor level were substantially small in number (< 6%) for evaluating 3000 scan sessions. Examples of each grading level are shown in Figure 6.
Table 3.
Visual investigation score for six structures of two large-scale studies: PREDICT-HD (Upper) and TRACK-HD (Bottom). Three experts rated segmentation quality on three level basis: 0 = poor, 1 = reasonable with minor intervention, and 3 = good quality of automated segmentations. Our method is applied on over 3000 scans and failure rate is less then 10% for all the structures.
| PREDICT-HD | accumben | caudate | putamen | globus | thalamus | hippocampus | ||||||
| score (n = 2228) | left | right | left | right | left | right | left | right | left | right | left | right |
| 0(poor) | *0.007 | *0.005 | *0.044 | *0.058 | *0.057 | *0.078 | *0.041 | *0.052 | *0.018 | *0.027 | *0.016 | *0.011 |
| 1 | 0.065 | 0.088 | 0.177 | 0.255 | 0.161 | 0.201 | 0.096 | 0.123 | 0.074 | 0.105 | 0.089 | 0.079 |
| 2(good) | 0.928 | 0.907 | 0.779 | 0.686 | 0.783 | 0.721 | 0.864 | 0.825 | 0.908 | 0.868 | 0.895 | 0.911 |
| Track On | accumben | caudate | putamen | globus | thalamus | hippocampus | ||||||
| score (n = 782) | left | right | left | right | left | right | left | right | left | right | left | right |
| 0(poor) | *0.028 | *0.040 | *0.030 | *0.052 | *0.058 | *0.091 | *0.095 | *0.085 | *0.009 | *0.015 | *0.037 | *0.033 |
| 1 | 0.103 | 0.119 | 0.077 | 0.122 | 0.136 | 0.143 | 0.183 | 0.187 | 0.005 | 0.022 | 0.078 | 0.081 |
| 2(good) | 0.869 | 0.841 | 0.893 | 0.827 | 0.806 | 0.766 | 0.722 | 0.729 | 0.985 | 0.963 | 0.885 | 0.886 |
Figure 6.
Subcortical segmentation examples from BRAINSCut. From top to bottom, images of each row corresponds to be rated as ‘poor’, ‘reasonable’, and ‘good’ via the visual inspection. As the figure shows, all the segmentations even at poor quality (< 10%) can be utilized for the further analysis with manual editing.
4. Discussion
This paper focused on two essential elements for achieving accurate and consistent segmentation for multicenter MR data: the choice of ML algorithm and the choice of intensity normalization strategy. The comparative experiments were conducted between eight ML algorithms and 11 normalization strategies.
We found that in virtually all cases, the choice of these two elements has significant impact on the quality of out-comes for the segmentation framework. The segmentation accuracy and generalizability were significantly increased by employing a random forest algorithm and the STAMP-based normalization strategies. Our study provides insight into the mechanisms underlying ML-based segmentation frameworks, and shows that the selection ML algorithm and the intensity normalization strategy can have profound impact on the performance characteristics of the resulting measurements. The results from the BRAINSCut framework applied to the large-scale multicenter MR data set are encouraging for using a ML-based segmentation tool as a cost- and time-efficient means to investigate volumetric changes in both neurodegenerative and healthy subjects. The reduction in operator time (6–10 hours for segmentation, vs 5–10 minutes for review and cleanup) makes the integration of computerized segmentation into a large-scale clinical data analysis very attractive. The result of this paper suggests that the automated segmentation framework, based on machine-learning techniques, operates robustly on large-scale multicenter MR data. Moreover, our study provides the straightforward comparative analysis framework for future studies to assess the performance characteristics of alternative approaches.
Our results are encouraging and should be explored with other available segmentation tools in the field, such as a label-fusion/propagation-based segmentation methods emerging approach in recent years [63, 64, 65, 66, 67, 68, 69, 70, 71, 72]. An extensive study of the available and promising techniques will guide us in further segmentation improvement. Future work will focus on the possibility of ML-based segmentation of the entire brain [73, 63]. The whole brain segmentation can benefit from accuracy by explicitly penalizing the possibility of mismatch between structures of interest and background tissues.
We have shown that our framework can be successfully applied to a wide range of brain MRI data to examine changes in subcortical volumes. In addition, this analytical approach could eventually lead to the identification of the complex mechanism of the ML-based segmentation framework for large amount of multicenter data processing. The the software implementation, including the trained model, is publicly available at github via the BRAINSTools package (https://github.com/BRAINSia/BRAINSTools).
5. Conclusion
This paper describes a segmentation framework, BRAINSCut, to delineate the brain subcortical structures consistently and effectively from large-scale multicenter MR data sets. Carefully designed comparative experiments reveal the relative benefits and failures of various ML-based segmentation framework choices. The excellent robustness and confirmed validity of BRAINSCut are achieved by employing 1) random forest, 2) a STAMP-based normalization, and 3) a series of validation studies that occurred repeatedly together with the software development to validate its robustness and reliability. Our study showed that judicious choice of ML and normalization methods can significantly enhance a ML-based segmentation framework in terms of accuracy and generalizability.
Figure 2.
Five correspondence measures between automated and manual delineations for thalamus in left hemisphere with asymmetry index between left and right structures in the far right side. The five measures include relative overlap (RO), dice index (DSC), Hausdorff distance (HD), and ICC of agreement (ICC(A)) and consistency (ICC(C)). The two left measures (light blue) in each graph are ANN trials with ℍ = 20 (ANN20) and ℍ = 60 (ANN60), and the right most three measures (light red) are random forest trial with 𝕋 = 10 (RF10), 𝕋 = 20 (RF25), and 𝕋 = 100 (RF100). The dashed red line at 0.75 in both ICC plots represents a bottom line suggested by Shrout et al. [62], where two independent traces, manual and automated, can be regarded as identical. The solid blue line with a star (⋆) mark shows the result of our previous study [39], which was highly optimized for single-sited study. It is hard to differentiate performance by RO, DSC, or HD (three measures from the left); but ICC measures generally suggest that random forest is superior. While ANN displayed over-fitting behavior as ℍ increases, the random forest model nicely converges as 𝕋 increases. Consistent asymmetry index for both structures could also be highlighted in that the result of right-side structure segmentation corresponds well to the left, which is shown here.
Figure 4.
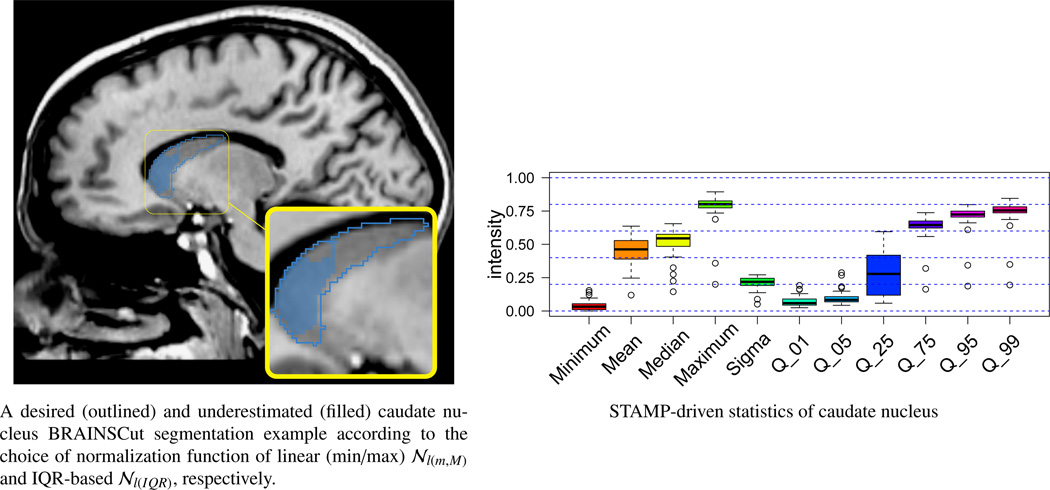
This figure shows two results of caudate nucleus segmentation (left) with corresponding STAMP-driven caudate nucleus region statistics (right). Two segmentation results were produced using linear (min/max) 𝒩l(m,M) and linear (IQR) 𝒩l(IQR) normalization for outlined blue and filled blue, respectively. Comparing filled and outline segmentations of caudate nucleus shows that segmentation is considerably improved choosing an adequate STAMP-based normalization technique. Underestimated segmentation (filled blue) is due to the instability (wide blue box in the box plot) of STAMP-driven statistics of q25.
Figure Amp.A1.
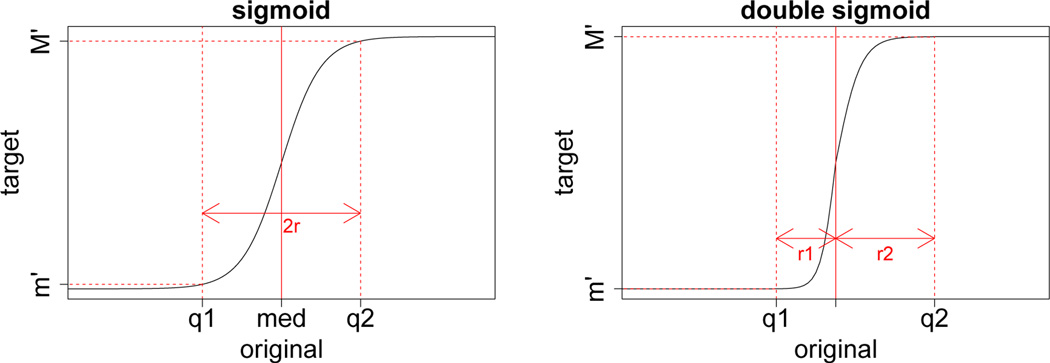
Contrasting shape of two sigmoidal shape normalization functions: 1) sigmoid (upper) and 2) double sigmoid (bottom). Double sigmoid function can deal with skewed data more effectively by taking account of two ranges from median, denoted as r1 and r2 in the graph, instead of one range r for entire data.
Figure Amp.A2.
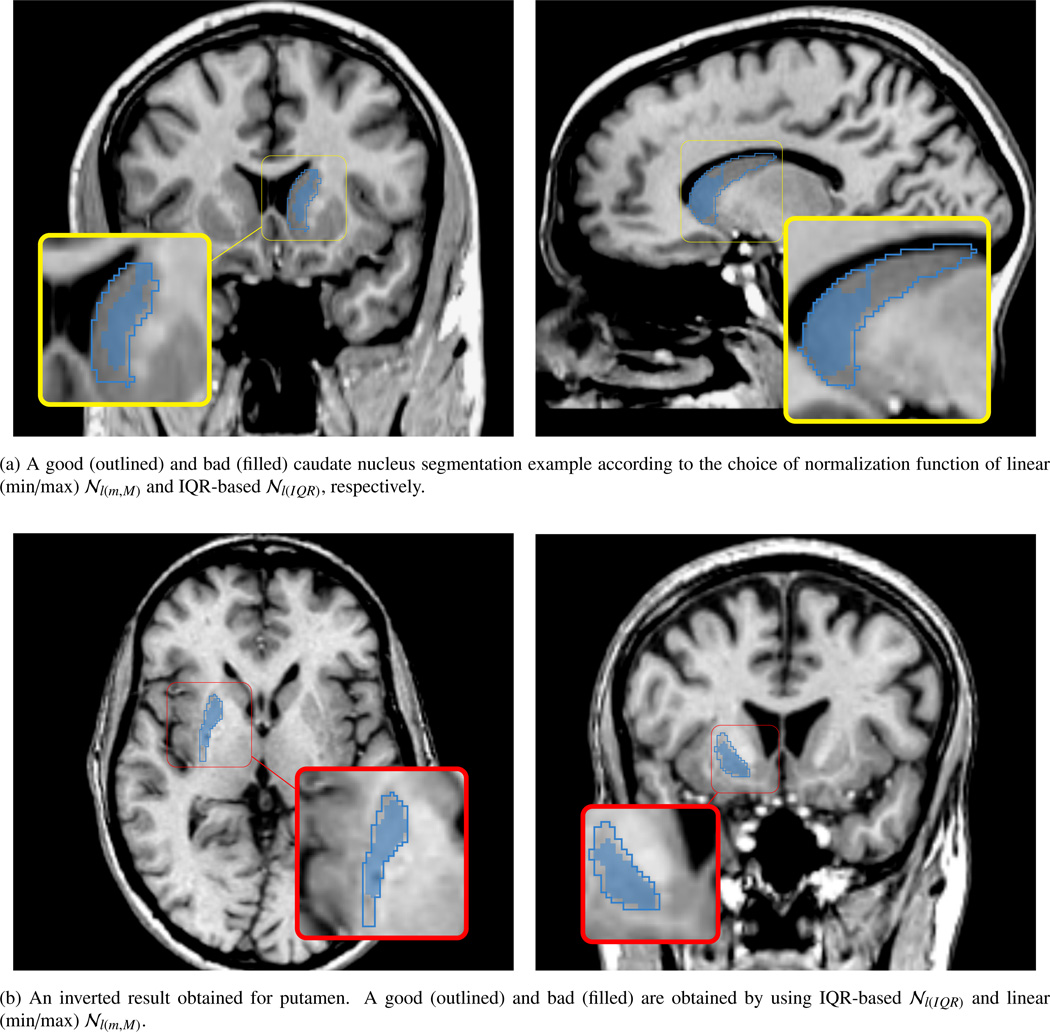
Contrasting segmentation failure (filled blue) and success (outlined blue) according to the choice of STAMP-based normalization for left caudate nucleus (a) and right putamen (b). Two normalization methods are utilized on this example: 1) linear (min/max) 𝒩l(m,M) and 2) IQR-based normalization 𝒩l(IQR). When linear (min/max) normalization is employed in the segmentation framework, the segmentation algorithm failed for the caudate nucleus but succeeded for the putamen. On the other hand, the segmentation framework with the IQR-based normalization showed an excellent segmentation accuracy for the putamen, but underestimated the caudate nucleus. From these cases, we clearly see that different normalization method have a substantial effect on the segmentation results visually and quantitatively. This visually unpleasant segmentation results did not observed in our 32 training data set, even with 10-fold cross validation study. This scan has randomly been tried and identified as a failure by visual inspection process.
Acknowledgment
This paper is funded by multiple grants: (R01 NS050568) - BRAINS Morphology and Image Analysis, (R01 EB000975) - Validation of Structural/Functional MRI Localization, (R01 EB008171) - 3D Shape Analysis for Computational Anatomy, (R01 NS040068) - Neurobiological Predictors of Huntington’s Disease, (R01 NS054893) - Cognitive and functional brain changes in preclinical Huntington’s disease (HD), (P41 RR015241) - Algorithms For Functional and Anatomical Brain Analysis; Algorithmic Methods For Anatomical Brain Analysis, (S10 RR023392) - Enterprise Storage In A Collaborative Neuroimaging Environment, (U54 EB005149) - Core 2b Huntington’s Disease - Driving Biological Project, (R03 EB008673) - NIPYPE: Neuroimaging in Python Pipelines and Interfaces.
Appendix
Amp.A. Normalization Functions
Amp.B. Multicenter Reliability Assessment through Traveling Human Phantom Data
To assess multicenter reliability, the automated segmentation tool is applied on eight independent subcortical structure segmentations of five subjects taken from eight sites.
Data: We have utilized traveling human phantom (THP) data for validation of our proposed tool. THP data consists of five subject scanned at eight sites repeatedly over a month period. Note that THP data was originally planned and collected to evaluate diffusion tensor imaging (DTI) process as reported in [43]. Eight sites participated in this multicenter image collection consists of two MR vendors of distinguished imaging histories: Siemens and Philips. The sites involved in this study had either a Siemens 3T TIM Trio scanner (gradient strength =45mT/m, slew rate = 200 T/m/sec) or Philips 3T Achieva scanner (gradient strength = 80mT/m, slew rate =200T/m/sec). Five healthy control subjects were recruited into this multicenter imaging study after informed consent was obtained in accordance with the Institutional Review Board at each of the imaging sites. All five subjects were imaged at the eight sites within a 30-day period. Collected data includes T1- and T2-weighted multi-modal MR images, acquired using using three-dimensional (3D) T1-weighted (MP-RAGE) and T2 (SPACE) sequences at each center.
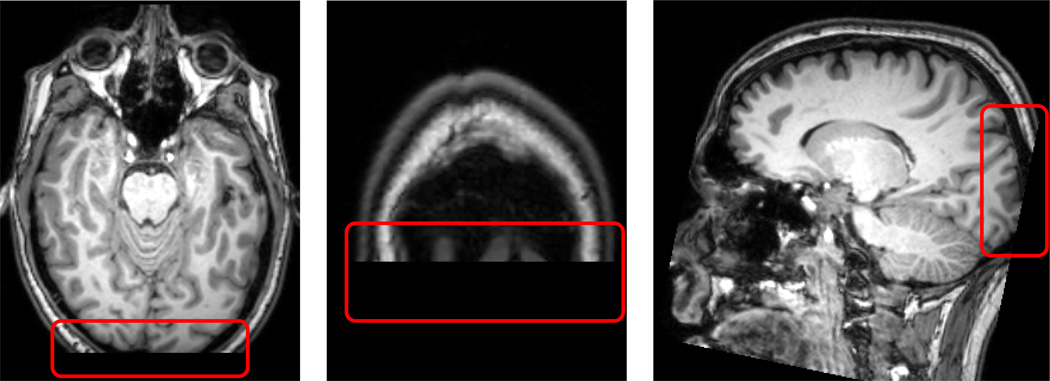
Each MRI anatomical volume was processed with the standard BAW procedure 2.2. After visual inspection stage based on our standard protocol, seven scan sessions are removed from further analysis due to the low quality of T1 images (Marked as (X) in Table Amp.B1). The common reason of low score was a insufficient coverage of whole brain region as shown in Figure Amp.B3, which, in turn, results in failure of spatial normalization of BAW process.
Table Amp.B1.
Quality report of THP data from the experts’ visual inspection. For each scans of a scan session, visual inspection score ranges from one to ten for the worst (1) and the best (10) image qualities. Each MR session can have multiple scans including more than one T1- and T2-weighted images. Score S is reported T1 first and followed by T2 separated by slash(/). If there is multiple scans that rated identical, the number of scans n are reported in parenthesis: [ST11, ST12 (n),… / ST21, ST22 (n),‥], where SI is a visual inspection score for scan I. The only data rated above > 5 is proceeded to the standard BAW procedure and acquired six subcortical structure segmentation results. Note that eight scans from UW and JHU are excluded from processing because no T1-weighted image is remained after the quality control.
| Center | Vendor | Visual Inspection Scores [T1s(repeat)/T2s] | ||||
|---|---|---|---|---|---|---|
| THP 1 | THP 2 | THP 3 | THP 4 | THP 5 | ||
| CCF | Siemens | 9/10 | 9/10 | 10/8 | 8/10 | 9/10 |
| IOWA | Siemens | 9,4/8 | 10/10 | 8/8 | 8/6 | 6/10 |
| MGH | Siemens | 10/7 | 10/10 | 8/8 | 8/9 | 10/8 |
| UCI | Siemens | 10/9 | 8/10 | 9/10 | 8/8 | 9/8 |
| UMN | Siemens | 7/7 | 8/8 | 10/9 | 10/8 | 8/8 |
| DART | Philips | 10(2),0/10,0 | 10(5)/8 | 8/8 | 10(2),0/10 | 10(3),8,8/9 |
| UW | Philips | 0/8 (X) | 8/8 | 0/8 (X) | 0/10 (X) | 8/10 |
| JHU | Philips | 0,0/8(X) | 10,8/8 | 0,0/10 (X) | 0,0/8 (X) | 0,0/5 (X) |
Figure Amp.B3.
Traveling Human Phantom Low Quality Scored MR Image Example: A Scan have an insufficient head region coverage to the posterior of brain. This insufficiency leads to the failure at the pre-processing stage while taking the scan into common AC-PC aligned space.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
PREDICT-HD Study official site: https://www.predict-hd.net/
TRACK-HD official site: http://www.track-hd.net
There are numerous references for each method, but we we have summarized and used notation primarily from [48].
For brevity only the results for the caudate are presented here. Results for other structures are available as supplemental materials.
Each of the extended experiments choose parameters based on either theoretical or previous [39] empirical studies.
Please see supplemental material for details on the search space explored.
A free software programming language and a software environment for statistical computing R package ‘samplesize’ is employed for the computation. The package is available at http://www.inside-r.org/packages/cran/samplesize/docs/n.ttest.
Derived six subcortical structures from BRAINSCut are rated by three independent experts for their accuracy. The rating is based on three levels: 0 = poor, 1 = reasonable, and 2 = good. (0) Poor indicates that manual tracing is required to use this measurement, (1) Reasonable indicates sufficient quality to use the measures, but would benefit from small edit, and (2) Perfect indicates that the segmentation would not benefit from manual tracing.
References
- 1.Rubin R. Principles of imaging in neuro-ophthalmology, Ophthalmology. Mosby, Philadelphia: 1999. pp. 943–949. [Google Scholar]
- 2.Styner M, Lieberman Ja, Pantazis D, Gerig G. Boundary and medial shape analysis of the hippocampus in schizophrenia. Medical image analysis. 2004;8(3):197–203. doi: 10.1016/j.media.2004.06.004. URL http://www.ncbi.nlm.nih.gov/pubmed/15450215. [DOI] [PubMed] [Google Scholar]
- 3.Szeszko PR, Narr KL, Phillips OR, McCormack J, Sevy S, Gunduz-Bruce H, Kane JM, Bilder RM, Robinson DG. Magnetic resonance imaging predictors of treatment response in first-episode schizophrenia. Schizophrenia bulletin. 2012;38(3):569–578. doi: 10.1093/schbul/sbq126. URL http://www.ncbi.nlm.nih.gov/pubmed/21084552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edland SD, Xu Y, Plevak M, O’Brien P, Tangalos EG, Petersen RC, Jack CR. Total intracranial volume: Normative values and lack of association with Alzheimer’s disease. Neurology. 2002;59(2):272–274. doi: 10.1212/wnl.59.2.272. [DOI] [PubMed] [Google Scholar]
- 5.Gerardin E, Chételat G, Chupin M, Cuingnet R, Desgranges B, Kim H-S, Niethammer M, Dubois B, Lehéricy S, Garnero L, Eustache F, Colliot O. Multidimensional classification of hippocampal shape features discriminates Alzheimer’s disease and mild cognitive impairment from normal aging. NeuroImage. 2009;47(4):1476–1486. doi: 10.1016/j.neuroimage.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schumann CM. The Amygdala Is Enlarged in Children But Not Adolescents with Autism; the Hippocampus Is Enlarged at All Ages. Journal of Neuroscience. 2004;24(28):6392–6401. doi: 10.1523/JNEUROSCI.1297-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scahill RI, Hobbs NZ, Say MJ, Bechtel N, Henley SMD, Hyare H, Langbehn DR, Jones R, Leavitt BR, Roos RAC, Dürr A, Johnson HJ, Lehéricy S, Craufurd D, Kennard C, Hicks SL, Stout JC, Reilman RR, Tabrizi SJ, et al. t. T.-H. Investigators. Clinical impairment in premanifest and early Huntington’s disease is associated with regionally specific atrophy. Human brain mapping 000. 2011 Jul; doi: 10.1002/hbm.21449. 2010 n/a–n/a. URL http://doi.wiley.com/10.1002/hbm.21449papers2://publication/doi/10.1002/hbm.21449http://www.ncbi.nlm.nih.gov/pubmed/22102212. [DOI] [PMC free article] [PubMed]
- 8.Younes L, Ratnanather JT, Brown T, Aylward EH, Nopoulos PC, Johnson HJ, Magnotta VA, Paulsen JS, Margolis RL, Albin RL, Miller MI, Ross CA, et al. Regionally selective atrophy of subcortical structures in prodromal HD as revealed by statistical shape analysis. Human brain mapping 00. 2012 Oct;:1–18. doi: 10.1002/hbm.22214. URL http://www.ncbi.nlm.nih.gov/pubmed/23281100. [DOI] [PMC free article] [PubMed]
- 9.Starkstein SE, Bylsma F, Peyser CI, Folstein M, Folstein SE. Neuroradiology Neuropsychological correlates of brain atrophy in Huntington’s disease : a magnetic resonance imaging study. Magnetic Resonance Imaging. 1992:487–489. doi: 10.1007/BF00598956. [DOI] [PubMed] [Google Scholar]
- 10.Paulsen JS, Magnotta Va, Mikos AE, Paulson HL, Penziner E, Andreasen NC, Nopoulos PC. Brain structure in preclinical Huntington’s disease. Biological psychiatry. 2006;59(1):57–63. doi: 10.1016/j.biopsych.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Douaud G, Gaura V, Ribeiro M-J, Lethimonnier F, Maroy R, Verny C, Krystkowiak P, Damier P, Bachoud-Levi aC, Hantraye P, Remy P. Distribution of grey matter atrophy in Huntington’s disease patients: a combined ROI-based and voxel-based morphometric study. NeuroImage. 2006;32(4):1562–1575. doi: 10.1016/j.neuroimage.2006.05.057. [DOI] [PubMed] [Google Scholar]
- 12.Aylward E, Anderson N, Bylsma F, Wagster M, Barta P, Sherr M, Feeney J, Davis A, Rosenblatt A, Pearlson G, et al. Frontal lobe volume in patients with Huntingtons disease. Neurology. 1998;50(1):252–258. doi: 10.1212/wnl.50.1.252. [DOI] [PubMed] [Google Scholar]
- 13.Paulsen JS, Langbehn DR, Stout JC, Langbehn, Aylward E, Ross Ca, Nance M, Guttman M, Johnson S, MacDonald M, Beglinger LJ, Duff K, Kayson E, Biglan K, Shoulson I, Oakes D, Hayden M, Predict-HD I. Detection of Huntington’s disease decades before diagnosis: the Predict-HD study. Journal of neurology, neurosurgery, and psychiatry. 2008;79(8):874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nopoulos P, Aylward E, Ross C. Cerebral cortex structure in prodromal Huntington disease. Neurobiology of diseaseof Disease. 2010;40:544–554. doi: 10.1016/j.nbd.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulsen J, Nopoulos P, Aylward E, Ross C. Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain research. doi: 10.1016/j.brainresbull.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nopoulos PC, Aylward EH, Ross CA, Mills JA, Langbehn DR, Johnson HJ, Magnotta VA, Pierson RK, Beglinger LJ, Nance MA, Barker RA, Paulsen JS. PREDICT-HD Investigators and Coordinators of the Huntington Study Group, Smaller intracranial volume in prodromal Huntington’s disease: evidence for abnormal neurodevelopment. Brain : a journal of neurology. 2011;134(Pt 1):137–142. doi: 10.1093/brain/awq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aylward EH, Liu D, Nopoulos PC, Ross Ca, Pierson RK, Mills Ja, Long JD, Paulsen JS. Striatal volume contributes to the prediction of onset of huntington disease in incident cases. Biological psychiatry. 2012;71(9):822–828. doi: 10.1016/j.biopsych.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan Z, Styner M, Muenzer J, Poe M, Escolar M. Correlation of automated volumetric analysis of brain MR imaging with cognitive impairment in a natural history study of mucopolysaccharidosis II. AJNR. American journal of neuroradiology. 2010;31(7):1319–1323. doi: 10.3174/ajnr.A2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabrizi SJ, Langbehn DR, Leavitt BR, Roos RAC, Dürr A, Craufurd D, Kennard C, Hicks SL, Fox NCNC, Scahill RI, Borowsky B, Tobin AJ, Rosas HD, Johnson HJ, Reilman RR, Landwehrmeyer GB, Stout JC, Durr A, Reilmann R, Landwehrmeyer B, et al. Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet neurology. 2009;8(9):791–801. doi: 10.1016/S1474-4422(09)70170-X. URL http://www.ncbi.nlm.nih.gov/pubmed/19646924http://dx.doi.org/10.1016/S1474-4422(09)70170-Xpapers2://publication/doi/10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TEJ, Bucholz R, Chang a, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath aC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder aZ, Xu J, Yacoub E. The Human Connectome Project: a data acquisition perspective. NeuroImage. 2012;62(4):2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3606888\&tool=pmcentrez\&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aylward EH. Change in MRI striatal volumes as a biomarker in preclinical Huntington’s disease. Brain research bulletin. doi: 10.1016/j.brainresbull.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 22.Aylward E, Rosenblatt a, Field K, Yallapragada V, Kieburtz K, McDermott M, Raymond L, Almqvist E, Hayden M, Ross C. Caudate volume as an outcome measure in clinical trials for Huntingtons disease: a pilot study. Brain Research Bulletin. 2003;62(2):137–141. doi: 10.1016/j.brainresbull.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Aylward E, Mills J, Liu D, Nopoulos P, Ross Ca, Pierson R, Paulsen JS. Association between Age and Striatal Volume Stratified by CAG Repeat Length in Prodromal Huntington Disease. PLoS currents. 2011;3 doi: 10.1371/currents.RRN1235. RRN1235. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3092625\&tool=pmcentrez\&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bendfeldt K, Hofstetter L, Kuster P, Traud S, Mueller-Lenke N, Naegelin Y, Kappos L, Gass A, Nichols TE, Barkhof F, Vrenken H, Roosendaal SD, Geurts JJG, Radue E-W, Borgwardt SJ. Longitudinal gray matter changes in multiple sclerosis–differential scanner and overall disease-related effects. Human brain mapping. 2012;33(5):1225–1245. doi: 10.1002/hbm.21279. URL http://www.ncbi.nlm.nih.gov/pubmed/21538703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mungas D, Harvey D, Reed BR, Jagust WJ, DeCarli C, Beckett L, Mack WJ, Kramer JH, Weiner MW, Schuff N, Chui HC. Longitudinal volumetric MRI change and rate of cognitive decline. Neurology. 2005;65(4):565–571. doi: 10.1212/01.wnl.0000172913.88973.0d. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1820871\&tool=pmcentrez\&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierson R, Johnson H, Harris G, Keefe H, Paulsen JS, Andreasen NC, Magnotta VA. Fully automated analysis using BRAINS: AutoWorkup. NeuroImage. 2011;54(1):328–336. doi: 10.1016/j.neuroimage.2010.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soneson C, Fontes M, Zhou Y, Denisov V, Paulsen J, Kirik D, Petersén A, et al. Early changes in the hypothalamic region in prodromal Huntington disease revealed by MRI analysis. Neurobiology of disease. 2010;40(3):531–543. doi: 10.1016/j.nbd.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolz R, Julkunen V, Koikkalainen J, Niskanen E, Zhang DP, Rueckert D, Soininen H, Lötjönen J. Multi-method analysis of MRI images in early diagnostics of Alzheimer’s disease. PloS one. 2011;6(10):e25446. doi: 10.1371/journal.pone.0025446. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3192759\&tool=pmcentrez\&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Younes L, Ratnanather JT, Brown T, Aylward E, Nopoulos P, Johnson H, Magnotta Va, Paulsen JS, Margolis RL, Albin RL, Miller MI, Ross Ca. Regionally selective atrophy of subcortical structures in prodromal HD as revealed by statistical shape analysis. Human brain mapping 00. 2012 Oct;:1–18. doi: 10.1002/hbm.22214. URL http://www.ncbi.nlm.nih.gov/pubmed/23281100. [DOI] [PMC free article] [PubMed]
- 30.Zijdenbos AP, Forghani R, Evans AC. Automatic “pipeline” analysis of 3-D MRI data for clinical trials: application to multiple sclerosis. IEEE transactions on medical imaging. 2002;21(10):1280–1291. doi: 10.1109/TMI.2002.806283. URL http://www.ncbi.nlm.nih.gov/pubmed/12585710. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J, Chan KL, Chong VF, Krishnan SM. Extraction of brain tumor from MR images using one-class support vector machine. Conference proceedings : … Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Conference. 2005;6:6411–6414. doi: 10.1109/IEMBS.2005.1615965. [DOI] [PubMed] [Google Scholar]
- 32.Akselrod-Ballin A, Galun M, Gomori MJ, Basri R, Brandt A. Atlas guided identification of brain structures by combining 3D segmentation and SVM classification. Medical Image Computing and Computer-Assisted Intervention. 2006;9(Pt 2):209–216. doi: 10.1007/11866763_26. [DOI] [PubMed] [Google Scholar]
- 33.Guo L, Liu X, Wu Y, Yan W, Shen X. Research on the segmentation of MRI image based on multi-classification support vector machine. Conference proceedings : … Annual International Conference of the IEEE Engineering in Medicine and Biology Society; IEEE Engineering in Medicine and Biology Society. Conference; 2007. pp. 6020–6023. 2007. [DOI] [PubMed] [Google Scholar]
- 34.Ruan S, Lebonvallet S. Multi-kernel SVM based classification for brain tumor segmentation of MRI multi-sequence; 2009 16th IEEE International Conference on Image Processing ICIP; 2009. pp. 3373–3376. [Google Scholar]
- 35.Morra JH, Tu Z, Apostolova LG, Green AE, Toga AW, Thompson PM. Comparison of AdaBoost and support vector machines for detecting Alzheimer’s disease through automated hippocampal segmentation. IEEE Transactions on Medical Imaging. 2010;29(1):30–43. doi: 10.1109/TMI.2009.2021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vrooman HA, Cocosco CA, Van Der Lijn F, Stokking R, Ikram MA, Vernooij MW, Breteler MMB, Niessen WJ. Multi-spectral brain tissue segmentation using automatically trained k-Nearest-Neighbor classification. NeuroImage. 2007;37(1):71–81. doi: 10.1016/j.neuroimage.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 37.Anbeek P, Vincken KL, Groenendaal F, Koeman A, van Osch MJP, van der Grond J. Probabilistic brain tissue segmentation in neonatal magnetic resonance imaging. Pediatric research. 2008;63(2):158–163. doi: 10.1203/PDR.0b013e31815ed071. [DOI] [PubMed] [Google Scholar]
- 38.Powell S, Magnotta Va, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC. Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures. NeuroImage. 2008;39(1):238–247. doi: 10.1016/j.neuroimage.2007.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim EY, Johnson H. Multi-structure segmentation of multi-modal brain images using artificial neural networks. Analysis. 2010;7623 76234B–76234B–12. [Google Scholar]
- 40.Vaidyanathan M, Clarke LP, Heidtman C, Velthuizen RP, Hall LO. Normal brain volume measurements using multispectral MRI segmentation. Magnetic Resonance Imaging. 1997;15(1):87–97. doi: 10.1016/s0730-725x(96)00244-5. [DOI] [PubMed] [Google Scholar]
- 41.Quddus A, Fieguth P, Basir O. Adaboost and Support Vector Machines for White Matter Lesion Segmentation in MR Images. Conference Proceedings of the International Conference of IEEE Engineering in Medicine and Biology Society. 2005;1:463–466. doi: 10.1109/IEMBS.2005.1616447. [DOI] [PubMed] [Google Scholar]
- 42.de Boer R, Vrooman Ha, Ikram MA, Vernooij MW, Breteler MMB, van der Lugt A, Niessen WJ. Accuracy and reproducibility study of automatic MRI brain tissue segmentation methods. NeuroImage. 2010;51(3):1047–1056. doi: 10.1016/j.neuroimage.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 43.Magnotta VA, Matsui JT, Liu D, Johnson HJ, Long JD, Bolster BD, Jr, Mueller BA, Lim KO, Mori S, Helmer KG, et al. Multi-Center Reliability of Diffusion Tensor Imaging. Brain …. 2012;2(6):345–355. doi: 10.1089/brain.2012.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghayoor A, Vaidya JG, Johnson HJ. Development of a novel constellation based landmark detection algorithm. SPIE Medical Imaging. 2013;8669 86693F–86693F–6. URL http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.2006471. [Google Scholar]
- 45.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young Kim E, Johnson HJ. Robust multi-site MR data processing: iterative optimization of bias correction, tissue classification, and registration. Frontiers in Neuroinformatics. 2013 Nov 7;:1–11. doi: 10.3389/fninf.2013.00029. URL http://www.frontiersin.org/Neuroinformatics/10.3389/fninf.2013.00029/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson HJ, Mccormick MM, Ibanez L, editors. I. S. Consortium. The ITK software guide. 3rd Edition. 2013. http://itk.org, URL http://itk.org/ItkSoftwareGuide.pdf. [Google Scholar]
- 48.Tan Pang-Ning, Steinbach Michael, Kumar Vipin. Introduction to Data Mining. 1 edition. Addison Wesley; 2005. [Google Scholar]
- 49.Cybenko G. Approximation by superpositions of a sigmoidal function. Mathematics of Control, Signals, and Systems (MCSS) 1989;2(4):303–314. [Google Scholar]
- 50.Hornik K, Stinchcombe M, White H. Multilayer feedforward networks are universal approximators. Neural networks. 1989;2(5):359–366. [Google Scholar]
- 51.Dietterich T. Ensemble methods in machine learning. Multiple classifier systems. URL http://link.springer.com/chapter/10.1007/3-540-45014-9\_1. [Google Scholar]
- 52.Mavandadi S, Feng S, Yu F, Dimitrov S, Nielsen-Saines K, Prescott WR, Ozcan A. A mathematical framework for combining decisions of multiple experts toward accurate and remote diagnosis of malaria using tele-microscopy. [doi:10.1371/journal.pone.0046192];PloS one. 2012 7(10):e46192. doi: 10.1371/journal.pone.0046192. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3469564\&tool=pmcentrez\&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iba W, Langley P. Induction of One-Level Decision Trees; Proceedings of the Ninth International Conference on …. [Google Scholar]
- 54.Freund Y, Schapire RE. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. Journal of Computer and System Sciences. 1997;55(1):119–139. [Google Scholar]
- 55.Hampel F. Robust statistics : A brief introduction and overview, Symposium; Robust Statistics and Fuzzy Techniques in Geodesy and GIS; URL http://scholar.google.com/scholar?hl=en\&btnG=Search\&q=intitle:Robust+statistics+:+A+brief+introduction+and+overview\#0. [Google Scholar]
- 56.Huber P. Robust statistics. 2011 URL http://library.mpib-berlin.mpg.de/toc/z2010\_703.pdfhttp://link.springer.com/content/pdf/10.1007/978-3-642-04898-2\_594.pdf. [Google Scholar]
- 57.Erceg-Hurn DM, Mirosevich VM. Modern robust statistical methods: an easy way to maximize the accuracy and power of your research. The American psychologist. 2008;63(7):591–601. doi: 10.1037/0003-066X.63.7.591. [DOI] [PubMed] [Google Scholar]
- 58.Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. 2nd Edition. San Francisco: Morgan Kaufmann; 2005. [Google Scholar]
- 59.Tabrizi SJ, Reilmann R, Roos RaC, Durr A, Leavitt B, Owen G, Jones R, Johnson H, Craufurd D, Hicks SL, Kennard C, Landwehrmeyer B, Stout JC, Borowsky B, Scahill RI, Frost C, Langbehn DR. Potential endpoints for clinical trials in premanifest and early Huntington’s disease in the TRACK-HD study: analysis of 24 month observational data. Lancet neurology. 2012;11(1):42–53. doi: 10.1016/S1474-4422(11)70263-0. URL http://www.ncbi.nlm.nih.gov/pubmed/22137354. [DOI] [PubMed] [Google Scholar]
- 60.Breiman L. Random forests. Machine learning. 2001:1–33. [Google Scholar]
- 61.Criminisi A, Shotton J, editors. Decision Forests for Computer Vision and Medical Image Analysis. London: Springer; 2013. [Google Scholar]
- 62.Shrout PE, Fleiss JL. Intraclass correlations: Uses in assessing rater reliability. Psychological Bulletin. 1979;86(2):420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 63.Sabuncu MR, Yeo BTT, Van Leemput K, Fischl B, Golland P. A generative model for image segmentation based on label fusion. IEEE transactions on medical imaging. 2010;29(10):1714–1729. doi: 10.1109/TMI.2010.2050897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jodoin P-M, Mignotte M, Rosenberger C. Segmentation framework based on label field fusion. IEEE Transactions on Image Processing. 2007;16(10):2535–2550. doi: 10.1109/tip.2007.903841. [DOI] [PubMed] [Google Scholar]
- 65.Mallar Chakravarty M, Steadman P, van Eede MC, Calcott RD, Gu V, Shaw P, Raznahan A, Louis Collins D, Lerch JP. Performing label-fusion-based segmentation using multiple automatically generated templates. Hum Brain Mapp. doi: 10.1002/hbm.22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H, Yushkevich P. Multi-atlas segmentation with joint label fusion and corrective learningan open source implementation. Frontiers in neuroinformatics. doi: 10.3389/fninf.2013.00027. URL http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3837555/ [DOI] [PMC free article] [PubMed]
- 67.Wang H, Suh JW, Das SR, Pluta J, Craige C, Yushkevich Pa. Multi-Atlas Segmentation with Joint Label Fusion. IEEE transactions on pattern analysis and machine intelligence. 2012;35(3):611–623. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang L, Wang X, Penwarden N, Ji Q. An Image Segmentation Framework Based on Patch Segmentation Fusion. EUSIPCO. 2006:1–5. Vol. 00, Ieee, 2006. [Google Scholar]
- 69.Daoqiang Zhang GWHJ, Shen D. LNCS 6893 - Confidence-Guided Sequential Label Fusion for Multi-atlas Based Segmentation. 2011:1–8. doi: 10.1007/978-3-642-23626-6_79. [DOI] [PubMed] [Google Scholar]
- 70.Heckemann Ra, Hajnal JV, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage. 2006;33(1):115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 71.Gousias IS, Hammers A, Counsell SJ, Srinivasan L, Rutherford Ma, Heckemann Ra, Hajnal JV, Rueckert D, Edwards aD. Magnetic resonance imaging of the newborn brain: automatic segmentation of brain images into 50 anatomical regions. PloS one. 2013;8(4):e59990. doi: 10.1371/journal.pone.0059990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage. 2011;56(3):907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khan AR, Beg MF. Multi-structure whole brain registration and population average; Conf Proc IEEE Eng Med Biol Soc; 2009. pp. 5797–5800. 2009. [DOI] [PubMed] [Google Scholar]