Abstract
Chromatin dynamics play an essential role in regulating the accessibility of genomic DNA for a variety of nuclear processes, including gene transcription and DNA repair. The posttranslational modification of the core histones and the action of ATP-dependent chromatin remodeling enzymes represent two primary mechanisms by which chromatin dynamics are controlled and linked to nuclear events. Although there are examples in which a histone modification or a remodeling enzyme may be sufficient to drive a chromatin transition, these mechanisms typically work in concert to integrate regulatory inputs, leading to a coordinated alteration in chromatin structure and function. Indeed, site-specific histone modifications can facilitate the recruitment of chromatin remodeling enzymes to particular genomic regions, or they can regulate the efficiency or the outcome of a chromatin remodeling reaction. Conversely, chromatin remodeling enzymes can also influence, and sometimes directly modulate, the modification state of histones. These functional interactions are generally complex, frequently transient, and often require the association of myriad additional factors.
Keywords: Chromatin remodeling, histone modifications, chromatin dynamics
1. A chromatin primer
Eukaryotic cells organize their genetic material into a DNA-protein complex, called chromatin. In general, chromatin assembly limits the accessibility of genomic sequences, and thus it creates inherent barriers for nuclear events such as transcription, DNA replication, and DNA repair. Consequently, chromatin structure must be dynamic or fluid, and local changes in chromatin structure are utilized to provide the cell with profoundly effective methods for fine-tuning DNA metabolism. Not too surprisingly, disruption of mechanisms that control chromatin dynamics can lead to aberrant gene expression, improper or nonexistent DNA repair, chromosomal translocations, inappropriate proliferation, developmental errors, onocogenesis, or even cell death.1
In its simplest form, chromatin consists of the nucleosome core particle, composed of 147 base pairs of DNA wrapped nearly twice around an octamer containing two copies each of the core histones H2A, H2B, H3, and H4.2 Nucleosomes are assembled into long, linear arrays in which each nucleosome is connected by 10–70 bp of linker DNA, with the length varying between species and cell types.3 The high affinity of the histone octamer for DNA ensures that nucleosome assembly is a significant barrier for enzymes requiring DNA access, and the folding or compaction of nucleosomal arrays can lead to additional constraints on nuclear processes.3 Consequently, this necessitates several methods for regulating the positioning and stability of nucleosomes, as well as means to disrupt nucleosomal array condensation. One method is the post-translational modification of histones, which can modulate chromatin folding4,5 and guide the binding of regulatory proteins.6,7 Non-allelic variants of the core histones, such as H2A.Z or H3.3, can also be incorporated into nucleosomes, and these variant nucleosomes can have altered stability and/or present novel opportunities for posttranslational modifications.8,9 The final, and perhaps most potent mechanism, is the use of ATP-dependent chromatin remodeling enzymes that can re-position, evict, or alter the composition of nucleosomes.10 Indeed, several chromatin remodeling enzymes use the energy of ATP hydrolysis to catalyze the deposition or removal of histone variants, and thus they play an integral role in regulating their chromosomal distributions. There are four subfamilies of ATP-dependent chromatin remodeling enzymes: SWI/SNF, INO80, ISWI, and CHD.1,10,11 Each family is defined by a characteristic ATPase subunit that is related to the DEAD/H superfamily of DNA helicases, but they also possess unique motifs that mediate binding to nucleosomes and individual complex subunits. As discussed in detail in this review, there exist numerous physical and functional interactions between chromatin remodeling enzymes and histone modifications, creating an intertwined network of mechanisms that control chromatin dynamics.
2. Targeting chromatin remodeling enzymes by histone marks
The majority of histone post-translational modifications occur within the 10–30 amino acid N-terminal domains of each of the histones (often called the histone “tails”). These domains extend from the nucleosomal surface, and although they do not directly contribute to the organization of nucleosomal DNA, the N-terminal tails provide interaction surfaces for a host of nucleosome binding proteins, and are essential for chromatin higher order folding.3,6 Histone modifications also occur within the globular domains that organize nucleosomal DNA, and given the remarkable number of histone modifications, it may well be that every solvent-exposed histone residue might be a target for modification. Common modifications include lysine acetylation, the mono-, di-, or tri- methylation of lysines, mono- or di-methylation of arginines, the phosphorylation of serines, threonines, or tyrosines, as well as lysine ubiquitylation and SUMOylation.6 These marks often occur in complex patterns, and there are numerous ongoing studies focused on understanding how such patterns are “read” by cellular machineries.7 Surprisingly, very few histone marks appear to affect chromatin structure dramatically by themselves (for an exception see Shogren-Knaak et al., 20064),6 with the large majority of histone modifications influencing either the binding or activity of other regulatory factors, such as ATP-dependent chromatin remodeling enzymes.
2.1. Bromodomain-acetylated lysine interactions
Since the early studies of Allfrey and colleagues, the acetylation of histone lysines, especially those in the H3 and H4 tails, has been described as a hallmark of transcriptionally active chromatin.12–14 Since the acetylation of a lysine neutralizes its positive charge, it was long believed that acetylated nucleosomes might be less stable or harbor less tightly bound DNA. These “looser” or “open” structures were proposed to facilitate transcription. Although this simple model is attractive, very little evidence supports this view. High levels of histone acetylation can disrupt chromatin condensation4,15 and slightly increase the mobility of nucleosomes on DNA,16 but nucleosomal DNA remains tightly bound to the octamer surface and nucleosome stability appears unchanged. A minor exception is the acetylation of H3-K56 that lies just beneath the first few turns of nucleosomal DNA. In the context of single nucleosomes, this mark slightly enhances the “breathing” of DNA at the very nucleosome edge, though this mark has little effect on the properties of nucleosomal arrays.17,18
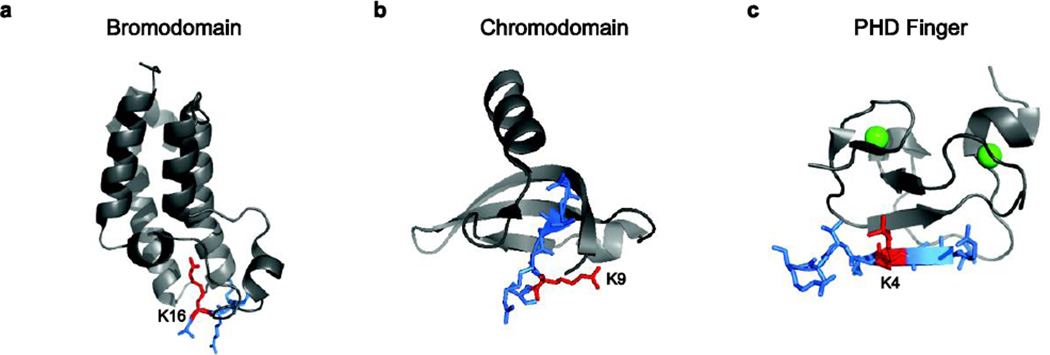
If histone acetylation is not generally sufficient to disrupt chromatin, then how might this type of mark generate active, open chromatin structures? One dominant mechanism appears to be the recruitment of ATP-dependent chromatin remodeling enzymes that contain subunits harboring acetyl-lysine recognition domains, known as bromodomains.19 Bromodomains are ~70 kDa and possess a hydrophobic binding pocket formed by an anti-parallel bundle of four α-helices (Fig. 1a).20,21 The side chain of an acetylated lysine residue inserts into this pocket and is anchored via interactions with bound water molecules and/or an internal asparagine side chain.20,21 Residues on the surface of the binding pocket may mediate sequence-specific interactions with nearby residues on the histone tail, conferring lysine specificity.21 Bromodomains often occur within proteins as tandem units, allowing for proteins that recognize multiple acetyl marks on the same or adjacent nucleosomes.7
Figure 1. Chromatin remodeling enzymes have specific domains that bind histone modifications.
(a) Bromodomains bind acetylated lysines. The bromodomain of Gcn5 bound to an H4 tail peptide acetylated at K16 (PDB: #1E6I). (b) Chromodomains bind methylated lysines. The chromodomain of HP1 protein Swi6 bound to an H3 tail peptide methylated at K9 (PDB: #1KNA). (c) PHD fingers bind methylated lysines. The PHD finger domain of ING2 bound to an H3 tail peptide methylated at K4 (PDB: #2G6Q).
The bromodomain was originally identified as a conserved sequence element of unknown function in a handful of genes including the Drosophila Brm and fsh genes, the S. cerevisiae swi2/snf2 and spt7 genes, and two human genes, ccg1 and ring3.22,23 Notably, BRM is the catalytic subunit of the Drosophila SWI/SNF remodeling enzyme, also called the PBAP complex. Jeanmougin and coworkers expanded the family and the motif, identifying 37 different members including a number of additional histone acetyltransferases (HATs), as well as the human (BRG1 and BRM) and budding yeast (Swi2/Snf2 and Sth1) homologs of BRM.24 Indeed, the presence of a bromodomain that is C-terminal to the conserved ATPase domain is a defining feature of the SWI/SNF family of remodeling enzymes.25
A functional relationship between histone acetylation and chromatin remodeling was first suggested by genetic experiments in yeast. First, the Gcn5 histone acetyltransferase and the SWI/SNF remodeling enzyme were found to regulate very similar sets of genes, and yeast strains lacking both enzymes were found to be extremely sick or inviable.26,27 Each of these chromatin enzymes also showed similar genetic interactions with other chromatin components,28 and chromatin immunoprecipitation studies indicated that the Gcn5 HAT is required for recruitment of the SWI/SNF enzyme to several inducible genes,29,30 presumably via the Swi2/Snf2 bromodomain. Likewise, a number of studies found that recruitment of the human SWI/SNF remodeling enzyme to a variety of target genes is also dependent on histone acetylation.31,32 Indeed, in an elegant study by Thanos and colleagues, it was found that the acetylation of lysines 8 and 12 of the histone H4 tail was specifically required for hSWI/SNF ATPase subunit BRG1’s recruitment to the IFN-β gene in vitro and in vivo, and the in vitro interaction could be ablated by the addition of excess competitor BRG1 bromodomain.33 Of course HATs and remodeling enzymes regulate processes other than transcription, and recent studies indicate that Gcn5-dependent histone acetylation also controls the recruitment of human SWI/SNF to DNA double strand breaks via the bromodomain on the BRG1 ATPase subunit (see additional detail below).34
Gcn5 is the catalytic subunit of several HAT complexes, including the SAGA complex, which acetylates histone H3-K4, 9, 14, 18, and 23 within the N-terminal tail.8,27 Two subunits of the SAGA complex, including Gcn5, contain bromodomains which may help to maintain the enzyme at target loci by interaction with its acetylated nucleosomal products.35 Workman and colleagues found that histone acetylation by the SAGA complex could promote SWI/SNF recruitment in vitro, and that SWI/SNF has a higher affinity for nucleosomes that contain tetra-acetylated H3 tails.36–38 Specifically, it was found that acetylated nucleosomes slowed the dissociation of SWI/SNF from arrays.36–38 These biochemical studies lend strong support to the view that histone marks, such as lysine acetylation, may “recruit” factors primarily by slowing their off-rate rather than promoting their initial association with a target locus. This view is supported by further genetic studies in yeast where removal of the Swi2/Snf2 bromodomain disrupts recruitment in vivo,38 though it is not entirely eliminated. In contrast, inactivation of SWI/SNF domains that mediate interactions with site-specific transcriptional activators has a much more severe effect on SWI/SNF function and recruitment.39,40 These studies suggest a unified view in which chromatin remodeling enzymes, such as SWI/SNF, are initially recruited to regulatory regions via interactions with site-specific DNA binding factors, and the subsequent interactions between bromodomains and acetylated histones stabilize chromatin associations, leading to stable nucleosome interactions that may be more productive for remodeling.
Whereas removal of the sole bromodomain within yeast SWI/SNF does not have dire consequences, the bromodomains within the related RSC remodeling enzyme play essential roles. Like SWI/SNF, mutations that affect the RSC complex are synthetically inviable in combination with inactivation of Gcn5, and RSC has also been shown to bind tetra-acetylated H3 nucleosomes in vitro.41,42 However, unlike the yeast SWI/SNF complex, the RSC complex and its fly and mammalian counterparts, PBAP and PBAF, have multiple bromodomains.43,44 Indeed, the namesake subunit of PBAP and BPAF is called Polybromo after its six bromodomains (Polybromo is also known as BAF180).45 This subunit appears to be an evolutionary fusion of the Rsc1, Rsc2, and Rsc4 subunits of yeast RSC which each have two bromodomains.46 Human SWI/SNF also appears to contain additional bromodomain-containing subunits, Brd7 and Brd9.46 Together with the single bromodomain within the catalytic subunit, these remodeling enzymes may contain between 4 and 10 bromodomains!
Why so many acetyl-lysine recognition modules? Some bromodomains may act in a redundant fashion, or they may bind different acetylated residues on adjacent nucleosomes or even on nearby transcription factors. Some bromodomains are clearly not redundant, as genetic studies have shown that one of the two bromodomains within both yeast Rsc1 and Rsc2 (which are mutually exclusive subunits of RSC) are essential for RSC function, and deletion of either tandem bromodomain within yeast Rsc4 also inactivates RSC function in vivo.47 One of the two bromodomains in Rsc4, and one of the six bromodomains in Drosophila Polybromo has been shown to bind specifically to H3-K14ac, and in vitro studies have demonstrate that H3-K14ac can recruit yeast RSC to nucleosomal substrates.16,41
Interestingly, work from the Cairns lab has shown that the multiple bromodomains within yeast RSC are involved in an autoregulatory mechanism that is controlled by the Gcn5 HAT.43 In addition to its typical substrate, the H3 tail, Gcn5 also acetylates K45 of Rsc4.43 The Rsc4-K45ac mark creates an intramolecular binding site for the first bromodomain of Rsc4, and this binding event blocks the second bromodomain from binding H3-K14.43 This implicates Gcn5 in both the positive and negative regulation of RSC, and suggests that bromodomain function is not confined simply to recruitment events.
Although bromodomains are generally a feature of the SWI/SNF family of remodeling enzymes, the yeast SWR-C also possesses two bromodomains within its subunit Bdf1.48 SWRC is a member of the INO80 family of remodeling enzymes that catalyzes the ATP-dependent exchange of H2A/H2B dimers for H2A.Z/H2B dimers, leading to targeted deposition of the H2A.Z histone variant within promoter proximal nucleosomes.49–51 This H2A.Z deposition reaction is greatly stimulated by histone H4 acetylation in vitro,48 and studies have indicated that the NuA4 HAT plays a key role in H2A.Z deposition by SWR-C in vivo.52 Recent studies from Wu and colleagues suggests a model in which SWR-C is targeted to nucleosome free regions upstream of RNA polymerase II promoters, and then it preferentially interacts with an adjacent, acetylated nucleosome via its Bdf1 subunit which then targets that nucleosome for efficient H2A.Z deposition.53 This model is consistent with a general theme that histone marks serve as a mechanism to fine-tune the positioning of a chromatin remodeling enzyme, rather than functioning as the primary method of recruitment.
2.2 Chromatin remodeling enzymes that bind methylated lysines
In contrast to lysine acetylation, the mono-, di-, or trimethylation of lysine residues does not neutralize their positive charge, and thus studies have long focused on identifying domains and proteins that recognize these marks rather than speculating on direct changes to chromatin structure.6 Furthermore, whereas lysine acetylation is generally a hallmark of transcriptionally active chromatin, lysine methylation appears in both active and inactive chromatin domains. For instance, repetitive DNA sequences that are packaged into condensed, heterochromatic domains are enriched in H3-K9 di- and tri-methylation, and developmentally silenced loci contain H3-K27 dimethylation.54 In contrast, actively transcribed genes contain nucleosomes that are di- and tri-methylated at H3-K4, K36, and K79.54 H3-K4me2/3 is enriched at the 5’ end of transcribed genes, whereas H3-K36me2/3 and K79me2/3 are enriched in gene bodies.6,54,55 As for acetyl lysine, both the “active” and “inactive” methyl marks are targets for chromatin remodeling enzymes.
The two most common methyl-lysine binding domains found in chromatin remodeling enzymes are plant homeodomain (PHD) fingers and chromodomains.7 A third common methyl lysine binding domain, called Malignant Brain Tumor (MBT) repeats, has not as yet been identified in remodeling enzymes, but to date this domain is restricted to factors involved in chromatin silencing (e.g. L3MBTL1).56–58 The first chromodomains were identified in the heterochromatin structural proteins, Polycomb and heterochromatin protein 1 (HP1).59 Chromodomains bind methyl-lysine side chains in a cage formed by aromatic residues along an anti-parallel β-sheet,60,61 whereas PHD finger domains consist of a two-stranded β-sheet and one or more α-helices coordinated by zinc ions (Fig. 1b–c).62 As in chromodomains, the methyl-lysine side chain binds within the β-sheet in a pocket lined by aromatic residues.7 In addition to the recognition of methylated lysines, some chromodomains can also bind to DNA and RNA, leading to the proposal that the chromodomain may have evolved from an ancestral fold with a wide-target specificity.63 Similarly, a PHD finger from the Msc1 subunit of the fission yeast SWR-C remodeling enzyme also exhibits E3 ubiquitin ligase activity, suggesting that PHD fingers can also have multiple functions.64
The CHD family of chromatin remodeling proteins is highly conserved from yeast to human. The distinguishing characteristic of this family is the presence of N-terminal, tandem chromodomains in addition to a C-terminal Swi2/Snf2 ATPase domain.1,65 There are three subfamilies of CHD proteins, the Chd1–2 family, the Chd3–4 family (also known as Mi-2α and Mi-2β), and the Chd5–9 family.1,65 The Chd1 and Chd2 proteins contain an additional DNA binding domain at their C-terminus, whereas Chd3–4 members contain one or more PHD fingers.1,65 The Chd5–9 family is more diverse and harbors other types of motifs, such as SANT domains that can interact with unmodified histone tails.66 The chromodomains of mouse and possibly Drosophila Chd1 are required for normal nuclear localization patterns and are essential for enzymatic activity,67–69 and the chromodomain and PHD fingers of Chd3/4 members are important for nucleosome binding and chromatin remodeling activities.69
Similar to SWI/SNF family members, some CHD remodeling enzymes are recruited to target loci by a combination of site-specific DNA binding proteins and histone modifications. For example, the NuRD complex is a multi-subunit remodeling enzyme conserved from flies to humans that harbors a Chd3/4 family member.65,70 NuRD enzymes are targeted by a variety of site-specific DNA binding proteins to repress transcription of genes during development. For example, the Drosophila NuRD complex is recruited by the hunchback (Hb) repressor to repress HOX gene transcription,71 and mammalian NuRD complexes interact with a variety of transcriptional repressors to regulate lineage commitment and differentiation of specific cell types.72–74 In addition to binding transcription factors, the PHD fingers of Chd3/4 members interact with H3 peptides methylated at either K9 or K36, and these interactions can be regulated by the methylation state of H3-K4.70,75 Like the case for SWI/SNF family members, it seems likely the recognition of histone marks by PHD fingers functions to stabilize a CHD family member at a chromatin locus following its initial recruitment.
Human Chd1 binds H3-K4me2/3 via one of its two chromodomains which helps to target it to active genes where it facilitates transcription.76,77 It is controversial whether yeast Chd1 is able to recognize H3-K4me, as its chromodomains lack one of the aromatic residues necessary for binding, and conflicting studies report on yChd1 interactions with the H3 tail.54,76–78 However, yeast Chd1 is known to associate with the SAGA and SLIK complexes, and thus yChd1 may be the factor that allows these HATs to target H3-K4me nucleosomes for H3 acetylation.76 Nevertheless, from yeast to mammals, Chd1 is recruited to the 5’ end of active genes via interactions with transcriptional initiators such as Mediator and PAF1, where its remodeling activity facilitates nucleosome turnover and gene expression.79,80 Intriguingly, recent evidence also suggests that yChd1 may work in conjunction with the ISWI family of remodeling enzymes to inhibit cryptic transcription in gene bodies by stabilizing nucleosomes harboring H3-K36me (see discussion in Section 5.2., below).
Several members of the ISWI family of remodeling enzymes contain subunits harboring PHD fingers that interact with H3-K4me3. For instance, the human NURF complex, a member of the ISWI family of ATP-dependent chromatin remodelers, recognizes H3-K4 methylation through its subunit BPTF.81 Interestingly, BPTF contains both a bromodomain and a PHD finger domain separated by an α-helical linker.82 When the PHD finger is bound to an H3-K4me3 nucleosome, the bromodomain selectively binds acetylated H4-K16 on the same nucleosome, and, in fact, this binding is cooperative.83 When both marks are bound, the linker region of BPTF can be positioned into the groove of DNA, a contact which may be involved in the remodeling mechanism.84 NURF is implicated in the transcriptional activation of genes during development, as its deletion leads to developmental defects in flies and frogs and is embryonic lethal in mice.85–87 Somewhat paradoxically, NURF’s specific association with nucleosomes requires active transcription, as H3-K4 methylation occurs during or immediately following elongation.54 However, since NURF also interacts with other transcriptional activators, it may be recruited to promoters by these other factors, where it can then assist in the remodeling of H3-K4me/H4-K16ac nucleosomes for rapid re-initiation. This theory is supported by NURF’s known role in the expression of inducible genes, such as heat shock protein genes in flies.85
3. Remodeling complexes that establish or remove histone marks
Although chromatin remodeling enzymes are defined by their ATP-dependent activities, several enzymes contain additional subunits with enzymatic activities that directly modulate histone modifications. As discussed below in four examples, these novel activities can regulate or target a chromatin remodeling event or further reinforce the desired transcriptional outcome.
3.1. Histone acetylation: The Tip60 complex
Tip60 was initially identified as a protein that interacted with the HIV transactivator, Tat,88 and functions as a transcriptional coactivator for a host of regulatory factors,89 including many nuclear hormone receptors.90 Subsequently Tip60 was shown to be one of the founding members of the MYST family of histone acetyltransferases that acetylate N-terminal lysine residues within histones H4 and H2A.91 Biochemical studies discovered that Tip60 is a subunit of a large protein complex that also contains a member of the INO80 family of ATPases, p400/domino, that is most highly related to yeast Swr1.92,93 Indeed, an elegant study from the Cote group demonstrated that the mammalian Tip60 complex is a physical merger of the essential yeast NuA4 HAT and SWR-C chromatin remodeling complexes that links histone acetylation to the ATP-dependent deposition of the H2A.Z histone variant.94 In this case, H4 acetylation not only enhances the binding of the complex to target nucleosomes via the bromodomain subunit, Bdf1 (Brd8 in humans), but acetylation also appears to directly stimulate the dimer exchange reaction.48 Interestingly the Tip60 HAT also harbors a chromodomain that appears to target this enzyme complex to histone H3-K9me2 at sites of DNA damage (Fig. 2a). Indeed, studies in Drosophila cells showed that Tip60 associates with DNA double strand breaks, where it acetylates the DNA damaged-induced, phosphorylated form of histone H2A.X, leading to its subsequent replacement with H2A.93 Recent studies of Tip60 have focused on embryonic stem cells, where the HAT activity of Tip60 plays a key role in stem cell maintenance and the repression of many developmental genes by the master regulator, Nanog.89
Figure 2. Chromatin remodeling enzymes can modify histone marks.
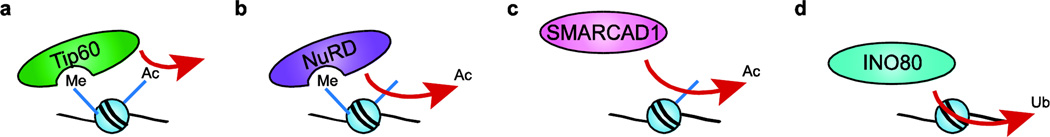
(a) Tip60 binds H3-K9me nucleosomes and acetylates H4 tails. (b) The NuRD complex binds H3-K9me nucleosomes and deacetylates them. (c) SMARCAD1 deacetylates newly-deposited nucleosomes during replication via association with subunits HDAC1 and HDAC2. (d) The Uch37 subunit of human INO80 has been recognized to posses DUB activity that may allow INO80 to target nucleosomes for deubiquitylation.
3.2. Histone deacetylation: NuRD and SMARCAD1
As discussed above, the NuRD complex is a highly conserved remodeling enzyme that functions in myriad transcriptional repression pathways during development. In addition to its Chd3/4 ATPase subunits that interact with H3-K9 or K36, the NuRD complex also contains two histone deacetylase subunits, HDAC1 and HDAC2, which target “active” histone acetyl-lysine marks for deacetylation (Fig. 2b).70 Interestingly, HDAC activity is stimulated by ATP hydrolysis, implicating nucleosome remodeling as an essential step in this process.95,96 A role for NuRD in establishing and maintaining repressive chromatin structures is further reinforced by the Mbd2 and Mbd3 subunits that interact with CpG methylated DNA.97,98 The ability of NuRD ability to modulate gene expression is important in regulating pluripotency and cell fate decisions in hematopoietic and embryonic stem cells.74 NuRD also associates with pericentric heterochromatin during S phase, where it forms higher-order chromatin structures necessary for S phase progression.99,100 Consistent with these activities, the loss of NuRD subunits is associated with both normal and premature aging.101
The mammalian SMARCAD1 remodeling enzyme also harbors the HDAC1 and HDAC2 deacetylase subunits.102 SMARCAD1 is brought to replicating regions via direct interaction with the replicative clamp, PCNA, where it deacetylates the newly deposited H3 and H4 histones (Fig. 2c).103 In combination with the subsequent methylation of H3-K9, these hypoacetylated nucleosomes facilitate an environment favorable to chromatin compaction and binding by heterochromatin proteins such as HP1, and thus SMARCAD1 functions to maintain heterochromatic regions during S phase.103 Congruently, inactivation of SMARCAD1 leads to hyperacetylation, chromosome segregation defects, and genome instability.103 Recently, SMARCAD1 and its budding yeast homolog Fun30 have also been implicated in promoting DNA processing at DNA double strand breaks,104,105 although the exact mechanism remains unknown.
3.3. Histone deubiquitylation: hINO80
The mono-ubiquitylation of lysine residues within the C-terminal tail domains of histones H2A and H2B occurs within both transcriptionally active chromatin domains and near sites of DNA double strand breaks.9 The mono-ubiquitylation of H2B-K123 is essential for establishing methylation of H3-K4 and H3-K79, and represents one of the best examples of histone modification crosstalk.106 Whereas histone deacetylases work in opposition to HATs, a large family of deubiquitylating enzymes (DUBs) functions to remove ubiquitin marks from proteins, including histones. Typically, DUBs physically interact with the proteasome machinery, and function to recycle ubiquitin molecules prior to ubiquitin-dependent degradation.107 Proteomic studies identified the DUB, Uch37, as an integral subunit of the human INO80 complex, where it associates with the DNA-binding subunit NFRKB.108 When present in hINO80, Uch37 is blocked from binding its ubiquitin substrate, but when Uch37 comes into contact with Rpn13, the 19S proteasome ubiquitin receptor, the DUB activity of Uch37 activity is activated.108 This interaction with Rpn13 is transient, occurs at the auto-inhibitory c-terminal tail of Uch37, and does not disrupt Uch37’s incorporation into INO80.108 Although the ubiquitylated substrate for Uch37 is unknown, its incorporation into the hINO80 complex makes a strong case that it may act on H2A-ub and/or H2B-ub (Fig. 2d).107 Current models propose that the DUB activity of hINO80 may play a key role in promoting transcriptional activation and DNA repair.
4. Histone marks that regulate remodeling enzyme activity
Chromatin remodeling enzymes make intimate contacts with their nucleosomal substrates, and thus it seems likely that they would take advantage of histone posttranslational modifications as a regulatory mechanism. Since the early genetic interactions were identified between SWI/SNF and the Gcn5 HAT,28 many studies have tested whether histone acetylation impacts ATP-dependent remodeling reactions. Generally, however, histone hyperacetylation does not have a dramatic impact on the ATPase or nucleosomal array remodeling activities of many enzymes, including yeast and human SWI/SNF, Xenopus NuRD (xMi-2), yeast RSC, or Drosophila CHRAC.11 Early work, though, showed that histone hyperacetylation inhibits the ability of ySWI/SNF to remodel multiple nucleosomal arrays (catalytic array remodeling), likely due to bromodomain effects on binding affinity.109 In contrast, several members of the ISWI family of enzymes are inhibited by histone hyperacetylation, such as Drosophila NURF,11 and yeast Chd1 and Isw2,16 due in part to acetylation of histone H4-K16 (see below).
Whereas early studies employed hyperacetylated histones isolated from cells treated with histone deacetylase inhibitors (e.g. butyrate), more recent studies have used native ligation methods to generate histones that are acetylated at defined locations.16,42 In this case, it was found that H4 acetylation, but not H3, altered the proportion of remodeled products by the yeast RSC enzyme, due to enhanced octamer transfer in trans.16 Furthermore, hyperacetylation of histone H3, but not H4, enhanced the ability of ySWI/SNF and yRSC to slide nucleosomes in cis and to evict H2A/H2B dimers.42 Interestingly, in the case of ySWI/SNF, these stimulatory effects required the Swi2/Snf2 bromodomain.42 Thus, these data indicate that bromodomainacetyl lysine interactions may not solely influence binding affinity, but also alter remodeling activity.
4.1. H4 K16 acetylation
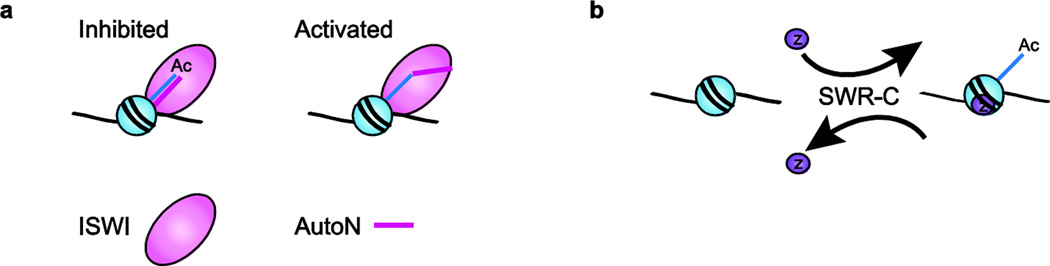
There are two clear examples where a single histone mark can impact a remodeling enzyme. The first is the inhibitory effect of H4-K16ac on the activity of ISWI family members. Genetic studies in Drosophila demonstrated that overexpression of the MOF acetyltransferase, the enzyme that catalyzes H4-K16ac, enhances the phenotype of an ISWI mutant, whereas inactivation of MOF alleviates the X-chromosome condensation phenotype of an ISWI mutant.110 Consistent with these genetic studies, the ISWI subfamily of remodeling enzymes requires the histone H4 N-terminal tail to mobilize nucleosomes in cis, and this activity is reduced ~4-fold by histone H4-K16ac.4 Recent studies indicate that this requirement for the H4 tail, and the inhibition by H4-K16ac, is due to an auto-inhibitory mechanism in which an ISWI domain (called AutoN) mimics the H4 tail and occludes the ATPase domain. This inhibitory domain is only released after engaging the H4 tail of a nucleosomal substrate (Fig. 3a).111,112 Indeed, removal of the ISWI AutoN domain creates an ISWI enzyme that no longer requires the H4 tail for activity.111–113 A similar auto-regulatory mechanism also appears to control the activity of yeast Chd1 which is also most active when bound to nucleosomes with an intact H4 tail.112,114
Figure 3. Histone modifications can regulate chromatin remodeling activity.
(a) ISWI contains a regulatory region homologous to the H4 tail, called AutoN, which binds the ATPase domain and inhibits activity. When ISWI binds the H4 tail, AutoN is displaced and ATPase activity resumes. However, H4 tails acetylated at K16 do not displace AutoN, and consequently inhibit ISWI function. (b) SWR-C exchanges H2A/H2B dimers for dimers containing H2A.Z. However, the presence of H3 K56ac alters the substrate specificity of SWR-C, allowing it to exchange dimers in both directions.
4.2. H3 K56 acetylation Histone
H3 is acetylated at K56 prior to being incorporated into nucleosomes, making it a mark of newly deposited nucleosomes.115,116 As a result, H3-K56ac is highly enriched within newly replicated chromatin and at gene promoters where nucleosomes are rapidly exchanged.117,118 Genetic studies have shown that the levels of H3-K56ac are important for maintenance of genome stability, and loss of H3-K56Ac also affects the regulation of a subset of inducible genes.119,120 H3-K56Ac was initially believed to be a histone mark that was unique to fungi, though more recent studies have found that this mark also occurs in mammalian cells,121 including embryonic stem cells.122 Unlike many histone marks, H3-K56 is located within the globular core of H3, on the surface of the nucleosome near the entry-exit point of DNA.2 H3-K56 is engaged in a water-mediated contact with the last turn of nucleosomal DNA, and its location led to the hypothesis that the loss of positive charge upon acetylation might reduce the interaction between histones and DNA, creating a less stable nucleosome.18 However, FRET experiments designed to measure nucleosomal breathing found only a modest effect of H3 K56ac, crystal structures bearing H3-K56Q or K56E substitutions showed no difference in nucleosome structure, and an H3-K56Q substitution did not disrupt 30 nm fiber compaction.17,18
Although H3-K56Ac does not seem to alter nucleosome structure, this mark does impact the functioning of several chromatin remodeling enzymes. First, it was found that H3-K56Ac has a modest effect on the rate of nucleosome remodeling by the yeast SWI/SNF and RSC enzymes (~1.4-fold increase),18 presumably due to the slight weakening of entry-exit DNA contact with the histone octamer. However, much more dramatic effects have been observed on the activity of INO80 family members.123 Remarkably, this single histone mark switches the substrate specificity of the yeast SWR-C complex, allowing it to catalyze the deposition of H2A/H2B dimers as well as H2A.Z/H2B dimers (Fig. 3b).123,124 H3-K56Ac also enhances the ability of the INO80 complex to evict H2A.Z/H2B dimers from nucleosomal substrates.123 Further mechanistic studies suggest that H3-K56Ac functionally interacts with the Swc2 subunit of SWR-C and that it alters the ability of the enzyme to discern a nucleosomal substrate from a remodeling product.123 Consequently, when SWR-C acts on an H3-K56Ac nucleosome, it appears to remove an H2A.Z/H2B dimer from its product, replacing it with an H2A/H2B dimer, creating a cycle of continuous dimer exchange.123 In vivo, constitutive H3-K56ac decreases the level of H2A.Z. at promoters throughout the yeast genome, consistent with H3-K56ac disrupting SWR-C activity.123
5. Multi-faceted interactions
The preceding sections have dealt with cases in which histone modifications and chromatin remodeling enzymes have direct, physical interactions that lead to functional consequences. In some other cases, however, the interactions are not direct, but rather the outcome of a chromatin remodeling event can alter a subsequent histone modification. One simple example that we will not discuss in detail would be a scenario in which a chromatin remodeling event promotes the binding of a site-specific DNA binding factor that subsequently recruits a histone modification enzyme (for example, see Cosma et al., 1999; Krebs et al., 1999).29,125
5.1. Chromatin remodeling and histone mark dynamics at DNA double-strand breaks
The formation of a DNA double-strand break (DSB) can be a catastrophic event during which the cell cycle is arrested and numerous proteins are recruited to mediate its repair, including many chromatin remodeling and histone-modifying enzymes. Although headway has been made in uncovering the roles of some of these proteins in the DSB response, the function of many chromatin modifiers remains unclear (for a review, see Papamichos-Chronakis and Peterson, 2013).9,126 One of the earliest signs of a DSB is the widespread phosphorylation of the histone variant H2A.X. at S129 in yeast and at S139 in mammals (often referred to as γ-H2A.X).127,128 Other histone marks associated with open chromatin structure also accumulate rapidly around DSBs, including H3-K4me3 and H2A and H2B ubiquitylation within their C-terminal domains.9
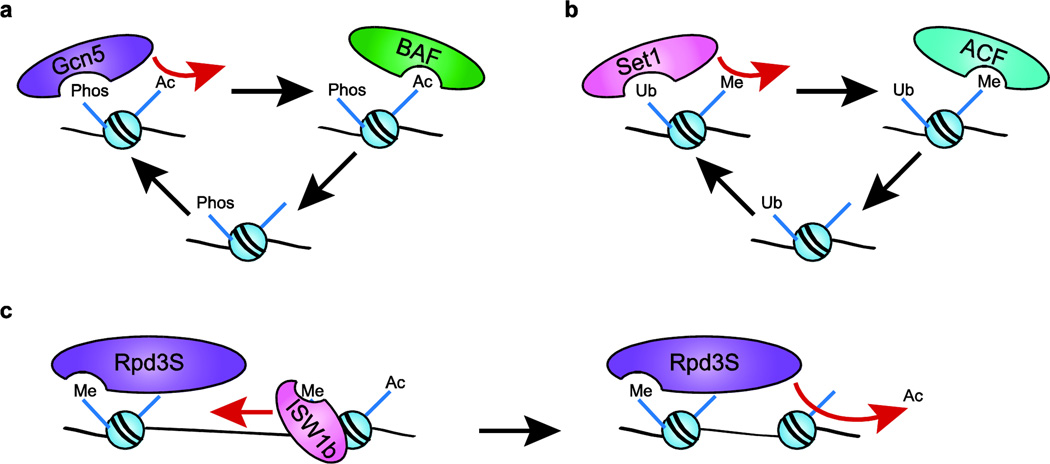
Some of the many chromatin remodeling complexes recruited to a DSB are SWI/SNF family members (ySWI/SNF and RSC in yeast or BAF in mammals). In mammalian cells, recruitment of the BAF complex requires γ-H2A.X, even though BAF does not appear to bind this mark directly.129 Furthermore, in the absence of the BAF complex, the domain of γ-H2A.X within DSB chromatin does not spread normally and it does not persist.34,129 Interestingly, Kwon and colleagues find that γ-H2A.X also recruits the human Gcn5 HAT and that the Gcn5-dependent acetylation of H3 promotes BAF recruitment via its associated bromodomain.34 In this case, the subsequent chromatin remodeling directed by BAF is proposed to increase the accessibility of neighboring nucleosomes, enhancing H2A.X phosphorylation, and as a result, more extensive H3 acetylation and BAF binding (Fig. 4a).34 In this way, γ-H2A.X formation leads to a cooperative feed forward loop in which it is able both to propagate itself and to initiate chromatin remodeling around DSBs.
Figure 4. Histone modifications and chromatin remodeling enzymes exhibit complex interactions.
(a) At DSB’s, γ-H2A.X recruits Gcn5 to nucleosomes, where it acetylates the H3 tail and consequently favors BAF binding. BAF then remodels nearby nucleosomes, leading to spreading of the γ-H2A.X mark and continuation of the cycle. (b) H2B ubiquitylation at a DSB recruits Set1, which methylates H3 K4, leading to binding by the ACF subunit SNF2H. The remodeling enzyme then remodels nearby nucleosomes in a way that favors spreading of the ubiquitin mark. (c) H3-K36me nucleosomes are targeted by Rpd3S for deacetylation in gene bodies. ISWI1b and CHD1 space methylated nucleosomes preferentially for Rpd3S binding to two nucleosomes at once.
A similar mechanism appears to control recruitment of the ISWI homolog, Snf2H, to mammalian DSBs. In this case the Rnf20/Rnf40 E3 ubiquitin ligase heterodimer ubiquitylates H2B-K120 at DSBs, and H2B-K120ub is required for efficient methylation of H3-K4 by the Set1 enzyme.130 H3-K4me then promotes the binding of SNF2H, a human homolog of Isw1 present in the ACF chromatin remodeling complex (Fig. 4b).130 Depletion of SNF2H prevents the formation of RPA, Rad51, and BRCA1 foci around a DSB,130 indicating that this histone modification-chromatin remodeling regulatory loop plays a key role in early recruitment of repair factors.
5.2. Chromatin dynamics during transcription elongation
During transcriptional elongation, nucleosomes are removed in front of the elongating RNA Polymerase II (RNAPII) and then rapidly reassembled after its passage.131 The Set2 methyltransferase is believed to travel with RNAPII whereby it methylates newly deposited nucleosomes at H3-K36; consequently, the level of H3 K36me2/3 within a gene body corresponds to its expression level.132–134 The re-assembly of nucleosomes following RNAPII passage is also associated with deposition of histones that harbor lysine acetylation marks typical of newly replicated chromatin.116,135 If allowed to persist, these acetyl marks appear to promote cryptic transcription within gene bodies.136,137 To counteract these deleterious effects, the Rpd3S deacetylase complex uses both the chromodomain in its subunit Eaf3 and the PHD finger in its subunit Rco1 to target HDAC activity to H3-K36me2/3 nucleosomes within gene bodies.138
Several lines of evidence have also suggested a role for the ISWI and Chd1 remodeling enzymes in controlling Rpd3S activity within gene bodies. Like Set2 methyltransferase, the Isw1 and Chd1 remodeling enzymes are required to inhibit histone acetylation and cryptic transcription within actively transcribed genes.139 The ISW1b complex, via its PHD-finger subunit, Ioc4, and Chd1 (presumably via its chromodomain) interact with H3-K36me3 nucleosomes, and ISW1b co-localizes with H3-K36me in vivo.139,140 Recently, the Li group has demonstrated that CHD and ISWI remodeling enzymes facilitate Rpd3S activity by spacing nucleosomes 30–40 base pairs apart, allowing Rpd3S to bridge two adjacent nucleosomes (Fig 4c).141 The ability of Rpd3S to bind two nucleosomes at once allows it to target both H3-K36me nucleosomes and their neighbors for deacetylation, allowing for the efficient stabilization of nucleosomes, even when chromatin domains are not saturated by the methylation mark.142 Similarly, in mammalian cells, Chd1 has been implicated in stabilizing nucleosome occupancy in gene bodies by mediating H2B ubiquitylation at K123 and K120 by the Rad6 and Bre1 enzymes.55,143 The ability of ISW1 and CHD family remodeling enzymes to slide nucleosomes to the preferred spacing of complexes that bind multiple nucleosomes (and, conversely, to move them outside the preferred linker length to inhibit binding) may be one of their primary functions in vivo.
6. Perspective
Since the identification of the first ATP-dependent remodeling enzyme144–146 and first nuclear histone acetyltransferase,147 there has been a wealth of both data and speculation on interactions between these two classes of chromatin enzymes. The number of chromatin remodeling enzymes and the diversity of histone modifying enzymes is now staggering, but we continue to see more examples of how these enzyme families functionally interface with each other. As illustrated throughout this review, histone marks can directly influence both the targeting and activity of remodeling enzymes, and chromatin remodeling enzymes can potentiate histone marks by both direct and indirect mechanisms. It seems apparent that these two families of enzymes have evolved to work together in the control of all nuclear events.
Highlights.
Chromatin modifications and remodeling act in concert to alter chromatin structure.
Remodeling enzymes have specialized binding domains for targeting histone marks.
Some remodeling complex subunits can establish or remove histone marks.
Histone modifications can regulate remodeling enzyme activity and specificity.
Many processes consist of a plethora of histone modification and remodeling events.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 5.Fierz B, Chatterjee C, McGinty K, Bar-dagan M, Muir W. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011;7:1–7. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013;20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 7.Patel DJ, Wang Z. Readout of epigenetic modifications. Annu. Rev. Biochem. 2013;82:81–118. doi: 10.1146/annurev-biochem-072711-165700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rando OJ, Winston F. Chromatin and transcription in yeast. Genetics. 2012;190:351–387. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papamichos-Chronakis M, Peterson CL. Chromatin and the genome integrity network. Nat. Rev. Genet. 2013;14:62–75. doi: 10.1038/nrg3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 11.Boyer L, et al. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
- 12.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pogo BG, Allfrey VG, Mirsky AE. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 1966;55:805–812. doi: 10.1073/pnas.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidali G, C BL, Bradbury EM, G AV. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc. Natl. Acad. Sci. U. S. A. 1978;75:2239–2243. doi: 10.1073/pnas.75.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of Higher-Order Folding by Core Histone Acetylation Dramatically Enhances Transcription of Nucleosomal Arrays by RNA Polymerase III. Mol. Cell. Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira H, Flaus A, Owen-Hughes T. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J. Mol. Biol. 2007;374:563–579. doi: 10.1016/j.jmb.2007.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe S, et al. Structural characterization of H3K56Q nucleosomes and nucleosomal arrays. Biochim. Biophys. Acta. 2010;1799:480–486. doi: 10.1016/j.bbagrm.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann H, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horn PJ, Peterson CL. The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front. Biosci. 2001;6:D1019–D1023. doi: 10.2741/horn. [DOI] [PubMed] [Google Scholar]
- 20.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 21.Owen DJ, et al. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haynes SR, et al. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamkun JW, et al. Brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 24.Jeanmougin F, Wurtz J-M, Le Douarin B, Chambon P, Losson R. The Bromodomain Revisited. Trends Biochem. Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 25.Dürr H, Flaus A, Owen-Hughes T, Hopfner K-P. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts SM, Winston F. Essential Functional Interactions of SAGA, a Saccharomyces cerevisiae Complex of Spt Ada, and Gcn5 Proteins, With the Snf/Swi and Srb/Mediator Complexes. Genetics. 1997;147:465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollard KJ, Peterson CL. Role for ADA / GCN5 Products in Antagonizing Chromatin- Mediated Transcriptional Repression. Mol. Cell. Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollard KJ, Peterson CL. Chromatin remodeling: a marriage between two families? Bioessays. 1998;20:771–780. doi: 10.1002/(SICI)1521-1878(199809)20:9<771::AID-BIES10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 29.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 30.Natarajan K, Jackson BM, Zhou H, Winston F, Hinnebusch AG. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell. 1999;4:657–664. doi: 10.1016/s1097-2765(00)80217-8. [DOI] [PubMed] [Google Scholar]
- 31.Serna IL. De MyoD Targets Chromatin Remodeling Complexes to the Myogenin Locus Prior to Forming a Stable DNA-Bound Complex MyoD Targets Chromatin Remodeling Complexes to the Myogenin Locus Prior to Forming a Stable DNA-Bound Complex. Mol. Cell Biol. 2005;10:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salma N, Xiao H, Mueller E, Imbalzano AN. Temporal Recruitment of Transcription Factors and SWI / SNF Chromatin-Remodeling Enzymes during Adipogenic Induction of the Peroxisome Proliferator-Activated Receptor γ Nuclear Hormone Receptor. Mol. Cell Biol. 2004;11:4651–4663. doi: 10.1128/MCB.24.11.4651-4663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 34.Lee H-S, Park J-H, Kim S-J, Kwon S-J, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29:1434–1445. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syntichaki P, Topalidou I, Thireos G. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature. 2000;404:414–417. doi: 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- 36.Hassan AH, Neely KE, Workman JL. Histone Acetyltransferase Complexes Stabilize SWI/SNF Binding to Promoter Nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 37.Chandy M, Gutiérrez JL, Prochasson P, Workman JL. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot. Cell. 2006;5:1738–1747. doi: 10.1128/EC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan AH, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 39.Prochasson P, Neely KE, Hassan AH, Li B, Workman JL. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Mol. Cell. 2003;12:983–990. doi: 10.1016/s1097-2765(03)00366-6. [DOI] [PubMed] [Google Scholar]
- 40.Ferreira ME, Prochasson P, Berndt KD, Workman JL, Wright APH. Activator-binding domains of the SWI/SNF chromatin remodeling complex characterized in vitro are required for its recruitment to promoters in vivo. FEBS J. 2009;276:2557–2565. doi: 10.1111/j.1742-4658.2009.06979.x. [DOI] [PubMed] [Google Scholar]
- 41.Kasten M, et al. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chatterjee N, et al. Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res. 2011;39:8378–8391. doi: 10.1093/nar/gkr535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.VanDemark AP, et al. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol. Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Charlop-Powers Z, Zeng L, Zhang Q, Zhou M-M. Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2. Cell Res. 2010;20:529–538. doi: 10.1038/cr.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicolas RH, Goodwin GH. Molecular cloning of polybromo, a nuclear protein containing multiple domains including five bromodomains, a truncated HMG-box, and two repeats of a novel domain. Gene. 1996;175:233–240. doi: 10.1016/0378-1119(96)82845-9. [DOI] [PubMed] [Google Scholar]
- 46.Middeljans E, et al. SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS One. 2012;7:e33834. doi: 10.1371/journal.pone.0033834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cairns BR, et al. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, bromodomains. Mol. Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 48.Altaf M, et al. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J. Biol. Chem. 2010;285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krogan NJ, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 50.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 51.Kobor MS, et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keogh M-C, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ranjan A, et al. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell. 2013;154:1232–1245. doi: 10.1016/j.cell.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sims RJ, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 55.Petty E, Pillus L. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet. 2013;29:621–629. doi: 10.1016/j.tig.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, et al. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell. 2007;28:677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonasio R, Lecona E, Reinberg D. MBT domain proteins in development and disease. Semin. Cell Dev. Biol. 2010;21:221–230. doi: 10.1016/j.semcdb.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trojer P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 59.Paro R, Hogness DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl. Acad. Sci. U. S. A. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fischle W, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Min J, Zhang Y, Xu R-M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J. Mol. Biol. 2000;304:723–729. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- 63.Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. Bioessays. 2004;26:133–140. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- 64.Qiu X, Dul BE, Walworth NC. Activity of a C-terminal plant homeodomain (PHD) of Msc1 is essential for function. J. Biol. Chem. 2010;285:36828–36835. doi: 10.1074/jbc.M110.157792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marfella CGa, Imbalzano AN. The Chd family of chromatin remodelers. Mutat. Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boyer La, Latek RR, Peterson CL. The SANT domain: a unique histone-tailbinding module? Nat. Rev. Mol. Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 67.Morettini S, et al. The chromodomains of CHD1 are critical for enzymatic activity but less important for chromatin localization. Nucleic Acids Res. 2011;39:3103–3115. doi: 10.1093/nar/gkq1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelley DE, Stokes DG, Perry RP. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase / helicase-like domain for proper association with chromatin. Chromosoma. 1999;108:10–25. doi: 10.1007/s004120050347. [DOI] [PubMed] [Google Scholar]
- 69.Bouazoune K, et al. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 2002;21:2430–2440. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Allen HF, Wade Pa, Kutateladze TG. The NuRD architecture. Cell. Mol. Life Sci. 2013;70:3513–3524. doi: 10.1007/s00018-012-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kehle J. dMi-2, a Hunchback-Interacting Protein That Functions in Polycomb Repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- 72.Kaji K, et al. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 73.Reynolds N, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yildirim O, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Musselman Ca, et al. Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem. J. 2009;423:179–187. doi: 10.1042/BJ20090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pray-Grant MG, Daniel Ja, Schieltz D, Yates JR, Grant Pa. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 77.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 78.Sims RJ, et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin JJ, et al. Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev. 2011;25:2198–2209. doi: 10.1101/gad.17554711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simic R, et al. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 82.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruthenburg AJ, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Badenhorst P, Voas M, Rebay I, Wu C. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 2002;16:3186–3198. doi: 10.1101/gad.1032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song H, Spichiger-Haeusermann C, Basler K. The ISWI-containing NURF complex regulates the output of the canonical Wingless pathway. EMBO Rep. 2009;10:1140–1146. doi: 10.1038/embor.2009.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Landry J, et al. Essential role of chromatin remodeling protein BPTF in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4:e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 89.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brady ME. Tip60 Is a Nuclear Hormone Receptor Coactivator. J. Biol. Chem. 1999;274:17599–17604. doi: 10.1074/jbc.274.25.17599. [DOI] [PubMed] [Google Scholar]
- 91.Smith ER, et al. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 93.Kusch T, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 94.Auger A, et al. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol. Cell. Biol. 2008;28:2257–2270. doi: 10.1128/MCB.01755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lys A, et al. Chromatin deacetylation by an ATP-dependent nucleosome remodeling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 96.Xue Y, et al. NURD, a Novel Complex with Both and Histone Deacetylase Activities. Mol. Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 97.Wade PA, et al. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 98.Zhang Y, et al. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Helbling Chadwick L, Chadwick BP, Jaye DL, Wade PA. The Mi-2/NuRD complex associates with pericentromeric heterochromatin during S phase in rapidly proliferating lymphoid cells. Chromosoma. 2009;118:445–457. doi: 10.1007/s00412-009-0207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sims JK, Wade PA. Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Mol. Biol. Cell. 2011;22:3094–3102. doi: 10.1091/mbc.E11-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pegoraro G, et al. Ageing-related chromatin defects through loss of the NURD complex. Nat. Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mermoud JE, Rowbotham SP, Varga-weisz PD. Keeping chromatin quiet: How nucleosome remodeling restores heterochromatin after replication. Cell Cycle. 2011;10:4017–4025. doi: 10.4161/cc.10.23.18558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rowbotham SP, et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol. Cell. 2011;42:285–296. doi: 10.1016/j.molcel.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 104.Costelloe T, et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489:581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen X, et al. The Fun30 nucleosome remodeller promotes resection of DNA doublestrand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Briggs SD, et al. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 107.Zediak VP, Berger SL. Hit and run: transient deubiquitylase activity in a chromatin-remodeling complex. Mol. Cell. 2008;31:773–774. doi: 10.1016/j.molcel.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yao T, et al. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol. Cell. 2008;31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Logie C, Tse C, Hansen JC, Peterson CL. The core histone N-terminal domains are required for multiple rounds of catalytic chromatin remodeling by the SWI/SNF and RSC complexes. Biochemistry. 1999;38:2514–2522. doi: 10.1021/bi982109d. [DOI] [PubMed] [Google Scholar]
- 110.Corona DFV, Clapier CR, Becker PB, Tamkun JW. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 2002;3:242–247. doi: 10.1093/embo-reports/kvf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Clapier CR, Cairns BR. Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature. 2012;492:280–284. doi: 10.1038/nature11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Manning BJ, Peterson CL. Releasing the brakes on a chromatin-remodeling enzyme. Nat. Struct. Mol. Biol. 2013;20:5–7. doi: 10.1038/nsmb.2482. [DOI] [PubMed] [Google Scholar]
- 113.Mueller-Planitz F, Klinker H, Ludwigsen J, Becker PB. The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat. Struct. Mol. Biol. 2013;20:82–89. doi: 10.1038/nsmb.2457. [DOI] [PubMed] [Google Scholar]
- 114.Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol. Cell. 2010;39:711–723. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peterson CL. Genome integrity: a HAT needs a chaperone. Curr. Biol. 2007;17:R324–R326. doi: 10.1016/j.cub.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 116.Ozdemir A, Masumoto H, Fitzjohn P, Verreault A, Logie C. Histone H3 lysine 56 acetylation: a new twist in the chromosome cycle. Cell Cycle. 2006;5:2602–2608. doi: 10.4161/cc.5.22.3473. [DOI] [PubMed] [Google Scholar]
- 117.Rufiange A, Jacques P-E, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 118.Kaplan T, et al. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 2008;4:e1000270. doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen C-C, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tan Y, Xue Y, Song C, Grunstein M. Acetylated histone H3K56 interacts with Oct4 to promote mouse embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11493–11498. doi: 10.1073/pnas.1309914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science. 2013;340:195–199. doi: 10.1126/science.1229758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Watanabe S, Peterson CL. Chromatin dynamics: Flipping the switch on a chromatin remodeling machine. Cell Cycle. 2013;12:2337–2338. doi: 10.4161/cc.25704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Krebs JE, Kuo MH, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bennett G, Papamichos-Chronakis M, Peterson CL. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat. Commun. 2013;4:2084. doi: 10.1038/ncomms3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shroff R, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Park J-H, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nakamura K, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 131.Williams SK, Tyler JK. Transcriptional regulation by chromatin disassembly and reassembly. Curr. Opin. Genet. Dev. 2007;17:88–93. doi: 10.1016/j.gde.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 132.Kizer KO, et al. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Krogan NJ, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li B, Howe L, Anderson S, Yates JR, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 135.Verreault A. De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev. 2000;14:1430–1438. [PubMed] [Google Scholar]
- 136.Li B, et al. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 138.Li B, et al. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 139.Smolle M, et al. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012;19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Maltby VE, et al. Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin. Mol. Cell. Biol. 2012;32:3479–3485. doi: 10.1128/MCB.00389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee C-H, Wu J, Li B. Chromatin remodelers fine-tune H3K36me-directed deacetylation of neighbor nucleosomes by Rpd3S. Mol. Cell. 2013;52:255–263. doi: 10.1016/j.molcel.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huh J-W, et al. Multivalent di-nucleosome recognition enables the Rpd3S histone deacetylase complex to tolerate decreased H3K36 methylation levels. EMBO J. 2012;31:3564–3574. doi: 10.1038/emboj.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lee J-S, et al. Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1. Genes Dev. 2012;26:914–919. doi: 10.1101/gad.186841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Côté J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 145.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 146.Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 147.Brownell JE, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]