Introduction
Chromatin signaling dynamics fundamentally regulate eukaryotic genomes. The reversible covalent post-translational modification (PTM) of histone proteins by chemical moieties such as phosphate, acetyl and methyl groups constitutes one of the primary chromatin signaling mechanisms. Modular protein domains present within chromatin-regulatory activities recognize or “read” specifically modified histone species and transduce these modified species into distinct downstream biological outcomes. Thus, understanding the molecular basis underlying PTM-mediated signaling at chromatin requires knowledge of both the modification and the partnering reader domains. Over the last ten years, a number of innovative approaches have been developed and employed to discover reader domain binding events with histones. Together, these studies have provided crucial insight into how chromatin pathways influence key cellular programs.
Here, we discuss approaches and limitations of the main methods currently used to define interactions between reader domains and histone post-translational modifications. We focus on lysine methylation as a model chromatin modification that can be used to illustrate the successes and challenges in the field. However, the principles of these approaches can be applied to study other modification systems. Lysine residues can be mono-, di- or tri-methylated, with the potential for at least one unique activity being coupled to the specific lysine residue and extent of methylation on that residue. Thus, methylation of lysine residues on a target protein can increase the signaling potential of the modified protein and as such lead to complex downstream signaling. The principal mechanism by which lysine methylation acts on histones is by mediating modular protein-protein interactions via reader proteins that are sensitive to methylated lysine. In this regard, the proteins that recognize a methylated lysine within a specific sequence context define the outcome of a lysine methylation event. To date, the dozens of methyl-lysine readers that have been discovered fall within ten distinct protein domain families: Chromodomain (CD), plant homeodomain (PHD) finger, Tudor, Malignant Brain Tumor (MBT), Proline-Tryptophan-Tryptophan-Proline (PWWP), Bromo Adjacent Homology (BAH), Ankryin repeats, WD40 repeats, ATRX-DNMT3A-DNMT3L (ADD), and zn-CW. Given the number of potential methylation sites and states on histone proteins and non-histone proteins and the observation that typically several readers exist for a single histone PTM site [1], it is virtually certain that large numbers of readers with important biological behaviors remain to be discovered.
Currently, there are three principal ways to screen for binding of a particular protein domain to a desired histone modification: 1) Hypothesis-driven pairwise screening between protein domains and methylated peptides, 2) High-throughput array-based screening where many protein domains or modified peptides can be probed in a single experiment, and 3) Identification of binding proteins isolated from nuclear extract by quantitative mass spectrometry. Each of these techniques has been utilized to characterize or identify binding interactions with varying degrees of success. Drawing on notable successful examples in the literature, we review the strengths and weakness of these approaches in their ability to identify and define the interaction between a protein domain and its associated methylated lysine.
Pairwise screening of protein domains or histone marks
The existence of methylated lysines on histones has been known for many decades [2]. However, until the discovery of the enzymes that modify histones, the function associated with this modification was largely unknown. The discovery in 2000 that SUV39H1 catalyzes H3K9 methylation fueled our understanding of the role of lysine methylation in the formation of heterochromatin and more broadly in regulating chromatin organization and function [3]. SUV39H1 interacts with the heterochromatin-associated protein HP1, which contains a CD module. Observations, including the proposal that recognition of acetylated lysine by bromodomain-containing proteins recruit the transcriptional machinery to target genes [4, 5] and the localization and activity of SUV39H1, HP1, and H3K9 methylation at heterochromatin, led the Kourzarides and Jenuwein labs to postulate that the CD of HP1 is a candidate H3K9 methyllysine binding domain. To test this hypothesis, peptides of the N-terminal H3 tails were synthesized incorporating various modifications including methylation at lysine 9. Peptide-binding assays with these reagents established a direct interaction between the HP1 CD and H3K9me3 peptides [6, 7]. These studies provided a paradigm for how methylated lysine acts at the molecular level and showed HP1 CD to be the first of many protein domains that function by binding to methylated lysines. Moreover, these two publications established a robust, productive, and straightforward method that has served as a blueprint for candidate-based testing of interactions between chromatin-associated domains and distinct modified histone peptides, of which several examples are described below.
The chromodomain is present in dozens of other proteins including polycomb group proteins. The finding that the HP1 CD can recognize H3K9me3 suggested that other CDs like the one in polycomb might share a similar function. For example, direct peptide-binding assays were performed to demonstrate that Drosophila Polycomb protein could bind H3K27me3 [8]. This work was further expanded to mammalian proteins where many orthologs of the Drosophila Polycomb exist. For example, mouse orthologs present in in the PRC1 complex, which include CBX2 and CBX7, are able to bind H3K27me3 [9]. The crystal structure of the unrelated PRC2 component EED led to the hypothesis that its WD40 propeller domain binds to H3K27me3, which was experimentally validated by candidate-based screening using peptide-binding assays [10, 11].
The chromodomain constitutes one of a few domain families that share structural homology. Other domains within this ‘Royal Family’ include the PWWP, MBT, agenet, and tudor domain. Among the proteins that contain the latter tudor domain, 53BP1 served as an early example of its capability as a methyl-lysine binding module. Our understanding of 53BP1 binding to H4K20me1/2 is rooted in genetic information from Schizosaccharomyces pombe. Specifically, the recruitment of the 53BP1 ortholog Crb2, a DNA damage response and tudor domain-containing protein, to double strand breaks was found to be dependent upon the H4K20 methyltransferase spSet9 [12]. From these data and protein array work from the Bedford lab, it was postulated that the tandem tudor domain of 53BP1 could bind methylated H4K20 [12, 13]. Further structural and biochemical data provided the support necessary to directly determine that the 53BP1 tandem tudor domain bound H4K20me1/2 [14].
Another example for how candidate-based screening of modified peptides was used to identify new methyl-lysine binding modules comes from the example of the PHD finger from the ING family of chromatin-regulatory proteins. The PHD finger of the ING family member ING2 was found to bind to nucleosomes purified from HeLa cells but not to recombinant nucleosomes, where the histones are individually expressed in bacteria and lack PTMs. This finding suggested that the ability of ING2 to interact with nucleosomes is dependent upon a PTM present on HeLa-purified nucleosomes. To determine the responsible modification, the ING2 PHD finger was screened against a large panel of modified histone peptides. This analysis revealed that the ING2 PHD finger was both necessary and sufficient for high affinity and specific binding to H3K4me3-containing peptides [15]. Several other PHD fingers from the ING family (ING1, ING3, ING4, ING5 and yeast YNG1, YNG2, and PHO23) were shown to have the same property [15]. In simultaneous work, the PHD finger of BPTF was also found to bind to H3K4me3 [16]. This study used H3K4me3 peptides to extract candidate domains from cellular extract rather than screening a domain against a panel of modified peptides. Ultimately in both cases, direct peptide pulldowns encompassing many methylated histone residues demonstrated the specificity of the PHD fingers from the INGs and BPTF for H3K4me3. The molecular and biophysical basis for this specificity was elucidated in accompanying publications describing the crystal structures of BPTF and ING2 complexed with H3K4me3 peptides [17, 18].
Each of the discoveries mentioned above provided great insight into our current understanding of protein methylation biology. However, in the absence of a clear and specific hypothesis to be tested, new high-throughput approaches have recently been developed to facilitate identification of novel reader domains and reader domain interactions with methylated proteins.
Array-based high-throughput screening
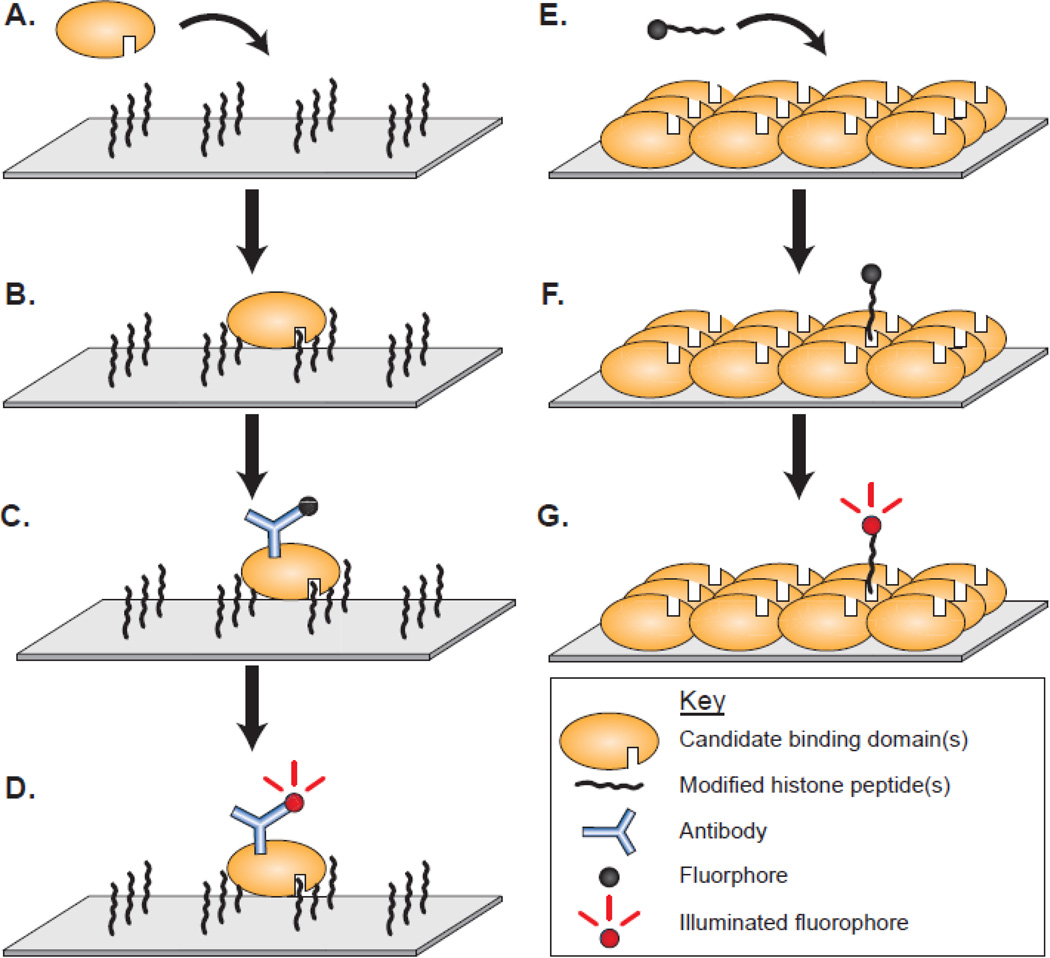
Advances in technology have allowed for higher throughput methods for screening domains and peptides against one another. Both modified peptides and protein domains of chromatin-associated proteins have been printed onto slide array platforms for screening. Each of these techniques has been useful in the discovery and definition of new protein interactions. Generally, these array platforms contain immobilized peptides or proteins upon which a query protein or peptide can be exposed (Figure 1). Common immobilization methods include direct peptide synthesis onto a substrate, biotin-streptavidin affinity, and glutathione-GST affinity, although theoretically any covalent or high affinity interaction could be utilized. Typical arrays can contain hundreds to thousands of individual spots that provide broad accessibility to comprehensive peptide and domain libraries that would otherwise be burdensome to test.
Figure 1.
Array-based screening of candidate binding domains and modified peptides. Peptide arrays contain many unique peptides that are probed with a candidate binding domain. A-B) Domains are incubated on the array surface and allowed interact with the peptides on the array. C) Antibodies are used to detect the domain either directly using fluorescently labeled anti-epitope antibody or indirectly through a fluorescently labeled secondary antibody. D) Binding interactions are visualized using a fluorescent array scanner. Conversely, protein domain arrays are spotted with several unique recombinantly expressed candidate protein domains to be probed with a query modified peptide. E) Fluorescently labeled peptides are applied to the immobilized protein domains. F) The query modified peptide is allowed to bind to the candidate domains. G) Peptides are then visualized as with a peptide array. The visualization schemes for both techniques are not limited to fluorescent visualization as techniques such as enhanced chemiluminescence has also been applied with effective results.
Peptide arrays
Synthesis of biotinylated peptides followed by high performance liquid chromatography (HPLC) purification allows for the production of high quality and pure peptides carrying a diverse set of histone modifications that can be immobilized onto streptavidin-coated slides. In the most straightforward form, a single protein domain can be incubated on a peptide array to act as an initial discovery tool (Figure 1a–d). To date, this approach has been highly productive and has led to the discovery of dozens of novel reader domains, including recent identification of tudor domain proteins that specifically bind to H3K36me3 to regulate PRC2 function and two that directly link disruption of the histone modification readout to human disease [19–24]. For example, a peptide array revealed that the non-canonical PHD finger of RAG2, an essential component of the RAG1/2 V(D)J recombinase that mediates antigen receptor gene assembly, could bind H3K4me3 peptides with great specificity [23]. This interaction was demonstrated to be critical for V(D)J recombination in vivo. Moreover, a residue essential for the interaction is mutated in patients suffering from Omenn's syndrome, an immunodeficiency disease, providing a molecular explanation for the mutation [23]. An array-based approach also led to the discovery that the BAH domain of ORC1 is a novel binding domain with specificity and affinity for H4K20me2 [24]. In this case, the ORC1 BAH domain bound to H4K20me2 peptides but not sixty other methylated peptides on the arrays. Thus, the approach of screening a candidate domain on peptide arrays had uncovered new mechanisms by which chromatin signaling impacts biology and human disease.
Array technology allows for the definition of a single protein domain in a very comprehensive manner. However, one can also perform the same analysis on a large number of domains to define characteristics of a larger protein family previously determined to have potential to recognize modified histones. One such study took on this challenge to examine several domains within the Royal Family of chromatin-associated domains as well as MRG, SWIRM, and BRK domains [25]. Many existing domain-peptide interactions were corroborated using this approach such as the HP1 CD interaction with H3K9me, but new interactions were also found such as the CD of MPP8 binding to methylated H3K9 [25, 26]. Interestingly, an independent peptide array experiment showed that MPP8 and HP1β could bind H3K9me and H3K23me, an interaction not seen in the previous array study [27]. While the array platforms and methodology were similar, the most likely cause of discrepancy between these experiments is the peptides on the array. In our experience, peptides are the most effective binding substrates when the modification of interest is centered within the peptide, which emphasizes the importance of peptide choices when defining binding events to modified histones.
In addition to individually synthesizing and purifying peptides for spotting on arrays with robots, a second approach of peptide array production referred to as SPOT synthesis (synthetic peptide arrays on membrane supports) has been used by multiple labs. On SPOT arrays, peptides are directly synthesized on cellulose membranes, anchored to the membrane by a chemical linker. Employing SPOT arrays has helped define the potential consensus sequence of certain lysine methyltransferases as well as characterize reader domains [28, 29]. For instance, the Royal Family member PWWP domain was found to bind H3K36me3 peptides present by SPOT array [30]. The advantage of SPOT arrays versus individual synthesis of peptides is the cost and versatility of this approach. The main disadvantage is that the integrity, purity, and spatial orientation of the peptides cannot be determined and/or controlled for in the same manner as with the individual synthesis approach.
The interplay between different histone modifications has proven to be an important consideration with regards to functional readout. To address combinatorial analysis, peptide arrays provide a powerful platform to perform this type of screening because of the large number of distinct peptides that can be tested simultaneously. This is highlighted by a recent elegant study dissecting the binding profile of the tandem tudor domain (TTD) of the E3 ubiquitin ligase UHRF1. The TTD of UHRF1 was known to be sufficient to specifically bind H3K9me2/3 [31, 32]. Notably, combinatorial peptides on an array revealed that the H3K9me3 binding occurs independent of phosphorylation at H3S10, a canonical chromatin signature of mitosis that excludes other H3K9me2/3 effectors [33]. UHRF2, the homolog of UHRF1, shares similar domain structure and in vitro functionality. However, defects in DNA methylation derived from mice lacking UHRF1 cannot be rescued by UHRF2 suggesting a non-redundant function for UHRF2 [34]. The UHRF1 TTD, therefore, provides a mechanism by which H3K9 methylation permits the faithful propagation of DNA methylation through mitosis [33]. The UHRF proteins contain a domain structure where its TTD is adjacent to a PHD finger. A larger construct spanning both the TTD and the PHD finger therefore represents a more complete binding module of UHRF1. Structural and molecular data indicated that the PHD finger binds unmodified H3 tails with most sensitivity to H3R2 methylation [35, 36]. When combined, both the TTD- and PHD finger-mediated binding events are required for efficient H3K9me2/3 recognition on a select number of peptides [37, 38]. Consistent with these data, a more exhaustive analysis of peptides further showed that a non-functional PHD finger was able to abrogate the many binding interactions of UHRF TTD-PHD [34, 39].
The expansive number of peptides on arrays can also enlighten more general phenomena described in previously published work. For example, the MBT domains of L3MBTL1 were found to directly interact with methylated p53K382, RbK860, and H4K20 [13, 40–44]. Structural analysis of these domains revealed that the middle MBT domain of the 3xMBT repeat contained a canonical aromatic pocket that was capable of binding methylated lysines in a manner that had little requirement for the surrounding sequence context [43, 44]. When probed on peptide arrays, the 3xMBT domain showed broad specificity for mono- and dimethylated lysines irrespective of the peptide sequence [45, 46]. The binding promiscuity of this naturally occurring domain allowed it to be utilized as a pan-specific mono- and dimethyl-lysine affinity reagent for pull-down and far western applications [45].
Another powerful application of peptide arrays is to characterize modification state-specific histone antibodies, which are extensively used in chromatin research. In an analysis of several commonly used antibodies, many antibodies were found to lack the advertised specificity [25, 47–49]. Inconsistency exists in the quality of modification-specific antibodies that are critical for understanding chromatin biology; therefore, care must be taken when interpreting data using these reagents. However, antibody quality can easily be determined using array technology.
Peptide arrays serve as a versatile and comprehensive approach for both identifying and defining interactions with modified histones, including antibody specificity. Nonetheless, like all techniques, peptide array platforms have limitations and drawbacks that should be taken into account. The majority of histone modifications occur within the N-terminus, which can be reasonably mimicked by an unstructured peptide, but some reader domain interactions with histone modifications are highly enhanced when presented in the native nucleosome form [50]. In addition, the finite number of peptides spotted on the surface of arrays naturally limits their power. While many different interactions and combinations can be spotted, not all interactions can feasibly be represented. This drawback is especially true for long-range interactions that are made difficult by the limitations of peptide synthesis.
Protein domain arrays
Peptide arrays are useful when asking whether a particular domain binds one of many histone peptides. Protein domain arrays can be used to perform the converse experiment where a modified peptide is allowed to bind to immobilized protein domains (Figure 1e–g). Many epigenetic binding domains families have been defined, which provide starting points for defining proteins with potential reader-type binding capacity. These types of domains have been expressed and immobilized onto an array to be probed with a peptide [13]. Protein domain arrays have been useful in defining binding partners on a broad scale for H3K4me, H3K9me, and H4K20me [13]. More targeted experiments helped identify 53BP1 tudor domain to dimethylated p53 at lysine 372 and the ankryin repeat of GLP as a binding domain that recognized the NF-κB subunit RelA when mono-methylated at lysine 310 [51, 52]. While modifications of p53 and RelA are not histone modifications, the principles used to define these interactions apply to histone modifications such as H3R17me2a and H4R3me2a recognition by the TDRD3 tudor domain [53].
Peptide screening by SILAC
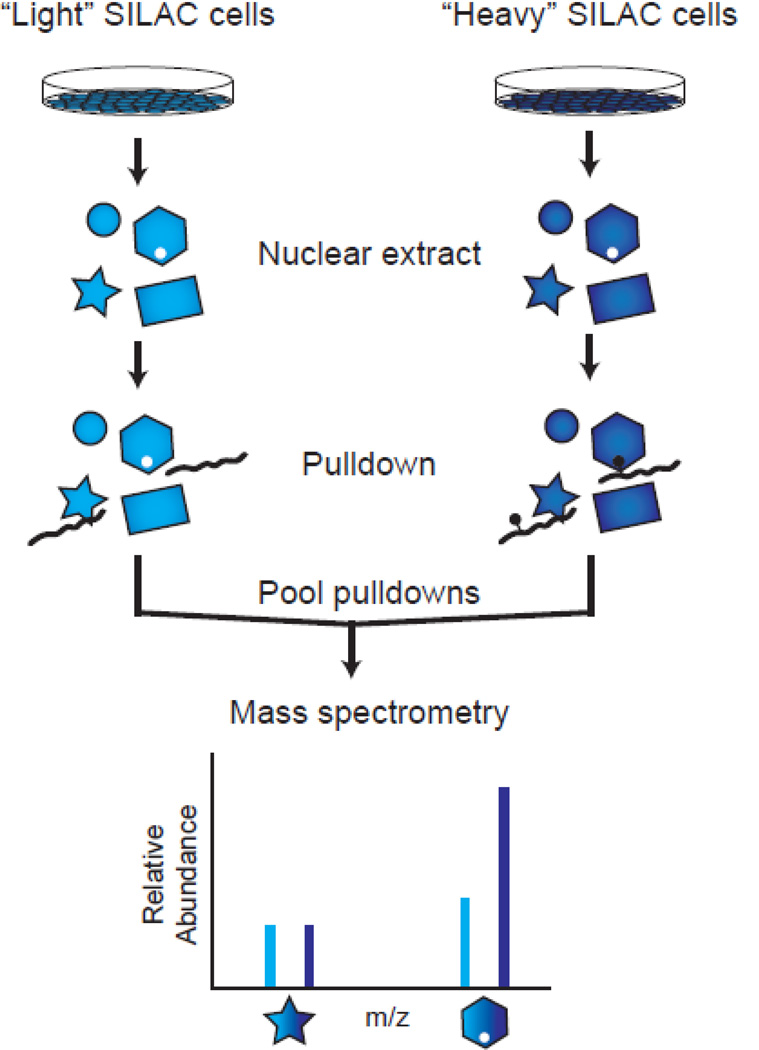
Candidate domain screening relies mostly on homology to previously characterized binding domains or directed experiments that are based on preliminary evidence and specific hypotheses. However, these criteria are not always met when beginning to characterize a binding event to methylated lysine. Thus, relatively unbiased methods have been developed to take advantage of SILAC (stable isotope labeling of amino acids in cell culture) and quantitative mass spectrometry to identify protein-protein interactions from cellular extracts. These extracts contain far greater numbers of proteins with potential to bind a particular modified peptide. SILAC provides one with the ability to enrich for proteins from the human proteome rather than a defined set of proteins. Pulldown experiments can then be performed with “heavy” and “light” isotopically labeled nuclear extracts and analyzed by quantitative mass spectrometry (Figure 2). Comparison of relative heavy-to-light ratios is used to identify proteins that are enriched in a modification-dependent manner.
Figure 2.
Peptide screening by SILAC. Cells are grown in tissue culture media that is isotopically labeled with “light” or “heavy” amino acids. Nuclear extracts are prepared from these cells to enrich for nuclear proteins that may bind histone modifications. Unmodified and modified peptides are used as bait in pulldowns from the light and heavy nuclear extracts, respectively. Beads containing the immobilized peptides and any bound proteins are pooled and boiled. The bound proteins can then be digested and analyzed by mass spectrometry. Heavy-to-light ratios are then compared to find proteins that are enriched or excluded due to the presence of the modification of interest.
The method of using SILAC-based quantitative mass spectrometry to identify methyl-lysine binding domains was validated when previously characterized binding domains were enriched from HeLa nuclear extracts: H3K4me3 interactions with BPTF, PHF8, and TFIID complex, H3K9me3 interactions with HP1α/β/γ, and H3K27me3 interactions with CDYL1/2 [54] and 53BP1 with H4K20me1 [55]. Additionally, proteins containing a PWWP domain, a Royal family member, were enriched in pulldowns for H3K36me3 [54]. Subsequent experiments showed that the interaction between MSH6 PWWP domain and H3K36me3 as important for recruitment of DNA mismatch repair machinery to chromatin [56]. Similarly, the PWWP domain of PSIP1/LEDGF was also shown to bind H3K36me3 [50]. SILAC pulldowns followed by mass spectrometry are biased toward higher abundance proteins because these binding events are easier to detect. Modifications to this procedure have been made to enrich for lower affinity or lower abundance interactions. For example, photo-crosslinking was effective at identifying the binding event of a lower abundance protein such as ING2 to H3K4me3 [57].
Beyond defining histone interactions with peptides
Peptides serve a convenient proxy to characterize binding interactions with the histone tails of the more physiologically relevant nucleosome. However, many modifications of importance occur within the globular domain of the nucleosome, such as H3K56 acetylation, H3K79 methylation, and H2BK120 ubiquitination. Even modifications on the histone tail, such as H3K36me, are located close to the nucleosome core where they may be influenced by the nucleosome structure as a whole. Evidence is beginning to support the idea that more physiologically relevant substrates may increase affinity or change identified substrates, even for residues toward the N-terminus of histones [50, 58–60].
A primary challenge for assembly of designer chromatin is the homogenous incorporation of post-translational modifications. Histones are too large to make using traditional peptide-synthesis methods, so other chemical techniques must be applied to get a modified substrate. These chemical manipulations can be broken down into two main categories: Methyl-lysine analogs (MLA) and protein ligation.
Methyl-lysine analogs (MLA) technology
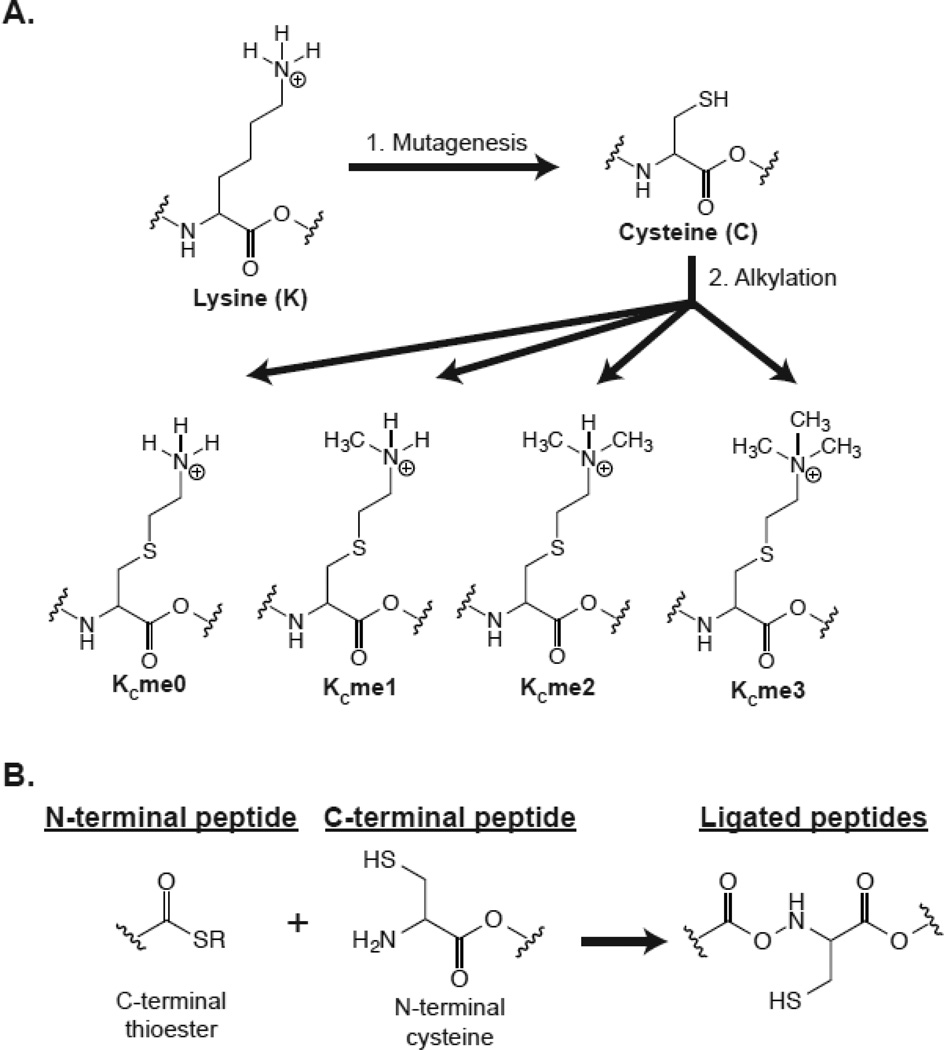
MLAs take advantage of the scarcity of cysteine resides in histones to perform alkylation reactions on a cysteine sulfhydryl group. Xenopus nucleosome core particles contain only a single cysteine residue at H3C110. H3C110A mutations do not severely impact DNA alignment around the nucleosome core particle and were used in the determination of the nucleosome core particle crystal structure [61, 62]. In this system, a lysine-to-cysteine mutation is made at the site of a desired MLA in a histone already lacking cysteines (e.g. an H3C110A background for H3 modifications). Subsequently, an alkylation reaction is performed on the cysteine sulfhydryl group of this lysine-to-cysteine mutant protein to make unmodified, mono-, di-, or trimethyllysine analogs (Figure 3a) [63]. MLA technology provides a relatively accessible and semi-genetically encoded method for representing a specific methyl modification within a whole protein. MLA histones can be used individually as free histones or can be incorporated into higher ordered structures such as octamers and nucleosomes. Nucleosome-incorporated MLAs of H3K36me3 were instrumental to the characterization of PWWP domains binding to nucleosomes, which indicated that the nucleosome DNA is important for their physiological affinity [50, 56].
Figure 3.
Chemical strategies to generate specifically modified histone proteins. A) Modified lysine analog construction. All cysteines must be removed from a candidate protein, such as making an H3C110A mutation. The desired modified lysine is then mutated to cysteine and expressed in Escherichia coli. Following expression and purification, alkylating reagents are applied to the cysteines to generate a specific methyl lysine state at that particular residue. B) Protein ligation. Two peptides can be ligated if they contain the correct chemical signatures. The N-terminal peptide must contain a C-terminal thioester moiety that can either be generated by peptide synthesis or by using natural iteins. The C-terminal peptide must contain an N-terminal cysteine residue, which can be installed by peptide synthesis or molecular techniques such as protease cleavage. The chemoselective reaction will occur spontaneously to generate a ligated protein through a native peptide bond.
Despite the promise and accessibility of this technique, MLAs may not behave exactly like a native methylated lysine because of the thioether generated from the alkylation reaction. For example, the TTD of 53BP1, CD of HP1β, and MBT domains of L3MBTL1 had notably decreased affinity for their cognate MLA-containing peptide relative to their affinity for the native methyl-lysine peptide [64]. Although, this phenomenon is not uniformly the case as the ING1 PHD finger and the UHRF1 TTD domain bound their MLA and native methyl-lysine substrates with comparable affinity [64]. Therefore when using MLAs as binding substrates, the structural changes caused by the thioether should be taken into consideration.
MLAs are also limited in their ability to test multiple different modifications in cis. In order to obtain a homogenous substrate, the same degree of methylation must be applied to all cysteines within a single alkylation reaction. If multiple methyl modifications are to be incorporated within the same histone, then they are required to have the same degree of methylation. Overall, MLA technology is a very powerful and accessible method that has been tremendously helpful in dissecting methyl-lysine biology.
Protein ligation technology
Protein ligation takes advantage of the ability to incorporate component histone peptides into full-length histones. Native chemical ligation (NCL) of two synthetic peptides can generate a native peptide bond between a peptide with a C-terminal thioester and a peptide with an N-terminal cysteine (Figure 3b) [65]. The related process of expressed protein ligation (EPL) uses the same chemical functionality except that one of the peptides is obtained from a biological source [66]. The flexibility of peptide synthesis allows for the potential to include a wide range of combinatorial modifications across the entire protein. Additionally, native modifications are preserved, which eliminates potential affinity issues sometimes observed with MLAs [64]. In the simplest usage EPL to generate a modified histone, a synthesized, modified peptide can be ligated to an unmodified portion expressed in bacteria. SILAC-based proteomics and EPL-generated mononucleosomes containing H3K4me3, H3K9me3, and H3K27me3 modifications were combined to identify canonical proteins associated with these marks [60]. More complicated syntheses have been implemented to make H3K56ac in a three-part ligation [67]. A residual cysteine will be created at any peptide junction, potentially resulting in structural problems. However, strategically placed cysteines can be desulfurized to alanine to get a native sequence if peptide junctures fall on an alanine [68].
Native chemical and expressed protein ligation provide the benefits of including native modifications within whole histone proteins. These reagents provide the most physiologically relevant substrates to define a binding event to histone modifications, especially when considering the potential importance of having a complete nucleosome structure. However, depending on the resources and expertise of the lab, these methods can be expensive, time consuming, and technically challenging to execute. Together, MLA technology and protein ligation technology have had a transformative impact on the chromatin biology and methyl lysine-signaling fields.
Conclusion
Although many protein modules have been identified to directly bind modified histones, our continued understanding of chromatin dynamics will be aided by the tools available to discover new binding proteins. Direct peptide pulldowns have had great historical significance in our foundational understanding of chromatin biology and will continue to be the most accessible method to test binding interactions. The popularization of designer chromatin reagents has provided a new perspective to ask outstanding questions, old and new. Integration of these reagents as time progresses will only expedite progress in identifying and characterizing reader domains. For example, modified chromatin reagents are also compatible with existing technologies such as slide arrays. Our ability to represent more specific chromatin environments and our ability to make these reagents will evolve. Work has been done to test the ability of proteins to bind trimethylated nucleosomes in the context of DNA methylation [60]. Also, nonsense-mediated suppression systems to incorporate modified, unnatural amino acids can be developed and improved [69]. Fully genetically encoded modifications will only facilitate the production and usage of customized chromatin. All of the aforementioned technologies have both strengths and weaknesses, which must carefully be considered when choosing and using reagents to interrogate the biology. Some of the most significant discoveries were made using the simplest methods. In the future, greater accessibility and development of these technologies will a boon to the work defining interactions with modified histones.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levy D, Gozani O. Decoding chromatin goes high tech. Cell. 2010;142:844–846. doi: 10.1016/j.cell.2010.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray K. The Occurrence of Epsilon-N-Methyl Lysine in Histones. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 3.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by sitespecific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 4.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 6.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 7.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 8.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones AE, MacKenzie F, Pan P, Li SS, Wang H, Min J. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) . Proc Natl Acad Sci U S A. 2010;107:19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ballare C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, Carlomagno T, Benitah SA, Di Croce L. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol. 2012;19:1257–1265. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai L, Rothbart SB, Lu R, Xu B, Chen WY, Tripathy A, Rockowitz S, Zheng D, Patel DJ, Allis CD, Strahl BD, Song J, Wang GG. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol Cell. 2013;49:571–582. doi: 10.1016/j.molcel.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde ME, Hong Z, Allen C, Roy S, Nunez JK, Nickoloff J, Kulesza CA, Yasui A, Cote J, Kutateladze TG. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol. 2012;19:1266–1272. doi: 10.1038/nsmb.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin S, Guo Y, Xu C, Bian C, Fu M, Gong S, Min J. Tudor domains of the PRC2 components PHF1 and PHF19 selectively bind to histone H3K36me3. Biochem Biophys Res Commun. 2013;430:547–553. doi: 10.1016/j.bbrc.2012.11.116. [DOI] [PubMed] [Google Scholar]
- 23.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier- Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, Casadio F, Bassi C, Amati B, Bedford MT, Guccione E, Gozani O. Epigenome microarray platform for proteomewide dissection of chromatin-signaling networks. PLoS One. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang Y, Horton JR, Bedford MT, Zhang X, Cheng X. Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding. J Mol Biol. 2011;408:807–814. doi: 10.1016/j.jmb.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Galka M, Iberg A, Wang Z, Li L, Voss C, Jiang X, Lajoie G, Huang Z, Bedford MT, Li SS. Systematic identification of methyllysine-driven interactions for histone and nonhistone targets. J Proteome Res. 2010;9:5827–5836. doi: 10.1021/pr100597b. [DOI] [PubMed] [Google Scholar]
- 28.Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhayalan A, Kudithipudi S, Rathert P, Jeltsch A. Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem Biol. 2011;18:111–120. doi: 10.1016/j.chembiol.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nady N, Lemak A, Walker JR, Avvakumov GV, Kareta MS, Achour M, Xue S, Duan S, Allali-Hassani A, Zuo X, Wang YX, Bronner C, Chedin F, Arrowsmith CH, Dhe-Paganon S. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem. 2011;286:24300–24311. doi: 10.1074/jbc.M111.234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte- Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH, Strahl BD. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Gao Q, Li P, Liu X, Jia Y, Wu W, Li J, Dong S, Koseki H, Wong J. S phasedependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 2011;21:1723–1739. doi: 10.1038/cr.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu L, Li Z, Wang P, Lin Y, Xu Y. Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell Res. 2011;21:1374–1378. doi: 10.1038/cr.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, Shi YG, Xu Y, Patel DJ, Shi Y. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng J, Yang Y, Fang J, Xiao J, Zhu T, Chen F, Wang P, Li Z, Yang H, Xu Y. Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J Biol Chem. 2013;288:1329–1339. doi: 10.1074/jbc.M112.415398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M, Ariyoshi M, Shirakawa M. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc Natl Acad Sci U S A. 2012;109:12950–12955. doi: 10.1073/pnas.1203701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 2013;27:1288–1298. doi: 10.1101/gad.220467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West LE, Roy S, Lachmi-Weiner K, Hayashi R, Shi X, Appella E, Kutateladze TG, Gozani O. The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression. J Biol Chem. 2010;285:37725–37732. doi: 10.1074/jbc.M110.139527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saddic LA, West LE, Aslanian A, Yates JR, 3rd, Rubin SM, Gozani O, Sage J. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010;285:37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, Wang YH, Reinberg D. L3MBTL1, a histonemethylation- dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 43.Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, MacKenzie F, Vedadi M, Arrowsmith CH. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol. 2007;14:1229–1230. doi: 10.1038/nsmb1340. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, Patel DJ. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell. 2007;28:677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore KE, Carlson SM, Camp ND, Cheung P, James RG, Chua KF, Wolf- Yadlin A, Gozani O. A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol Cell. 2013;50:444–456. doi: 10.1016/j.molcel.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nady N, Krichevsky L, Zhong N, Duan S, Tempel W, Amaya MF, Ravichandran M, Arrowsmith CH. Histone recognition by human malignant brain tumor domains. J Mol Biol. 2012;423:702–718. doi: 10.1016/j.jmb.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 47.Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol. 2011;21:53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levy D, Liu CL, Yang Z, Newman AM, Alizadeh AA, Utz PJ, Gozani O. A proteomic approach for the identification of novel lysine methyltransferase substrates. Epigenetics Chromatin. 2011;4:19. doi: 10.1186/1756-8935-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothbart SB, Lin S, Britton LM, Krajewski K, Keogh MC, Garcia BA, Strahl BD. Poly-acetylated chromatin signatures are preferred epitopes for site-specific histone H4 acetyl antibodies. Sci Rep. 2012;2:489. doi: 10.1038/srep00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Nuland R, van Schaik FM, Simonis M, van Heesch S, Cuppen E, Boelens R, Timmers HM, van Ingen H. Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenetics Chromatin. 2013;6:12. doi: 10.1186/1756-8935-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, Espejo A, Zee BM, Liu CL, Tangsombatvisit S, Tennen RI, Kuo AY, Tanjing S, Cheung R, Chua KF, Utz PJ, Shi X, Prinjha RK, Lee K, Garcia BA, Bedford MT, Tarakhovsky A, Cheng X, Gozani O. Lysine methylation of the NF-kappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-kappaB signaling. Nat Immunol. 2011;12:29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 53.Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, Bedford MT. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40:1016–1023. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 55.Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNAdependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Foley EA, Molloy KR, Li Y, Chait BT, Kapoor TM. Quantitative chemical proteomics approach to identify post-translational modification-mediated protein-protein interactions. J Am Chem Soc. 2012;134:1982–1985. doi: 10.1021/ja210528v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nikolov M, Stutzer A, Mosch K, Krasauskas A, Soeroes S, Stark H, Urlaub H, Fischle W. Chromatin affinity purification and quantitative mass spectrometry defining the interactome of histone modification patterns. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.005371. M110 005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Musselman CA, Ramirez J, Sims JK, Mansfield RE, Oliver SS, Denu JM, Mackay JP, Wade PA, Hagman J, Kutateladze TG. Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proc Natl Acad Sci U S A. 2012;109:787–792. doi: 10.1073/pnas.1113655109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flaus A, Luger K, Tan S, Richmond TJ. Mapping nucleosome position at single basepair resolution by using site-directed hydroxyl radicals. Proc Natl Acad Sci U S A. 1996;93:1370–1375. doi: 10.1073/pnas.93.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 63.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seeliger D, Soeroes S, Klingberg R, Schwarzer D, Grubmuller H, Fischle W. Quantitative assessment of protein interaction with methyl-lysine analogues by hybrid computational and experimental approaches. ACS Chem Biol. 2012;7:150–154. doi: 10.1021/cb200363r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 66.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci U S A. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shimko JC, North JA, Bruns AN, Poirier MG, Ottesen JJ. Preparation of fully synthetic histone H3 reveals that acetyl-lysine 56 facilitates protein binding within nucleosomes. J Mol Biol. 2011;408:187–204. doi: 10.1016/j.jmb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wan Q, Danishefsky SJ. Free-radical-based, specific desulfurization of cysteine: a powerful advance in the synthesis of polypeptides and glycopolypeptides. Angew Chem Int Ed Engl. 2007;46:9248–9252. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- 69.Wang YS, Wu B, Wang Z, Huang Y, Wan W, Russell WK, Pai PJ, Moe YN, Russell DH, Liu WR. A genetically encoded photocaged Nepsilon-methyl-L-lysine. Mol Biosyst. 2010;6:1557–1560. doi: 10.1039/c002155e. [DOI] [PubMed] [Google Scholar]