Abstract
A novel approach to personalizing postgrafting immunosuppression in hematopoietic cell transplant (HCT) recipients is evaluating inosine monophosphate dehydrogenase (IMPDH) activity as a drug-specific biomarker of mycophenolic acid (MPA)-induced immunosuppression. This prospective study evaluated total MPA, unbound MPA, and total MPA glucuronide plasma concentrations and IMPDH activity in peripheral blood mononuclear cells (PMNC) at five time points after the morning dose of oral mycophenolate mofetil (MMF) on day +21 in 56 nonmyeloablative HCT recipients. Substantial interpatient variability in the pharmacokinetics and pharmacodynamics was observed and accurately characterized by the population pharmacokinetic/dynamic model. IMPDH activity decreased with increasing MPA plasma concentration, with maximum inhibition coinciding with maximum MPA concentration in most patients. The overall relationship between MPA concentration and IMPDH activity was described by a direct inhibitory Emax model with an IC50 = 3.23 mg/L total MPA and 57.3 ng/mL unbound MPA. The day +21 IMPDH area under the effect curve (AUEC) was associated with cytomegalovirus reactivation, non-relapse mortality, and overall mortality. In conclusion, a pharmacokinetic/dynamic model was developed that relates plasma MPA concentrations with PMNC IMPDH activity after an MMF dose in HCT recipients. Future studies should validate this model and confirm that day +21 IMPDH AUEC is a predictive biomarker.
Keywords: Mycophenolic acid, therapeutic drug monitoring, population pharmacokinetics, limited sampling schedule, hematopoietic cell transplantation, covariates, albumin, cyclosporine
INTRODUCTION
The availability of allogeneic hematopoietic cell transplant (HCT) has expanded with the development of lower-dose nonmyeloablative conditioning regimens, which depend on achieving a delicate balance between recipient and donor cells to obtain immunosuppression of the recipient, optimal anti-tumor effect, and minimal toxicity.(1) Nonmyeloablative HCT recipients often receive mycophenolate mofetil (MMF) and a calcineurin inhibitor (cyclosporine or tacrolimus) as postgrafting immunosuppression, which aims to facilitate allogeneic engraftment and control graft-versus-host disease (GVHD).(2, 3)
There is considerable variability in the clinical outcomes of these patients. Some of the interpatient variability in clinical outcomes could, in part, reflect differences in each recipient’s sensitivity to MMF. The pharmacokinetics and drug-specific pharmacodynamics of mycophenolic acid (MPA), the therapeutically active metabolite of MMF, are potential sources of this interindividual variability (IIV). MMF is rapidly hydrolyzed to MPA in the gastrointestinal (GI) tract. After rapid absorption, MPA undergoes hepatic metabolism by various UDP-glucuronosyltransferase (UGT) isoenzymes to form MPA glucuronide (total MPAG).(4) After oral MMF administration, there is considerable between-patient variability in total and unbound MPA area under the concentration-time curves (AUCs).(5, 6) The available pharmacodynamic data in allogeneic HCT recipients suggest a relationship between MPA AUC and clinical outcomes.(7) Although some HCT centers have proposed personalizing MMF doses based on MPA AUC(8), there is an ongoing debate regarding the benefits of such therapeutic drug monitoring in solid organ transplantation.(9)
MPA is a selective, reversible, and noncompetitive inhibitor of inosine monophosphate dehydrogenase (IMPDH).(10) IMPDH is the rate-limiting enzyme involved in the de novo synthesis of guanosine nucleotides; IMPDH catalyzes the oxidation of inosine 5’-monophosphate (IMP) to xanthosine 5’-monophosphate (XMP) by a nicotinamide adenine dinucleotide (NAD)+-dependent reaction.(11) Characterizing the pharmacodynamic relationship between MPA and IMPDH activity is critical to understanding the potential benefit of alternative MMF dosing strategies in nonmyeloablative HCT recipients. Thus, we sought to characterize the pharmacokinetic/dynamic relationship between total and unbound MPA plasma concentrations and ex vivo IMPDH activity in peripheral blood mononuclear cells (PMNC) in nonmyeloblative HCT recipients receiving MMF as postgrafting immunosuppression.
METHODS
Patient characteristics
Between November 2008 and February 2012, 105 patients participated in a prospective ancillary biomarker study in nonmyeloablative allogeneic HCT recipients. Study participation influenced neither the conditioning regimen nor postgrafting immunosuppression. Patients (age >18 y) receiving fludarabine monophosphate (Fludara®) and total body irradiation (TBI) conditioning, a related or unrelated donor GCSF-mobilized PMNC graft, and postgrafting immuosuppression with a calcineurin inhibitor (cyclosporine or tacrolimus) and MMF were eligible for recruitment in this study. One participant received both PMNC and bone marrow because of inadequate PMNC yield from the donor’s apheresis. The choice and kinetics-based dose targeting of the calcineurin inhibitor were determined by the HCT protocol. In addition to cyclosporine or tacrolimus, some participants also received sirolimus as part of their postgrafting immunosuppression. Exclusion criteria included: diagnosis of an immunodeficiency disorder or scheduled to receive immunosuppression in addition to fludarabine/TBI (e.g., alemtuzumab, thymoglobulin) during HCT conditioning to day +28 post graft infusion. This protocol was approved by the Institutional Review Board at the Fred Hutchinson Cancer Research Center (FHCRC Protocol 1980, Clinicaltrials.gov #NCT00764829). Written informed consent was obtained from all patients prior to study participation.
The MMF dose and administration frequency were specified by the HCT protocol. No participants received oral enteric-coated MPA (Myfortic®). MMF doses were based on body weight and rounded to the nearest 250 mg dose, as previously described.(6) MMF doses were not adjusted based on MPA plasma concentrations or IMPDH determinations.
This cohort was divided into two separate datasets, the development dataset and the validation dataset. The development dataset was used to develop the pharmacokinetic-dynamic model relating plasma MPA concentrations with PMNC IMPDH activity and to validate our previous population pharmacokinetic model.(5, 6) The validation dataset was used to validate the pharmacokinetic-dynamic model relating plasma MPA concentrations and PMNC IMPDH. The patient characteristics of the development and validation datasets are shown in Table 1.
Table 1.
Participant characteristicsa
| Development dataset |
Validation dataset |
Overall | |
|---|---|---|---|
| Number of participants | 34 | 22 | 56 |
| Sex (female/male, % male) | 14/20 (59%) | 7/15 (68%) | 21/35 (63%) |
| Recipients’ age (year) | 63 (28 – 72) | 58 (26 – 73) | 62 (26 – 73) |
| Adjusted ideal body weight (kg) | 70.6 (44.3 – 88.8) | 71.2 (44.4 – 93.4) | 70.7 (44.3 – 93.4) |
| Height (cm) | 172 (149 – 184) | 173 (147 – 194) | 172 (147 – 194) |
| Body surface area (m2) | 1.96 (1.40 – 2.60) | 1.95 (1.50 – 2.47) | 1.95 (1.40 – 2.60) |
| Donor type (related/unrelated, % related) | 6/28 (18%) | 5/17 (23%)b | 11/45 (20%) |
| Female donor to male recipient | 7 (21%) | 8 (36%) | 15 (27%) |
| Kahl relapse risk | |||
| Low | 13 (38%) | 6 (27%) | 19 (34%) |
| Standard | 13 (38%) | 13 (59%) | 26 (46%) |
| High | 8 (24%) | 3 (14%) | 11 (20%) |
| Covariates associated with MPA clearance | |||
| Concomitant calcineurin inhibitorc | |||
| Cyclosporine | 25 (74%) | 18 (82%) | 43 (77%) |
| Tacrolimus | 9 (26%) | 4 (18%) | 13 (23%) |
| Serum creatinine (mg/dL) | 1.1 (0.6 – 2.0) | 1.2 (0.7 – 2.2) | 1.1 (0.6 – 2.2) |
| Serum albumin (g/dL) | 3.4 (2.1 – 4.2) | 3.5 (2.4 – 4.1) | 3.4 (2.1 – 4.2) |
| Pharmacokinetic sampling around morning MMF dose on day +21d | |||
| Before and 1, 2, 2.5, 6 he | 29 (85%) | 0 | 29 (52%) |
| Before and 1.25, 2, 3, 4 hf | 5 (15%) | 22 (100%) | 27 (48%) |
| Day +28 donor T-cell chimerism | 85% (35–100) | 95% (61–100) | 89% (35–100) |
Categorical data presented as number (percentage) of participants meeting stated criteria; continuous data presented as median (min–max);
three patients (one related, two unrelated donor) had antigen-level mismatch;
concomitant sirolimus in eight cyclosporine and three tacrolimus patients (see Patient Characteristics in Methods);
modified to improve patient adherence (see Sample Collection in Methods),
November 2008 – March 2011
April 2011 – February 2012.
Sample collection
Peripheral blood samples (8 mL drawn into EDTA vacutainers, Supplemental Fig. 1) were obtained on day +21. The total MPA, unbound MPA, and total MPAG plasma concentrations and IMPDH activity in PMNC cells were quantitated in each sample. All assays were performed on each of the samples. Of the 56 AUCs, two were collected on day +19, 17 on day +20, 32 on day +21, three on day +22, one on day +23 and one on day +25. Samples were drawn before and 1, 2, 2.5, and 6 h after oral MMF for 29 patients (November 2008 to March 2011; development and validation cohorts) and before and 1.25, 2, 3, and 4 h after oral MMF for 27 patients (April 2011 to February 2012, validation cohort). These patients were primarily treated in the ambulatory clinic, and therefore limited sampling schedules were used to maximize adherence. From November 2008 to March 2011, adherence with pharmacokinetic samples was 63% (41 of 65 participants); this decreased to 50% (6 of 12 participants) when participants were paid per AUC obtained. In April 2011, the sampling schedule was shortened to 4 h after the oral MMF dose based on a prior analysis.(6) With this change, the adherence improved to 76% (26 of 34 participants). In 50 of the 56 participants, all five samples were collected. Among the remaining participants, two had one sample collected, one had three samples collected, two had four samples collected, and one had six samples collected. IMPDH activity on day +2 after HCT was planned; this could not be conducted, however, due to myelosuppression resulting from the conditioning regimen.
Table 1 lists the participant characteristics, including biochemistry values and concomitant medications associated with MPA pharmacokinetic parameters. Our previous population pharmacokinetic model after IV or oral MMF administration indicated that MPA clearance was significantly increased (by 33.8%) with concomitant cyclosporine and negatively correlated with albumin concentration.(6)
Reagents and Chemicals
All nucleotides used as substrates for the enzymatic assay or as chromatographic standards were obtained from Sigma (St. Louis, MO). NAD was also purchased from Sigma. Acetonitrile, ammonium acetate, methanol, sodium hydroxide, sodium phosphate monobasic, ammonium acetate, ammonium hydroxide, and potassium chloride were all purchased from Thermo Fisher (Waltham, MA). Dulbecco’s Phosphate Buffered Saline (PBS) was purchased from Invitrogen Grand Island, NY). Ficoll Hypaque solution (density 1.077g/mL) was obtained from GE Healthcare (Uppsala, Sweden).
Quantitation of MPA and Total MPAG
Each plasma sample was analyzed for total MPA, unbound MPA, and total MPAG plasma concentrations using reverse-phase high performance liquid chromatography (HPLC) with mass spectrometry (MS) detection. For total MPA and total MPAG quantitation, plasma samples (100µL) with the internal standard (20µL mycphenolic acid-d3 and mycophenolic acid β-D-glucuronide-d3) were combined with 50µL methanol and 1000µL acetonitrile, vortexed, and subsequently centrifuged. The supernatant (1µL) was injected onto an LC/MS running a gradient of 2.0 mM ammonium formate (pH 3.3) and acetonitrile through an Agilent C18 column (2.1mm × 150mm × 5µ, Agilent Technologies, Palo Alto, CA). Monitored ions included m/z 321 for the (M+H+) ion of MPA, m/z 324 for the (M+H+) ion of MPA-d3 (internal standard), m/z 495.2 for the (M+H+) ion of total MPAG, and m/z 498.1 for the (M+H+) ion of total MPAG-d3 (internal standard). The dynamic range was 0.3 to 15.5 mg/L for MPA and 0.21.5 to 215 mg/L for total MPAG. The inter-day coefficient of variation was less than 10.7%.
Unbound MPA plasma concentrations were determined by equilibrium dialysis using Pierce RED (Rapid Equilibrium Dialysis) Devices (Thermo Fisher Scientific, Waltham, MA). Following incubation and processing of plasma samples according to Pierce RED manufacturer instructions, samples were analyzed using LC/MS as above with slight modifications (i.e., mobile phase was an isocratic mixture of 55% 2.0 mM ammonium formate (pH 3.3) and 45% acetonitrile, and the total run time was 5 min). The percentage of unbound (free) MPA was calculated as follows: unbound MPA = 100 × (1-bound MPA).
Isolation of human PMNC and IMPDH activity assay
PMNC were isolated within 6 h of collection by diluting blood in PBS at a 1:1 v:v ratio and subsequently layering on top of Ficoll as previously described.(12) In brief, PMNC were collected, diluted to 10 mL with PBS to wash, and centrifuged. The cell counts were quantitated using a Horiba Diagnostics ABX Micro 60 (Irving, CA), and distilled water was added to the supernatant to adjust the cell concentration to 0.5 × 106 cells/mL lysate. This PMNC cell count was used to standardize the IMPDH activity measurement. After storage at −80°C, the IMPDH activity was determined from the conversion of inosine monophosphate (IMP) to xanthosine 5′-monophosphate (XMP) according to procedures adapted from Glander et al.(13) and Daxecker et al.(14) The incubation reaction mixture included NaH2PO4, KCl, IMP (0.8 mL of 6.0 mmol/L), and NAD (0.8mL of 4.5 mmol/L, made fresh each day). The incubation reaction was started via the addition of the reaction mixture to 50 µL pre-warmed cell lysate (standard concentration of 0.5 × 106 cells/mL). For each incubation, the enzymatic reaction was terminated after 2.5 h incubation by the addition of methanol followed by internal standard (8-Bromo adenosine 5’-monphosphate, BMP), processed, and injected on the LC/MS running a gradient of 0.1 M ammonium acetate (pH=8.5) and acetonitrile through a Thermo Scientific Hypercarb column (2.0mm × 100mm × 5µ, part no. 35005-102130, Thermo Scientific, Bellefonte, PA). An Agilent G1946D MSD (Agilent Technologies, Palo Alto, CA) API-ES in positive ion mode was used. The MSD was run in the selected ion monitoring (SIM) mode. Monitored ions included m/z 365 for the (M+H+) ion of XMP, m/z 348 for the (M+H+) ion of AMP, and m/z 426 for the (M+H+) ion of BMP, the internal standard. Typical retention times were 4.3 min for XMP, 5.1 min for IMP, and 7.0 min for BMP. The limit of quantification (signal to noise >60 and CV <2%) was 58 pmol.
A quality control pooled lysate obtained from the PMNC of healthy subjects was run in triplicate with every incubation. The coefficient of variation for the control lysate was 6.2 % over 39 incubations. Each PMNC sample was run in triplicate; the average of the triplicates is reported as the IMPDH activity. IMPDH activity is standardized to the ex vivo PMNC count determined after Ficoll isolation and expressed as pmol XMP/106 cells/h.
MPA population pharmacokinetic-dynamic analysis
Pharmacokinetic (total MPA, unbound MPA, and total MPAG plasma concentrations) and pharmacodynamic (XMP formation in PMNC to provide IMPDH activity) data were available at each concentration-time point. The pharmacokinetic-dynamic models were developed in a sequential manner: the pharmacokinetic model was developed first, then each participant’s pharmacokinetic parameters were fixed for the creation of the pharmacodynamic model.
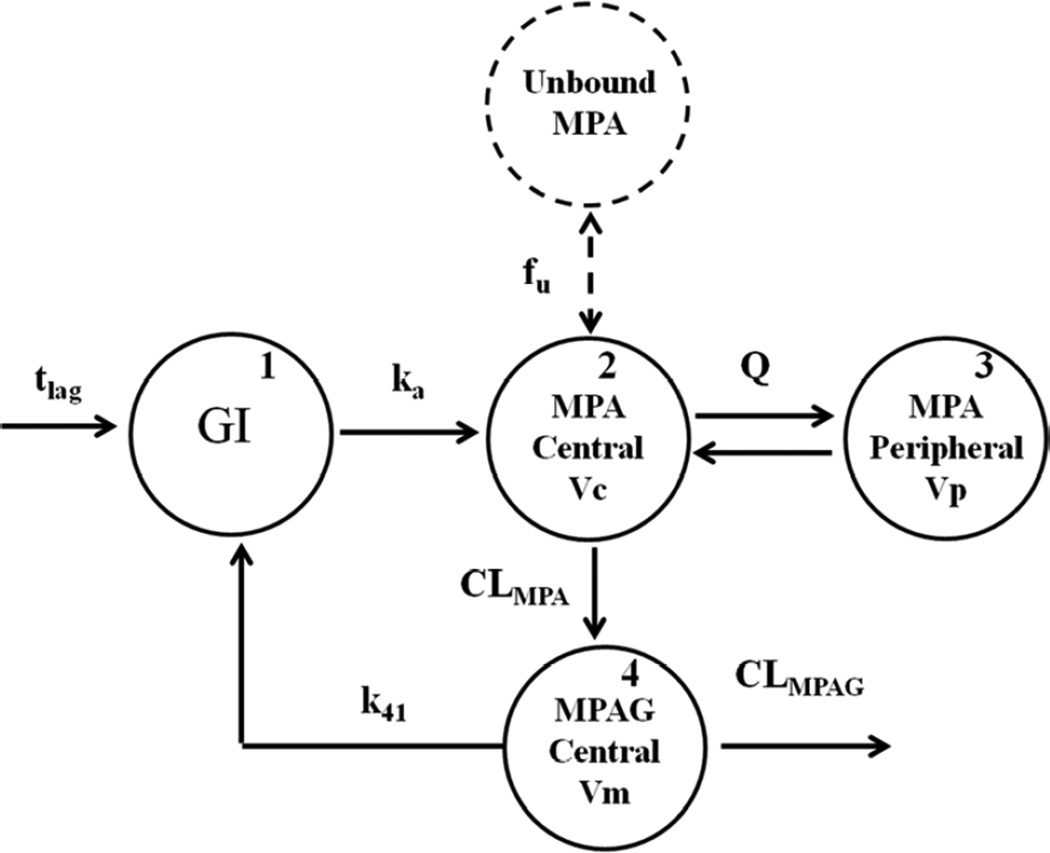
The initial pharmacokinetic model of Li et al.(6), which was developed in 402 HCT recipients, was modified to include total MPAG (as described by Musuamba et al(15)) and unbound MPA concentrations. Briefly, total MPA pharmacokinetics was described using a two-compartmental model with first-order elimination and first-order absorption with a lag time (Fig. 1). The model was parameterized as clearance (CL), volume of the central compartment (Vc), volume of the peripheral compartment (Vp), and intercompartment clearance (Q). Total MPAG pharmacokinetics was described using a one-compartmental model with first-order elimination. It was assumed that MPA is metabolized to total MPAG by a first-order process and that 100% of MPA is converted to total MPAG because urinary MPA data was not available. Collection of urinary excretion data was not possible since the participants were treated in the ambulatory clinic. Therefore, total MPAG CL and Vd are apparent clearance and volume of distribution (i.e., CL/fm and V/fm). Enterohepatic recirculation (EHC) of total MPAG was represented as a first-order process between the total MPAG central compartment and the GI tract. Unbound MPA concentrations were modeled as: [MPAunbound] = [MPAtotal] × fu, with [MPAunbound], [MPAtotal], and fu representing unbound MPA concentration, total MPA concentration, and the unbound fraction, respectively.
Figure 1.
Final pharmacokinetic model characterizing total MPA, unbound MPA, and total MPAG concentrations following oral MMF administration. tlag = lag time; GI = gastrointestinal tract; ka = first-order absorption rate constant; fu= fraction unbound of MPA; Vc = volume of central compartment of total MPA; Q = inter-compartment clearance of total MPA; Vp = volume of peripheral compartment of total MPA; CLMPA = clearance of total MPA; CLtotal MPAG = clearance of total MPAG; Vm = volume of central compartment of total MPAG; k41 = first-order rate constant of enterohepatic recirculation (EHC).
We also sought to validate the covariate effect of the calcineurin inhibitor (cyclosporine or tacrolimus) on MPA clearance.(5, 6) MPA clearance was eventually defined by the following equation:
| (Equation 1) |
with body weight in kg, albumin in g/dL, and CSP = 1 for cyclosporine and 0 for tacrolimus.
The Bayesian total MPA AUC0–8h of 27 participants who received MMF every 8 h was calculated. The AUCs of patients receiving concomitant tacrolimus were compared to those of patients taking cyclosporine. In addition, this simulation was executed using population mean parameter estimates to compare total MPA AUC0–12h in patients receiving either concomitant cyclosporine or tacrolimus. Both Bayesian estimations and simulations were performed using NONMEM VII (Icon Development Solutions, LLC, Ellicott City, MD).
Subsequent to finalization of the population pharmacokinetic model, the relationship between MPA concentrations and IMPDH activity was explored graphically and modeled using an inhibitory Emax model:
| (Equation 2) |
with E0 as baseline IMPDH activity (minimal inhibition), Imax as maximal IMPDH inhibition, Cp as the MPA plasma concentration, IC50 as the MPA concentration that causes 50% of maximal IMPDH inhibition, and γ as the Hill coefficient that governs the slope of MPA concentration vs. IMPDH activity.
Population pharmacokinetic-dynamic analysis was performed using nonlinear mixed effects modeling (NONMEM® VII .1.0; Icon Development Solutions, LLC, Ellicott City, MD). The Monte Carlo Important Sampling expectation maximization (EM) method was used throughout the modeling process. Pharmacokinetic models were simultaneously fitted to total MPA, unbound MPA, and total MPAG plasma concentrations. MMF dose and total MPAG concentration were multiplied by 0.739 and 0.594, respectively, to convert these values to their equivalent MPA values.(16) Clearance and volume of distribution were allometrically scaled to body weight with exponents fixed to 0.75 and 1, respectively. Between-subject variability was modeled using an exponential error model. Pharmacokinetic data were log-transformed and an additive error model was applied to describe residual error. A proportional error model was used for pharmacodynamic residual error.
Stepwise forward selection and backward elimination, based on the likelihood ratio test and a pre-specified alpha level, were applied across the base, intermediate and final models.(5) A covariate was included in the intermediate model when a decrease of at least 6.6 in the objective function value (OFV, p<0.01) occurred. A covariate was retained in the final model when an increase of at least 10.8 in OFV (p<0.001) occurred when this covariate was removed. The tested covariates included age, body surface area (BSA), serum albumin, blood urea nitrogen (BUN), serum creatinine (Scr), creatinine clearance, alkaline phosphatase (ALK), alanine aminotransferase (ALT), aspartate aminotransferase (AST), lactate dehydrogenase (LDH), total bilirubin, and direct bilirubin. All biochemistry values were obtained within 7 days before the pharmacokinetic-dynamic sampling.
Model validation
The pharmacokinetic-dynamic model was validated internally and externally. Goodness-of-fit (GOF) and visual predictive check (VPC) plots were used for internal validation. GOF plots included observed versus predicted concentrations as well as conditional weighted residuals (CWRES) versus model predictions and time post dose. For VPC plots, five hundred datasets were simulated from parameter estimates of the final model, and the 5th, 50th, and 95th percentiles of simulated data were compared with observed data.
External validation was carried out using two methods: 1) predicting observed pharmacokinetic-dynamic data in the validation dataset (n=22 participants); and 2) comparing results of Monte Carlo simulations to the observed data. Using the population model developed from the development dataset, predicted concentrations (both population and Bayesian predictions) were calculated for each participant in the validation dataset, using the known MMF dosing history and available pharmacokinetic sampling times. Predictions were obtained by setting POSTHOC and MAXEVAL=0 options in the NONMEM $ESTIMATION command. Bias in model prediction was assessed by calculating the percent mean prediction error (MPE%) as follows:
| (Equation 3) |
| (Equation 4) |
where PRED and IPRE are population prediction and individual (Bayesian) predictions, respectively, and OBS represents the observed data of the participants in the validation dataset. The Monte Carlo simulation approach generated a total of 100 studies of 22 participants. Simulations combined estimated pharmacokinetic-dynamic parameters from the development dataset with participant characteristics, dosing, and sampling information from the validation dataset. The median, 5th and 95th percentiles of simulated data were plotted alongside the corresponding median, 5th and 95th percentiles of observed data. This simulation was performed in NONMEM VII. Plots and statistical analysis were performed in S-PLUS 8.0 (Insightful Corp, Seattle, WA) or the open-source statistical software R (version 2.10.0).
Clinical outcomes
The area under the effect–time curve (AUEC, pmol × 106 cells) for IMPDH activity was calculated for the statistical analysis with clinical outcomes. Specifically, the participant’s predose value for IMPDH activity was considered as baseline, and then the area below the baseline was calculated using noncompartmental analysis to provide the AUEC. Because the latest assessment of IMPDH AUEC occurred on day +25, analysis of clinical outcomes was restricted to events taking place on day +26 or later.
Clinical outcomes of interest were toxicity to MMF (i.e., cytomegalovirus (CMV) reactivation), efficacy of MMF (i.e., day +28 donor T-cell chimerism, acute and chronic GVHD), and overall HCT outcomes (i.e., relapse, non-relapse mortality, and overall survival). Day +28 T-cell chimerism was not evaluated because too few participants had low donor chimerism: 9% had donor chimerism less than 50% and 32% had donor chimerism less than 75%. The median (range) for day +28 donor T-cell chimerism was 89% (35–100%). Similarly, only one participant experienced graft rejection, so this was not evaluated.
Of the 56 participants, 12 (21%) were CMV positive with a CMV positive donor, 21 (38%) were CMV positive with a CMV negative donor, 8 (14%) were CMV negative with a CMV positive donor, and 15 (27%) were CMV negative with a CMV negative donor. When either the donor or recipient was CMV positive, the CMV antigenemia assay to detect CMV pp65 antigen was performed on a weekly basis for the first three months following HCT. Twenty-two participants experienced CMV reactivation post-transplant. Acute GVHD was graded as previously described.(17) Hematological diseases were classified as low, standard, or high risk of relapse per the Kahl criteria to evaluate relapse rate in a consistent manner.(18) We defined disease relapse or disease progression as disease recurrence following complete remission or progression of persistent disease. Clinical endpoints were measured to the time of last clinical follow-up. The median time to last clinical follow-up was 1.4 years (range, 0.3 – 3.3 years).
Statistical analysis
IMPDH AUEC was treated as a fixed covariate. Cumulative incidence curves for acute GVHD were estimated using methods previously described.(19) Cox regression analysis was used to model the impact of IMPDH AUEC on time-to-event endpoints. Death and relapse were treated as competing risks for analysis of acute and chronic GVHD. Relapse was treated as a competing risk for the analysis of non-relapse mortality. The effects of IMPDH AUEC on hazard ratios (HRs) were expressed as the effect per doubling of IMPDH AUEC. All reported p-values are two-sided, and those estimated from regression models are derived from the Wald test. No adjustments were made for multiple comparisons.
RESULTS
The characteristics of the 56 participants are summarized in Table 1. The dose, administration route, and administration frequency of MMF were determined by the participant’s HCT transplant protocol.
Population pharmacokinetic model
A total of 167 pharmacokinetic samples (total MPA, unbound MPA, and total MPAG) were used for population pharmacokinetic model building. A previously developed MPA pharmacokinetic model was modified to include total MPAG EHC (Fig. 1). Total MPAG was assumed to be excreted to the GI tract by a continuous first-order process. It was estimated that 29.5% of MPA (i.e., EHC%=k41/(k41+CLTOTAL MPAG/Vm)) underwent EHC. Data fitting was significantly improved when EHC was only integrated with concomitant tacrolimus. The current dataset did not support a more physiologically-based EHC model.(20) Of note, the lag time (Tlag) could not be estimated for each participant since their food intake was not available. The population mean of Tlag was fixed to a previous estimate(5) since the sampling schedule was inadequate to estimate this parameter. Due to convolution of the absorption and distribution phases of MPA, and the absence of IV data, Vp could not be identified and was fixed to a previously determined value.(5) Pharmacokinetic parameters were well estimated, as shown in Table 2. The VPCs and the time courses of total MPA concentration, unbound MPA concentration, and total MPAG concentration are shown in Supplemental Fig. 2.
Table 2.
Population pharmacokinetic analysis for total MPA, unbound MPA and total MPA glucuronide (total MPAG) plasma concentrations
| Pharmacokinetic Model | Parameter Estimates [RSE%] | ||
|---|---|---|---|
| Base Model | Final Model | ||
| Parameter | Explanation | ||
| ka (h−1) | First-order rate constant representing both formation and absorption process | 0.913 [11.7] | 0.916 [12.4] |
| Tlag (h) | Lag time of oral absorption | 0.228 FIXED | 0.228(21) FIXED |
| CLMPA (L/h/70kg)a | Clearance of total MPA | 31.4 [5.6] | 31.4 [5.7] |
| CLTOTAL MPAG (L/h/70kg)b | Clearance of total MPAG | 1.29 [7.2] | 1.32 [5.9] |
| QMPA (L/h/70kg) | Intercompartmental clearance of total MPA | 11.6 [29.8] | 11.5 [27.7] |
| k41 (L/h/70kg) | First-order rate constant representing total MPAG enterohepatic recirculation (EHC) | 0.0503 [45.3] | 0.0558 [44.1] |
| Vc (L/70kg) | Volume of central compartment of total MPA | 25.3 [13.0] | 26.5 [12.9] |
| Vp (L/70kg) | Volume of peripheral compartment of total MPA | 247(21) FIXED | 247(21) FIXED |
| Vm/fm (L/70kg) | Volume of central compartment of total MPAG | 9.91 [8.9] | 9.91[8.8] |
| fu (%) | Fraction unbound of MPA | 1.76 [2.6] | 1.76 [2.7] |
| θcreatinineb | Power coefficient of creatinine covariate effect on total MPAG clearance | Not estimated (NE) | −0.919 [19.5] |
| Interindividual variabilityc | |||
| ka (CV%) | 55.0 [35.1] | 55.0 [42.2] | |
| Tlag (CV%) | 133.4 [24.7] | 132.7 [25.9] | |
| CLMPA (CV%) | 38.2 [21.2] | 38.5 [21.4] | |
| CLTOTAL MPAG (CV%) | 53.7 [18.5] | 44.0 [18.5] | |
| Q (CV%) | 162.8 [35.3] | 161.2 [29.5] | |
| k41 (CV%) | 118.3 [70.7] | 110.5 [65.7] | |
| Vc (CV%) | 47.1 [46.8] | 47.0 [79.6] | |
| Vp (CV%) | 84.2 [49.5] | 86.4 [109.2] | |
| Vm/fm (CV%) | 44.4 [34.3] | 45.1 [32.1] | |
| fu (CV%) | 14.9 [31.8] | 15.0 [33.0] | |
| Residual Variabilityd | |||
| Total MPA | 0.20 [11.3] | 0.20 [11.8] | |
| Unbound MPA | 0.20 [11.2] | 0.20 [11.8] | |
| Total MPAG | 0.09 [11.9] | 0.09 [12.1] | |
Typical CL values were calculated as described in Equation 1. Body weight calculations described in Patient Characteristics in Methods;
CLTOTAL MPAG = CLpop × (Scr/1.12)−0.919;
Bioavailability was fixed to 1.
Additive residual error was on a natural logarithmic-scale.
Abbreviations: CV = coefficient of variation; RSE = relative standard error
Covariate analysis identified Scr as a significant covariate of total MPAG CL. Inclusion of Scr in the model resulted in a 24-unit decrease in OFV and a 9.7 % decrease in IIV of total MPAG CL. The final pharmacokinetic model included both cyclosporine and serum creatinine as model covariates. The GOF plots are presented in Supplemental Fig. 3a–c.
Cyclosporine inhibits multidrug resistance-associated protein 2 (MRP2)-mediated EHC of total MPAG and thus results in decreased MPA AUC and increased MPA clearance (Supplemental Fig. 4). Median total MPA AUC0–8h was increased by 33% in participants receiving concomitant tacrolimus. In addition, simulations using population means demonstrated that the MPA AUC0–12h in participants taking tacrolimus was, on average, 1.38-fold higher than in patients taking cyclosporine. Overall, these results confirmed our previous findings(6) and can be considered as an external validation for Equation 1 where MPA CL is increased by 33.8% with concomitant cyclosporine.
Population pharmacokinetic-dynamic model
For the pharmacodynamic analysis, 267 PMNC samples were available for ex vivo IMPDH activity quantitation. Of these, 263 samples had three replicates for IMPDH activity and four samples had two replicates. Within-sample variability was calculated by dividing the lowest XMP formation rate by the highest rate within each sample (i.e. minimum/maximum). The within-sample variability ranged from 74% to 100%. The majority of replicates (257, 96%) had within-sample variability greater than 90%. For pharmacodynamic model building, there were 166 IMPDH activity measurements in the development dataset and 101 IMPDH activity measurements for the validation dataset.
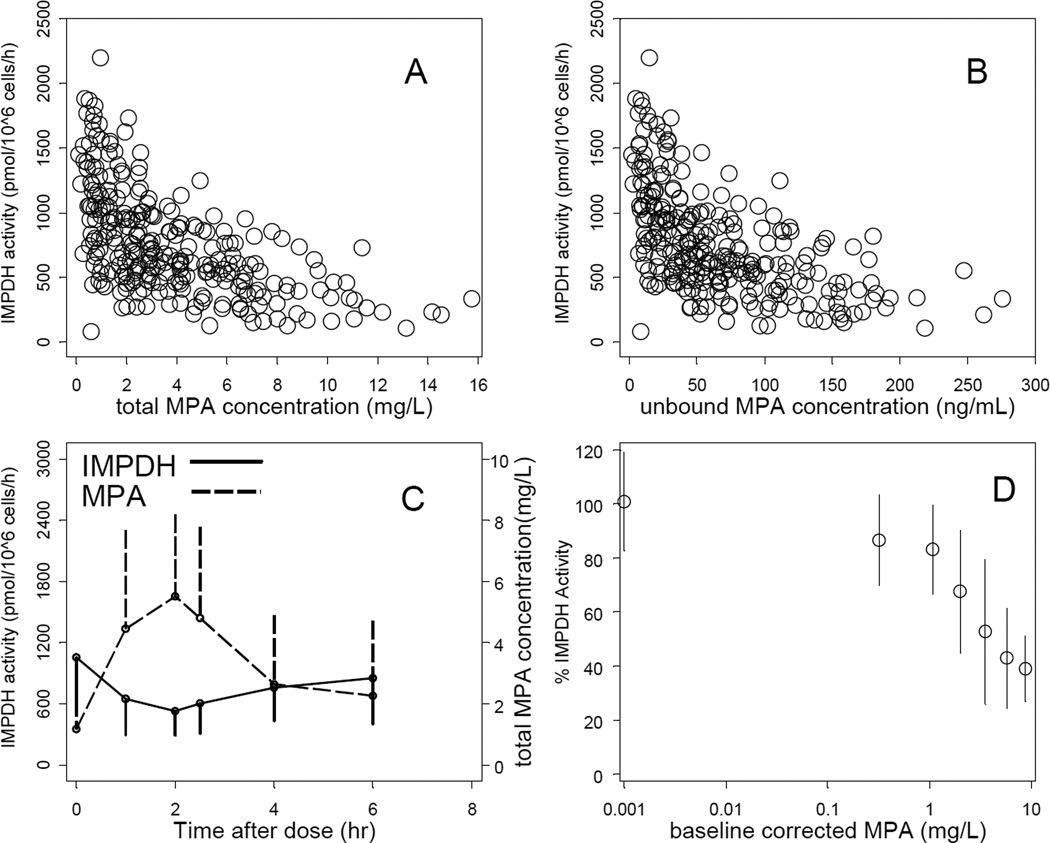
The concentration-response relationships between MPA concentration (either total or unbound) and IMPDH activity are shown in Fig. 2. IMPDH activity was inversely related to total MPA (Fig. 2A) and unbound MPA (Fig 2B) concentrations. The greatest level of inhibition was associated with the greatest MPA concentration, after which IMPDH activity returned to near pre-dose values. This association is shown for the entire population (Fig. 2C) as mean plus standard deviation and also using each participant’s pre-dose MPA concentration and IMPDH activity as their own controls (Fig. 2D). Maximum inhibition coincided with maximum MPA concentration, indicating a direct effect relationship. When analyzing all 267 pharmacokinetic-dynamic data points in the development dataset, the MPA concentration – IMPDH activity relationship could be well described by an inhibitory direct effect Emax model (Eq. 2). The final model-based IC50 estimate was 3.23 mg/L (RSE 10.7%; IIV 53.1%CV) for total MPA and 57.3 ng/mL (RSE 11.2%; IIV 56.3%CV) for unbound MPA. Maximum inhibition (Imax) could not be estimated from the observed data and was therefore fixed to 1. The Hill coefficient was estimated to be close to 1 and was fixed to 1 in the final parameter estimation. Table 3 summarizes the estimated and fixed population pharmacodynamic parameters.
Figure 2.
Interpatient variability in IMPDH activity in PMNC on day +21. Concentration-response of IMPDH activity by total MPA (A), and unbound MPA (B) plasma concentrations. Time course of IMPDH activity and total MPA concentration (mean±SD) (C). Percentage of IMPDH activity (mean±SD) vs. total MPA corrected for baseline (i.e., pre-dose) values (D).
Table 3.
Population pharmacodynamics parameters describing relationship between total or unbound MPA concentrations and IMPDH activity on day +21 in nonmyeloablative HCT recipients taking MMF
| Parameter | Explanation | Estimatesa [RSE%]b for | |
|---|---|---|---|
| IMPDH activity (pmol XMP/106 cells/h) | |||
| E0 | Baseline IMPDH activity (immediately before day +21 MMF dose) | 1370 [5.6] | |
| Total MPA (mg/L) | Unbound MPA (ng/mL) | ||
| IC50 | MPA concentration causing 50% maximal inhibition | 3.23 [10.7] | 57.3[11.2] |
| IIV_E0 (CV%) | Interindividual variability of E0 | 27.6 [30.8] | 27.3 [31.4] |
| IIV_IC50 | (CV%) Interindividual variability of IC50 | 53.1 [34.2] | 56.3 [33.4] |
| Proportional residual error (%) | 0.20 [19.3] | 0.20 [19.3] | |
Maximum inhibition (Imax) could not be estimated based on observed data and, therefore, was fixed to 1. Hill coefficient was estimated close to 1 and was fixed to 1 in the final parameter estimation.
The base model is the final model, because none of the evaluated covariates met the criteria for inclusion in the pharmacodynamic model.
Abbreviations: CV = coefficient of variation; IIV = interindividual variability; RSE = relative standard error.
The final pharmacodynamic model did not include any of the available covariates. Age was negatively correlated with E0, with a Pearson correlation of 0.25 (p-value=0.06). Inclusion of age as a model covariate did not result in a significant improvement in model fitting, and age was thus not included in the final model. Notably, the calcineurin inhibitor and graft source (i.e., related vs. unrelated donor) did not affect the IC50 or E0 (Supplemental Fig. 5).
The final pharmacokinetic-dynamic parameter estimates were updated using a combination of both the model-building and validation datasets (Tables 2 and 3). Both VPC and GOF plots demonstrated that the final model precisely described the observed pharmacokinetic and pharmacodynamic data (Supplemental Fig. 2–3).
Validation of the population pharmacokinetic-dynamic model
The participant characteristics were similar between the model-building and validation datasets (Table 1). Two methods were applied to the validation dataset to externally corroborate the developed population pharmacokinetic-dynamic model. The results from both methods demonstrated that the population pharmacokinetic-dynamic model accurately predicted the observed MPA pharmacokinetics and IMPDH activity in a seperate cohort of HCT participants. For the population prediction, the MPE% for total MPA, unbound MPA, total MPAG, and IMPDH activity were 28.9%, 10.6%, 27.1%, and −1.85%, respectively. For individual Bayesian predictions, MPE% for total MPA, unbound MPA, total MPAG, and IMPDH activity were 4.85%, −2.0%, 0.4%, and −0.97%, respectively. As shown in Supplemental Fig. 6, the majority of validation data fall within the 95% CI of the predicted values that were derived from the pharmacokinetic-dynamic parameters estimated from the development dataset.
Pharmacodynamic Relationships
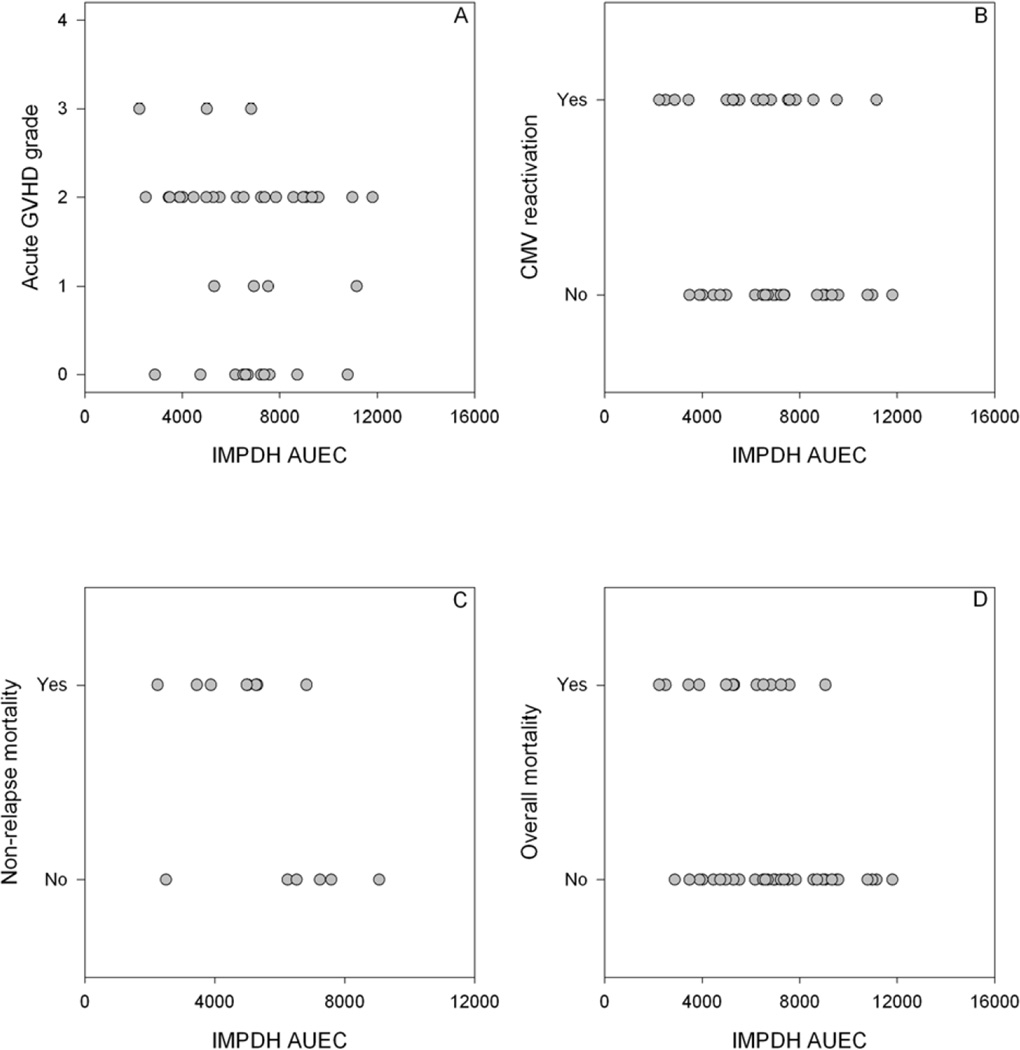
We also sought to evaluate if clinical outcomes could be predicted by the day +21 IMPDH AUEC. The analysis was adjusted for Kahl relapse risk category, female donor to male recipient, and donor type (unrelated vs. related). As shown in Table 4, Fig. 3, and Supplemental Fig. 7, day +21 IMPDH was associated with CMV reactivation (p=0.003), non-relapse mortality (p=0.04), and overall survival (p=0.03).
Table 4.
Association of day +21 IMPDH AUEC with HCT outcomes
| Eventsa | OR/HRb (95% CI) | P-value | |
|---|---|---|---|
| Day +28 T-cell chimerism ≥95% | 23 | 0.96 (0.4–2.6) | 0.96 |
| Grade 2–4 acute GVHD | 33 | 0.72 (0.4–1.3) | 0.26 |
| Grade 3–4 acute GVHD | 4 | 0.13 (0.0–1.0) | 0.05 |
| Extensive chronic GVHD | 30 | 1.38 (0.7–2.9) | 0.38 |
| Relapse | 10 | 0.95 (0.3–3.2) | 0.93 |
| CMV reactivation | 21 | 0.29 (0.1–0.7) | 0.003 |
| Non-relapse mortality | 9 | 0.23 (0.1–1.0) | 0.04 |
| Overall mortality | 17 | 0.40 (0.2–0.9) | 0.03 |
Events on or after day +26.
Day +28 T-cell chimerism analyzed as binary endpoint (OR), all others as time-to-event endpoint (HR). OR and HR are effects per doubling of IMPDH AUEC. All analyses adjusted for Kahl relapse risk category (low, standard, high), donor-recipient gender (female to male, other), and donor (related, unrelated).
Figure 3.
Association of IMPDH AUEC (pmol × 106 cells) with acute GVHD (A), CMV reactivation (B), non-relapse mortality (C), and overall mortality (D) in 45 patients receiving an unrelated donor graft. Only events on or after day +26 are included.
DISCUSSION
The translational relevance of this work is that we created a pharmacokinetic-dynamic model that characterizes the relationship between IMPDH activity with plasma MPA concentrations after oral MMF administration in nonmyeloablative HCT patients. Our key results are: 1) weight-based dosing of MMF results in considerable interpatient variability in the inhibition of IMPDH activity; 2) the validation of a previous MPA population pharmacokinetic model in HCT recipients(6); 3) the development and validation of a population pharmacokinetic–dynamic model of MPA concentrations with IMPDH activity. No clinical covariates were found for the pharmacodynamic parameters (Supplemental Fig. 6), indicating that IMPDH activity should be directly measured and that further research is needed to explain pharmacodynamic variability. IMPDH AUEC on day +21 was associated with CMV reactivation, non-relapse mortality, and overall mortality (Table 4). These findings should be confirmed in a larger patient population, with the long-term goal of using IMPDH activity in PMNC as a predictive biomarker to improve survival.
The immunosuppressant MMF is an integral component of postgrafting immunosuppression after HCT. HCT recipients receiving MMF dosed by body weight have varying clinical outcomes.(21) We recently constructed MPA population pharmacokinetic models after IV MMF(5) or oral MMF(6) administration that revealed considerable interindividual variability in MPA pharmacokinetics. We validated this pharmacokinetic model and refined it by adding concentration-time data of unbound MPA and total MPAG. Oral MMF dosed based on body weight still results in considerable interpatient variability in the ex vivo IMPDH activity in PMNC isolated from participants receiving the same 15 mg/kg dose of oral MMF (Fig. 2 and Fig. 4). An inhibitory Emax model adequately described the inhibition of IMPDH activity in PMNC by MPA. The IIV of the pharmacodynamic parameters varied from 27.3% to 56.3% (Table 3). This IIV is greater than the IIV of many pharmacokinetic parameters with the notable exception of the IIV of the volume estimates, which range from 47 to 86.4% (Table 2). The large IIV in the volume estimates could be attributed to the inability to accurately characterize the maximum MPA concentration due to the use of a limited sampling schedule, which is a necessity in this patient population treated in the ambulatory clinic.
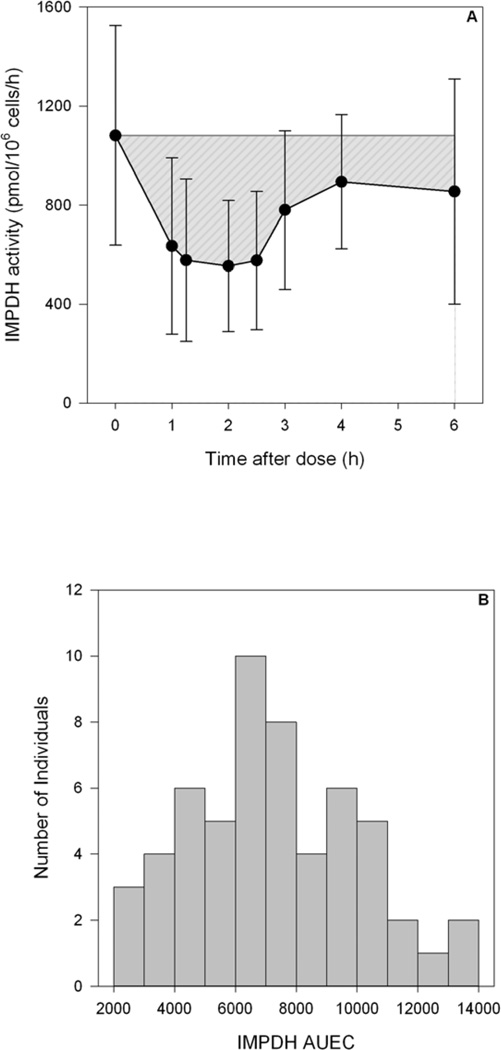
Figure 4.
Visual representation of IMPDH AUEC (gray area, in units of pmol × 106 cells) after a single dose of MMF at steady-state (A) and distribution of IMPDH AUEC in all patients (B).
We hypothesized that estimating its drug-specific pharmacodynamics (IMPDH activity in PMNC) could be a predictive measure of an individual’s response to MMF. Determining IMPDH activity may provide a direct quantitative method to evaluate the degree of MMF-induced immunosuppression and subsequent clinical efficacy and toxicity. Notably, on day +21, the PMNC can be derived from the recipient, a mixture of donor and recipient cells, or solely the donor cells. The day +28 donor T-cell chimerism is presented in Table 1; most patients had >95% T-cell chimerism. Variability in the cellular uptake and activation of MPA in PMNC cells may account for this differential cellular sensitivity. Notably, concomitant calcineurin inhibitor and graft source were not associated with the IC50 and Emax of IMPDH inhibition, suggesting these factors do not influence sensitivity to MMF (Supplemental Fig. 5).
To our knowledge, this publication is the first to evaluate the association of IMPDH activity after MMF administration in nonmyeloablative HCT recipients. Within HCT recipients, Laverdière (11) recently characterized IMPDH activity in 19 HCT recipients whose conditioning regimen, graft source, and MMF regimen were not detailed. These investigators reported a 5.3- fold variability in IMPDH activity; our results show a 10-fold variability in IMPDH activity (Fig. 2A and 2B) and 6-fold variability in IMPDH AUEC (Fig. 4B). Although we developed a validated LC-MS method to quantitate IMPDH activity, we could not quantitate IMPDH activity on day +2 due to extremely low white blood cell counts, predominantly due to the fludarabine/TBI conditioning.
In conclusion, we have shown that adequate number of PMNC can be isolated from HCT recipients on day +21 to quantitate XMP, and thus IMPDH activity, using our highly sensitive assay. We presented an integrated population-based model of total MPA, unbound MPA, and total MPAG plasma concentrations and the associated degree of immunosuppression, as quantified by IMPDH activity in PMNC. The final model captured the central tendencies and IIV well; there were, however, no clinical covariates associated with the pharmacodynamic parameters. Such a model provides an approach towards individualized oral MMF dosing and a firm rationale for further studies investigating whether dosing MMF on the basis of IMPDH activity can improve clinical outcomes. Subsequent translational studies will be necessary to evaluate whether IMPDH activity after MPA provides a novel biomarker to predict an individual’s sensitivity and response to MMF, with the long-range goal of individualizing postgrafting immunosuppression and/or MMF doses to improve the efficacy and/or decrease the toxicity of nonmyeloablative HCT.
Supplementary Material
Acknowledgements
The authors are very grateful to the patients who participated in this study. The authors also wish to thank all physicians, nurses, and support personnel for their care of patients on this study.
This work is supported in part by grants: HL91744, CA18029, CA78902, HL36444, HL093294.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors report no additional disclosures.
REFERENCES
- 1.Deeg HJ, Maris MB, Scott BL, Warren EH. Optimization of allogeneic transplant conditioning: not the time for dogma. Leukemia. 2006;20:1701–1705. doi: 10.1038/sj.leu.2404327. [DOI] [PubMed] [Google Scholar]
- 2.Maris MB, Niederwieser D, Sandmaier BM, et al. HLA-matched unrelated donor hematopoietic cell transplantation after nonmyeloablative conditioning for patients with hematologic malignancies. Blood. 2003;102:2021–2030. doi: 10.1182/blood-2003-02-0482. [DOI] [PubMed] [Google Scholar]
- 3.Nash RA, Johnston L, Parker P, et al. A phase I/II study of mycophenolate mofetil in combination with cyclosporine for prophylaxis of acute graft-versus-host disease after myeloablative conditioning and allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2005;11:495–505. doi: 10.1016/j.bbmt.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Staatz CE, Tett SE. Clinical pharmacokinetics and pharmacodynamics of mycophenolate in solid organ transplant recipients. Clin Pharmacokinet. 2007;46:13–58. doi: 10.2165/00003088-200746010-00002. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Mager DE, Bemer MJ, et al. A Limited Sampling Schedule to Estimate Mycophenolic Acid Area Under the Concentration-Time Curve in Hematopoietic Cell Transplantation Recipients. J Clin Pharmacol. 2011;52:1654–1664. doi: 10.1177/0091270011429567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Mager DE, Sandmaier BM, Maloney DG, Bemer MJ, McCune JS. Population pharmacokinetics and dose optimization of mycophenolic acid in HCT recipients receiving oral mycophenolate mofetil. J Clin Pharmacol. 2013;53:393–402. doi: 10.1002/jcph.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDermott CL, Sandmaier BM, Storer B, et al. Nonrelapse Mortality and Mycophenolic Acid Exposure in Nonmyeloablative Hematopoietic Cell Transplantation. Biol Blood Marrow Transplant. 2013;19:1159–1166. doi: 10.1016/j.bbmt.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haentzschel I, Freiberg-Richter J, Platzbecker U, et al. Targeting mycophenolate mofetil for graft-versus-host disease prophylaxis after allogeneic blood stem cell transplantation. Bone Marrow Transplant. 2008;42:113–120. doi: 10.1038/bmt.2008.85. [DOI] [PubMed] [Google Scholar]
- 9.Kuypers DR, Le Meur Y, Cantarovich M, et al. Consensus report on therapeutic drug monitoring of mycophenolic acid in solid organ transplantation. Clin J Am Soc Nephrol. 2010;5:341–358. doi: 10.2215/CJN.07111009. [DOI] [PubMed] [Google Scholar]
- 10.Allison AC, Eugui EM. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology. 2000;47:85–118. doi: 10.1016/s0162-3109(00)00188-0. [DOI] [PubMed] [Google Scholar]
- 11.Laverdière I, Caron P, Couture F, Guillemette C, Lévesque E. Liquid chromatography-coupled tandem mass spectrometry based assay to evaluate inosine-5'-monophosphate dehydrogenase activity in peripheral blood mononuclear cells from stem cell transplant recipients. Analytical chemistry. 2012;84:216–223. doi: 10.1021/ac202404y. [DOI] [PubMed] [Google Scholar]
- 12.Bemer MJ, Sorror M, Sandmaier BM, O'Donnell PV, McCune JS. A pilot pharmacologic biomarker study in HLA-haploidentical hematopoietic cell transplant recipients. Cancer Chemother Pharmacol. 2013 doi: 10.1007/s00280-013-2232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glander P, Sombogaard F, Budde K, et al. Improved assay for the nonradioactive determination of inosine 5'-monophosphate dehydrogenase activity in peripheral blood mononuclear cells. Ther Drug Monit. 2009;31:351–359. doi: 10.1097/FTD.0b013e31819c3f3d. [DOI] [PubMed] [Google Scholar]
- 14.Daxecker H, Raab M, Muller MM. Influence of mycophenolic acid on inosine 5'-monophosphate dehydrogenase activity in human peripheral blood mononuclear cells. Clinica chimica acta; international journal of clinical chemistry. 2002;318:71–77. doi: 10.1016/s0009-8981(01)00801-4. [DOI] [PubMed] [Google Scholar]
- 15.Musuamba FT, Rousseau A, Bosmans JL, et al. Limited sampling models and Bayesian estimation for mycophenolic acid area under the curve prediction in stable renal transplant patients co-medicated with ciclosporin or sirolimus. Clin Pharmacokinet. 2009;48:745–758. doi: 10.2165/11318060-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 16.Bullingham RE, Nicholls AJ, Kamm BR. Clinical pharmacokinetics of mycophenolate mofetil. Clin Pharmacokinet. 1998;34:429–455. doi: 10.2165/00003088-199834060-00002. [DOI] [PubMed] [Google Scholar]
- 17.Przepiorka D, Weisdorf D, Martin P, et al. 1994 Consensus Conference on Acute GVHD Grading. Bone Marrow Transplant. 1995;15:825–828. [PubMed] [Google Scholar]
- 18.Kahl C, Storer BE, Sandmaier BM, et al. Relapse risk in patients with malignant diseases given allogeneic hematopoietic cell transplantation after nonmyeloablative conditioning. Blood. 2007;110:2744–2748. doi: 10.1182/blood-2007-03-078592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng HY, Liu WB, Huang X, et al. [Efficacy and safety of low-dose cyclophosphamide plus corticosteroids for type I/II myasthenia gravis] Zhonghua Yi Xue Za Zhi. 2012;92:2323–2326. [PubMed] [Google Scholar]
- 20.Pharmacokinetic and Pharmacodynamic Data Analysis: Concepts and Applications. Swedish Pharmaceutical Press; [Google Scholar]
- 21.Li H, Mager DE, Bemer MJ, Sandmaier BM, Maloney D, McCune JS. Population Pharmacokinetics and Dose Optimization of Mycophenolic Acid in HCT Recipients Receiving Oral Mycophenolate Mofetil. [accepted for publication August 2, 2012];J Clin Pharmacol. 2012 doi: 10.1002/jcph.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.