Abstract
Why Helicobacter pylori colonizes only gastric tissue is unknown. It is found on gastric mucus-secreting cells and in the overlying gastric mucus but not deep in gastric glands. This localization mirrors the expression of trefoil factor 1, TFF1. We hypothesized that H. pylori interacting with TFF1 could explain the tropism of this bacteria for gastric tissue. Recombinant human TFF1 expressed in Escherichia coli was purified by affinity chromatography, ion-exchange chromatography, and gel filtration. Binding of H. pylori was assessed by using flow cytometry and the BIAcore system, which allows real-time monitoring of molecular interactions. In flow cytometry, H. pylori bound to the TFF1 dimer, but Campylobacter jejuni strains and the laboratory strain of E. coli, HB101, did not bind. When the BIAcore system was used, H. pylori bound strongly to TFF1-coated dextran chips compared with uncoated chips. Binding was inhibited by a TFF1 monoclonal antibody and by soluble TFF1. H. pylori bound to porcine gastric mucin only if it was pretreated with TFF1. In conclusion, H. pylori interacts avidly with the dimeric form of TFF1, and this interaction enables binding to gastric mucin, suggesting that TFF1 may act as a receptor for the organism in vivo. This interaction may underline the previously unexplained tropism of this organism for gastric tissue and its colocalization with the gastric mucin MUC5AC.
Helicobacter pylori infection causes gastritis and duodenal ulcer disease in humans (1, 2). Infection is associated with gastric carcinoma, and H. pylori has been designated a class 1 carcinogen by the World Health Organization. Infection is thought to be acquired in early childhood (3), and unless it is eradicated with antimicrobials, it can persist for life. A striking characteristic of H. pylori is that it exhibits strict host and tissue specificity. It colonizes humans and nonhuman primates but does not colonize other species naturally. H. pylori colonizes only gastric tissue. The majority of organisms are found swimming in the mucous gel layer of the stomach (4). A small percentage (approximately 2%) are adherent to the gastric epithelium. The organisms are found in the gastric pits but rarely if ever deep in the glands (5). Tropism of the organism for the stomach is unlikely to be explained by environmental conditions in the stomach, as areas of gastric metaplasia in the duodenum and gastric tissue in Meckels diverticulum can be colonized (6, 7). Receptors for H. pylori that have been identified include the Lewis b (Leb) (8) and Lewis x (Lex) (9) blood group antigens. Binding to Leb and Lex is mediated by the H. pylori adhesins BabA and SabA, respectively. However, other adhesins such as the AlpA and AlpB proteins have been identified, but no receptor for these proteins has been identified (10), indicating that other receptors exist on the gastric mucosa. In addition, neither Leb nor Lex is expressed solely in the stomach, and H. pylori also binds in vitro to Leb expressed on duodenal tissue (11), which is never observed in vivo. In contrast, areas of gastric metaplasia in the duodenum can be colonized in vivo, indicating that there is a receptor for H. pylori specific for gastric tissue. To date, no receptor for H. pylori has been identified to explain the tropism the organism exhibits for gastric tissue.
The trefoil peptides TFF1, TFF2, and TFF3 are a group of small proteins belonging to the trefoil factor family (TFF) (12). Trefoil peptides are localized within mucous granules in mucous-secreting cells and are secreted from the cells. They play a role in the process of restitution after mucosal injury and protect the integrity of the epithelial barrier (13, 14). They are expressed in a site-specific manner in gastrointestinal epithelium. TFF1 is expressed in foveolar epithelial surface cells of the stomach (15). TFF2 is expressed in the deeper glandular epithelium in the distal stomach but not in the surface mucosa and pits. It is also present in the acini of Brunners glands in the duodenum (16, 17). TFF3 is expressed by goblet cells in the large and small intestine (18).
In normal gastrointestinal mucosal structures, there is a general correlation between mucin glycoproteins and trefoil peptide gene expression (19). TFF1 segregates with MUC5AC in fovelar pit cells of the gastric body and superficial regions of the antral glands, TFF2 with MUC6 in the fundus and deep antral glands of the stomach, and TFF3 and MUC2 are coexpressed throughout the large and small bowel mucosa. It has been shown that H. pylori associates with gastric mucous-secreting cells that produce MUC5AC and not with cells deeper down in the glands that produce MUC6 (20). In addition, H. pylori has been shown to preferentially colonize the mucus gel layer of surface mucous cell-type mucins (21). H. pylori is not found in association with areas of intestinal metaplasia unless they have an incomplete phenotype, which expresses MUC5AC (22). The expression of TFF1 and MUC5AC in the gastrointestinal tract mirrors the localization of H. pylori. We therefore aimed to test the hypothesis that TFF1 can interact with H. pylori, possibly explaining why H. pylori colonizes only the stomach and colocalizes with MUC5AC.
Methods
Bacteria. The H. pylori type strain NCTC 11637 and Escherichia coli strain HB101 were obtained from the Public Health Laboratory Service, now renamed as Health Protection Agency (Colindale, England). H. pylori strain SS1 was obtained from Jani O'Rourke (University of New South Wales, Sydney), strain N6 was obtained from Agnes Labigne (Institut Pasteur, Paris). Strains PU4 and PU44 were cultured from gastric biopsy specimens obtained from children undergoing endoscopy. Campylobacter jejuni strains 81116 and F164 were derived from patients with campylobacteriosis. Bacteria were grown at 37°C on Columbia blood agar plates containing 7% (vol/vol) defibrinated horse blood. H. pylori and C. jejuni strains were incubated under microaerophillic conditions for 48-72 h.
Immunohistochemical and Immunofluorescent Staining. Gastric biopsy tissue was taken from children undergoing endoscopy at Our Lady's Hospital for Sick Children, Dublin. Ethical approval for this study was obtained from the Ethics Committee in the hospital. Biopsy tissue was checked for infection with H. pylori by Campylobacter-like organism (CLO) testing and by culture and histology. For immunofluorescent staining, specimens were prefixed in Zamboni solution for 60 min, rinsed in PBS with 10% (wt/vol) sucrose, embedded in OCT compound, and frozen in liquid nitrogen. Specimens were cut and stained with polyclonal H. pylori antibody (DAKO), and anti-rabbit IgG conjugated to Texas red (Molecular Probes). They were subsequently stained with a monoclonal TFF1 antibody and anti-mouse IgG conjugated to FITC (DAKO). Specimens were mounted in fluorescent mounting medium (DAKO) and viewed under a fluorescence microscope. For immunohistochemical staining, formalin-fixed, paraffin-embedded tissue was used. Cut sections were deparaffinated. Tissue that was stained for H. pylori and TFF1 was treated for 5 min with proteinase K (DAKO). Endogenous peroxidase was blocked. H. pylori organisms were detected by using the DAKO Envision+, rabbit peroxidase system. Polyclonal anti-H. pylori (DAKO) was applied, followed by labeled polymer conjugated to horseradish peroxidase. Enzyme activity was detected with diaminobenzidine, resulting in a brown color. Monoclonal TFF1 antiserum was applied, followed by a secondary biotinylated goat anti-mouse IgG (DAKO). Sections were incubated with ABcomplex conjugated to alkaline phosphatase (Vector Laboratories), and enzyme activity was detected with a Vector blue alkaline phosphatase substrate kit, resulting in a blue color. Tissue stained for TFF1 and TFF2 was heated in a pressure cooker in 0.01M sodium citrate, pH 6.0, for 1 min. Endogenous peroxidase was blocked. TFF1 was detected by using the DAKO Envision+, mouse peroxidase system (DAKO). Monoclonal TFF1 antiserum was applied, followed by labeled polymer conjugated to horseradish peroxidase. Enzyme activity was detected with diaminobenzidine, resulting in a brown color. Subsequently, monoclonal TFF2 antiserum was applied, followed by a secondary biotinylated goat anti-mouse IgG (DAKO). Sections were incubated with ABcomplex conjugated to alkaline phosphatase (Vector Laboratories), and enzyme activity was detected with a Vector blue alkaline phosphatase substrate kit, resulting in a blue color. Sections were mounted in Glycergel (DAKO) and viewed under a light microscope.
Production and Purification of Recombinant TFF1. TFF1 is a small secreted protein of 60 aa that contains a single trefoil domain. It has seven cysteine residues. Six are involved in maintaining the structure of the trefoil domain. The seventh (Cys-58) does not form part of the trefoil domain and is located three residues from the C terminus. TFF1 has been shown to be able to dimerize through Cys-58 by formation of a disulfide bond. An analogue of TFF1 in which Cys-58 has been replaced by serine is unable to dimerize and is found only in the monomeric form. Recombinant monomeric and dimeric human TFF1, which were expressed in E. coli, were purified by a combination of affinity chromatography, ion-exchange chromatography, and gel filtration as previously described (23).
Coating of Latex Beads with Trefoil Peptides and Mucin. Latex beads (Polysciences) (200 μl) were pelleted by centrifugation, resuspended in 500 μl of a 100 μg/ml solution of trefoil peptide in 0.1 M boric acid, pH 8.5, followed by end-over-end rotation overnight at 20°C. Beads were washed in boric acid, and unreacted sites were blocked by incubation of the beads in a 2% (wt/vol) solution of BSA in 0.1 M boric acid overnight at room temperature. Beads were washed in 0.1 M sodium phosphate buffer, pH 7.4, and resuspended and stored in 500 μl of sodium phosphate buffer containing 5% (vol/vol) glycerol and 1% (wt/vol) BSA at 4°C in the dark. Adsorption of the TFF1 monomer and dimer to the beads was determined by staining the beads with a rabbit polyclonal antibody raised against a synthetic peptide corresponding to the C-terminal 30 aa of TFF1 and an anti-mouse IgG conjugated to FITC. Reactivity of the beads with the antibody was assessed by flow cytometry. Beads coated with either monomeric TFF1 or dimeric TFF1 gave a positive fluorescence signal compared with the fluorescence of BSA-coated beads, which were used as a negative control (results not shown).
Porcine mucin purchased from Sigma was dissolved in 0.1 M boric acid, pH 8.5 (5 mg/ml), and coated onto latex beads as described above.
Flow Cytometric Adherence Assay. Adherence of H. pylori to beads coated with monomeric TFF1, dimeric TFF1, mucin, or BSA was assessed by using a flow cytometric assay. Five microliters of beads was pelleted in an Eppendorf tube; 10 μl of bacteria (OD600 = 0.2) was added, and beads and bacteria were incubated together at 37°C for 30 min. The contents of the tube were stained with rabbit-raised anti-H. pylori antibody (24) and a goat-raised anti-rabbit IgG conjugated to FITC. Adherent bacteria were detected by flow cytometry. Using a dot-plot display of forward and right-angle scatter, the flow cytometer (Becton Dickinson) was gated to include single beads and to exclude nonadherent bacteria, debris, and clumped beads. A total of 10,000 gated events were collected. Analysis of the data was performed with the cellquest software program from Becton Dickinson. The ability of a monoclonal antibody against TFF1 to neutralize the interaction of H. pylori with the TFF1 dimer-coated beads was tested by preincubation of the beads with the anti-TFF1 antibody for 30 min. The specificity of the interaction between H. pylori and the TFF1 dimer on the TFF1-coated latex beads was tested by incubation of H. pylori with TFF1 dimer (50 μg/ml) for 30 min before incubation of the bacteria with the TFF1-coated beads. Adherence of C. jejuni and E. coli to latex beads coated with TFF1 dimer was assessed as described above for H. pylori. Beads were stained with an in-house rabbit-raised C. jejuni antibody or E. coli antibody (DAKO).
Surface Plasmon Resonance Analysis. The BIAcore system based on the principle of surface plasmon resonance (25), which uses sensor chip chemistry and an integrated flow system to allow real-time monitoring of molecular interactions at the sensor surface (26), was used to further characterize the interaction of H. pylori with TFF1. The BIAcore sensor chip consists of a glass slide with a thin layer of gold deposited on one side, which is covered with a covalently bound carboxymethylated (CM) dextran matrix attached by a hydroxyalkyl thiol linker layer. Surface plasmon resonance is generated through the interaction of the light energy with the gold film, and this resonance is used to monitor concentrations of analyte on the surface of the chip. It can also monitor changes resulting from binding interactions between ligands of cells and proteins immobilized on the chip.
The CM-dextran matrix was activated by mixing equal volumes of 100 mM N-hydroxysuccinimide and 400 mM N-ethyl-N-(dimethylaminopropyl) carbodiimide hydrochloride and injecting the mixture over the sensor chip surface. TFF1 dimer was dissolved in 10 mM acetate buffer, pH 4.0, and injected over the surface for 30 min at a flow rate of 2 μl/min. The unreacted sites on the sensor chip surface were then capped by injection of 1 M ethanolamine, pH 8.5.
The bacteria (OD600 = 0.390) were diluted in phosphate-buffered saline solution (Dulbecco A PBS; 8 mM sodium phosphate, pH 7.3/0.15 M NaCl). They were initially characterized for nonspecific interactions to the CM-dextran surface.
TFF1 Preparation for Use in Inhibitive Assay. A 2 mg/ml solution of dimeric TFF1 was prepared. Standards were prepared from the stock solution in PBS. Each standard was incubated separately with an equal volume of the appropriate bacterial dilution, allowed to reach equilibrium, and then passed over the surface of the chip.
Results
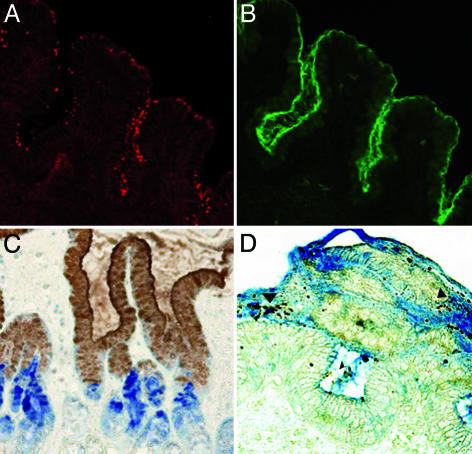
Colocalization of H. pylori and TFF1 in Human Gastric Biopsy Specimens. Colocalization of H. pylori and TFF1 in the gastric mucosa of infected humans was demonstrated by double immunofluorescent and immunohistochemical staining. H. pylori colocalized with TFF1 in the surface epithelium and pits of the gastric mucosa and within the overlying gastric mucus. TFF2 was expressed in the deeper glandular epithelium (Fig. 1).
Fig. 1.
Localization of H. pylori, TFF1, and TFF2 in the antrum of the human stomach. (A) Immunofluorescent staining of H. pylori in the antrum of the human stomach. (B) The same field is shown, but the staining of TFF1 is demonstrated. (C) The distribution of TFF1 and TFF2 in the human stomach. TFF1 (brown) is expressed in the surface mucosa of the gastric pits and in the overlying mucus, whereas TFF2 (blue) is found in the deeper glandular epithelium. (D) The majority of H. pylori organisms (brown, arrowhead) are visible within TFF1-containing gastric mucus (blue). (A, B, and D, original magnification ×200; C, original magnification ×100).
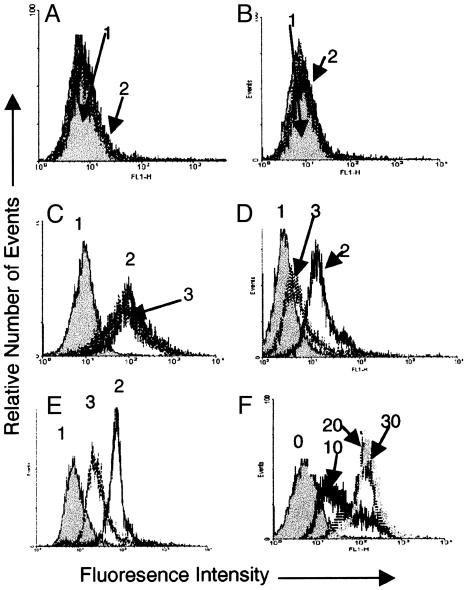
The Interaction of H. pylori with Monomeric and Dimeric TFF1. H. pylori strain PU4, a CagA-positive clinical isolate, bound to latex beads coated with the dimeric form of human TFF1. It did not bind to beads coated with the monomeric TFF1 or to BSA-coated beads (Fig. 2 A-C). Binding of H. pylori to TFF1 dimer-coated beads was detected within 10 min, and maximal binding was detected at 20 min (Fig. 2F). The binding of H. pylori strain PU4 to TFF1 dimer-coated beads could be inhibited by a monoclonal antibody to TFF1, and preincubation of H. pylori with soluble recombinant TFF1 resulted in inhibition of the subsequent binding of H. pylori to TFF1-coated beads (Fig. 2 D and E). Microscopy confirmed that H. pylori adhered to TFF1 dimer and not to BSA-coated beads (Fig. 3).
Fig. 2.
Interaction of H. pylori with monomeric and dimeric TFF1. (A) Binding of H. pylori to BSA-coated latex beads. Peak 1 (gray) represents BSA-coated beads that have not been incubated with the bacteria; it has been overlaid with a histogram representing binding to beads that have been incubated with bacteria (peak 2, speckled lines). The two histograms are almost identical, showing that bacteria did not adhere to the BSA. (B) Binding of H. pylori to TFF1 monomer-coated latex beads. Peak 1 (gray) represents coated latex beads, and it has been overlaid with a histogram representing beads that have been incubated with bacteria (peak 2, speckled lines). The two histograms are very similar, indicating that minimal bacterial adherence is occurring. (C) Binding of H. pylori to TFF1 dimer-coated latex beads. Peak 1 (gray) represents coated beads, and the clear histograms (peaks 2 and 3, speckled lines) represent coated beads that have been incubated with H. pylori. The positive shift in fluorescence indicates that the bacteria have bound to the beads. (D) The effect of a monoclonal antibody against TFF1 on binding of H. pylori to TFF1 dimer-coated latex beads. Peak 1 represents coated beads, peak 2 represents coated beads that have been incubated with the bacteria, and peak 3 represents coated beads that were treated with a monoclonal antibody against TFF1 before incubation with the bacteria. (E) The effect of soluble dimeric TFF1 on binding of H. pylori to TFF1 dimer-coated latex beads. Peak 1 represents coated beads, peak 2 represents coated beads that have been incubated with the bacteria, and peak 3 represents coated beads that been incubated with bacteria that were pretreated with TFF1 dimer. (F) Binding of H. pylori strain PU4 to latex beads coated with the dimeric form of TFF1. Beads were incubated with bacteria for 10, 20, and 30 min. All experiments were done at least three times. Results shown are from one representative experiment.
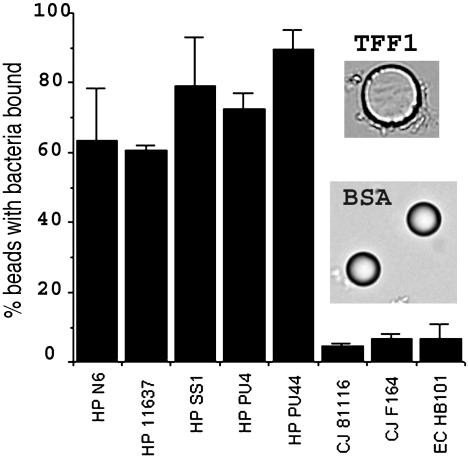
Fig. 3.
Binding of different bacterial strains to TFF1 dimer-coated latex beads. HP, H. pylori; CJ, C. jejuni; EC, E. coli. Adherence of the bacteria to the beads was assessed by flow cytometry. Results are expressed as the mean results of three experiments ± SD of the mean. Also shown are light micrographs of TFF1 dimer and BSA-coated beads that have been incubated with H. pylori. The diameter of the beads is 10 μm.
Four other strains of H. pylori tested bound to the TFF1 dimer. They were the type strain NCTC 11637 (CagA positive, with a functional cag pathogenicity island), strain N6 isolated from a patient with gastritis (CagA positive), the mouse-adapted strain SS1 (CagA positive but lacks a functional cag pathogenicity island), and strain PU44 (CagA negative). Two strains of C. jejuni and E. coli HB101 did not adhere to beads coated with the dimeric form of TFF1 (Fig. 3).
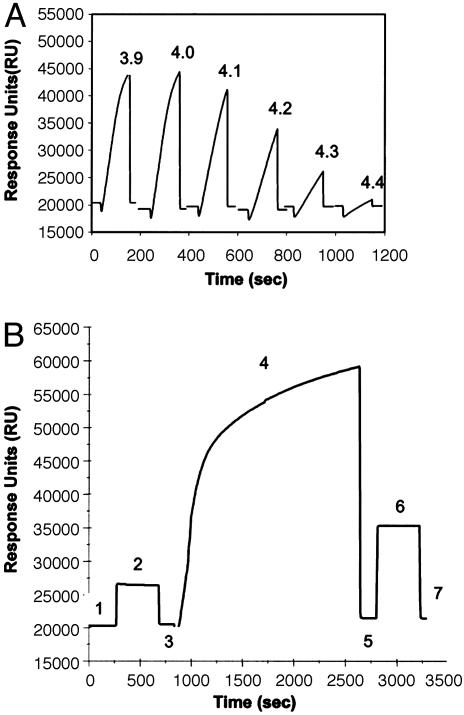
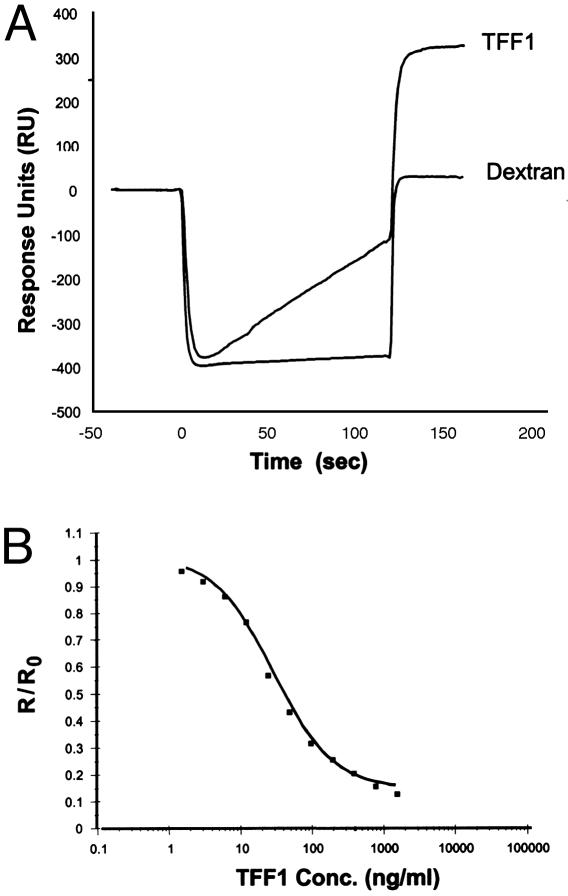
Surface Plasmon Resonance Analysis of the Interaction of H. pylori with the Dimeric Form of TFF1. The optimal pH determined for immobilization of dimeric TFF1 onto the surface of a CM5 sensor chip was pH 4.0 (Fig. 4A). All subsequent immobilizations of TFF1 were carried out by preparing the conjugate at this pH. Fig. 4B shows a typical sensorgram for the immobilization of TFF1 onto the surface of a chip. Immobilization of TFF1 resulted in ≈880 response units (RU) binding to the surface of the chip, corresponding to a protein concentration on the chip of ≈0.88 ng/mm2. H. pylori were passed over the surface of the TFF1-coated chip and bound specifically to the TFF1-coated chip (320 RU). Nonspecific interactions of the cells with the dextran matrix were found to be negligible (26 RU) (Fig. 5A).
Fig. 4.
Coating of a CM-dextran chip with recombinant dimeric human TFF1. (A) Preconcentration of TFF1 peptide onto the surface of a CM-dextran chip. Solutions of 100 μg/ml TFF1 in 10 mM sodium acetate buffer from pH 3.9 to 4.4 were passed over the same unactivated chip surface at 2 μl/min for 2 min. The optimal pH for immobilization was 4.0. (B) Typical sensorgram for the immobilization of TFF1 onto the surface of a sensor chip. Hepes-buffered saline (HBS) is passed over the sensor chip (line 1). A solution of 0.05 M N-hydroxysuccinimide and 0.2M N-ethyl-N-(dimethylaminopropyl) carbodiimide hydrochloride is passed over the sensor chip surface, resulting in activation of the CM-dextran on the chip surface (line 2). HBS is passed over the surface, and the baseline returns to normal with a small increase in response units, which can be attributed to the binding of N-hydroxysuccinimide esters to the surface (line 3). A solution of 100 μg/ml TFF1 prepared in 10 mM acetate buffer, pH 4.0, was passed over the surface for 30 min (line 4). The amount of peptide bound to the surface of the chip is recorded in line 5. Deactivation and capping of the surface N-hydroxysuccinimide esters was carried out with 1 M ethanolamine hydrochloride (pH 8.5) (line 6), which removes weakly bound peptide (line 7). The value recorded here is the amount of peptide bound to the surface of the chip, and this figure is presented as response units (RU).
Fig. 5.
Binding of H. pylori to an immobilized dimeric TFF1 surface. (A) Overlay plot demonstrating the binding of H. pylori strain PU4 to an immobilized TFF1 surface and also over a blank dextran surface. This resulted in negligible binding to the control dextran surface. The bacteria bound specifically to the TFF1 surface. (B) Inhibition assay carried out for free dimeric TFF1 by using a bacterial optical density of 0.391 on a dimeric TFF1 immobilized chip surface. The results shown are the average of triplicate results. The binding response at each TFF1 concentration (R) was divided by the cell binding response determined in the absence of TFF1 (R0) to give a normalized binding response (R/R0).
We determined the concentration of unbound TFF1 that is required to inhibit the bacteria binding to a TFF1-coated chip. The IC50 value for soluble TFF1 to inhibit binding of H. pylori to TFF1 bound to the surface of a sensor chip was 30.5 ng/ml, demonstrating that H. pylori has a high affinity for TFF1. A concentration of 1.56 μg/ml soluble TFF1 abolished binding of H. pylori to the coated chip (Fig. 5B).
Lack of Correlation Between Binding of H. pylori to TFF1 and to Leb. The best-characterized receptor for H. pylori to date is the Lewis b (Leb) blood group antigen. We used the BIAcore system to assess binding of the five strains of H. pylori to a sensor chip coated with Leb coupled to human serum albumin (Isosep, Sweden). The concentration immobilized onto the surface of the chip was ≈13,029 RU, which corresponds to a concentration of 13 ng/mm2, and the pH used for immobilization was 4.1. Only one of the five strains, strain NCTC 11637, bound to the Leb-coated chip (140 RU). The other four strains did not bind at all or binding was minimal compared with binding to uncoated dextran chips. Therefore, we could find no correlation between binding of the bacteria to TFF1 and binding to Leb.
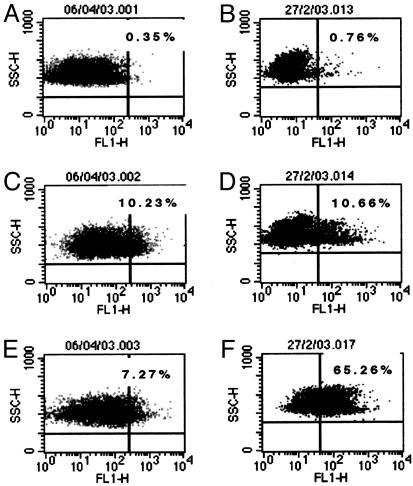
The Effect of TFF1 on Binding of H. pylori to Porcine Gastric Mucin. We wanted to determine whether binding of H. pylori to TFF1 had any effect on binding of the organism to gastric mucin. First, we tested binding of H. pylori on its own to porcine gastric mucin. We could not detect any appreciable binding of H. pylori to porcine gastric mucin. However, when the bacteria were preincubated with TFF1 before incubation with porcine mucin-coated beads, H. pylori acquired the ability to bind to the beads. TFF1 did not enhance binding of H. pylori to BSA-coated beads (Fig. 6).
Fig. 6.
Binding of H. pylori to porcine gastric mucin. H. pylori strain PU4 was incubated with the dimeric form of recombinant TFF1 for 30 min at 37°C, before incubation with latex beads coated with either BSA or porcine gastric mucin. Control bacteria were incubated with PBS before incubation with the beads. The percentage of beads that stained positive for bacteria adherent are shown in the top right corners of each dot plot. (A, C, and E) Results obtained with BSA-coated beads. (B, D, and F) Results obtained with mucin-coated beads. (A and B) Results obtained for beads with no bacteria attached. (C and D) Results obtained with beads incubated with bacteria that were preincubated with PBS. (E and F) Results obtained with beads incubated with bacteria that were preincubated with TFF1. Results shown are typical results obtained from one representative experiment.
Discussion
The reason why H. pylori specifically colonizes gastric tissue has not been identified to date. Our finding that H. pylori binds to the dimeric form of TFF1 could explain the unique ability of this organism to colonize only gastric tissue in vivo. This finding could also explain why H. pylori colocalizes with MUC5AC. The TFF1 dimer is strongly associated with the gastric mucin MUC5AC (27). In addition, it has been shown that TFF1 dimers are biologically more active than TFF1 monomers both in vitro and in vivo (28).
Binding of H. pylori to the dimeric form of TFF1 occurs rapidly. Binding seems to be a very specific event in that H. pylori colocalizes with TFF1 in infected human gastric biopsies. Furthermore, antibodies to TFF1 blocked H. pylori adherence to purified TFF1, and preincubation of H. pylori with recombinant dimeric TFF1 markedly diminished adherence. All H. pylori strains tested bound the dimeric form of TFF1, indicating that binding of H. pylori to TFF1 seems to be a trait widely expressed in strains. Strains of C. jejuni, which colonizes the small bowel of humans, and E. coli HB101 did not bind TFF1. Of interest, TFF1 expression is up-regulated in AGS cells upon infection with H. pylori (29) and in mice infected with Helicobacter heilmannii (30). In addition, recent work from the laboratory of S. Falkow has found that TFF1 expression is strongly up-regulated in polarized T84 monolayers in response to H. pylori infection, and this up-regulation is CagA dependent (D. Scott Merrell, personal communication). Increased H. pylori density in vivo has been associated with CagA status (31). Our results suggest that up-regulation of TFF1 by CagA-positive strains in vivo should confer on the organisms an enhanced ability to colonize the gastric mucosa.
It has been clearly demonstrated that the majority of H. pylori organisms are found within the gastric mucous layer (21), suggesting that all H. pylori isolates must have the ability to colonize gastric mucus. The only receptor that has been identified for H. pylori in MUC5AC is the Leb blood group antigen (32). However, only strains of H. pylori that express the BabA adhesin are capable of binding to Leb (33). Furthermore, H. pylori can bind to MUC5AC in the absence of Leb (11). In our study, there was no correlation between the ability of the H. pylori strains to bind Leb and to bind TFF1, as all strains bound to TFF1 and only one bound to Leb. There must be a receptor more universally present in gastric mucin that the bacteria can interact with. Binding to TFF1 may explain why H. pylori interacts only with gastric tissue and MUC5AC because, unlike Leb, which is expressed throughout the gastrointestinal tract, TFF1 is expressed only in the stomach in association with MUC5AC.
In this study, we could detect no interaction of H. pylori with porcine gastric mucin unless the organism was preincubated with TFF1, in which case the organism bound extremely well to mucin. It is unlikely that the reason for the lack of binding of untreated H. pylori is because the mucin is porcine, as one of the first and the best animal model of H. pylori infection is infection of the gnotobiotic pig (34). There are variable reports in the literature of H. pylori binding to gastric mucin. Some studies found that H. pylori did bind in vitro to human gastric mucin (35, 36). Linden et al. (32) reported no binding of H. pylori to MUC5AC for Leb-binding-negative isolates. A reason for the variation in binding may be the heterogeneity of the mucin samples from individual laboratories. Purification of mucins presents technical difficulties due to the size of mucin molecules, their high molecular weight, their polydispersity, and the overall complexity of the sample. Complete purification of mucin would remove dimeric TFF1 from the preparation. Our findings suggest that purified gastric mucin would have significantly less affinity for H. pylori than mucin with TFF1 present. The purity of the porcine mucin preparation used in this study is not likely to be a concern because H. pylori did not bind to it in the absence of TFF1.
There is some evidence that TFF1 interacts with MUC5AC through binding with the VWFC1 and VWFC2 (von Willebrand factor C) cysteine-rich domains of the mucin (37). The two putative receptor ligand recognition domains of the disulfide-linked homodimer of TFF1 have been shown to be separated at opposite ends of a flexible linker (38). The variable distance and orientation of the TFF1 binding sites offer versatility, which is needed for TFF1 to act as an adapter molecule binding to receptor binding sites on two different proteins and bringing them together. It is plausible to suggest that such an interaction may occur when H. pylori binds to TFF1 and gastric mucus.
Whereas the mouse model of H. pylori infection has been extensively studied, TFF1-deficient mice, which have been constructed (39), are unlikely to be suitable for studies on the role of TFF1 in gastric colonization by H. pylori. The antral and pyloric mucosa of TFF1-null mice is dysfunctional and exhibits severe hyperplasia and dysplasia. The antral and pyloric epithelial cells are improperly differentiated and almost entirely devoid of mucus, and inactivation of TFF1 can also suppress expression of TFF2 in the stomachs of some mice (39).
In conclusion, H. pylori interacts with the dimeric form of TFF1, and this interaction enables binding to gastric mucin, suggesting that TFF1 may act as a receptor for the organism in vivo. This interaction may underline the previously unexplained tropism of this organism for gastric tissue and its colocalization with MUC5AC. There is a need now to develop reliable cell culture and animal models to study the interaction of H. pylori with the gastric mucus layer.
Acknowledgments
We thank Dr. Michael McDermot and David Grehan for help with immunohistochemical staining. This work was supported by a grant from the Children's Medical and Research Foundation, Crumlin, Ireland (to M.C. and B.D.) and by grants from The Wellcome Trust and Cancer Research U.K. (to F.E.B.M. and B.R.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TFF1, trefoil factor 1; CM, carboxymethylated; RU, response units.
References
- 1.Marshall, B. J. & Warren, J. R. (1984) Lancet i, 1311-1315. [DOI] [PubMed] [Google Scholar]
- 2.NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease (1994) J. Am. Med. Assoc. 272, 65-69. [PubMed] [Google Scholar]
- 3.Rowland, M. (2000) Lancet 355, 332-333. [DOI] [PubMed] [Google Scholar]
- 4.Thomsen, L. L., Gavin, J. B. & Tasman-Jones, C. (1990) Gut 31, 1230-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falk, P., Roth, K. A., Boren, T., Westblom, T. U., Gordon, J. I. & Normark, S. (1993) Proc. Natl. Acad. Sci. USA 90, 2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyatt, J. I., Rathbone, B. J., Sobala, G. M., Shallcross, T., Heatley, R. V., Axon, A. T. & Dixon, M. F. (1990) J. Clin. Pathol. 43, 981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ackerman, Z., Peston, D. & Cohen, P. (2003) Dig. Dis. Sci. 48, 1068-1072. [DOI] [PubMed] [Google Scholar]
- 8.Boren, T., Falk, P., Roth, K. A., Larson, G. & Normark, S. (1993) Science 262, 1892-1895. [DOI] [PubMed] [Google Scholar]
- 9.Mahdavi, J., Sonden, B., Hurtig, M., Olfat, F. O., Forsberg, L., Roche, N., Angstrom, J., Larsson, T., Teneberg, S., Karlsson, K. A., et al. (2002) Science 297, 573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odenbreit, S., Till, M., Hofreuter, D., Faller, G. & Haas, R. (1999) Mol. Microbiol. 31, 1537-1548. [DOI] [PubMed] [Google Scholar]
- 11.Van de Bovenkamp, J. H., Mahdavi, J., Korteland-Van Male, A. M., Buller, H. A., Einerhand, A. W., Boren, T. & Dekker, J. (2003) Helicobacter 8, 521-532. [DOI] [PubMed] [Google Scholar]
- 12.Wright, N. A., Hoffmann, W., Otto, W. R., Rio, M. C. & Thim, L. (1997) FEBS Lett. 408, 121-123. [DOI] [PubMed] [Google Scholar]
- 13.Dignass, A., Lynch-Devaney, K., Kindon, H., Thim, L. & Podolsky, D. K. (1994) J. Clin. Invest. 94, 376-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kindon, H., Pothoulakis, C., Thim, L., Lynch-Devaney, K. & Podolsky, D. K. (1995) Gastroenterology 109, 516-523. [DOI] [PubMed] [Google Scholar]
- 15.Rio, M. C., Bellocq, J. P., Daniel, J. Y., Tomasetto, C., Lathe, R., Chenard, M. P., Batzenschlager, A. & Chambon, P. (1988) Science 241, 705-708. [DOI] [PubMed] [Google Scholar]
- 16.Hanby, A. M., Poulsom, R., Singh, S., Elia, G., Jeffery, R. E. & Wright, N. A. (1993) Gastroenterology 105, 1110-1116. [DOI] [PubMed] [Google Scholar]
- 17.Hanby, A. M., Poulsom, R., Elia, G., Singh, S., Longcroft, J. M. & Wright, N. A. (1993) J. Pathol. 169, 355-360. [DOI] [PubMed] [Google Scholar]
- 18.Podolsky, D. K., Lynch-Devaney, K., Stow, J. L., Oates, P., Murgue, B., De-Beaumont, M., Sands, B. E. & Mahida, Y. R. (1993) J. Biol. Chem. 268, 6694-6702. [PubMed] [Google Scholar]
- 19.Longman, R. J., Douthwaite, J., Sylvester, P. A., Poulsom, R., Corfield, A. P., Thomas, M. G. & Wright, N. A. (2000) Gut 47, 792-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van den Brink, G. R., Tytgat, K. M., Van der Hulst, R. W., Van der Loos, C. M., Einerhand, A. W., Buller, H. A. & Dekker, J. (2000) Gut 46, 601-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hidaka, E., Ota, H., Hidaka, H., Hayama, M., Matsuzawa, K., Akamatsu, T., Nakayama, J. & Katsuyama, T. (2001) Gut 49, 474-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genta, R. M., Gurer, I. E., Graham, D. Y., Krishnan, B., Segura, A. M., Gutierrez, O., Kim, J. G. & Burchette, J. L., Jr. (1996) Gastroenterology 111, 1206-1211. [DOI] [PubMed] [Google Scholar]
- 23.Chadwick, M. P., Westley, B. R. & May, F. E. B. (1997) Biochem. J. 327, 117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clyne, M. & Drumm, B. (1993) Infect. Immun. 61, 4051-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinn, J. & O'Kennedy, R. (1999) Anal. Lett. 7, 1475-1517. [Google Scholar]
- 26.Jonsson, U., Fagerstam, L., Ivarsson, B., Johnsson, B., Karlsson, R., Lundh, K., Lofas, S., Persson, B., Roos, H., Ronnberg, I., et al. (1991) BioTechniques 11, 620-627. [PubMed] [Google Scholar]
- 27.Newton, J. L., Allen, A., Westley, B. R. & May, F. E. B. (2000) Gut 46, 312-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchbank, T., Westley, B. R., May, F. E. B., Calnan, D. P. & Playford, R. J. (1998) J. Pathol. 185, 153-158. [DOI] [PubMed] [Google Scholar]
- 29.Guillemin, K., Salama, N. R., Tompkins, L. S. & Falkow, S. (2002) Proc. Natl. Acad. Sci. USA 99, 15136-15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller, A., O'Rourke, J., Grimm, J., Guillemin, K., Dixon, M. F., Lee, A. & Falkow, S. (2003) Proc. Natl. Acad. Sci. USA 100, 1292-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atherton, J. C., Tham, K. T., Peek, R. M., Jr., Cover, T. L. & Blaser, M. J. (1996) J. Infect. Dis. 174, 552-556. [DOI] [PubMed] [Google Scholar]
- 32.Linden, S., Nordman, H., Hedenbro, J., Hurtig, M., Boren, T. & Carlstedt, I. (2002) Gastroenterology 123, 1923-1930. [DOI] [PubMed] [Google Scholar]
- 33.Ilver, D., Arnqvist, A., Ogren, J., Frick, I. M., Kersulyte, D., Incecik, E. T., Berg, D. E., Covacci, A., Engstrand, L. & Boren, T. (1998) Science 279, 373-377. [DOI] [PubMed] [Google Scholar]
- 34.Krakowka, S., Eaton, K. A., Rings, D. M. & Morgan, D. R. (1991) Rev. Infect. Dis. 13, S681-S685. [DOI] [PubMed] [Google Scholar]
- 35.Tzouvelekis, L. S., Mentis, A. F., Makris, A. M., Spiliadis, C., Blackwell, C. & Weir, D. M. (1991) Infect. Immun. 59, 4252-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirmo, S., Artursson, E., Puu, G., Wadstrom, T. & Nilsson, B. (1999) J. Microbiol. Methods 37, 177-182. [DOI] [PubMed] [Google Scholar]
- 37.Tomasetto, C., Masson, R., Linares, J. L., Wendling, C., Lefebvre, O., Chenard, M. P. & Rio, M. C. (2000) Gastroenterology 118, 70-80. [DOI] [PubMed] [Google Scholar]
- 38.Williams, M. A., Westley, B. R., May, F. E. & Feeney, J. (2001) FEBS Lett. 493, 70-74. [DOI] [PubMed] [Google Scholar]
- 39.Lefebvre, O., Chenard, M. P., Masson, R., Linares, J., Dierich, A., LeMeur, M., Wendling, C., Tomasetto, C., Chambon, P. & Rio, M. C. (1996) Science 274, 259-262. [DOI] [PubMed] [Google Scholar]