Abstract
The response regulator CtrA, which silences the Caulobacter origin of replication and controls multiple cell cycle events, is specifically proteolyzed in cells preparing to initiate DNA replication. At the swarmer-to-stalked cell transition and in the stalked compartment of the predivisional cell, CtrA is localized to the cell pole just before its degradation. Analysis of the requirements for CtrA polar localization and CtrA proteolysis revealed that both processes require a motif within amino acids 1-56 of the CtrA receiver domain, and neither process requires CtrA phosphorylation. These results strongly suggest that CtrA polar localization is coupled to its cell cycle-regulated proteolysis. The polarly localized DivK response regulator promotes CtrA localization and proteolysis, but it does not directly recruit CtrA to the cell pole. Mutations in the divJ and pleC histidine kinases perturb the characteristic asymmetry of CtrA localization and proteolysis in the predivisional cell. We propose that polar recruitment of CtrA evolved to ensure that CtrA is degraded only in the stalked half of the predivisional cell, perhaps by localizing a proteolytic adaptor protein to the stalked pole. This is an example of controlled proteolysis of a cytoplasmic protein that is associated with its active recruitment to a specific subcellular address.
Keywords: Caulobacter, CtrA, ClpXP
In Caulobacter crescentus, DNA replication occurs once per cell division, and the phases of the cell cycle are linked to observable morphological changes (reviewed in refs. 1 and 2). Motile swarmer cells (Fig. 1) in the G1 phase of the cell cycle develop into stalked cells (Fig. 1) and initiate chromosome replication. During the swarmer-to-stalked cell (SW-ST) (or G1-S) transition, each cell sheds its polar flagellum and builds a stalk at the same site. As DNA replication proceeds, stalked cells elongate and become predivisional cells with distinct poles. A new flagellum is built at the pole opposite the stalk, so that each cell division produces a motile swarmer cell and a stalked cell. Before cell separation, a diffusion barrier is established between the swarmer and stalked compartments of the predivisional cell so that they contain distinct sets of signal transduction proteins that yield progeny with different replicative fates (3): the stalked progeny can initiate DNA replication immediately, whereas the swarmer cell must first differentiate into a new stalked cell.
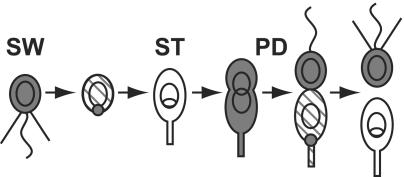
Fig. 1.
Polar localization and proteolysis of CtrA during the Caulobacter cell cycle. Motile swarmer cells (SW) have a polar flagellum (wavy line) and pili (straight lines) and cannot initiate DNA replication (closed circular chromosome). As the swarmer cell differentiates into a stalked cell (ST), the flagellum and pili are lost and CtrA (gray shading) is localized to the incipient stalked pole and then proteolyzed. As DNA replication proceeds (theta structure), the cell elongates, resynthesizes CtrA, and builds a flagellum at the pole opposite the stalk. In late predivisional cells (PD), CtrA is localized and degraded specifically in the stalked compartment of the cell to produce a replication-competent stalked cell devoid of CtrA and a swarmer cell containing CtrA.
A key signal transduction protein that regulates Caulobacter cell cycle progression is the essential response regulator CtrA (4). CtrA directly induces or represses the transcription of ≈55 operons in the Caulobacter genome, including genes needed for flagellum and pili biosynthesis, DNA methylation, cell division, chemotaxis, and metabolism (5, 6). However, CtrA also represses chromosome replication by binding to five sites within the replication origin (7). Because CtrA has multiple functions, its activity is tightly controlled by three mechanisms: cell cycle-regulated transcription (8), cell cycle-regulated phosphorylation (9, 10), and rapid proteolysis at the SW-ST transition and in the stalked compartment of the predivisional cell (9). These processes ensure that CtrA activity is present in swarmer and predivisional cells, but absent in stalked cells preparing to enter S phase.
Previous studies have shown that regulated proteolysis of CtrA requires a bipartite motif consisting of hydrophobic residues at the extreme C terminus and residues within amino acids 1-56 of the receiver domain (9, 11). The proteolytic motif in the CtrA receiver domain is within amino acids 26-44, which lie on the β2-α2 face of the receiver domain (11). The ATP-dependent ClpXP protease is necessary for regulated degradation of CtrA (12). The C-terminal amino acids of CtrA resemble one type of recognition signal for ClpX, the ssrA tag (13-15), which binds directly to ClpX (16). We therefore proposed that the hydrophobic residues at the C terminus of CtrA bind directly to ClpX. In this model, the role of the proteolytic determinant in the receiver domain would be to interact with an additional regulatory factor that specifies the correct cell cycle timing of CtrA degradation. A final component known to promote CtrA proteolysis is the essential single-domain response regulator DivK (17). A conditional divK mutant fails to degrade CtrA, and the cells arrest in G1 with one chromosome (18). The precise role of DivK is unknown: DivK may interact directly with CtrA, or it may be part of a signal transduction pathway that promotes a variety of events during the SW-ST transition.
Fusion of either the entire CtrA protein (this study) or the receiver domain and C terminus of CtrA to the yellow fluorescent protein (YFP) (11) revealed that CtrA transiently accumulates at the cell pole just before proteolysis, both at the SW-ST transition and in the stalked compartment of the predivisional cell (Fig. 1). The aims of this study are to determine the requirements for CtrA accumulation at the cell pole and to analyze the relationship between CtrA polar localization and proteolysis. We report here that polar accumulation of CtrA requires the proteolytic motif in the first half of the CtrA receiver domain. The C-terminal proteolytic determinant (and thus CtrA turnover) is not necessary for CtrA localization at the cell pole. divK mutant cells, which cannot degrade CtrA, are also impaired in CtrA polar localization. These results strongly suggest that recruitment of CtrA to the cell pole is an integral part of the proteolytic mechanism. Although DivK and CtrA are colocalized at the cell pole, DivK does not act as a polar binding site for CtrA. We propose that DivK acts in a signaling pathway that provides temporal and spatial control over CtrA polar localization and proteolysis.
Methods
Bacterial Strains, Media, and Plasmids. Unless noted otherwise, Caulobacter strain NA1000 (19), a synchronizable wild-type derivative of CB15, was used in all experiments. Caulobacter cells were grown in PYE or M2G medium (20) supplemented with 1 μg/ml chloramphenicol, 5 μg/ml kanamycin, or 1 μg/ml oxytetracycline. Derivatives of plasmids pJS14 (ref. 21 and J. Skerker, personal communication), pMR10, and pMR20 (22) were mobilized into Caulobacter from Escherichia coli strain S17-1 (23) by bacterial conjugation (20). To induce expression of genes from the xylose-inducible promoter Pxyl (24), PYE was supplemented with 0.03% xylose, and M2G was supplemented with 0.3% xylose. Expression of transgenes from Pxyl was induced 1-2 h before cells were synchronized or photographed. YFP fusion genes were created by restriction enzyme-mediated subcloning of sequence-verified constructs (11). Plasmids and strains used in this study are listed in the supporting information, which is published on the PNAS web site.
Synchronization, Immunoblotting, and Fluorescence-Activated Cell Sorter (FACS) Analysis. We used Ludox density centrifugation (4) to isolate G1-phase swarmer cells from mixed cultures undergoing exponential growth. FACS analysis of DNA content (25) and immunoblotting (11) were performed as described. Anti-GFP antiserum (P. Viollier, personal communication) was diluted 1:5,000.
Fluorescence and Differential Interference Contrast (DIC) Microscopy. Cells were immobilized on agarose pads composed of 1% (wt/vol) agarose in M2G medium containing 0.3% xylose. Images were acquired as described (11), with 2-s exposure time for YFP. A developing swarmer cell was considered to have polarly localized YFP if the signal intensity at one pole was increased at least 50% over the intensity at the opposite pole. (For predivisional cells, the intensities at the stalked pole and the opposite end of the stalked compartment were compared.) In cells with diffuse YFP signal, the mean intensity difference between two poles was 11%, and the intensity difference never exceeded the 50% cutoff value.
Results
CtrA Accumulates at the Cell Pole Just Before Proteolysis. Cell cycle-regulated proteolysis of CtrA requires residues within the first 56 aa of the receiver domain and residues at the extreme C terminus of the protein (11). When the receiver domain and last 15 aa of CtrA are fused to the C terminus of YFP (YFP-RD+15), CtrA proteolysis can be visualized during the Caulobacter cell cycle by using time-lapse fluorescence microscopy. When cells expressing YFP-RD+15 were observed at 30-min intervals during synchronous growth, YFP-RD+15 was localized to the incipient stalked pole in ≈1/3 of developing swarmer cells and to the stalked pole in ≈1/10 of predivisional cells, just before the fusion protein was proteolyzed (Fig. 1 and ref. 11). To determine whether polar foci occur in all cells proteolyzing CtrA, we isolated wild-type swarmer cells expressing YFP-RD+15 and collected time-lapse images every 6 min to detect transient foci. During the SW-ST transition, 82% (n = 181) of developing cells displayed a polar YFP-RD+15 signal before degradation of the fusion protein. Thus, YFP-RD+15 accumulates at one pole of almost all developing swarmer cells. Because we must immobilize the cells to obtain fluorescence images, we could not detect a temporal relationship between the appearance of YFP-RD+15 polar foci and the loss of motility, which also occurs during the SW-ST transition.
To quantify polar foci in predivisional cells, we harvested wild-type swarmer cells expressing YFP-RD+15, allowed them to grow into the early predivisional stage, and collected time-lapse images every 6 min until the cells had divided. In the 60 min leading up to cell division, 51% (n = 158) of predivisional cells displayed a focus of YFP-RD+15 at the stalked pole. It may be that YFP-RD+15 does not accumulate at the stalked pole of every predivisional cell before its proteolysis. However, it is possible that polar foci of YFP-RD+15 occur in all predivisional cells but are shorter-lived and therefore more difficult to detect.
To confirm that the receiver domain and C terminus of CtrA have the same intracellular dynamics as the full-length protein, we expressed a YFP-CtrA fusion protein by using the inducible Pxyl promoter (24) on a low-copy number plasmid (22) in wild-type Caulobacter. We observed mixed populations of cells at a single time point by using DIC and fluorescence microscopy to determine the percentage of swarmer, stalked, and predivisional cells containing a polar YFP signal. YFP-CtrA foci were present in 16% of swarmer cells, no stalked cells, and 15% of predivisional cells (defined as cells possessing both a stalk and a pinch at the presumptive division site, Fig. 2A and Table 1). In a similar experiment using unsynchronized cells, YFP-RD+15 was located at the pole in 16% of swarmer cells, no stalked cells, and 15% of predivisional cells (Fig. 2B and Table 1). These results imply that fusion proteins containing only the receiver domain and C terminus of CtrA faithfully reproduce the dynamics of the full-length CtrA protein in vivo.
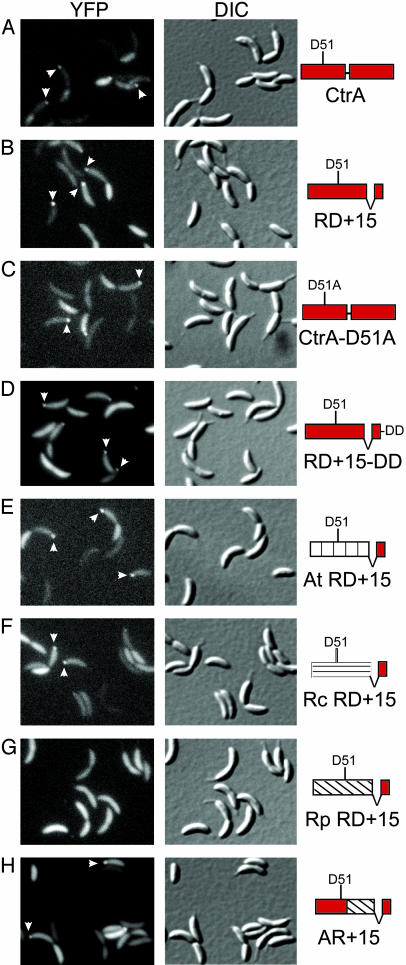
Fig. 2.
Amino acids 1-56 of the receiver domain are required for polar localization of CtrA. Fluorescence and DIC microscopy of CB15N cells expressing YFP fused to the indicated CtrA variants. Each YFP fusion was expressed from the Pxyl promoter on the low-copy plasmid pMR10 except for YFP-Rp RD+15, which was carried on the high-copy plasmid pJS14 to achieve expression levels comparable with the other YFP fusions. Arrowheads indicate polar foci of YFP fusion proteins.
Table 1. Incidence of polar foci of YFP fusions.
| SW cells
|
ST cells
|
PD cells
|
||||
|---|---|---|---|---|---|---|
| Protein fused to YFP | % with focus | n | % with focus | n | % with focus | n |
| CtrA | 16 | 406 | 0 | 196 | 15 | 325 |
| RD+15 | 16 | 522 | 0 | 229 | 15 | 303 |
| CtrA-D51A | 17 | 586 | 0 | 319 | 10 | 373 |
| RD+15-DD | 26 | 384 | 3 | 151 | 15 | 244 |
| At RD+15 | 16 | 377 | 0 | 211 | 11 | 350 |
| Rc RD+15 | 16 | 498 | 0 | 117 | 6 | 175 |
| Rp RD+15 | 0 | 359 | 0 | 134 | 0 | 260 |
| AR+15 | 16 | 514 | 0 | 148 | 7 | 217 |
SW, swarmer; ST, stalked; PD, predivisional.
Polar Accumulation of CtrA Does Not Require CtrA Phosphorylation or Proteolysis. To analyze the relationship between CtrA polar accumulation and proteolysis, we fused CtrA variants with known proteolytic phenotypes to YFP. We expressed each fusion protein from the Pxyl promoter on a low-copy number plasmid in wild-type Caulobacter and examined mixed populations of cells with DIC and fluorescence microscopy (Fig. 2 and Table 1). We used Western blot analysis with antibodies against YFP and CtrA to ensure that all fluorescent signal is created by full-length fusion proteins and not breakdown products (data not shown). CtrA-D51A, a full-length CtrA protein with a point mutation at the site of phosphorylation, cannot be phosphorylated and is inactive (9), but is properly degraded during the cell cycle (11). Polar foci of YFP-CtrA-D51A are present in 17% of swarmer cells, no stalked cells, and 10% of predivisional cells (Fig. 2C and Table 1). Thus, phosphorylation of CtrA at D51 is not essential for polar accumulation or proteolysis. Changing the final two residues of YFP-RD+15 from AA to DD creates a fusion protein that lacks the C-terminal degradation motif and does not undergo cell cycle-regulated proteolysis (11). However, polar foci of YFP-RD+15-DD are present in 26% of swarmer cells, 3% of stalked cells, and 15% of predivisional cells (Fig. 2D and Table 1). Polar accumulation and proteolysis of CtrA can therefore be separated, and the C-terminal degradation motif in CtrA is not required for polar localization. The presence of YFP-RD+15-DD foci in stalked cells and their increased frequency in swarmer cells, as compared to variants that are degraded, suggest that CtrA proteolysis normally accelerates the dissipation of polar foci. In time-lapse experiments, polar foci of YFP-RD+15-DD occur transiently in developing swarmer and predivisional cells, even though the fusion protein is not specifically proteolyzed (data not shown). Together, these results imply that polar foci are formed by cell cycle-regulated recruitment of CtrA to the cell pole, rather than by stabilizing CtrA at the pole, whereas it is degraded elsewhere in the cytoplasm.
A Motif in Amino Acids 1-56 of the CtrA Receiver Domain Is Required for Polar Localization. We previously fused homologs of the CtrA receiver domain from other α-proteobacteria to the C terminus of Caulobacter CtrA and assessed their turnover to locate the receiver domain determinant for cell cycle-regulated proteolysis (11). Only the receiver domain of CzcR, the Rickettsia prowazekii (Rp) CtrA homolog, failed to specify proteolysis at the SW-ST transition in Caulobacter. In this study, we fused YFP to each of these constructs to determine which homologs support transient polar localization. The CtrA receiver domains from Agrobacterium tumefaciens (At) and Rhodobacter capsulatus (Rc) both specify turnover during the SW-ST transition of the Caulobacter cell cycle, and when fused to YFP, both receiver domains form polar foci in swarmer and predivisional cells (Fig. 2 E and F and Table 1). In contrast, a construct containing the CzcR receiver domain is stable during the Caulobacter cell cycle, and the YFP-Rp RD+15 fusion protein does not form polar foci in any Caulobacter cell type (Fig. 2G and Table 1). We used chimeras of Caulobacter CtrA and CzcR to locate a determinant necessary for cell cycle-regulated proteolysis within amino acids 1-56 of the CtrA receiver domain (11). Here we found that a YFP fusion protein containing amino acids 1-56 of CtrA, amino acids 57-117 of CzcR, and the CtrA C terminus forms transient polar foci in 16% of swarmer cells, no stalked cells, and 7% of predivisional cells (Fig. 2H and Table 1). These results strongly suggest that the same molecular signal within amino acids 1-56 of the CtrA receiver domain is necessary both for cell cycle-regulated proteolysis and for transient polar localization.
DivK Signals both the Proteolysis and Polar Localization of CtrA. The essential single-domain response regulator DivK is required for CtrA proteolysis at the SW-ST transition (18). In a conditional divK loss-of-function mutant (divKcs), Caulobacter cells arrest in the G1 phase of the cell cycle with one chromosome. The cells elongate and grow stalks but do not degrade CtrA or initiate DNA replication (18). Because polar localization and proteolysis are closely linked in time and appear to use the same sequence motif in the CtrA receiver domain, we asked whether DivK is required for CtrA localization as well as for degradation.
We created a divKcs strain that expresses YFP-RD+15 from the inducible Pxyl promoter. We grew both wild-type and divKcs cells at the permissive temperature of 33°C and induced expression of YFP-RD+15 for 2 h before isolating G1-phase swarmer cells. We then released the swarmer cells of each strain into medium containing xylose at 20°C, the nonpermissive temperature for divKcs. At intervals, we withdrew samples from each culture for microscopy, Western blot analysis of CtrA levels, and FACS analysis of DNA content.
At 20°C, wild-type cells in culture take 4 h to divide (18). We observed synchronized wild-type and divKcs cells growing at 20°C for 3 h, to focus on the time in which CtrA is normally proteolyzed and DNA replication and stalk biogenesis begin. In the wild-type culture, CtrA and YFP-RD+15 are degraded during the SW-ST transition and are later resynthesized (Fig. 3B). FACS analysis shows that wild-type cells initiate DNA replication and proceed synchronously into S phase (data not shown). To assess morphological development, we determined the percentage of cells at each time point with a visible stalk: after 3 h at 20°C, >50% of wild-type cells have a polar stalk (Fig. 3A). At the first time point, 40% of wild-type cells have already entered the SW-ST transition and possess a polar focus of YFP-RD+15 (Fig. 3A), en route to degrading CtrA and YFP-RD+15 (Fig. 3B). The number of cells containing a polar focus of YFP decreases as the experiment progresses because of proteolysis of the fusion protein.
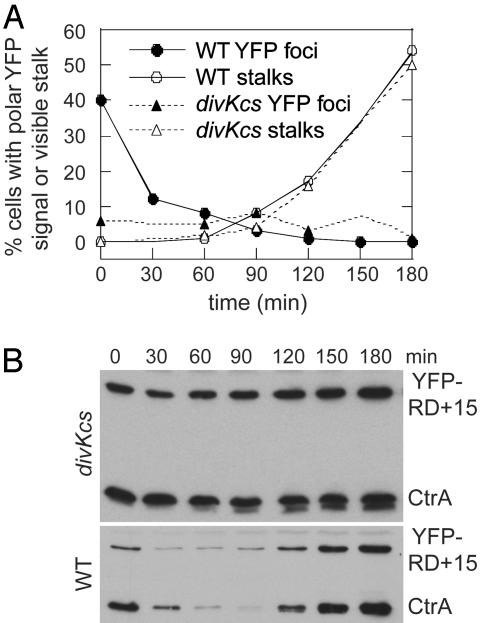
Fig. 3.
divKcs cells are impaired in CtrA localization and proteolysis. CB15N and divKcs swarmer cells expressing YFP-RD+15 were isolated from mixed cultures grown at 33°C and then incubated in a shaking water bath at 20°C. At the indicated times, samples were withdrawn from the liquid culture for DIC and fluorescence microscopy (A), Western blotting (B), and FACS analysis (not shown). Cultures were observed for 180 min, which corresponds to the first 3/4 of the cell cycle. (A) Graph showing the percentage of CB15N or divKcs cells at each time point displaying a polar focus of YFP-RD+15 or a polar stalk. Each time point represents >500 cells. (B) Western blots of CB15N or divKcs cells expressing YFP-RD+15 probed with anti-CtrA antiserum. Each lane contains protein extract from an equal number of cells.
In agreement with previous results (18), synchronized divKcs cells held at 20°C do not degrade CtrA or YFP-RD+15 (Fig. 3B), and they fail to initiate DNA replication (data not shown). divKcs cells build stalks at the same time as wild-type cells (Fig. 3A). However, in contrast to the early burst of YFP foci seen in wild-type cells, divKcs cells contain polar foci of YFP-RD+15 at a low frequency throughout the incubation at 20°C (Fig. 3A). DivK thus appears to regulate the temporally controlled accumulation of CtrA at the cell pole in addition to promoting CtrA degradation.
Polar Localization of DivK Is Not Required for CtrA Recruitment to the Pole. DivK, a cytoplasmic protein, is dynamically localized during the Caulobacter cell cycle (27). In the swarmer cell, DivK is diffuse, but during the SW-ST transition, DivK accumulates at the incipient stalked pole. As the cell grows and prepares to divide, a second focus of DivK appears at the pole opposite the stalk. Around the time of division, DivK is released from the pole in the swarmer compartment and remains at the stalked pole of the stalked compartment. Because CtrA accumulates at sites where DivK is also localized, we asked whether DivK is directly responsible for recruiting CtrA to the pole by observing YFP-RD+15 in two mutant strains that mislocalize DivK, but do not block the functions of DivK that are essential for viability.
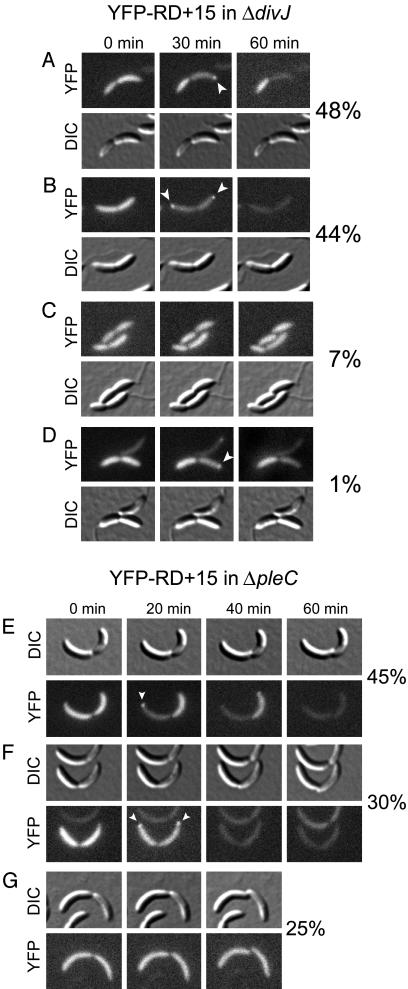
DivJ is a histidine kinase that phosphorylates DivK in vitro (17) and in vivo (28). Substitution of divJ with an antibiotic resistance cassette is not lethal, but the stereotypic asymmetry of Caulobacter cell division is perturbed (29). Motile and piliated cells are present in a ΔdivJ mutant population, but many cells have bipolar stalks or a stalk misplaced along the side of the cell (28). ΔdivJ cells contain reduced levels of DivK∼P (28), and all cells fail to localize DivK to the poles (27). Despite their inability to localize DivK, ΔdivJ swarmer (data not shown) and predivisional cells (Fig. 4 A, B, and D) can still localize and degrade YFP-RD+15. Thus, DivK localization to the pole is not essential for transient accumulation of YFP-RD+15 at the same site. However, time-lapse microscopy revealed that the normal asymmetry of CtrA proteolysis and polar localization is abolished in at least half of ΔdivJ predivisional cells. Only 48% of ΔdivJ predivisional cells proteolyze CtrA specifically in one compartment before division (Fig. 4A), as compared to 100% of wild-type predivisional cells. In ΔdivJ predivisional cells with a single polar stalk, we found a few instances (1% of all cells examined) where CtrA localization and proteolysis occurred in the compartment opposite the stalk, a reversal of the wild-type CtrA polarity (Fig. 4D). In 44% of ΔdivJ predivisional cells, YFP-RD+15 foci appear at both poles, and the fusion protein is degraded in both halves of the cell before division (Fig. 4B). Conversely, in 7% of ΔdivJ predivisional cells, division proceeds without any localization or degradation of YFP-RD+15 (Fig. 4C). The ΔdivJ mutation thus impairs the normal asymmetry of CtrA localization and proteolysis in many predivisional cells. However, the physical localization of DivK at the cell pole is not essential for polar recruitment of CtrA.
Fig. 4.
ΔdivJ and ΔpleC mutants that mislocalize DivK can localize and degrade CtrA. Time-lapse DIC and fluorescence microscopy showing the patterns of localization and proteolysis of YFP-RD+15 in ΔdivJ (A-D) or ΔpleC (E-G) predivisional cells. Arrowheads indicate polar foci of YFP-RD+15. The percentage of predivisional cells displaying each pattern is reported at the right of each series of photographs.
PleC is a second histidine kinase that can phosphorylate DivK in vitro (17), but which reduces the level of DivK∼P in vivo (28). The ΔpleC mutant is viable but lacks a stalk and pili and has bipolar, nonfunctional flagella (30, 31). ΔpleC cells contain elevated levels of DivK∼P (28) and cannot release localized DivK from the swarmer cell pole (27). To determine whether constitutive polar localization of DivK affects the normal pattern of CtrA localization and proteolysis, we observed YFP-RD+15 in ΔpleC cells by using time-lapse microscopy (Fig. 4 E-G). In 45% of ΔpleC predivisional cells, normal CtrA polarity is preserved in that YFP-RD+15 is proteolyzed in one half of the cell before division (Fig. 4E). In contrast, 30% of ΔpleC predivisional cells localize and degrade YFP-RD+15 in both halves of the cell before division (Fig. 4F), and 25% fail to localize or degrade the fusion protein before dividing (Fig. 4G). Because ΔpleC cells are stalkless, we could not determine whether any cells displayed reversed polarity of CtrA localization and proteolysis. divJ and pleC mutations therefore impair the asymmetric localization and proteolysis of CtrA in the predivisional cell. We propose that DivJ and PleC affect these processes via DivK, but we cannot rule out the involvement of additional signaling pathways. It is clear, however, that polar localization of CtrA does not require DivK to be localized at the same pole. Thus, DivK does not function as the physical factor that recruits CtrA to the cell pole, but rather serves in a signal transduction pathway that activates CtrA localization and proteolysis.
Discussion
The CtrA receiver domain and C-terminal 15 aa together are sufficient for temporally regulated degradation of a YFP fusion protein. A mutation that converts the two C-terminal alanine residues (AA) of CtrA to two aspartates (DD) abolishes CtrA degradation but preserves transient polar localization. This result distinguishes between two possible mechanisms for the generation of CtrA foci. In one model, CtrA appears concentrated at the pole only because it is being rapidly degraded elsewhere in the cell, whereas in the second model CtrA is actively recruited to the pole from the cytoplasm. Because the nondegradable variant YFP-RD+15-DD forms transient polar foci, we conclude that the foci are generated by an active process of recruitment.
Analysis of YFP fusions to other CtrA variants showed that the ability of a receiver domain to specify cell cycle-regulated proteolysis correlates with its ability to form polar foci. CtrA-D51A is both proteolyzed (11) and localized to the pole, indicating that phosphorylation at D51 is dispensable for both processes. The CtrA receiver domains from A. tumefaciens and R. capsulatus are degraded at the SW-ST transition, and both are polarly localized in developing swarmer and predivisional cells. Conversely, the receiver domain from Rickettsia prowazekii CzcR is neither proteolyzed at the SW-ST transition nor localized to the cell pole. A YFP fusion encoding a chimera of the CtrA and CzcR receiver domains revealed that the signal for polar localization lies in amino acids 1-56 of Caulobacter CtrA, which is also needed for cell cycle-regulated degradation.
We deduced from sequence analysis of the degradable and nondegradable CtrA homologs and chimeras that one determinant of cell cycle-regulated CtrA proteolysis is within a surface-exposed group of amino acids on the β2-α2 face of the receiver domain (11). Because the ability of a receiver domain to be degraded correlates with its ability to form polar foci, and because we have found no CtrA variants that prevent polar localization but leave proteolysis intact, we propose that the same molecular signal in the receiver domain is used both for both processes. In this model, polar localization is an intrinsic mechanistic step in CtrA proteolysis. The receiver domain proteolytic determinant may be recognized by an adaptor protein that promotes the interaction of CtrA with ClpXP at a specific time in the cell cycle. We believe that such a regulatory factor exists because ClpX and ClpP are essential proteins present at all times of the cell cycle (12) and because purified ClpXP does not degrade CtrA in vitro (11). The interaction between the receiver domain and the proposed adaptor protein might recruit CtrA to the cell pole as well (Fig. 5). A direct link between polar localization and proteolysis may have evolved to ensure that CtrA is degraded only in the stalked half of the predivisional cell, whereas ClpXP is present and active in both compartments. In immunofluorescence experiments, ClpX is found throughout the cell in all Caulobacter cell types (data not shown).
Fig. 5.
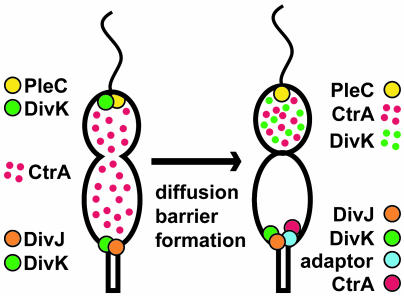
Model of asymmetric CtrA proteolysis in the predivisional cell. (Left) In the predivisional cell, PleC is located at the swarmer pole, DivJ is located at the stalked pole, and DivK is bipolar. CtrA is diffuse throughout the cell, and DivK∼P levels are equal in the two halves of the cell. (Right) After formation of a cytoplasmic diffusion barrier, DivK∼P levels in swarmer compartment fall, and DivK is released from the pole. In response to the asymmetry of DivK∼P levels, CtrA remains diffuse and stable in the swarmer compartment, but is localized to the pole and degraded in the stalked compartment. We propose that an adaptor protein located specifically at the stalked pole of the cell binds to the β2-α2 face of the CtrA receiver domain to directly mediate the polar recruitment and degradation of CtrA.
Although DivK is required for CtrA proteolysis (18) and DivK itself is localized to the nascent stalked pole of the cell at the SW-ST transition (27), DivK does not directly recruit CtrA to the pole. In the ΔdivJ mutant, where DivK is delocalized, and in the ΔpleC mutant, where DivK is constitutively located at the cell poles (27), half of the predivisional cells localize and degrade CtrA with normal asymmetry. Thus, DivK localization to a pole is neither necessary nor sufficient for CtrA recruitment to the same pole. However, DivK exerts some control over CtrA localization. Polar foci of CtrA form in synchronized divKcs cells, but the timing of polar recruitment is disturbed, and CtrA is not subsequently degraded. These and other data suggest that DivK plays a general role in triggering events of the SW-ST transition. DivK regulates the ClpXP-dependent proteolysis of the chemotaxis receptor McpA at the SW-ST transition (18). Furthermore, G1 arrest cannot be achieved only by preventing CtrA preoteolysis (9, 18). The divKcs allele must also prevent the cell cycle-regulated dephosphorylation of CtrA at the SW-ST transition, or it must affect an unknown pathway that combines with CtrA stability to prevent the initiation of DNA replication.
The normal asymmetry of CtrA localization and proteolysis is lost in half of ΔdivJ and ΔpleC predivisional cells, indicating that these kinases participate in generating asymmetry of CtrA content in the progeny swarmer and stalked cells. We propose that DivJ and PleC act through their effects on DivK phosphorylation (Fig. 5), but they may affect CtrA polarity through a distinct, unknown pathway. The PleC kinase reduces DivK∼P levels and is located at the swarmer pole of the predivisional cell, whereas the DivJ kinase increases the cellular amount of DivK∼P and is located at the stalked pole (28, Fig. 5 Left). After a barrier to cytoplasmic diffusion is formed between the swarmer and stalked halves of the predivisional cell (3), levels of DivK∼P can begin to diverge, rising in the stalked compartment and falling in the swarmer compartment (Fig. 5 Right). Evidence that DivK activity levels are different in the swarmer and stalked compartments comes from the release of DivK specifically from the swarmer pole around the time of cell division (27, 32). Because phosphorylation of DivK is required for its polar localization (32), the asymmetric release of DivK argues that DivK∼P levels fall only in the swarmer compartment. It may be that a difference in DivK signaling activity is needed for asymmetric CtrA proteolysis, with high DivK∼P levels activating CtrA proteolysis or low DivK∼P levels preventing it. This model is attractive because it suggests a molecular mechanism by which the cell can detect the formation of a cytoplasmic diffusion barrier and couple it to the rapid, asymmetric proteolysis of CtrA.
The control of CtrA proteolysis is a critical factor in normal cell cycle progression and in asymmetric division of Caulobacter cells. There are several examples of regulated, Clp-mediated proteolysis in which an adaptor protein promotes the degradation of a specific substrate or group of substrates (33-35), but this is an instance in which active recruitment of a cytoplasmic substrate to a specific subcellular address is associated with its proteolysis.
Supplementary Material
Acknowledgments
We thank members of the Shapiro laboratory for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM32506 and Department of Energy Grant DE-FG03-01ER63219.
Abbreviations: SW-ST, swarmer-to-stalked cell; YFP, yellow fluorescent protein; FACS, fluorescence-activated cell sorter; DIC, differential interference contrast.
References
- 1.Ausmees, N. & Jacobs-Wagner, C. (2003) Annu. Rev. Microbiol. 57, 225-247. [DOI] [PubMed] [Google Scholar]
- 2.McAdams, H. H. & Shapiro, L. (2003) Science 301, 1874-1877. [DOI] [PubMed] [Google Scholar]
- 3.Judd, E. M., Ryan, K. R., Moerner, W. E., Shapiro, L. & McAdams, H. H. (2003) Proc. Natl. Acad. Sci. USA 100, 8235-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quon, K. C., Marczynski, G. T. & Shapiro, L. (1996) Cell 84, 83-93. [DOI] [PubMed] [Google Scholar]
- 5.Laub, M. T., McAdams, H. H., Feldblyum, T., Fraser, C. M. & Shapiro, L. (2000) Science 290, 2144-2148. [DOI] [PubMed] [Google Scholar]
- 6.Laub, M. T., Chen, S. L., Shapiro, L. & McAdams, H. H. (2002) Proc. Natl. Acad. Sci. USA 99, 4632-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quon, K. C., Yang, B., Domian, I. J., Shapiro, L. & Marczynski, G. T. (1998) Proc. Natl. Acad. Sci. USA 95, 120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domian, I. J., Reisenauer, A. & Shapiro, L. (1999) Proc. Natl. Acad. Sci. USA 96, 6648-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domian, I. J., Quon, K. C. & Shapiro, L. (1997) Cell 90, 415-424. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs, C., Ausmees, N., Cordwell, S. J., Shapiro, L. & Laub, M. T. (2003) Mol. Microbiol. 47, 1279-1290. [DOI] [PubMed] [Google Scholar]
- 11.Ryan, K. R., Judd, E. M. & Shapiro, L. (2002) J. Mol. Biol. 324, 443-455. [DOI] [PubMed] [Google Scholar]
- 12.Jenal, U. & Fuchs, T. (1998) EMBO J. 17, 5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flynn, J. M., Neher, S. B., Kim, Y. I., Sauer, R. T. & Baker, T. A. (2003) Mol. Cell 11, 671-683. [DOI] [PubMed] [Google Scholar]
- 14.Karzai, A. W., Roche, E. D. & Sauer, R. T. (2000) Nat. Struct. Biol. 7, 449-455. [DOI] [PubMed] [Google Scholar]
- 15.Keiler, K. C., Waller, P. R. & Sauer, R. T. (1996) Science 271, 990-993. [DOI] [PubMed] [Google Scholar]
- 16.Flynn, J. M., Levchenko, I., Seidel, M., Wickner, S., Sauer, R. T. & Baker, T. A. (2001) Proc. Natl. Acad. Sci. USA 98, 10584-10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hecht, G. B., Lane, T., Ohta, N., Sommer, J. M. & Newton, A. (1995) EMBO J. 14, 3915-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung, D. & Shapiro, L. (2002) Proc. Natl. Acad. Sci. USA 99, 13160-13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evinger, M. & Agabian, N. (1977) J. Bacteriol. 132, 294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ely, B. (1991) Methods Enzymol. 204, 372-384. [DOI] [PubMed] [Google Scholar]
- 21.Kovach, M. E., Phillips, R. W., Elzer, P. H., Roop, R. M. & Peterson, K. M. (1994) BioTechniques 16, 800-802. [PubMed] [Google Scholar]
- 22.Roberts, R. C., Toochinda, C., Avedissian, M., Baldini, R. L., Gomes, S. L. & Shapiro, L. (1996) J. Bacteriol. 178, 1829-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon, R., Prieffer, U. & Puhler, A. (1983) Bio/Technology 1, 784-790. [Google Scholar]
- 24.Meisenzahl, A. C., Shapiro, L. & Jenal, U. (1997) J. Bacteriol. 179, 592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winzeler, E. & Shapiro, L. (1995) J. Mol. Biol. 251, 346-365. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 27.Jacobs, C., Hung, D. & Shapiro, L. (2001) Proc. Natl. Acad. Sci. USA 98, 4095-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wheeler, R. T. & Shapiro, L. (1999) Mol. Cell 4, 683-694. [DOI] [PubMed] [Google Scholar]
- 29.Ohta, N., Lane, T., Ninfa, E. G., Sommer, J. M. & Newton, A. (1992) Proc. Natl. Acad. Sci. USA 89, 10297-10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sommer, J. M. & Newton, A. (1989) J. Bacteriol. 171, 392-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, S. P., Sharma, P. L., Schoenlein, P. V. & Ely, B. (1993) Proc. Natl. Acad. Sci. USA 90, 630-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam, H., Matroule, J.-Y. & Jacobs-Wagner, C. (2003) Dev. Cell 5, 149-159. [DOI] [PubMed] [Google Scholar]
- 33.Levchenko, I., Seidel, M., Sauer, R. T. & Baker, T. A. (2000) Science 289, 2354-2356. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, Y., Gottesman, S., Hoskins, J. R., Maurizi, M. R. & Wickner, S. (2001) Genes Dev. 15, 627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dougan, D. A., Reid, B. G., Horwich, A. L. & Bukau, B. (2002) Mol. Cell 9, 673-683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.