Abstract
Whole-genome expression profiling revealed Escherichia coli MG1655 genes induced by growth on mucus, conditions designed to mimic nutrient availability in the mammalian intestine. Most were nutritional genes corresponding to catabolic pathways for nutrients found in mucus. We knocked out several pathways and tested the relative fitness of the mutants for colonization of the mouse intestine in competition with their wild-type parent. We found that only mutations in sugar pathways affected colonization, not phospholipid and amino acid catabolism, not gluconeogenesis, not the tricarboxylic acid cycle, and not the pentose phosphate pathway. Gluconate appeared to be a major carbon source used by E. coli MG1655 to colonize, having an impact on both the initiation and maintenance stages. N-acetylglucosamine and N-acetylneuraminic acid appeared to be involved in initiation, but not maintenance. Glucuronate, mannose, fucose, and ribose appeared to be involved in maintenance, but not initiation. The in vitro order of preference for these seven sugars paralleled the relative impact of the corresponding metabolic lesions on colonization: gluconate > N-acetylglucosamine > N-acetylneuraminic acid = glucuronate > mannose > fucose > ribose. The results of this systematic analysis of nutrients used by E. coli MG1655 to colonize the mouse intestine are intriguing in light of the nutrient-niche hypothesis, which states that the ecological niches within the intestine are defined by nutrient availability. Because humans are presumably colonized with different commensal strains, differences in nutrient availability may provide an open niche for infecting E. coli pathogens in some individuals and a barrier to infection in others.
Escherichia coli is the predominant facultative anaerobe in the gastrointestinal tracts of mammals (1). It is arguably the best understood of all model organisms, yet the essence of how E. coli colonizes its mammalian hosts is not understood (2). In fact, very little is known about the acquisition and metabolism of nutrients that underpin the growth and persistence of colonized microorganisms.
The intestinal biota is remarkably diverse, containing an estimated 500 different bacterial species (1, 3-5). Freter's nutrient-niche theory postulates that numerous ecological niches within the intestine are defined by nutrient availability (6, 7). According to this hypothesis, individual species have a preference for one, or very few, of the nutrients that come from ingested food, epithelial and bacterial cell debris, and the mucus layer lining the epithelium. The diversity of available nutrients is thought to result in a balanced ecosystem in which each nutrient-defined niche is occupied by an individual bacterium, with individual population sizes determined by the available concentration of the preferred nutrient.
The majority of intestinal bacteria require a fermentable carbohydrate for growth, and fermentation is assumed to be the mode of metabolism used by most species (8). Most carbohydrate in the colon is in the form of mucosal polysaccharides, which are degraded by a few anaerobes that dominate the intestinal biota (9-11). The monosaccharides released from mucin and other mucosal glycoproteins support the growth of many intestinal bacteria such as E. coli, which does not make polysaccharide-degrading enzymes (10). Because the population of individual species is proportional to the concentration of its preferred nutrient(s), the relatively low population of E. coli in the intestine indicates that the concentration of its preferred nutrient(s) is low.
A large and growing body of evidence indicates that commensal E. coli grows in the intestine on nutrients acquired from mucus. Fluorescence in situ hybridization of intestinal thin slices showed that E. coli BJ4 (12) and MG1655 (13) are dispersed in the mucus layer. E. coli BJ4 grows rapidly in the mouse intestine, with a generation time of 40-80 min (14). In vitro, rapid growth (30-min generation time) occurs in intestinal mucus, but not in luminal contents (15, 16). Among mutants unable to colonize the mouse intestine are those that fail to penetrate mucus (13, 17), have difficulty surviving in mucus (18), or have difficulty growing on mucus (16, 18, 19). Thus, the ability of E. coli to grow and survive in mucus appears critical for intestinal colonization.
It is important to understand how gastrointestinal pathogens acquire the nutrients necessary to infect their hosts and initiate the disease process. This understanding of host-pathogen interactions should build on knowledge of model systems. Thus, we conducted a genome-wide search for metabolic pathways used by E. coli K-12 to colonize the mouse intestine. Here we present the gene expression profile of E. coli MG1655 grown in vitro on mucus as the sole source of carbon and energy, conditions that mimic nutrient availability in the mouse intestine. The induced pathways were systematically eliminated by gene knockouts and the mutants were tested for their relative colonization fitness. The results are discussed in terms of the nutrient-niche hypothesis and the prediction that the commensal E. coli flora serves as a first line of defense against E. coli intestinal pathogens.
Methods
Bacterial Strains and Growth Conditions. Strains derived from E. coli MG1655 StrR (streptomycin-resistant) and E. coli MG1655 StrR NalR (nalidixic acid-resistant) were used throughout this work (13). For DNA array experiments, cells were grown in triplicate at 37°C without shaking in 18-mm test tubes containing 5 ml of the Mops-based culture medium designed by Neidhardt et al. (20). Mouse cecal mucus was prepared from streptomycin-treated CD-1 mice (not colonized with E. coli), as described previously (13). Cultures were grown in triplicate to early (OD600 = 0.1) or late (OD600 = 0.3) logarithmic phase in 5 or 10 mg/ml lyophilized mucus. For in vitro nutrient preference experiments, cultures were grown in 96-well microtiter plates containing 100 μl of Mops minimal medium supplemented with the indicated carbon sources, incubated in a plate reader (FLUOstar Optima, BMG Labtechnologies, Offenburg, Germany) at 37°C, and the OD600 was measured every 10 min. Enzymatic BioAnalysis sugar test kits were obtained from Boehringer Mannheim.
Mutant Constructions. Knockout mutants were constructed by allelic replacement with kanamycin- or chloramphenicol-resistance cassettes, as described by Datsenko and Wanner (21). Primers used to construct deletion mutants were designed according to the E. coli MG1655 genome sequence (22). Mutants were confirmed for their growth phenotype and sequenced for genotype confirmation.
DNA Array Analysis of E. coli MG1655 in Mouse Cecal Mucus. Isolation of total RNA, labeling of samples, and duplicate hybridization to matched Panorama E. coli membrane arrays (Sigma-GenoSys, Woodlands, TX) were described previously (23-25). Raw data were imported into an Oracle database, normalized by expressing individual spot intensities as a fraction of the sum of all gene-specific spot intensities in each image, and analyzed as described previously (24). Clustering algorithms were executed in Spotfire decisionsite for functional genomics software (Spotfire, Somerville, MA). The data and protocols are available on our web site (www.ou.edu/microarray).
Mouse Colonization Experiments. The relative fitness of E. coli strains for colonization of the mouse intestine was assayed as described (13, 16, 19). Briefly, three male CD-1 mice (5-8 weeks old) were given drinking water containing streptomycin sulfate (5 g/liter) for 24 h to eliminate resident facultative bacteria (26). After 18 h of starvation for food and water, the mice were fed 1 ml of 20% sucrose containing 105 to 106 colony-forming units (cfu) of both the wild-type and mutant strains. After ingestion of the bacterial suspension, food and streptomycin/water were returned to the mice. Fecal plate counts were obtained after 5 h, 24 h, and on odd-numbered days thereafter. The NalR and antibiotic resistance markers introduced by allelic replacement had no effect on colonization in control experiments (19). The colonization experiments are available on the Internet (www.ou.edu/microarray).
Results
Gene Expression Profiles of Mucus-Grown E. coli MG1655. We determined the gene expression profile of E. coli MG1655 under conditions designed to mimic nutrient availability in the intestine. In the first “low-mucus” experiment, E. coli MG1655 was cultured on 5 mg/ml mouse cecal mucus and compared with cells grown on glucose (Fig. 1). Significantly induced genes directed our attention to pathways for catabolism of amino sugars: N-acetylglucosamine (NAG), N-acetylneuraminic (sialic) acid, and glucosamine; pentoses: fucose and ribose; amino acids: tryptophan, threonine, serine, and aspartate; and phosphatidylethanolamine. The low-mucus data set also indicated elevated expression of the tricarboxylic acid (TCA) cycle, gluconeogenesis, and anaerobic respiration (data not shown).
Fig. 1.
Expression profiles of E. coli MG1655 genes involved in catabolism of carbohydrates found in mucus (15). Genes are represented in rows, as indicated to the right, and are grouped by the corresponding sugar degradation pathway. PH1, ratio of early logarithmic phase (OD600 = 0.1) cells grown on 10 mg/ml mucus vs. minimal glucose (0.2%); PH2, from the same culture grown to late logarithmic phase (OD600 = 0.3); Low, ratio of cells grown to late logarithmic phase (OD600 = 0.3) on 5 mg/ml mucus vs. minimal glucose; Cont, ratio of cells grown to late logarithmic phase on chemically defined rich medium (20) vs. minimal glucose. Ratios are displayed colorimetrically: bright red indicates genes with ≥2.5-fold higher expression (≈2 standard deviations) and bright green indicates ≤2.5-fold lower expression in the experimental condition compared with minimal glucose; the colors darken to black to indicate no change in expression. GlcNAc, N-acetylglucosamine; GalNAc, N- acetylgalactosamine; NANA, sialic acid (N-acetylneuraminic acid); GlcNH4, glucosamine. See text for further details.
Surprisingly, there was no evidence in the low-mucus experiment for induction of genes involved in catabolism of preferred nutrients such as gluconate and mannose, which are known components of mucus (15). Therefore, the experiment was repeated with a higher concentration (10 mg/ml) of mucus (Fig. 1 and Table 1). Induction of genes for catabolism of sugar acids such as gluconate occurred in early logarithmic phase (phase 1) (Fig. 1). The relative induction ratios for the sugar acid genes declined at the later time point (phase 2), explaining why induction of these genes was not detected in the first experiment. Comparison of phases 1 and 2 revealed other subtleties of growth on mucus: genes involved in catabolism of gluconate, glucuronate, sialic acid, NAG, mannose, maltose, and ribose were induced at the early time point, whereas genes involved in degradation of ethanolamine and fucose, anaerobic respiration, and the TCA cycle were induced at the later time point (Fig. 1). Significantly induced genes other than those involved in metabolism are shown in Table 1, but they were not considered further.
Table 1.
Genes significantly* induced by mucus
| Functional group | Phase 1 | Phase 2 |
|---|---|---|
| Amino acid biosynthesis | gcvH ilvG | |
| Carbon compound catabolism | ansB gntK hycI nanA rbsK tnaA uxaA uxaB uxaC uxuA | ansB eutJ eutM eutN eutP eutQ eutT fucA fucO gabT tnaA |
| Cell processes | cspE flaG flgB flgC flgD flgE flgF flgG flgK flgL fliC fliL htpG motB mukB | cspD cspE flgB flgC flgE flgG flgK flgL fliC glgS katG motB |
| Central intermediary metabolism | aphA aspA fruK glpK glpQ pck | agp appA aspA glpK glpQ pck sdhB sdhD |
| DNA replication, repair | dksA | helD |
| Energy metabolism | atpC frdA frdB frdC glpA glpB glpC pta | fdoG fdoH fdoI frdA frdB frdC glpA glpB glpC hyfD |
| Hypothetical, unclassified, unknown | b 1937 fliZ yaiN ygaZ yhbU yjeE yjiY yjjI ymfJ | b 1937 ybhO ychH ydaA yfcZ yhbS yhbT yqeC |
| Nucleotide biosynthesis, metabolism | deoB deoC | |
| Phage, transposon, or plasmid | pspE | |
| Putative cell structure | ynaF | |
| Putative enzymes | hybC mipB ybiW ycgT ygjH | gatZ paaH pepB prpC ydfI yeiT ygbD ygeX ygeY yqeF |
| Putative factors | yraI | yliH |
| Putative membrane protein | ompW | ompW |
| Putative regulatory proteins | ybjN yfiA | |
| Putative transport proteins | yidE | ftnB ygbN |
| Regulatory function | asnC cspA glnD tar tsr | gatR 3 glnD phoH tar |
| Transcription, RNA processing | rhoL | |
| Translation, posttranslational modification | rplC rplD rplF rplP rplV rplW rpmC rpmD rpsC rpsN rpsQ rpsS rpsT rpsU | rplD rplK |
| Transport and binding proteins | exuT lamB malE manX manZ rbsB rbsD secG uhpT | dcuA gatA gatC glpT |
Significantly induced genes: log ratio ≥2 standard deviations from mean of log ratios and P < 0.0002.
To determine whether E. coli can consume building blocks directly from mucus, we examined the expression profile of biosynthesis genes, assuming that exogenous amino acids and nucleosides would cause repression. Thirty of 97 amino acid and 6 of 66 nucleotide biosynthetic genes were significantly repressed in cells grown on mucus compared with glucose (data not shown). Supplementation of mucus medium with amino acids and nucleosides had no effect (i.e., additional biosynthetic genes were not repressed). These results suggest that E. coli can acquire purines, pyrimidines, and amino acids from mucus, thus increasing the partitioning of sugars into energy-yielding pathways as opposed to biosynthesis.
In summary, the gene expression profiles of mucus-grown E. coli MG1655 identified genes involved in catabolism of NAG, sialic acid, glucosamine, fucose, ribose, glucuronate, galacturonate, gluconate, and maltose. There was also evidence for catabolism of phospholipids and amino acids, which require the TCA cycle and gluconeogenesis. These data were the starting point for testing the roles of individual nutrients in supporting E. coli colonization of the mouse.
Genes, Pathways, and Nutrients That Contribute to Colonization. The streptomycin-treated mouse is a useful model for determining the relative fitness of enteric bacteria for intestinal colonization (27-30). The way to find out what an individual strain grows on in the intestine is to simultaneously feed low numbers (105 cfu per streptomycin-treated mouse) of the wild-type parent and an otherwise isogenic mutant that is blocked in a specific metabolic pathway. Mutant strains that do not compete effectively for nutrients involved in initiation (5 h to 3 days after feeding) will fail to reach the same high numbers as the wild type. Mutants that do not compete effectively for nutrients involved in maintenance (day 7 after feeding and beyond) will decline in numbers relative to the wild type. Cocolonization during the maintenance stage indicates the strains are able to use limiting nutrients equally well or they are not competing for the same limiting nutrient(s).
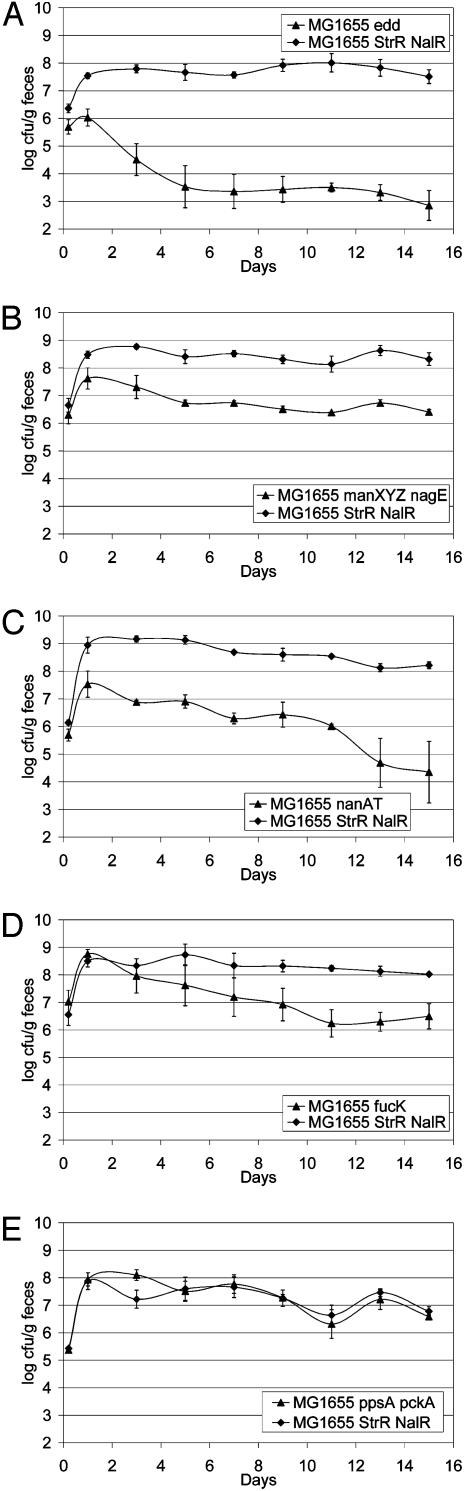
The mutant with a defect in the Entner-Doudoroff pathway (Δedd), the primary route for gluconate catabolism, had a major colonization defect characterized by failure to grow rapidly during initiation and a subsequent reduction to less than 103 cfu/g of feces during maintenance (Fig. 2A). The mutant with a defect in transport of NAG and glucosamine (ΔnagE ΔmanXYZ) had a significant defect in initiation, but no apparent defect in maintenance, as indicated by its cocolonization with the wild-type parent at a 2 log lower population (Fig. 2B). The mutant with a defect in transport and catabolism of sialic acid (ΔnanAT) had a significant defect in initiation and possibly maintenance (Fig. 2C).
Fig. 2.
Colonization of the mouse intestine by E. coli MG1655 StrR NalR (wild-type) and mutant derivatives of E. coli MG1655 StrR, as follows. Sets of three mice were fed one of the following: 105 cfu each of wild-type and MG1655 StrR Δedd::KmR (KmR, kanamycin-resistant) (A); 105 cfu each of wild-type and MG1655 StrR ΔmanXYZ::KmR ΔnagE (B); 105 cfu each of wild-type and MG1655 StrR ΔnanAT::KmR (C); 105 cfu each of wild-type and MG1655 StrR ΔfucK::KmR (D); or 105 cfu each of wild-type and MG1655 StrR ΔppsA ΔpckA::Cm (E). At the indicated times, fecal samples were homogenized, diluted, and plated as described (13). Bars represent standard error of the log10(mean cfu/g of feces) for each set of three mice.
Mutants with defects in catabolism of fucose (ΔfucK, Fig. 2D and ΔfucAO, not shown) initiated colonization as well as the wild-type parent, but each had a significant defect during maintenance characterized by a population level 1½ to 2 logs lower than the wild-type parent. The mutant with a defect in catabolism of mannose (ΔmanA, not shown) had a significant defect (≈2-3 logs lower) in maintenance. The mutant with a defect in catabolism of ribose (ΔrbsK, not shown) had a minor defect (≈1-2 logs lower) in maintenance. The mutant with a defect in catabolism of hexuronic acids (ΔuxaC, not shown) had a minor defect in maintenance (1-2 logs lower). It appears that the colonization defect is in glucuronate catabolism, because the defect in galacturonate catabolism (ΔuxaB, not shown) resulted in no loss of colonizing ability. Genes and corresponding nutrients that did not appear to be used by E. coli MG1655 in the mouse intestine, despite being induced when grown on mucus, included ethanolamine (ΔeutBC), glycerol 3-phosphate released from phospholipids (ΔglpTQ), glycerol (ΔglpK), fructuronate (ΔgntP), glucuronate released from oligosaccharides by β-glucuronidase (ΔuidA), tryptophan (ΔtnaA), and gluconeogenic nutrients (ΔppsA ΔpckA, shown as an example in Fig. 2E).
Order of in Vitro Nutrient Preference. We tested the hypothesis that the relative importance of these nutrients for colonization is correlated with the genetic program for nutrient preference. The order of nutrient choice was determined by measuring growth on combinations of carbon sources in laboratory cultures. Using standard microbiology methods, we looked for the presence of diauxie, and where it was not present we used biochemical assays to confirm the timing of the disappearance of individual sugars from the mixture. The relative impact of the metabolic lesions on colonization fitness does, in fact, parallel the order of nutrient preference in vitro for E. coli MG1655: gluconate > NAG > sialic acid = glucuronic acid > mannose > fucose > ribose (Table 2).
Table 2. Growth and nutrient preference of E. coli MG1655.
| Sugar | In vitro growth rate, μ | Order in vitro | Stage in vivo |
|---|---|---|---|
| Gluconate | 0.59 ± 0.04 | 1 | I, M |
| NAG | 0.35 ± 0.01 | 2 | I |
| Sialic acid | ND | 3 | I |
| Glucuronate | 0.35 ± 0.04 | 3 | M |
| Mannose | 0.35 ± 0.03 | 5 | M |
| Fucose | 0.31 ± 0.05 | 6 | M |
| Ribose | 0.19 ± 0.03 | 7 | M |
Standing cultures: Mops minimal medium with 0.2% sugar. μ is expressed in generations per hour; ND, not determined; I, initiation; M, maintenance.
Colonization of Central Metabolism Mutants. Given the essential role of intermediary metabolism, it can be hypothesized that any mutant in central pathways will be defective in colonization because of a diminished growth rate, even on carbon sources that are catabolized via different routes. If this is true, the major effect of the edd mutation on colonization could be ascribed to its centrality rather than its specificity for hexonate catabolism.
To test this hypothesis, we constructed mutants that are defective in glycolysis (pgi, phosphoglucose isomerase), the pentose phosphate pathway (gnd, 6-phosphogluconate dehydrogenase), the TCA cycle (sdhB, succinate dehydrogenase), and gluconeogenesis (ppsA pckA, phosphoenolpyruvate synthase and phosphoenolpyruvate carboxykinase, respectively). A gnd mutation causes an 11% decrease in the growth rate on gluconate and a 15% decrease in the growth rate on glucose by comparison to the wild type; a pgi mutant grows 35% slower on gluconate than does the wild type (31). An edd mutant grows on glucose with a rate identical to that of the wild type, but grows 60-70% more slowly on gluconate (32).
Colonization of the MG1655 edd and ppsA pckA mutants was described above (Fig. 2 A and E). The former had a major effect and the latter had no effect on colonization. The pgi mutation had a major effect on initiation and maintenance (Table 3). The sdhB and gnd mutations had no effect on colonization (Table 3). Thus, the hypothesis that lesions in central metabolism decreased fitness of E. coli MG1655 for colonization was not supported. The colonization defects of the pgi and edd mutants were likely caused by defects in catabolism of specific nutrients, and not simply because they are steps in central carbon metabolism. Because the TCA cycle and gluconeogenesis mutants were fit for colonization, this ruled out a major role for catabolism of lipids, dicarboxylates, and amino acids in colonization by E. coli MG1655.
Table 3. Colonization by E. coli MG1655 central metabolism mutants.
| Mutation | Pathway | Colonization defect* | Phase of colonization† |
|---|---|---|---|
| edd | Entner-Doudoroff | Major | I, M |
| pgi | Glycolysis | Major | I, M |
| sdhB | TCA cycle | None | |
| gnd | Pentose phosphate | None | |
| ppsA pckA | Gluconeogenesis | None |
Major, >3 logs between wild type and mutant; none, <1 log between wild type and mutant.
I, initiation (5 h to day 3); M, maintenance (after day 7).
Discussion
In this study, we conducted a genome-wide search for metabolic pathways that allow E. coli K-12 (MG1655) to colonize the mouse intestine. In the absence of a suitable in vivo model, we designed in vitro conditions to mimic nutrient availability in the mammalian intestine. The bacteria were cultured on minimal medium with mucus, which contains 50% mucin in addition to a complex mixture of glycoproteins and glycolipids, as well as cell debris (15). Cultivation without shaking results in partially anaerobic conditions, which most closely reflects the limited oxygen availability of mucosal tissues in the mouse intestine (33). A disadvantage of this in vitro model is that it does not mimic the growth parameters or microbial competition that exists in the animal intestine. This model did allow us to use whole-genome expression profiling to reveal genes induced by growth on mucus. Most were nutritional genes corresponding to catabolic pathways for nutrients found in mucus (Table 1). We systematically knocked out these pathways and tested the mutants for their relative fitness for colonization in competition with their wild-type parent. More than half of the metabolic lesions tested had no effect on the ability of E. coli MG1655 to colonize the mouse intestine (e.g., Fig. 2E). We found that only mutations in sugar pathways affected colonization, not mutations in phospholipid and amino acid catabolism, gluconeogenesis, the TCA cycle, and the pentose phosphate pathway (Tables 2 and 3).
Rapid growth of E. coli during the initiation stage likely depends on utilization of nonlimiting nutrients made available by removing the streptomycin-sensitive facultative microflora, i.e., nutrients that the anaerobic flora apparently does not use. In fact, E. coli could not grow from 105 to 108 cfu per gram of feces if nutrients were limiting (Fig. 2 A). While it might be argued that rapid growth during initiation is an artifact of streptomycin treatment, it is for this reason that the streptomycin-treated mouse is so useful: nutrient consumption during initiation reflects the in vivo nutrient preference. Conventionally colonized mice (i.e., not streptomycin-treated) are resistant to colonization because the preferred nutrients are not available (34). Thus, the maintenance stage is more akin to persistent colonization of the intestine (it is at this stage that mice become colonization resistant). During maintenance, the growth rate of colonized strains must match the turnover rate of the mucosal contents (6), and persistence most likely depends on nutrient(s) that become limiting after E. coli has grown to high numbers. One microbial strategy for persistence during maintenance might be to switch to cometabolism of several limiting nutrients.
In the laboratory, E. coli K-12 can grow on 11 of the 13 known sugar constituents of mucosal glycoproteins and glycolipids (15). In vivo, only a subset of these sugars appeared to be involved in colonization. Gluconate was the major carbon source used by E. coli MG1655 to colonize, having an impact on both the initiation and the maintenance stages (Fig. 2 A). The effect of the edd mutation on colonization could be attributed to its specificity for gluconate catabolism, because the hypothesis was not supported that lesions in central metabolism decreased fitness for colonization. NAG and sialic acid were involved in initiation, but not maintenance (Fig. 2 B and C). Glucuronate, mannose, fucose (Fig. 2D), and ribose were involved in maintenance, but not initiation. The order of preference for these seven sugars in the laboratory paralleled the relative impact of the corresponding metabolic lesions on E. coli MG1655 colonization (Table 2). The results are intriguing, in light of the nutrient-niche hypothesis: colonization of the mouse intestine by E. coli MG1655 is supported, to various degrees, by (at least) seven mucus-derived sugars.
Two sugars that E. coli K-12 cannot grow on because it lacks the pathway genes, galactosamine and N-acetylgalactosamine, can be used by other E. coli strains (35). Differences in metabolic potential due to the presence or absence of certain pathways could provide a nutritional basis for cocolonization. This possibility has not been tested, but if true, it could be extended to include differences in the order of consumption of preferred nutrients from complex mixtures such as mucus.
The nutrient-niche hypothesis (6) can be reconsidered in light of what we have learned about the nutrients used by E. coli to colonize the mouse intestine. The hypothesis is oversimplified if considered from the perspective that each of the 500 species present grows best on a single simple nutrient; there simply are not that many nutrients available in the ecosystem. Instead, it seems prudent to consider that the complex microbial community of the intestine is established by competition for diverse nutrients that are generated in the intestine through mucosal polysaccharide degradation to release free sugars and distinct oligosaccharides with various carbohydrate compositions (10).
For bacteria in the mammalian intestine, as in a chemostat, the growth rate in the initiation stage must exceed the washout rate and the growth rate during the maintenance stage must equal the washout rate (6, 36). Two different microorganisms with a preference for the same growth-limiting nutrient cannot coexist in chemostats; one will eventually out-compete the other (37). However, if two microorganisms use different growth-limiting nutrients, they can maintain stable populations in a chemostat (38). Thus the mammalian intestine can be thought of as a chemostat in which several hundred species of bacteria are in equilibrium, competing for resources from a mixture of limiting nutrients (6, 39). The chemostat allows for simultaneous utilization of several limiting nutrients by an individual species (40). It remains to be established whether this observation extends to in vivo nutrient utilization by E. coli. The fact that seven, or more, nutrients support E. coli colonization suggests that cometabolism of available nutrients in the intestine is worthy of study.
The systematic analysis of E. coli MG1655 described here suggests that there is a nutritional basis for the steady progression of commensal strains in human hosts (34). Further studies are needed to address the hypothesis that preferential utilization of different nutrients and/or the ability to switch to alternative nutrition is the basis for cocolonization of different E. coli strains and that direct competition for the same nutrient(s) leads to decreased fitness of some strains. The results of such studies may explain how various commensal strains affect nutrient availability, in some cases providing an open niche for infecting E. coli pathogens and in other cases preventing infection. For example, it is known that certain E. coli strains can prevent Shigella flexneri (41) and Salmonella typhimurium (42) infection in mice. Hence, individual differences in the E. coli flora could be the reason for differences in human susceptibility to gastroenteritis (43). If so, there is the opportunity to build a nutritional framework for understanding how commensal strains serve as the first line of defense against intestinal infections.
Acknowledgments
We gratefully acknowledge the support of the Korea Science and Engineering Foundation (D.-E.C.) and National Institutes of Health Grants RO1-AI48945-01 and RR-01-005. W.E.N. was supported by Grant H-401 from the University of Rhode Island Experiment Station.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: StrR, streptomycin-resistant; NalR, nalidixic acid-resistant; cfu, colony-forming units; NAG, N-acetylglucosamine; TCA, tricarboxylic acid.
References
- 1.Finegold, S. M., Sutter, V. L. & Mathisen, G. E. (1983) in Human Intestinal Microflora in Health and Disease, ed. Hentges, D. J. (Academic, New York), pp. 3-31.
- 2.Relman, D. A. & Falkow, S. (2001) Trends Microbiol. 9, 206-208. [DOI] [PubMed] [Google Scholar]
- 3.Borrelio, S. P. (1986) in Microbial Metabolism in the Digestive Tract, ed. Hill, M. J. (CRC, Boca Raton, FL), pp. 2-16.
- 4.Bryant, M. P. (1974) Am. J. Clin. Nutr. 27, 1313-1319. [DOI] [PubMed] [Google Scholar]
- 5.Hill, M. J. (1995) in Role of Gut Bacteria in Human Toxicology and Pharmacology, ed. Hill, M. J. (Taylor and Francis, London), pp. 3-18.
- 6.Freter, R. (1983) in Human Intestinal Microflora in Health and Disease, ed. Hentges, D. J. (Academic, New York), pp. 33-54.
- 7.Freter, R. (1988) in Virulence Mechanisms of Bacterial Pathogens (Am. Soc. Microbiol., Washington, DC), pp. 45-60.
- 8.Salyers, A. A. & Leedle, J. A. Z. (1983) in Human Intestinal Microflora in Health and Disease, ed. Hentges, D. J. (Academic, New York), pp. 129-146.
- 9.Corfield, A. P., Wagner, S. A., Clamp, J. R., Kriaris, M. S. & Hoskins, L. C. (1992) Infect. Immun. 60, 3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoskins, L. C., Agustines, M., McKee, W. B., Boulding, E. T., Kriaris, M. & Niedermeyer, G. (1985) J. Clin. Invest. 75, 944-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoskins, L. C. & Boulding, E. T. (1981) J. Clin. Invest. 67, 163-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulsen, L. K., Lan, F., Kristensen, C. S., Hobolth, P., Molin, S. & Krogfelt, K. A. (1994) Infect. Immun. 62, 5191-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moller, A. K., Leatham, M. P., Conway, T., Nuijten, P. J., de Haan, L. A., Krogfelt, K. A. & Cohen, P. S. (2003) Infect. Immun. 71, 2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulsen, L. K., Licht, T. R., Rang, C., Krogfelt, K. A. & Molin, S. (1995) J. Bacteriol. 177, 5840-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peekhaus, N. & Conway, T. (1998) J. Bacteriol. 180, 3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweeney, N. J., Laux, D. C. & Cohen, P. S. (1996) Infect. Immun. 64, 3504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCormick, B. A., Klemm, P., Krogfelt, K. A., Burghoff, R. L., Pallesen, L., Laux, D. C. & Cohen, P. S. (1993) Microb. Pathog. 14, 33-43. [DOI] [PubMed] [Google Scholar]
- 18.Newman, J. V., Kolter, R., Laux, D. C. & Cohen, P. S. (1994) Microb. Pathog. 17, 301-311. [DOI] [PubMed] [Google Scholar]
- 19.Sweeney, N. J., Klemm, P., McCormick, B. A., Moller-Nielsen, E., Utley, M., Schembri, M. A., Laux, D. C. & Cohen, P. S. (1996) Infect. Immun. 64, 3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neidhardt, F. C., Bloch, P. L. & Smith, D. F. (1974) J. Bacteriol. 119, 736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datsenko, K. A. & Wanner, B. L. (2000) Proc. Natl. Acad. Sci. USA 97, 6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blattner, F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453-1474. [DOI] [PubMed] [Google Scholar]
- 23.Chang, D. E., Smalley, D. J. & Conway, T. (2002) Mol. Microbiol. 45, 289-306. [DOI] [PubMed] [Google Scholar]
- 24.Conway, T., Kraus, B., Tucker, D. L., Smalley, D. J., Dorman, A. F. & McKibben, L. (2002) BioTechniques 32, 110, 112-114, 116, 118-119. [DOI] [PubMed] [Google Scholar]
- 25.Tao, H., Bausch, C., Richmond, C., Blattner, F. R. & Conway, T. (1999) J. Bacteriol. 181, 6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, C. P. & Bohnhoff, M. (1963) J. Infect. Dis. 113, 59-66. [DOI] [PubMed] [Google Scholar]
- 27.Cohen, P. S., Rossoll, R., Cabelli, V. J., Yang, S. L. & Laux, D. C. (1983) Infect. Immun. 40, 62-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nevola, J. J., Laux, D. C. & Cohen, P. S. (1987) Infect. Immun. 55, 2884-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wadolkowski, E. A., Laux, D. C. & Cohen, P. S. (1988) Infect. Immun. 56, 1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wadolkowski, E. A., Burris, J. A. & O'Brien, A. D. (1990) Infect. Immun. 58, 2438-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraenkel, D. G. & Vinopal, R. T. (1973) Annu. Rev. Microbiol. 27, 69-100. [Google Scholar]
- 32.Bausch, C., Peekhaus, N., Utz, C., Blais, T., Murray, E., Lowary, T. & Conway, T. (1998) J. Bacteriol. 180, 3704-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He, G., Shankar, R. A., Chzhan, M., Samouilov, A., Kuppusamy, P. & Zweier, J. L. (1999) Proc. Natl. Acad. Sci. USA 96, 4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apperloo-Renkema, H. Z., Van der Waaij, B. D. & Van der Waaij, D. (1990) Epidemiol. Infect. 105, 355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brinkkotter, A., Kloss, H., Alpert, C. & Lengeler, J. W. (2000) Mol. Microbiol. 37, 125-135. [DOI] [PubMed] [Google Scholar]
- 36.Lee, A. (1985) Adv. Microb. Ecol. 8, 115-162. [Google Scholar]
- 37.Slater, J. H. (1988) in Microorganisms in Action: Concepts and Applications in Microbial Ecology, eds. Lynch, J. M. & Hobbie, J. E. (Blackwell Scientific, Oxford), pp. 51-74.
- 38.Taylor, P. A. & Williams, P. J. (1975) Can. J. Microbiol. 21, 90-98. [DOI] [PubMed] [Google Scholar]
- 39.Tilman, D. (1982) Monogr. Popul. Biol. 17, 1-296. [PubMed] [Google Scholar]
- 40.Lendenmann, U. & Egli, T. (1995) Microbiology 141, 71-78. [DOI] [PubMed] [Google Scholar]
- 41.Freter, R. (1972) Infect. Immun. 6, 134-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hudault, S., Guignot, J. & Servin, A. L. (2001) Gut 49, 47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neill, M. A. (1998) in Escherichia coli O157:H7 and Other Shiga Toxin-Producing E. coli Strains, eds. Kaper, J. B. & O'Brien, A. D. (Am. Soc. Microbiol., Washington, D.C.), pp. 357-363.