Abstract
Entry of herpes simplex virus (HSV) 1 into cells requires the interaction of HSV gD with herpesvirus entry mediator or nectin1 receptors, and fusion with cell membrane mediated by the fusion glycoproteins gB, gH, and gL. We report that the gD ectodomain in soluble form (amino acids 1-305) was sufficient to rescue the infectivity of a gD-null HSV mutant, indicating that gD does not need to be anchored to the virion envelope to mediate entry. Entry mediated by soluble gD required, in addition to the receptor-binding sites contained within residues 1-250, a discrete downstream portion (amino acids 261-305), located proximal to the transmembrane segment in full-length gD. We named it as profusion domain. The pro-fusion domain was required for entry mediated by virion-bound gD, because its substitution with the corresponding region of CD8 failed to complement the infectivity of gD-/+ HSV. Furthermore, a receptor-negative gD (gDΔ6-259) inhibited virus infectivity when coexpressed with wild-type gD; i.e., it acted as a dominant-negative gD mutant. The pro-fusion domain is proline-rich, which is characteristic of regions involved in protein-protein interactions. P291L-P292A substitutions diminished the gD capacity to complement gD-/+ HSV infectivity. We propose that gD forms a tripartite complex with its receptor and, by way of the proline-rich pro-fusion domain, with the fusion glycoproteins, or with one of them. The tripartite complex would serve to recruit/activate the fusion glycoproteins and bring them from a fusion-inactive to a fusion-active state, such that they execute fusion of the virion envelope with cell membrane.
Herpes simplex virus (HSV) enters cells through the coordinated action of four essential glycoproteins; gD, gB, gH, and gL, that act after the binding of gC and gB to the glycosaminoglycans of cell-surface proteoglycans (1, 2). Of the four glycoproteins required for entry, gD is the receptor-binding glycoprotein. It interacts with two alternative protein receptors, HVEM (herpesvirus entry mediator) and nectin1, that belong to the tumor necrosis factor receptor family (3), and to a growing family of intercellular adhesion molecules with Ig structure, respectively (4-8). The four essential glycoproteins required for HSV entry are required and are also sufficient to induce fusion of cells that express a gD receptor (9). The gD-binding site on HVEM maps mainly to the N-terminal cysteine-rich domain 1, with a hot spot at Y23 (10, 11). For nectin1, the N-terminal V domain, in particular its CC′C″ ridge (amino acids 64-104) is sufficient to mediate HSV entry (12-15). Critical residues were located in the 69-75 region and at positions 77 and 85 (16, 17). Insertion and deletion mutants in gD were the first mutants used to define functional regions (18). Subsequently, the x-ray crystal structure of the first 259 residues of gD [of the 315 that compose the ectodomain] was solved (19). The gD ectodomain is composed of an Ig-folded core (residues 56-184), with N- and C-terminal extensions. The latter folds back toward the Ig core so that it lies proximal to the N terminus. The gD contact with HVEM resides exclusively in N-terminal residues 1-34. The residues critical for the interaction with nectin1 appear to be more widespread on gD molecule and are included within amino acids 34-243 (20-22). Cumulatively, the receptor-binding sites therefore lie between residues 1-243. Studies on the binding affinity of a panel of truncated forms of gD with soluble receptors showed that gD243t binds with low affinity, whereas gD250t and gDs truncated downstream of residue 250 bind with high affinity; the highest affinity is exhibited by gD275t, gD285t, and gDΔ290-299t. gD306t exhibits a 100-fold lower affinity than gDΔ290-299t (20, 23).
To understand the molecular basis of HSV entry into cells, a key question that remains to be addressed is how the receptor recognition by gD triggers fusion of the virion envelope with cell membrane. At least two scenarios can be envisioned for gD. In one case, gD serves the typical function of a receptor-binding glycoprotein, i.e., it brings the virion envelope and the cell membrane into juxtaposition. The subsequent involvement of gB, gH, and gL executes fusion. Because gD lacks a fusion peptide, it is not expected to directly penetrate the target membrane. Alternatively, gD does more than merely bring the virion envelope and the cell membrane close. One can envision that its interaction with receptor induces conformational changes to gD that allow the recruitment/activation of gB, gH, and gL, or of cellular factors required for fusion. The folding of the gD N terminus to form a hairpin after binding to HVEM argues that gD does indeed undergo at least one conformational modification when it interacts with its receptor (19).
To discriminate between these two possibilities, we asked whether soluble gD can substitute for virion-bound gD and rescue the infectivity of a gD-null HSV. Surprisingly, we found that soluble ectodomain was the sole gD region essential for HSV entry. We then mapped the regions necessary for entry mediated by soluble gD as well as by virion-bound gD, and found that, in addition to the domain that carries the receptor-binding sites, the gD ectodomain requires a discrete proline-rich domain located proximal to the transmembrane (TM) sequence, here named the pro-fusion domain. Its substitution or mutation of some of the prolines diminished the capacity of full-length gD to complement the infectivity of gD-/+ HSV mutant. We propose that, after the interaction of gD with its receptor, the pro-fusion domain recruits the fusion glycoproteins gB, gH, and gL (or a subset of them), leading to the formation of a tripartite complex (receptor-gD-fusion glycoproteins) that triggers fusion.
Materials and Methods
Cells and Viruses. Cells were grown in DMEM containing 5-10% FCS. J cells expressing nectin 1 (nectin1-J) were described (8). J cells expressing HVEM were derived by transfection with pBEC10 DNA (3). The gD- F-gDβ and RR1097 viruses (24, 25) were grown in gD-expressing R6 cells (26) (gD-/+ stock), or rabbit skin cells (gD-/- stock). Extracellular virions were used.
Abs. MAbs H170 and HD1 were directed to gD, mAb H1817 to gB (Goodwin Institute, Plantation, FL), and polyclonal Ab ZC15 to the gD cytoplasmic tail (C-tail) (27).
Plasmids. pEA99 carries the HSV-1 gD gene in pcDNA3.1(-) (28). pEA101 carries the HSV-1 UL26.5 promoter in place of CMV promoter. pEA102 carries the gD gene in pEA101 (26). For construction of gDΔ6-259 (pCF21), two Asp-718 sites were inserted at nucleotide positions 90 and 852 in pEA99, by site-directed mutagenesis (29), with primer 5′-CAA ATA TGC CTT GGC GGT ACC CTC TCT CAA GCT GG-3′, and 5′-CCG GAG CTG TCC GAG GTA CCC AAC GCC ACG CAG-3′, generating pCF20, which was collapsed with Asp-718. For construction of gD1-260-CD8 chimera, the portion of CD8 gene encoding 52 amino acids upstream of TM, the TM, and the C-tail (48 amino acids) was PCR-amplified with primers gDCD8RevEcoRI: 5′-GCA TTT AGG GAA TTC TAT AGA ATA GGG-3′ and chimgD260CD8Forw: 5′-CTG CCC CCG GAG CTG TCC GAG ACC GTC TTC CTG CCA GCG AAG CCC ACC-3′. The portion of gD up to amino acid 260 was PCR-amplified with the primers gDCD8ForwXhoI: 5′-GGT CTC TTT TGT CTC GAG CGT TCC GGT ATG GGG G-3′ and chimgD260CD8Rev: 5′-GGT GGG CTT CGC TGG CAG GAA GAC GGT CTC GGA CAG CTC CGG GGG CAG-3′. The chimeric construct was obtained by mixing the two fragments in equimolar amounts and 20 cycles of denaturation, annealing, and extension. The EcoRI-XhoI-digested sequence was cloned into pcDNA3.1(-), generating pgD-CD8. All constructs were sequenced for accuracy.
Proline Mutagenesis. Mutagenesis was performed in pEA102, as detailed (29), with the following primers: PQ 313/314 LA Asp-718: 5′-GGA GGA CCC CGT GGG TAC CGT GGC GCT GGC AAT CCC ACC AAA CTG-3′; TP 329/330 AL PstI: 5′-GTC GAT CCA GGA CGC TGC AGC GCT TTA CCA TCC CCC GGC-3′; PP 316/317 LA BglII: 5′-GAC GGT GGC GCC GCA GAT CTT ACT AAA CTG GCA CAT ACC-3′; and QD 325/326 VA PvuII: 5′-GGT AAA GCG CTG CAG CTG CCA CGA TCG ACG GTA TGT-3′. The primers were designed with silent mutations that yielded the indicated restriction sites.
Rescue of gD-/- Infectivity by Soluble Forms of gD. Except when otherwise stated, aliquots of pelleted gD-/- virions (3-10 μl) were mixed with soluble gDs, prepared as described (20, 30), and added to the cells, grown in 48-well dishes. After 150 min adsorption at 37°C, the inoculum was removed and cells were overlaid with DMEM containing 1% FBS. Infection was monitored at 16 h by 5-bromo-4-chloro-3-indolyl β-d-galactoside staining (3, 8).
Infectivity Complementation Assay. Cells were transfected by means of Polyfect (Qiagen, Valencia, CA) with the indicated plasmids; 4 h later they were infected with gD-/+ FgDβ (3 pfu per cell). The viral inoculum was then removed, adsorbed virus was inactivated with pH 3 citrate buffer, cells were overlaid with DMEM containing 1% FBS, and were frozen 24 h after transfection. Progeny virus was titrated in R6 cells.
Cell-Cell Fusion Assay. The assay was performed essentially as described (31) by using expression plasmids encoding gD1-260CD8 or wild-type (wt)-gD, plus gB, gH, or gL (80 ng of each plasmid DNA/2 cm2 well).
Western Blot Analysis of Virion gD. Virions were pelleted from the medium of BHK cells cotransfected with pEA99 plus gDΔ6-259 or pEA99 plus pcDNA3.1(-) (12 plus 18 μg of plasmid DNA/150 cm2 flask), and were subjected to SDS/PAGE. Separated proteins were transferred to a Hybond ECL nitrocellulose membrane (Amersham Pharmacia, Milan), and probed with anti-gD and anti-gB mAbs.
Results
Soluble gD Ectodomain (gDΔ290-299t) Rescues the Infectivity of gD-/- HSV. To determine whether soluble gD can substitute for virion-bound gD, gDΔ290-299t was mixed with the gD-null virus F-gDβ (24), grown in noncomplementing cells (gD-/- or gD-null stock) during adsorption to BHK cells. Infection was quantified by β-galactosidase activity of the reporter gene engineered in the virus genome. Fig. 1A shows that gD-/- HSV was able to infect BHK cells in the presence of gDΔ290-299t, but not in the presence of an irrelevant glycoprotein, fetuin (Fig. 1B).
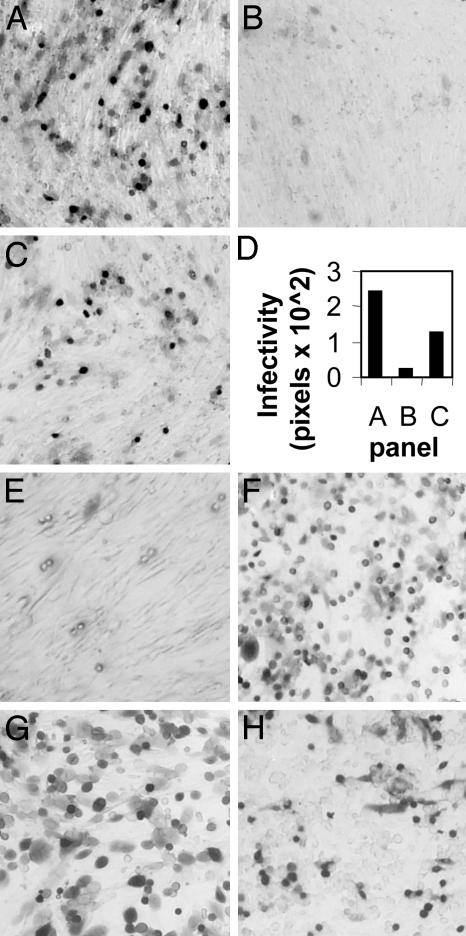
Fig. 1.
Rescue of gD-/- HSV infectivity by gDΔ290-299t (300 nM). (A and B) Cells infected with gD-/- HSV (3 μl) in the presence of gDΔ290-299t (A) or fetuin (B). Virions were mixed with glycoprotein during 150 min adsorption at 37°C. (C) The same amount of gD-/- HSV used in A was preincubated with gDΔ290-299t for 1 h at 4°C, and was then overlaid on BHK cells for 150 min. (D) Quantification of the results. Digital micrographs were taken with a 1.5× objective. The green areas (corresponding to infected cells) were quantified by the histogram program of Photoshop, and expressed in pixels. (E) A total of 200 μl of gD-/- virions were preincubated with 300 nM gDΔ290-299t for 1 h at 4°C, and were pelleted by centrifugation at 215,000 × g for 40 min. Virions were resuspended in 150 μl of DMEM and 100 μl were used to infect BHK cells. A-E, BHK cells. COS (F), nectin1-J (G), and J cells expressing HVEM (H) were infected as in A. J cells expressing HVEM were not cloned. All cells stained with 5-bromo-4-chloro-3-indolyl β-d-galactoside.
We determined whether preincubation of gD-/- virions with gDΔ290-299t for 1 h at 4°C, or at 37°C, before adsorption to BHK cells, affected the efficiency of infectivity rescue. The highest efficiency of infection was achieved when soluble gD and virions were applied simultaneously to cells (compare Fig. 1 C and A and quantification in D). Subsequent experiments were performed by simultaneously mixing virions, gD and cells, except where otherwise stated.
We next asked whether the incubation of gD-/- virions with gDΔ290-299t, followed by virion centrifugation to remove the unbound gD, and subsequent adsorption to cells, was sufficient to rescue the gD-/- virus infectivity. gD-/- virions were preincubated with gDΔ290-299t for 1 h at 4°C, centrifuged, and overlaid onto cells. This treatment did not confer any infectivity to gD-null virions (Fig. 1E). Furthermore, preexposure of cells to 300 nM gDΔ290-299t, followed by its removal before addition of virus, did not result in a significant infectivity (data not shown). These control experiments indicate that, in order to rescue the infectivity of gDnull HSV, gD has to be present at the time of virus addition to the cells.
gDΔ290-299t rescued the infectivity of gD-/- HSV in different cell lines (COS and rabbit skin), as well as in nectin1-J or J cells expressing HVEM (representative results in Fig. 1 F-H). We also checked whether gDΔ290-299t rescued the infectivity of the gD-null virus RR1097. In contrast with F-gDβ, which has a deletion of gD and of a portion of glycoprotein I, RR1097 is deleted only for the gD gene. gDΔ290-299t rescued the infectivity of RR1097 (data not shown), indicating that the effect was due to the deletion of gD, and not to other genetic alterations present in F-gDβ. Cumulatively, the results indicate that the ability of soluble gD to rescue the infectivity of gD-/- HSV is an intrinsic property of gD, and is independent of the type of receptor, cell line, and deleted virus used.
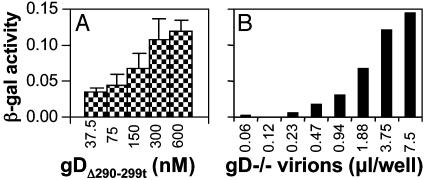
To determine whether the rescue of gD-/- HSV infectivity was dose-dependent, BHK cells were infected with replicate aliquots of gD-/- HSV mixed with increasing amounts of gDΔ290-299t, from 37.5 to 600 nM. A plateau level was achieved at 300 nM gD (Fig. 2A), and therefore this concentration was used in subsequent experiments. (Fig. 2B). When increasing amounts of virions were mixed with 300 nM gDΔ290-299t, the number of infected cells increased as a function of virus concentration. Thus, infectivity was also dose-dependent with respect to virus titer. It should be noted that, in a cell-cell fusion assay (9), gDΔ290-299t did not substitute for membrane-bound gD. The reasons are unclear at present.
Fig. 2.
Rescue of gD-/- HSV infectivity by gDΔ290-299t is dose-dependent with respect to gD (A) and to virus (B). (A) Triplicate aliquots (3 μl) of gD-/- HSV were mixed with the indicated concentrations of gDΔ290-299t and were overlaid on BHK cells. (B) Increasing amounts of gD-/- virions were mixed with 300 nM gDΔ290-299t and were overlaid on BHK cells. β-galactosidase activity was measured at 16 h after infection. Bars denote ± SE.
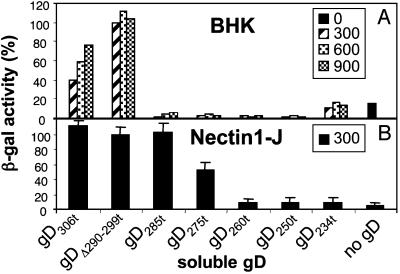
Residues 261-305 of Soluble gD Are Required to Rescue the Infectivity of gDnull HSV. To determine which regions of the ectodomain of gD were required to rescue the infectivity of gDnull HSV, we made use of a panel of soluble forms of gD. Each was truncated at a different site downstream of the receptor-binding sites, and displayed different affinities for receptors (20). We found that only gDΔ290-299t and gD306t rescued gD-/- virus infectivity in BHK cells (Fig. 3A). Because these cells do not express the human receptors, we repeated this experiment with nectin 1-J cells (Fig. 3B). In this case, we found that forms of gD truncated as far as 285 were as effective as gD306t. Shorter forms were much less effective. The differences cannot be explained solely on the basis of affinity because the affinity of gD250t for nectin 1 is higher than that of gD306t, and yet, gD250t is ineffective at rescue. Likewise, the affinity of gDΔ290-299t for nectin1 is 100-fold higher than that of gD306t, and both have similar ability to rescue the gD-null virus. Remarkably, the gD domain that carries the receptor-binding sites (residues 1-250) was not sufficient to rescue the infectivity of the gD-null virus on either cell, and a downstream region was additionally required. This mapped to residues 261-285 for nectin1-J cells, and extended to residue 306 for BHK cells. The difference may be due to a higher level of nectin1 expression in the nectin1-J cells, or properties of the hamster receptors present in BHK cells. Cumulatively, the additional domain was encoded within residues 261-305.
Fig. 3.
Effect of different gDs, truncated at different residues in the ectodomain, on rescue of gD-/- HSV infectivity. BHK (A), or nectin1-J (B) cells were exposed to replicate aliquots of gD-/- HSV mixed with the indicated soluble gDs. gD was present at the indicated nM concentrations in A, and at 300 nM in B. The value obtained with 300 nM gDΔ290-299t is 100%. Each column represents the average of duplicates in A, and of triplicates in B. Bars denote ± SE.
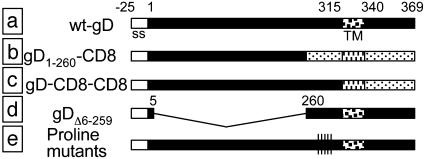
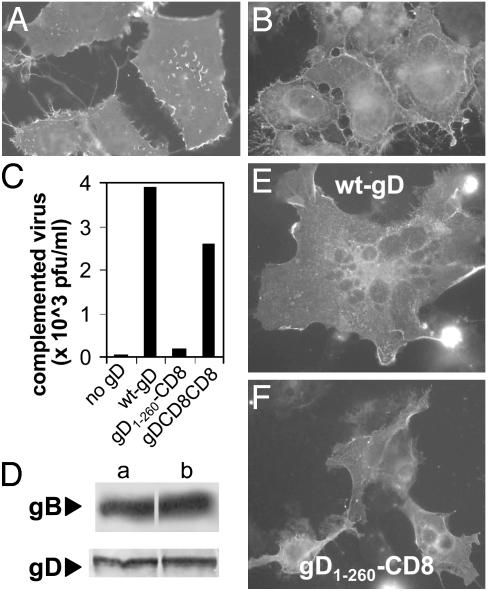
Substitution in Membrane-Bound gD of the Membrane-Proximal Domain Reduces HSV Infectivity and Cell-Cell Fusion Activity. In the following experiments, we asked whether the 261-305 region represents a functional domain in membrane-bound gD. Three types of constructs were derived, as illustrated in Fig. 4. In the gD1-260-CD8 chimera, the entire portion downstream of residue 260 (membrane-proximal, TM, and C-tail) was substituted with the corresponding regions of CD8. When this chimera was expressed in BHK cells, it retained reactivity to mAbs H170 (Fig. 5A) and HD1, a neutralizing Ab that recognizes a discontinuous epitope involved in entry (data not shown). These transfected cells were also able to bind a soluble form of nectin 1 (Fig. 5B). These assays assured that the gD1-260-CD8 chimera was properly folded, maintained its ability to interact with the receptor, and was expressed at the cell surface. To determine whether this chimera could function in infectivity complementation, a gD-/+ HSV stock was obtained by growing F-gDβ in a cell line that expresses gD from a transgene; the virus stock carries gD in the envelope and is infectious for one replicative cycle. BHK cells were transfected with the plasmid encoding gD1-260-CD8, superinfected with gD-/+ HSV, and progeny virus was titered 24 h later. gD1-260-CD8 was unable to complement the infectivity of the gD-/+ virus (Fig. 5C). In contrast, a chimera in which the entire gD ectodomain (1-315) was fused to the TM and C-tail of CD8 (gD-CD8-CD8) was functional in complementation, as reported (32). As expected, full-length gD complemented gD-/+ HSV infectivity, whereas a plasmid carrying no gD gene did not. The inability of gD1-260-CD8 to complement was not due to a defect in gD incorporation into virions, because the same amount of gD was present in complemented virions grown in cells transfected with gD1-260-CD8 or with wt-gD (Fig. 5D).
Fig. 4.
Schematic representation of the gD constructs used in this study. (a) wt-gD. Empty bar, signal sequence (ss) cleaved in mature gD. The TM coordinates are marked. (b) gD1-260-CD8 chimera. (c) gD-CD8-CD8 from ref. 32. (d) Receptor-negative gDΔ6-259, the region between residues +6 and +259 was collapsed. (e) Proline mutants.
Fig. 5.
(A)gD1-260-CD8 expression in COS cells, detected by immunofluorescence with mAb H170. (B) Binding of a soluble nectin1 chimera made of nectin1 ectodomain fused to the constant fragment of human Ig (ref. 12; 1.6 ng/μl) to paraformaldehyde-fixed gD1-260-CD8-expressing COS cells, detected by means of FITC-conjugated anti-human IgG Ab. (C) Infectivity complementation by gD1-260-CD8, gD-CD8-CD8, wt-gD, or no gD. Four h after transfection, BHK cells were infected with gD-/+ HSV (3 pfu per cell), and were then rinsed with pH 3 citrate buffer. Complemented virus was titrated at 24 h in R6 cells. (D) Quantification of gD, and of gB as control, present in complemented virions produced in C. Virions made in cells expressing (Da) wt-gD and (Db) gD1-260-CD8. The equal amounts of gB in the two virion preparations denote equal amounts of virions loaded in the gel. (E and F) Cell-cell fusion induced in COS cells by cotransfection of wt-gD or gD1-260-CD8 with plasmids encoding gB, gH, and gL, and stained with mAb H170 to gD. In E, a giant multinucleated syncytium is shown. In F, cells contain either one or two nuclei.
The gD1-260-CD8 chimera was also defective in the cell-cell fusion assay, as observed in COS cells cotransfected with expression plasmids encoding gB, gH, gL, and either wt-gD (Fig. 5E) or gD1-260-CD8 (Fig. 5F). The cell-cell fusion in BHK cells gave similar results (data not shown). Altogether, the experiments define the membrane-proximal region of full-length gD (residues 261-305) as a functional domain critical for HSV infectivity and cell-cell fusion (the pro-fusion domain).
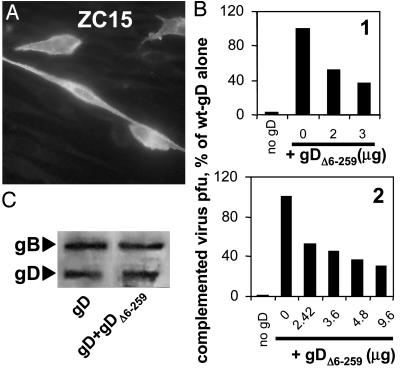
A Receptor-Negative gD (gDΔ6-259) Behaves as a Dominant-Negative Mutant. To gain further evidence for a functional role of the pro-fusion domain, we reasoned that a gD in which the receptor-binding sites were deleted and the pro-fusion domain was maintained, should behave as a dominant-negative mutant. We hypothesized that this form of gD, when coexpressed with full-length gD, should compete with and inhibit the activity of the pro-fusion domain of full-length gD. Accordingly, we generated the construct gDΔ6-259, which lacks residues 6-259 (corresponding to the receptor-binding sites) and maintains residues 260-369, i.e., pro-fusion domain plus the TM and C-tail. The signal sequence (residues -25 to 0) up to residue +5 were maintained to enable proper translocation to the endoplasmic reticulum. gDΔ6-259 expression and intracellular localization was checked by immunofluorescence with the anti-C-tail polyclonal Ab ZC15 (Fig. 6A). As expected, gDΔ6-259 alone was unable to complement the gD-/+ virus (data not shown).
Fig. 6.
(A) Expression of the receptor-negative gDΔ6-259 in methanol-fixed BHK cells, stained with polyclonal Ab ZC15 to the C-tail. (B) Effect of coexpression of wt-gD plus gDΔ6-259 on infectivity complementation of gD-/+ HSV. A fixed amount of wt-gD (2 μg per T25 flask for experiment 1, and 1.2 μg for experiment 2) was cotransfected with the indicated amounts of gDΔ6-259, in the range from 0- to 8-fold the amount of wt-gD. The amounts of transfected DNAs were made equal by addition of empty vector. Further details of the complementation assay were as in Fig. 5B. Experiments 1 and 2 are two independent experiments. (C) Quantification of gD, and of gB as control, present in complemented virions of B, experiment 1, wt-gD plus 3 μggDΔ6-259. Details as in Fig. 5D.
We then coexpressed gDΔ6-259 plus full-length gD in BHK cells, in relative amounts ranging from 1:1 to 8:1 (experiments 1 and 2 of Fig. 6B). Cells were superinfected with gD-/+ HSV 4 h later; progeny virus was harvested at 24 h. The presence of the receptor-negative gDΔ6-259 reduced HSV infectivity in a dose-dependent manner. Inhibition was not due to a reduced incorporation of full-length gD into virions, because the complemented virions made in the presence or absence of gDΔ6-259 contained approximately the same amount of full-length gD (Fig. 6C). Thus, the receptor-negative gDΔ6-259 behaved as a dominant-negative mutant.
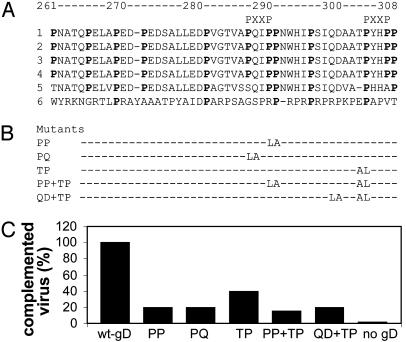
Mutational Analysis of Prolines Present in the Pro-Fusion Domain. Fig. 7A shows the alignment of pro-fusion domains from a number of HSV-1, HSV-2, and pseudorabies virus gDs. Overall gD is highly conserved among different type-1 and type-2 HSV strains, and conservation extends to this region. The most characteristic feature of the sequence is the high content in prolines, which exhibit defined spacings. Some spacings are conserved in pseudorabies virus gD. Two minimal motifs (PXXP) that can bind proteins carrying SH-3 motifs are also present (33). Bioinformatic analysis did not predict other motifs.
Fig. 7.
(A) Alignment of gD sequences from HSV-1, strains F (1), KOS (2), Patton (3), ANG (4), HSV-2 strain HG52 (5), and pseudorabies virus strain Kaplan (6). Proline residues are bold. In pseudorabies virus, conserved prolines are bold. Minimal motifs (PXXP) able to bind SH3 domains are marked. (B) Designation of proline mutants and engineered substitutions. (C) Infectivity complementation of gD-/+ HSV by gD proline mutants. Transfections, infections, and virion titrations were as detailed in Fig. 5C.
To investigate the role of prolines, some of them were substituted either with leucine, one of the least conservative substitutions for proline, or alanine. We concentrated site-directed mutagenesis on the region downstream of residue 285, which was the richest in prolines. The substitutions are indicated in Fig. 7B. BHK cells were transfected with the plasmids encoding the proline mutants, wt-gD or no gD, as positive and negative controls, and were superinfected with the gD-/+ HSV. All substitutions decreased infectivity (Fig. 7C). The highest reduction was observed with the double P316L-P317A plus T329L-P330A mutant. This mutant did not display any reduction in HSV-inducible expression, as judged by immunofluorescence (data not shown). The results indicate that prolines are critical residues in the pro-fusion domain.
Discussion
The results presented in this report form the basis of two fundamental conclusions with broad conceptual implications regarding the entry of HSV-1 into susceptible cells.
The first conclusion concerns the functional structure of gD. Earlier studies (1, 2, 9) established that gD attaches to one of three different classes of cell-surface receptors and also indicated that it plays a role in the fusion of the envelope with the cell membrane. It was also established that the region that interacts with the receptors maps to the ectodomain. The TM and C-tail regions can be exchanged with those of another glycoprotein (CD8), or gD ectodomain may be glycosylphosphatidylinositol-anchored to the virion envelope or fused to the gH ectodomain, and virus infectivity is still maintained (32, 34). In this report, we show that we can differentiate three discrete domains within gD, each with a defined function. Specifically, gD consists of a domain that harbors the receptor-binding sites, a domain critical for the fusion of the envelope to the cell membrane, and finally, a domain consisting of the TM and C-tail of the protein. Several important concepts have emerged from our studies. First, we have identified a discrete sequence of gD required for fusion of the envelope to the cell membrane that is located entirely in the ectodomain of gD, in proximity of the TM sequence (the pro-fusion domain). Second, we found that the soluble gD ectodomain (receptor-binding region plus the profusion domain) is both necessary and sufficient for virus entry. Our studies further show that the TM and C-tail regions have no demonstrable function. The only function that can be attributed to them at this time is that they ensure the gD is delivered to the gD receptor along with the virion.
Another concept to emerge from our studies concerns the interaction of gD with other virion glycoproteins. The published data to date clearly indicate that gD plays a role in the fusion of the envelope with the cell membrane, in that it is an essential component of all HSV fusion assays reported to date (9). An implication of those studies is that gD interacts in some fashion with the fusion glycoproteins gB, gH, and gL, or with a subset of them. In this report, we were unable to rescue the infectivity of gDnull HSV by preincubation of the virions with soluble gD, implying that gD does not bind to other virion glycoproteins. In addition, we found that preexposure of cells to the gD ectodomain failed to make these cells susceptible to the gD-null virus. How do we explain these seemingly contradictory results? We propose that the role of gD in entry is to assemble a tripartite complex, in which gD interacts by means of the N-terminal domain with its receptor and by means of the pro-fusion domain with fusion glycoproteins simultaneously, or in sequence, but only in the presence of each component of the tripartite complex. The end result is the recruitment of the fusion glycoproteins, and their switch from a fusion-inactive to a fusion-active state. Our results do not exclude the participation of additional cellular proteins with this tripartite complex.
The Pro-Fusion Domain of gD and Its Proposed Role in Virus Entry. Two series of experiments mapped the gD pro-fusion domain to the membrane-proximal region encoding amino acids 261-305. First, the soluble forms of gD that were able to rescue the gD-null virus infectivity contained the receptor-binding sites (amino acids 1-243) plus the downstream region (amino acids 261-305). Second, substitution of this region in conjunction with the TM and C-tail regions with the corresponding portions of CD8-abolished infectivity. By contrast, a gD-CD8-CD8 chimera that carries the entire gD ectodomain fused to the TM and C-tail of CD8 conferred infectivity (32). The mapping studies were supported by the properties of a receptor-negative gD mutant, gDΔ6-259. When coexpressed in virions together with wt-gD, it reduced HSV infectivity; i.e., it behaved as a dominant-negative gD mutant. Consistent with current findings, a number of earlier observations suggested that gD carries a domain in this region that functions at a postreceptor-binding step. Thus, mAb DL6 directed to an epitope located at residues 272-279 blocks virus infectivity, but does not interfere with the binding of gD to its receptors (35). The gDΔ277-305 mutant was defective in infectivity complementation and in cell-cell fusion, but not in binding to nectin1 (18, 21). Virions carrying a gD insertion mutant (gD▾243) were impaired in rate of entry, but were unimpaired in binding affinity to both receptors (22).
The most striking feature of the pro-fusion domain is its high content of prolines, which exhibit defined spacings. Substitution of some prolines reduced HSV infectivity in a complementation assay, indicating that prolines represent critical residues in the pro-fusion domain. The observation that proline-rich regions are often in cassettes that function in protein-protein interactions (36) supports the proposal that the pro-fusion domain interacts with target proteins.
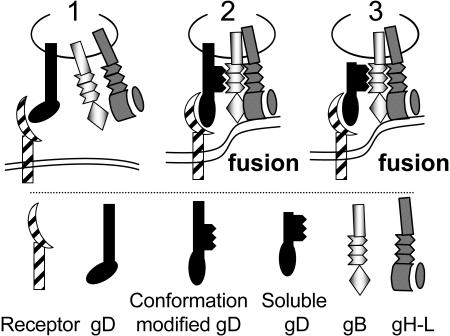
Taking into account that HVEM recognition induces conformational changes to gD (19), we propose that when gD interacts with its receptor, its conformation is modified (Fig. 8). These changes in conformation act to “cis-signal” the receptor recognition to the fusion glycoproteins (or to cellular factors required to carry out fusion), and thus link receptor recognition to the triggering of fusion. The proline-rich pro-fusion domain is responsible for the cis-signaling activity, and for assembly of a tripartite complex. This complex may be a stable, or, more likely, a transient complex, such that gD is not part of the ultimate structure that executes fusion. Future studies will focus on the composition and structure of the tripartite complex and the mechanism through which this complex activates the fusion capabilities of gB, gH, and gL.
Fig. 8.
Proposed role of gD in HSV entry (1). gD interacts with is receptor. A conformational modification ensues, such that (2) gD interacts with/recruits one of the fusion glycoproteins through the proline-rich pro-fusion domain (black zigzag lines). A tripartite complex (receptor-gD-fusion glycoprotein) is assembled. The fusion glycoproteins execute fusion. The respective role of gB and gH-gL is not differentiated here (3). Soluble gD can substitute for full-length gD, leading to formation of the tripartite complex and to fusion.
Acknowledgments
We thank Drs. P. Spear, A. Minson, D. Johnson, and R. Roller for gifts of plasmids and viruses, and Elisabetta Romagnoli for invaluable assistance with cell cultures. This work was supported by the Fondo per gli Investimenti della Ricerca di Base-Autonomous Project, the Fondo per gli Investimenti della Ricerca di Base-Coordinated Project, Cofin-Ministero dell'Istruzione, dell'Università e della Ricerca 40% 2002 and 2003; Consiglio Nazionale delle Ricerche-Functional Genomics, University of Bologna 60%; and by U.S. Public Health Service Grants AI-18289 (to G.H.C. and R.J.E.) and AI-56045 and NS-36731 (to R.J.E. and G.H.C.).
Abbreviations: C-tail, cytoplasmic tail; HVEM, herpesvirus entry mediator; TM, transmembrane; wt-gD, wild-type gD; nectin 1-J, J cells expressing nectin 1.
References
- 1.Campadelli-Fiume, G., Cocchi, F., Menotti, L. & Lopez, M. (2000) Rev. Med. Virol. 10, 305-319. [DOI] [PubMed] [Google Scholar]
- 2.Spear, P. G., Eisenberg, R. J. & Cohen, G. H. (2000) Virology 275, 1-8. [DOI] [PubMed] [Google Scholar]
- 3.Montgomery, R. I., Warner, M. S., Lum, B. J. & Spear, P. G. (1996) Cell 87, 427-436. [DOI] [PubMed] [Google Scholar]
- 4.Eberlé, F., Dubreuil, P., Mattei, M. G., Devilard, E. & Lopez, M. (1995) Gene 159, 267-272. [DOI] [PubMed] [Google Scholar]
- 5.Lopez, M., Eberlé, F., Mattei, M. G., Gabert, J., Birg, F., Bardin, F., Maroc, C. & Dubreuil, P. (1995) Gene 155, 261-265. [DOI] [PubMed] [Google Scholar]
- 6.Geraghty, R. J., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Spear, P. G. (1998) Science 280, 1618-1620. [DOI] [PubMed] [Google Scholar]
- 7.Takai, Y. & Nakanishi, H. (2003) J. Cell Sci. 116, 17-27. [DOI] [PubMed] [Google Scholar]
- 8.Cocchi, F., Menotti, L., Mirandola, P., Lopez, M. & Campadelli-Fiume, G. (1998) J. Virol. 72, 9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner, A., Bruun, B., Minson, T. & Browne, H. (1998) J. Virol. 72, 873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connolly, S. A., Landsburg, D. J., Carfi, A., Wiley, D. C., Eisenberg, R. J. & Cohen, G. H. (2002) J. Virol. 76, 10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly, S. A., Landsburg, D. J., Carfi, A., Wiley, D. C., Cohen, G. H. & Eisenberg, R. J. (2003) J. Virol. 77, 8127-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cocchi, F., Lopez, M., Menotti, L., Aoubala, M., Dubreuil, P. & Campadelli-Fiume, G. (1998) Proc. Natl. Acad. Sci. USA 95, 15700-15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krummenacher, C., Rux, A. H., Whitbeck, J. C., Ponce-de-Leon, M., Lou, H., Baribaud, I., Hou, W., Zou, C., Geraghty, R. J., Spear, P. G., et al. (1999) J. Virol. 73, 8127-8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocchi, F., Lopez, M., Dubreuil, P., Campadelli-Fiume, G. & Menotti, L. (2001) J. Virol. 75, 7987-7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krummenacher, C., Baribaud, I., Ponce De Leon, M., Whitbeck, J. C., Lou, H., Cohen, G. H. & Eisenberg, R. J. (2000) J. Virol. 74, 10863-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menotti, L., Cocchi, F. & Campadelli-Fiume, G. (2002) Virology 301, 6-12. [DOI] [PubMed] [Google Scholar]
- 17.Martinez, W. M. & Spear, P. G. (2002) J. Virol. 76, 7255-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang, H. Y., Cohen, G. H. & Eisenberg, R. J. (1994) J. Virol. 68, 2529-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carfi, A., Willis, S. H., Whitbeck, J. C., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Wiley, D. C. (2001) Mol. Cell 8, 169-179. [DOI] [PubMed] [Google Scholar]
- 20.Whitbeck, J. C., Muggeridge, M. I., Rux, A. H., Hou, W., Krummenacher, C., Lou, H., van Geelen, A., Eisenberg, R. J. & Cohen, G. H. (1999) J. Virol. 73, 9879-9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou, G., Avitabile, E., Campadelli-Fiume, G. & Roizman, B. (2003) J. Virol. 77, 3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milne, R. S., Hanna, S. L., Rux, A. H., Willis, S. H., Cohen, G. H. & Eisenberg, R. J. (2003) J. Virol. 77, 8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krummenacher, C., Nicola, A. V., Whitbeck, J. C., Lou, H., Hou, W., Lambris, J. D., Geraghty, R. J., Spear, P. G., Cohen, G. H. & Eisenberg, R. J. (1998) J. Virol. 72, 7064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ligas, M. W. & Johnson, D. C. (1988) J. Virol. 62, 1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rauch, D. A., Rodriguez, N. & Roller, R. J. (2000) J. Virol. 74, 11437-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou, G., Galvan, V., Campadelli-Fiume, G. & Roizman, B. (2000) J. Virol. 74, 11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Showalter, S. D., Zweig, M. & Hampar, B. (1981) Infect. Immun. 34, 684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menotti, L., Lopez, M., Avitabile, E., Stefan, A., Cocchi, F., Adelaide, J., Lecocq, E., Dubreuil, P. & Campadelli Fiume, G. (2000) Proc. Natl. Acad. Sci. USA 97, 4867-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menotti, L., Casadio, R., Bertucci, C., Lopez, M. & Campadelli-Fiume, G. (2002) J. Virol. 76, 5463-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rux, A. H., Willis, S. H., Nicola, A. V., Hou, W., Peng, C., Lou, H., Cohen, G. H. & Eisenberg, R. J. (1998) J. Virol. 72, 7091-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avitabile, E., Lombardi, G. & Campadelli-Fiume, G. (2003) J. Virol. 77, 6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browne, H., Bruun, B., Whiteley, A. & Minson, T. (2003) J. Gen. Virol. 84, 1085-1089. [DOI] [PubMed] [Google Scholar]
- 33.Khan, I. H., Sawai, E. T., Antonio, E., Weber, C. J., Mandell, C. P., Montbriand, P. & Luciw, P. A. (1998) J. Virol. 72, 5820-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cairns, T. M., Milne, R. S., Ponce-de-Leon, M., Tobin, D. K., Cohen, G. H. & Eisenberg, R. J. (2003) J. Virol. 77, 6731-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicola, A. V., Ponce de Leon, M., Xu, R., Hou, W., Whitbeck, J. C., Krummenacher, C., Montgomery, R. I., Spear, P. G., Eisenberg, R. J. & Cohen, G. H. (1998) J. Virol. 72, 3595-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kay, B. K., Williamson, M. P. & Sudol, M. (2000) FASEB J. 14, 231-241. [PubMed] [Google Scholar]