Abstract
Most mitochondrial proteins are synthesized on cytosolic ribosomes and must be imported across one or both mitochondrial membranes. There is an amazingly versatile set of machineries and mechanisms, and at least four different pathways, for the importing and sorting of mitochondrial precursor proteins. The translocases that catalyze these processes are highly dynamic machines driven by the membrane potential, ATP, or redox reactions, and they cooperate with molecular chaperones and assembly complexes to direct mitochondrial proteins to their correct destinations. Here, we discuss recent insights into the importing and sorting of mitochondrial proteins and their contributions to mitochondrial biogenesis.
Introduction
Eukaryotic cells are divided into numerous organelles, compartments that are surrounded by membranes. The vast majority of proteins, however, are synthesized in a single compartment, the cytosol. About half of these proteins have to be transported into or across at least one cellular membrane to reach their functional destination (Schnell and Hebert, 2003; Wickner and Schekman, 2005; Neupert and Herrmann, 2007). For example, the budding yeast Saccharomyces cerevisiae synthesizes about 6000 different proteins with about 3000 of these proteins directed to the various cell organelles, raising several fundamental questions about how the precursor proteins are directed to the correct target organelle, how they are translocated across the hydrophobic membranes, and how they are sorted and assembled into their functional forms.
Mitochondria are crucial for numerous cellular processes, including energy metabolism, programmed cell death (apoptosis), signaling, and metabolic pathways involving lipids, amino acids, and iron (Figure 1A). These organelles have an outer membrane and an inner membrane with folded cristae that give rise to two aqueous compartments, the intermembrane space and the matrix. Mitochondria contain about 1000 (yeast) to 1500 (human) different proteins (Sickmann et al., 2003; Perocchi et al., 2006; Pagliarini et al., 2008). Of these, about 1% are synthesized on ribosomes in the matrix (Figure 1A), and the remainder are synthesized on ribosomes in the cytosol. Mitochondrial proteins synthesized in the matrix were inherited from the prokaryotic ancestor of mitochondria and represent crucial membrane components of the oxidative phosphorylation machinery. But mitochondria also contain hundreds of proteins derived from prokaryotes that are synthesized in the cytosol, because the genes encoding them were transferred to the nucleus. Mitochondrial proteins that were “invented” during the evolution of eukaryotic cells also are encoded by nuclear genes and are synthesized in the cytosol.
Figure 1. Principles of Mitochondrial Protein Biogenesis.
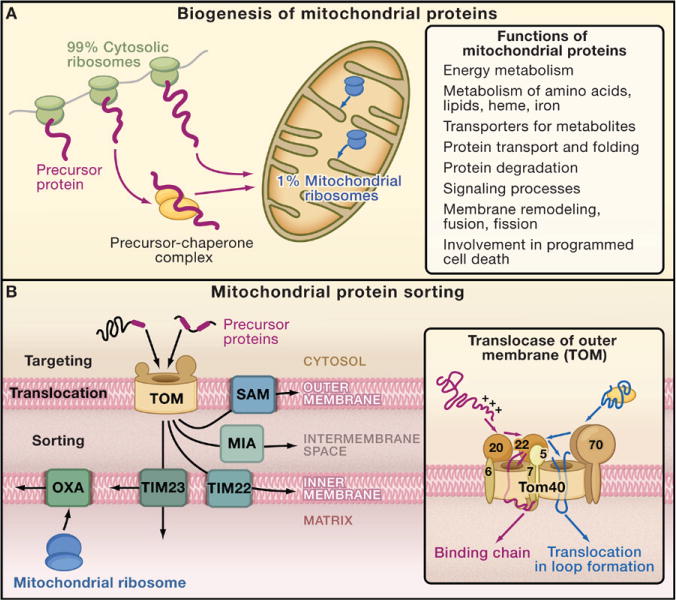
(A) Cytosolic and mitochondrial protein synthesis. Most mitochondrial proteins are synthesized on cytosolic ribosomes and are imported into the organelle. About 1% of mitochondrial proteins are synthesized inside the organelle.
(B) Sorting pathways of mitochondria. The translocase of the outer membrane (TOM complex) is the main entry gate into mitochondria. Subsequently, the precursor proteins follow different sorting pathways. MIA, mitochondrial intermembrane space assembly; OXA, insertase/export machinery of the inner membrane; SAM, sorting and assembly machinery; TIM22 complex, carrier translocase of the inner membrane; TIM23 complex, presequence translocase of the inner membrane. (Inset) The TOM complex consists of seven different subunits. The receptors Tom20, Tom22, and Tom70 recognize precursor proteins and transfer them to the central component, the channel-forming Tom40. Three small Tom proteins, Tom5, Tom6, and Tom7, are involved in the assembly and dynamics of the TOM complex. (Presequence-carrying precursor proteins, red; hydrophobic precursors with internal targeting signals, blue.)
Despite the central role of mitochondria in bioenergetics, the respiratory chain proteins and the mitochondrial ATP synthase are not strictly essential for life, at least in organisms that are capable of fermentation. However, the mitochondrial protein import and folding machineries and the matrix machinery for the biosynthesis of iron-sulfur clusters are strictly essential for cell viability (Neupert and Herrmann, 2007; Lill and Mühlenhoff, 2008).
Originally, it was believed that all mitochondrial precursor proteins are imported via one main pathway and mechanism, the so-called presequence pathway. This pathway involves an outer and inner membrane translocase, with amino-terminal presequences in precursor proteins acting as classical targeting signals directing proteins into mitochondria. The demonstration that the precursors of metabolite carrier proteins of the mitochondrial inner membrane use different signals and a different sorting route suggested that there were two routes for mitochondrial import: the presequence pathway and the carrier pathway. Then, 5 years ago, our view of mitochondrial protein biogenesis changed very rapidly because of the identification of numerous new import components and (at least) two new import pathways (Table 1). New mechanistic principles, such as redox-regulated import, formation of supercomplexes with the respiratory chain, and two-membrane coupling of translocases, were discovered. It also became clear that the precursor protein (preprotein) translocases are composed of modular units that cooperate with each other in a highly dynamic manner. This Review discusses recent advances that provide important insights into the biogenesis of mitochondria and the principles of intracellular protein sorting. This Review highlights the machineries and mechanisms of the four principal pathways that direct proteins to their intramitochondrial destination: the presequence pathway to the matrix and inner membrane, the carrier protein pathway to the inner membrane, the redox-regulated import pathway into the intermembrane space, and the β-barrel pathway (and possibly other routes) into the outer membrane.
Table 1.
Components of the Mitochondrial Protein Import and Sorting Machineries
| Name | Proposed Function | Essential | Year | Aliases |
|---|---|---|---|---|
| Translocase of the Outer Mitochondrial Membrane (TOM) | ||||
| Tom20 | Initial receptor for preproteins with a presequence | – | 1989 | Mas20, Mom19, Pom13, Rir16 |
| Tom70 | Initial receptor for hydrophobic precursor proteins (carriers) | – | 1990 | Mas70, Mom72, Omp1 |
| Tom71 | Homolog of Tom70 (low abundance) | – | 1996 | Tom72 |
| Tom40 | Channel-forming protein of TOM complex, β-barrel topology | Yes | 1989 | Isp42, Mom38 |
| Tom22 | Receptor at cis and trans side of the outer membrane, organizer of TOM | – | 1993 | Mas17, Mas22, Mom22 |
| Tom5 | Transfer of preproteins from receptors to channel, assembly of TOM | – | 1997 | Mom8, Mom8a |
| Tom6 | Assembly of TOM complex | – | 1993 | Isp6, Mom8b |
| Tom7 | Disassembly and dynamics of TOM complex | – | 1996 | Mom7, Yok22 |
| Sorting and Assembly Machinery of the Outer Mitochondrial Membrane (SAM) | ||||
| Sam50 | Central component of SAM complex, β-barrel topology, channel | Yes | 2003 | Omp85, Tob55 |
| Sam37 | Subunit of SAM complex, promotes release of precursors | – | 2003 | Mas37, Pet3027, Tom37, metaxin 1 |
| Sam35 | Binding of β signal, partner of Sam50 | Yes | 2004 | Fmp20, Tob38, Tom38, metaxin 2 |
| Mdm10 | Involved in β-barrel assembly, associates with SAM and Mdm12/Mmm1 | – | 2004 | Fun37 |
| Mim1 | Involved in biogenesis of α-helical proteins, interaction with SAM | – | 2004 | Tom13 |
| Mitochondrial Intermembrane Space Import and Assembly (MIA) | ||||
| Mia40 | Receptor in IMS, transfers disulfide bonds to IMS precursors | Yes | 2004 | Fmp15, Tim40 |
| Erv1 | Oxidation of and cooperation with Mia40 in transfer of disulfide bonds | Yes | 2005 | ALR |
| Hot13 | Promotes oxidation of Mia40 by Erv1, binding of zinc ions | – | 2004 | |
| Intermembrane Space Chaperones | ||||
| Tim9–Tim10 | Chaperone complex, transfer of hydrophobic proteins through IMS | Yes | 1998 | Mrs11 (Tim10) |
| Tim8–Tim13 | Chaperone complex, transfer of hydrophobic proteins through IMS | – | 1999 | DDP1 (Tim8) |
| Carrier Translocase of the Inner Mitochondrial Membrane (TIM22) | ||||
| Tim22 | Core of TIM22 complex, channel-forming (twin-pore) | Yes | 1996 | |
| Tim54 | Membrane protein with domain in IMS, binds Tim9-Tim10-Tim12 complex | – | 1997 | |
| Tim18 | Involved in assembly of TIM22 complex | – | 2000 | |
| Tim9–Tim10–Tim12 | Membrane-bound chaperone, tethering of carrier precursors to TIM22 | Yes | 1998 | Mrs5 (Tim12) |
| Presequence Translocase of the Inner Mitochondrial Membrane (TIM23) | ||||
| Tim23 | Channel-forming subunit of TIM23 complex | Yes | 1993 | Mas6, Mim23, Mpi3 |
| Tim17 | Tight association with Tim23, involved in lateral sorting of preproteins | Yes | 1994 | Mim17, Mpi2, Sms1 |
| Tim50 | Intermembrane space receptor, gating of Tim23 channel | Yes | 2002 | |
| Tim21 | Modulator of TIM23, interaction with TOM complex and respiratory chain | – | 2005 | |
| Presequence Translocase-Associated Motor (PAM) | ||||
| mtHsp70 | Molecular chaperone, binds preproteins, hydrolyzes ATP, protein folding | Yes | 1990 | Ssc1, Ens1 |
| Mge1 | Mitochondrial nucleotide-exchange factor for mtHsp70 | Yes | 1994 | Yge1 |
| Tim44 | Membrane anchor for mtHsp70, preprotein binding | Yes | 1992 | Isp45, Mim44, Mpi1 |
| Pam18 | J protein, stimulates ATPase activity of mtHsp70 | Yes | 2003 | Tim14 |
| Mdj2 | Homolog of Pam18 (low abundance) | – | 2005 | |
| Pam16 | J-like protein, forms a module with and controls Pam18 activity | Yes | 2004 | Mia1, Tim16 |
| Pam17 | Involved in integrity of Pam18-Pam16 module, binds to Tim23 | – | 2005 | Fmp18 |
| Mitochondrial Processing Peptidases | ||||
| MPP α/β | Mitochondrial processing peptidase, heterodimer of Mas1 (β) and Mas2 (α) | Yes | 1988 | Mif1, PEP (Mas1), Mif2 (Mas2) |
| Oct1 | Matrix intermediate peptidase, completes maturation of some preproteins | – | 1992 | MIP |
| IMP | Inner membrane peptidase complex consisting of Imp1, Imp2 and Som1 | – | 1991 | |
| m-AAA | Inner membrane protease complex consisting of Yta12 and Yta10 | – | 2002 | Rca1 (Yta12), Yta10 (Afg3) |
| i-AAA | Inner membrane protease complex consisting of Yme1, Mgr1 and Mgr3 | – | 2006 | Osd1, Yta11 (Yme1), Fmp24 (Mgr3) |
| Pcp1 | Rhomboid intramembrane protease, processing of Mgm1 and Ccp1 | – | 2002 | Mdm37, Rbd1 |
| Mitochondrial Export Machinery | ||||
| Oxa1 | Insertion of proteins into the IM, interacts with mitochondrial ribosome | – | 1997 | |
| Mba1 | Cooperates with Oxa1 in the membrane insertion of COX subunits | – | 2001 | |
| Mdm38 | Membrane integration, interacts with mitochondrial ribosome | – | 2006 | Mkh1 |
| Cox18 | Homolog of Oxa1 with export function for Cox2 subunit of COX | – | 2002 | Oxa2 |
| Pnt1 | Role in export of mitochondrially encoded Cox2, interacts with Cox18 | – | 1999 | |
| Mss2 | Role in export of mitochondrially encoded Cox2, interacts with Cox18 | – | 2001 | |
| Folding in Matrix | ||||
| Mdj1 | J protein, cooperates with mtHsp70 | – | 1994 | |
| Hsp60/Hsp10 | Chaperonin, protein folding, hydrolyzes ATP | Yes | 1989 | Cpn60, Mif4, Mna2/Cpn10 |
| Hsp78 | Role in prevention of protein aggregation, disaggregation, ATP-dependent | – | 1993 | |
| Zim17 | Zinc finger heat shock protein, chaperone for mtHsp70 | – | 2005 | Hep1, Tim15 |
Many mitochondrial proteins are required for protein import, sorting, processing, and folding. A number of components have more than one name. The Table lists the standard name of the Saccharomyces genome database (SGD) unless another name has become the principal name in the literature, e.g., mtHsp70. mtHsp70 and its cochaperone Mge1 are listed under their import function (presequence translocase-associated motor), but they also function in protein folding in the matrix. (Essential, the protein is strictly essential for cell viability, that is, deletion of the gene blocks cell growth under all conditions tested; –, not strictly essential. The year in which the role of the component in mitochondrial protein import and sorting was defined is shown. COX, cytochrome c oxidase; Hsp, heat shock protein; IMS, intermembrane space.)
TOM—The Mitochondria’s Portal
Nearly all mitochondrial preproteins are imported via the general entry gate, the translocase of the outer membrane or TOM complex (Figure 1B). The central component of TOM is Tom40, an integral membrane protein with a β-barrel structure that forms the channel for preprotein translocation across the outer membrane. Tom40 is organized as an oligomer that forms two to three channels per TOM complex (Ahting et al., 2001; Model et al., 2008). Three receptor proteins function as part of the TOM complex (Figure 1B). Tom20 is the initial recognition site for preproteins with presequences (Saitoh et al., 2007) and transfers the preproteins to the central receptor Tom22. Tom70 forms the initial recognition site for precursors of inner membrane metabolite carrier proteins, which carry multiple internal targeting signals; Tom70 transfers these precursor proteins to Tom22 (Kiebler et al., 1993; van Wilpe et al., 1999). From here, the precursors are inserted into the Tom40 channel.
Though TOM is the common import site for precursor proteins, at least two different mechanisms for translocation through the channel have been observed (Figure 1B). In the first, presequence-carrying preproteins are translocated as loosely folded linear polypeptide chains with the amino-terminal presequence going first. The presequence sequentially binds to a series of binding sites: to Tom20 and Tom22, then with the help of Tom5 to Tom40, and eventually to Tom7 and the intermembrane space domain of Tom22. This chain of presequence binding sites continues with Tim proteins of the translocase of the inner membrane (binding chain hypothesis) (Komiya et al., 1998; Kanamori et al., 1999; Esaki et al., 2003, 2004; Chacinska et al., 2005). In the second mechanism, the precursors of carrier proteins are not translocated as linear chains but traverse the outer membrane in a loop formation such that both termini are still on the cytosolic surface while a middle portion has already reached the intermembrane space (Wiedemann et al., 2001). Potentially, there may exist a third mechanism of preprotein import via TOM that does not use the interior of the channel. In this model, some precursors of outer membrane proteins would be inserted at the protein-lipid interface (Gabriel et al., 2001; Ahting et al., 2005). This mechanism would solve a major conceptual problem with regard to the β-barrel channel and protein insertion into the lipid phase of the outer membrane. The Tom40 channel likely forms a very stable β-barrel that probably cannot be pulled apart for energetic reasons as too many hydrogen bonds would have to be broken, and thus a lateral exit of precursor proteins from the channel interior to the outside is unlikely (Ahting et al., 2001; Gabriel et al. 2001; Gentle et al., 2004; Kutik et al., 2008b). It is possible that some α-helical proteins of the outer membrane may use a pathway at the protein-lipid interface, although direct experimental demonstration of this third pathway is lacking so far.
Tom22 not only provides specific binding sites for preproteins on the cytosolic side and on the intermembrane space side, but its membrane domain also plays a crucial role in the oligomeric organization of the 450 kDa TOM complex. In the absence of Tom22, the complex dissociates into smaller Tom40-containing units, and the biogenesis of mitochondria is strongly impaired (van Wilpe et al., 1999; Wiedemann et al., 2003; Meisinger et al., 2004). The three small Tom proteins are involved in the assembly and stability of the TOM complex. Tom6 and Tom7 play antagonistic roles. Tom6 stabilizes the large TOM complex, whereas Tom7 favors its dissociation, thus supporting a dynamic organization of the TOM complex. Tom5 promotes assembly of the TOM complex and participates in preprotein transfer from Tom22 to the Tom40 channel (Dietmeier et al., 1997; Model et al., 2001; Wiedemann et al., 2003).
Mitochondrial Targeting Signals
Cleavable presequences are the classical type of mitochondrial targeting signals. However, numerous types of noncleavable targeting and sorting signals, which are located in the mature regions of mitochondrial proteins, have also been described (Figure 2), reflecting the different sorting routes for proteins within mitochondria.
Figure 2. Targeting and Sorting Signals of Mitochondrial Precursor Proteins.
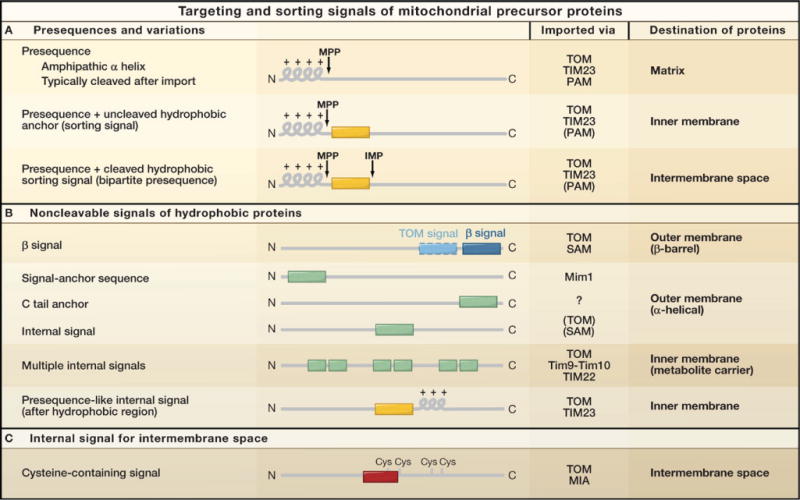
Mitochondrial precursor proteins contain cleavable or noncleavable targeting signals. The three main classes of signals are shown. The signals direct the precursor proteins to different sorting machineries. Presequences are usually cleaved off by the mitochondrial processing peptidase (MPP) in the matrix. Additional hydrophobic sorting signals can be removed by the inner membrane peptidase complex (IMP). The putative signal directing β-barrel precursors through the TOM complex is shown in light blue. The mitochondrial intermembrane space signal (MISS) for the MIA pathway is shown in red; MISS includes a cysteine residue of the precursor (shown here for the first cysteine). MIA, mitochondrial intermembrane space assembly; Mim1, putative insertase of the mitochondrial outer membrane; PAM, presequence translocase-associated motor; SAM, sorting and assembly machinery; Tim9–Tim10, chaperone complex in the intermembrane space; TIM22, carrier translocase of the inner membrane; TIM23, presequence translocase of the inner membrane; TOM, translocase of the outer membrane.
Presequences and Variations
Presequences are located at the amino terminus of precursor proteins and form positively charged amphipathic α helices (Figure 2A). They have a length of typically 15–50 amino acid residues, although presequences shorter than ten residues and up to ~100 residues have been described. Upon import into the matrix, most presequences are proteolytically removed by the dimeric mitochondrial processing peptidase (MPP). Presequences apparently contain at least three different types of information as revealed by their differential mode of interaction with binding sites. The amphipathic α helix possesses a hydrophobic surface that is recognized by Tom20 and a positively charged surface recognized by Tom22 (Moberg et al., 2004; Saitoh et al., 2007; Yamano et al., 2008b). The high-resolution structures of presequence-Tom20 and presequence-MPP complexes surprisingly revealed that presequences radically change their conformation during import into mitochondria. Whereas presequences adopt an α-helical conformation when in a complex with Tom20 (Saitoh et al., 2007), they sit in an extended conformation in the binding site of the enzyme MPP (Taylor et al., 2001), indicating that the specific recognition of the cleavage site in an extended conformation represents a third type of information. Moreover, the positive net charge of presequences is the critical determinant for the electrophoretic effect of the membrane potential during translocation across the inner membrane (Shariff et al., 2004; van der Laan et al., 2006, 2007; Krayl et al., 2007).
Presequences direct the attached proteins into the matrix; however, a number of preproteins additionally contain a hydrophobic sorting signal that is located after the matrix-targeting signal (Figure 2A). This sorting signal arrests translocation in the mitochondrial inner membrane with the preproteins being released laterally into the lipid phase (Glick et al., 1992). In the case of inner membrane proteins, the sorting signal typically remains part of the mature protein and anchors the protein in the membrane. For several intermembrane space proteins, the sorting signal is cleaved off by the inner membrane peptidase (IMP) on the outer surface of the inner membrane, and thus the mature protein is released into the intermembrane space (Figure 2A; Table 1) (Glick et al., 1992; Jan et al., 2000; Gakh et al., 2002). In a few cases, other inner membrane proteases were also shown to cleave mitochondrial presequences (Table 1). These include the AAA proteases that contain their active center either on the matrix side (m-AAA) or intermembrane space side (i-AAA), and the rhomboid protease Pcp1 that is critical for cleaving the precursor of a mitochondrial morphology protein within the membrane (Nolden et al., 2005; Rainey et al., 2006; Duvezin-Caubet et al., 2007). Thus, different proteases contribute to the specificity of mitochondrial protein sorting. Additional cleavage events for some proteins imported into the matrix can occur after cleavage by MPP. For example, the mitochondrial intermediate peptidase Oct1 removes an additional octapeptide sequence, though the functional reason for this second cleavage is not yet known (Gakh et al., 2002).
A multitude of Internal Targeting Signals
A large number of mitochondrial proteins are not synthesized with cleavable presequences but contain targeting information within regions of the mature protein (Figures 2B and 2C). This includes all mitochondrial outer membrane proteins, the majority of intermembrane space proteins, numerous multispanning inner membrane proteins, and a few matrix proteins. In the case of these matrix proteins and a few outer and inner membrane proteins, presequence-like sequences at the amino terminus are used for targeting but are not cleaved. However, the majority of noncleavable precursor proteins contain quite distinct targeting signals at various positions within their primary structure. The exact nature and targeting mechanisms of these internal signals are the subject of current research.
Mitochondrial outer membrane proteins of the β-barrel type contain a β signal in their carboxy-terminal region that bears no similarity to the α-helical presequences. The signal, identified recently, is formed by the last β strand of the proteins and is specifically recognized by the sorting and assembly machinery (SAM) of the outer membrane (Kutik et al., 2008b) (Figure 2B). An additional TOM signal for the initial recognition by mitochondria is located at an internal position, yet its exact nature has not been defined. Many outer membrane proteins, however, are of the α-helical type. Targeting sequences have been found at the amino terminus (signal anchor sequence), at the carboxy terminus (tail anchor). and in the middle of the proteins (Beilharz et al., 2003; Setoguchi et al., 2006; Otera et al., 2007; Stojanovski et al., 2007; Kemper et al., 2008). These signals typically consist of an α-helical transmembrane segment and are often flanked by positively charged residues. The signals do not follow a unique insertion pathway into the outer membrane; we will discuss below four different possibilities for the insertion of α-helical outer membrane proteins.
Inner membrane proteins carrying noncleaved targeting signals include the large class of metabolite carriers, with the ADP/ATP carrier and phosphate carrier as typical representatives. The carrier proteins usually contain six transmembrane segments, do not have homologs in prokaryotes, and seem to have been “invented” by eukaryotic cells. They are found in all classes of eukaryotes, from trypanosomes and yeast to humans. The targeting information in carrier proteins is present in several sequence elements, each of about 10 amino acid residues, that can be distributed across the length of the protein (Figure 2B). The noncontiguous carrier signals probably cooperate in the recruitment of several import receptors and in the translocation of carrier precursors across the outer membrane in a loop formation (Wiedemann et al., 2001; Curran et al., 2002a; Vasiljev et al., 2004). Signals in the carboxy-terminal portion of the carriers are important for their insertion into the inner membrane (Brandner et al., 2005). Some inner membrane proteins contain an internal positively charged “presequence-like” signal that is often preceded by a hydrophobic sequence. These precursor proteins are also thought to be translocated in a loop formation (Neupert and Herrmann, 2007).
An additional, recently identified targeting signal directs noncleavable precursors into the intermembrane space (Figure 2C). The mitochondrial intermembrane space signal (MISS) includes a cysteine residue that forms a mixed disulfide bond with the intermembrane space receptor Mia40 (Milenkovic et al., 2007, 2009; Sideris and Tokatlidis, 2007). In addition to the transient covalent interaction, a hydrophobic residue in the signal is important for the specific recognition of precursor proteins by the hydrophobic binding cleft of the receptor (Banci et al., 2009; Milenkovic et al., 2009).
The multitude of different mitochondrial targeting and sorting signals demonstrates the versatility of the protein import apparatus of this organelle. Despite this complexity, three principles can be distinguished in the targeting signals for mitochondrial proteins: (1) single linear targeting signals at the amino terminus or internal positions, (2) multiple noncontiguous signals that direct translocation in a loop formation, and (3) redox-regulated signals that undergo transient covalent interaction with the corresponding import receptor.
Presequence Pathway and Translocase Coupling
The presequence translocase of the inner membrane (TIM23 complex) and its associated motor (PAM), which transport presequence-carrying precursor proteins across the inner membrane and into the matrix, are the most complicated translocase and sorting machinery of mitochondria. The reaction cycle of the TIM23 complex involves cooperation with the TOM complex of the outer membrane, the respiratory chain of the inner membrane, and the motor of the matrix, as well as the utilization of two different energy sources, the electrochemical membrane potential and ATP. The molecular mechanism of action of the TIM23-PAM machinery is still under debate.
Presequence Translocase of the Inner membrane
The core of the TIM23 complex is formed by three essential inner membrane proteins: Tim50 with a receptor function in the intermembrane space (Mokranjac et al., 2009; Tamura et al., 2009), the channel-forming protein Tim23 (Meinecke et al., 2006; Alder et al., 2008), and Tim17, which is involved in motor recruitment and lateral sorting of preproteins (Chacinska et al., 2005; Martinez-Caballero et al., 2007) (Figure 3A, left panel). Tim50 and Tim23, as well as the fourth subunit, Tim21, expose domains to the intermembrane space that are involved in the transient interaction of the TIM23 complex with the TOM complex and thus facilitate preprotein translocation from the outer to the inner membrane (Chacinska et al., 2005; Mokranjac et al., 2009; Tamura et al., 2009). Tim21 also participates in the transient coupling of the TIM23 complex with the respiratory chain complexes III and IV, and thus supports the membrane potential (Δψ)-driven import step (van der Laan et al., 2006; Wiedemann et al., 2007; Dienhart and Stuart, 2008). The membrane potential plays a dual role in preprotein import: Δψ activates the Tim23 channel and exerts an electrophoretic effect on the positively charged presequences (Δψ is negative on the matrix side) (Shariff et al., 2004; Meinecke et al., 2006; Krayl et al., 2007; van der Laan et al., 2007). Reconstitution experiments with the purified translocase inserted into liposomes revealed that the minimal unit for preprotein integration into the inner membrane is the four-subunit TIM23 complex, a cardiolipin-rich membrane, and a membrane potential (van der Laan et al., 2007) (cardiolipin is the characteristic dimeric phospholipid of mitochondria). Preproteins following this insertion pathway into the inner membrane contain a hydrophobic sorting signal behind the matrix-targeting signal. The sorting signal arrests translocation in the inner membrane and causes a lateral release into the lipid phase of the membrane (stop-transfer mechanism) (Glick et al., 1992). Although the details of the mechanism are not understood, the release requires a lateral opening of the Tim23 channel and involves the activity of Tim17 (Chacinska et al., 2005).
Figure 3. The Presequence Pathway to the Mitochondrial Inner Membrane and Matrix.
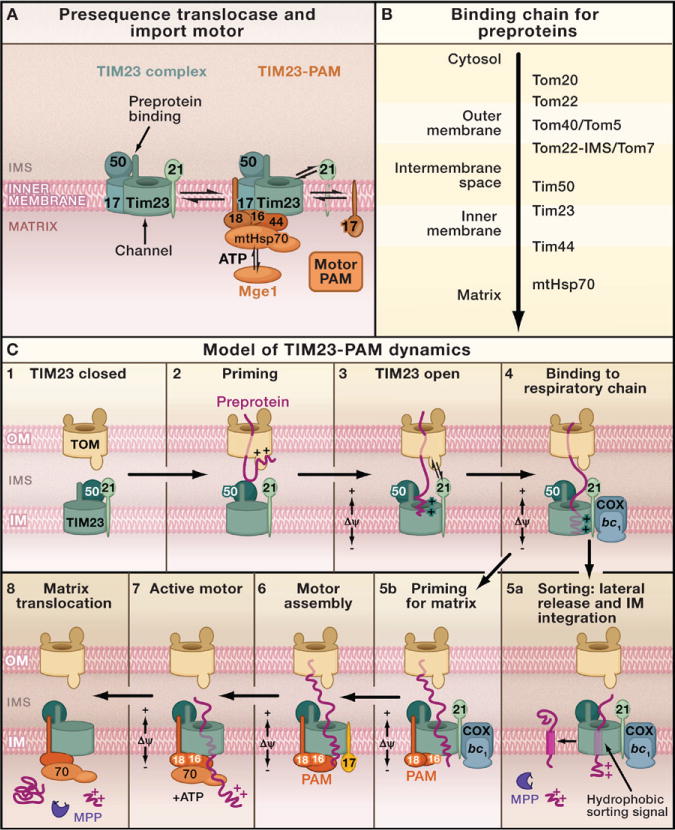
(A) Forms of the presequence translocase of the inner membrane (TIM23 complex). (Left image) Motor-free TIM23 complex that can insert preproteins into the inner membrane. (Right image) TIM23 complex associated with PAM (presequence translocase-associated motor). The central chaperone that binds to preproteins is mtHsp70. The function of mtHsp70 is regulated by the cochaperones Pam16-Pam18, Tim44, Pam17, and the nucleotide exchange factor Mge1. The function of the individual components is described in Table 1.
(B) Presequence-carrying preproteins are directed into the matrix by a sequential chain of binding sites.
(C) Hypothetical model for the dynamic cooperation of the TIM23 complex of the inner membrane (IM) with the TOM complex of the outer membrane (OM), the respiratory chain complexes III (bc1-complex) and IV (cytochrome c oxidase, COX), and the motor PAM. The membrane potential (Δψ) activates the Tim23 channel and exerts an electrophoretic effect on the positively charged presequences of preproteins. ATP drives the action of mtHsp70. The mitochondrial processing peptidase (MPP) removes the presequences.
Multisubunit Import motor
The central subunit of the matrix-exposed import motor PAM is the molecular chaperone heat shock protein 70 (mtHsp70) (Figure 3A, right panel). The mtHsp70 chaperone binds to a translocating polypeptide chain and drives its movement into the matrix in a reaction cycle that requires hydrolysis of ATP. The nucleotide exchange factor Mge1, a homolog of bacterial GrpE, promotes the release of ADP from mtHsp70 and thus stimulates a new round of ATP binding and preprotein translocation. The most remarkable feature of PAM is the existence of four membrane-bound cochaperones that direct and regulate the activity of mtHsp70 at the exit site of the Tim23 channel (Figure 3A; Table 1). Tim44 provides a dynamic (ATP-sensitive) binding site for mtHsp70 close to the channel. Moreover, Tim44 itself also binds to the preprotein in transit and is the first motor subunit that contacts the preprotein emerging on the matrix side of the Tim23 channel (Krayl et al., 2007; Slutsky-Leiderman et al., 2007). Pam18 contains a J domain that stimulates the ATPase activity of mtHsp70. Pam16, a J-related protein, forms a stable complex with Pam18 and controls its activity (Kozany et al., 2004; Li et al., 2004; Mokranjac et al., 2006). Pam17 is involved in the organization of the TIM23-PAM interaction (Hutu et al., 2008). Pam17 exerts its function before the preprotein is translocated through the channel and is then released from the motor (Popov-Celeketic et al., 2008a). The motor subunits interact with several sites on the TIM23 complex: Pam18 binds to Tim17, and Pam17 binds to Tim23 (Chacinska et al., 2005; D’Silva et al., 2008; Hutu et al., 2008; Popov-Celeketic et al., 2008a). The mitochondrial import motor is one of the most complex Hsp70 systems known.
Functions of TIm23-PAm
The sequential chain of binding sites for presequences, described above for preprotein transfer across the TOM complex, continues with the TIM23-PAM complex. Presequences are recognized by Tim50 and Tim23 on the intermembrane space side and, after translocation through the channel, are bound by Tim44 and mtHsp70. The motor components likely just recognize the unfolded state of the polypeptide chain and are not specific for presequences. The binding chain hypothesis thus includes about eight sequential sites for presequence transfer from the cytosol into the matrix (Figure 3B) (Komiya et al., 1998; Chacinska et al., 2005; Saitoh et al., 2007; Yamano et al., 2008b; Mokranjac et al., 2009; Tamura et al., 2009). Together with the membrane potential, the binding chain is crucial for directing the presequences into mitochondria. Although the import of preproteins is largely unidirectional when the overall transfer rates are considered, retrograde transport has been demonstrated for some preproteins. A remarkable case is the biogenesis of fumarase: the mitochondrial and cytosolic forms of the enzyme are encoded by one gene and are synthesized as one presequence-carrying preprotein (Karniely et al., 2006). All of the precursor molecules are partially imported into mitochondria such that the preproteins span both TOM and TIM23 complexes. When the presequence reaches the matrix, it is cleaved off. A fraction of the precursor molecules are imported completely to become the mitochondrial form of the enzyme. The remainder are released from mitochondria into the cytosol by retrograde movement to generate a presequence-free, cytosolic form.
There are three major controversies about the presequence translocase and motor that have stimulated debate in the mitochondrial import field. (1) “Stop transfer versus conservative sorting.” For many years, it has been debated whether presequence-carrying preproteins with a hydrophobic sorting signal behind the matrix-targeting signal are arrested during translocation in the TIM23 complex and laterally released (stop transfer), or whether the preproteins are completely imported into the matrix and exported into the inner membrane in a route conserved from the bacterial ancestor of mitochondria (conservative sorting). Current evidence indicates that both mechanisms exist. Preproteins with a cleavable presequence and a typical hydrophobic sorting signal use the stop transfer mechanism for lateral sorting into the inner membrane (Glick et al., 1992; Chacinska et al., 2005; Neupert and Herrmann, 2007). A conserved mechanism of export into the inner membrane has been demonstrated with the identification of Oxa1 as a core component of the mitochondrial protein export machinery (Hell et al., 1998). Oxa1 is homologous to the bacterial export component YidC (Preuss et al., 2005) and together with several more components (Mba1, Mdm38, Cox18, Pnt1, and Mss2) is responsible for inner membrane insertion of proteins synthesized in the mitochondrial matrix (Table 1). In addition, a few nuclear-encoded mitochondrial proteins seem to follow the conservative sorting route after import into the matrix (Neupert and Herrmann, 2007).
(2) “Trapping versus pulling.” Two different models were proposed for how mtHsp70 drives protein translocation into the matrix. In the trapping or Brownian ratchet model, mtHsp70 molecules bind to the unfolded precursor polypeptide in transit and thus prevent its back-sliding. When, by diffusion, a further segment of the preprotein reaches the matrix, an additional mtHsp70 molecule binds and thus favors an inward-directed movement of the preprotein (Okamoto et al., 2002; Liu et al., 2003; Yamano et al., 2008a). In the pulling or power stroke model, mtHsp70 that is bound to the membrane via Tim44 changes its conformation and thus actively drives the preprotein into the matrix. In this model, several mtHsp70 molecules and power strokes will be needed to drive the import of the preprotein (Chauwin et al., 1998). Functional studies with mutants of mtHsp70 indicated that the motor function cannot easily be explained by a single mechanism, but suggested that a combination of both trapping and pulling operates during preprotein import (Krayl et al., 2007). The folding state of the translocating polypeptide dictates requirements: for preproteins with folded domains, an active pulling function of mtHsp70 contributes to the unfolding of the domains as a prerequisite for their insertion into the import channels, whereas the import of largely unfolded preproteins can be explained by a trapping mechanism (Wilcox et al., 2005; Krayl et al., 2007). The recent identification of three PAM proteins (Pam16, Pam17, and Pam18) that regulate the architecture of the motor complex and the function of mtHsp70 at the TIM23 channel (Li et al., 2004; Mokranjac et al., 2006; D’Silva et al., 2008; Hutu et al., 2008) revealed that the mitochondrial import motor has a much higher complexity than anticipated, consistent with this composite model of regulated trapping and pulling.
(3) “Modular TIM23-PAM machinery versus single-entity translocase.” The most recent discussion on the function of the presequence translocase addresses the composition of the translocase at different stages of preprotein import. It has been proposed that at least two different forms exist (Chacinska et al., 2005; van der Laan et al., 2006, 2007; Wiedemann et al., 2007): a motor-free TIM23 complex that contains Tim21 and can mediate preprotein insertion into the inner membrane, and a TIM23-PAM complex (lacking Tim21) that is responsible for preprotein translocation into the matrix (Figure 3A). Alternatively, it has been proposed that the subunits of TIM23 and PAM are permanently associated, forming a single-entity translocase (Figure 3A, right panel) (Popov-Celeketic et al. 2008a). A detailed comparison of both models suggests that they are not mutually exclusive, and, as in the two previous controversies, a combination of the models may represent the best description of the underlying mechanism. In both models, Tim21 and Pam17 do not interact with the TIM23 core complex at the same time, but play an antagonistic role (Chacinska et al., 2005; Hutu et al., 2008; Popov-Celeketic et al., 2008a). Moreover, in the experiments favoring a single-entity translocase, only a fraction of TIM23 and PAM subunits were found in one complex (Popov-Celeketic et al., 2008a). We propose that the different models and results represent different “snapshots” of the multistep TIM23-PAM reaction cycle. In Figure 3C, we present a dynamic model that combines both views in a sequential interaction of preproteins and import components.
Dynamic Coupling of the Presequence Translocase
The model starts with the inactive TIM23 complex in the absence of preprotein (Figure 3C, step 1). Here, the Tim23 channel has to be in the closed state to prevent a massive leakage of ions across the mitochondrial inner membrane, a situation that would be deleterious to the energetic state of a cell. The intermembrane space domain of Tim50 plays a crucial role in keeping the Tim23 channel closed (Meinecke et al., 2006). When a preprotein is translocated through the TOM channel, Tim50 is the first inner membrane protein that binds directly to the preprotein and promotes its binding to the intermembrane space domain of Tom22 (step 2) (Chacinska et al., 2005; Mokranjac et al., 2009; Tamura et al., 2009). Then Tim21 binds transiently to Tom22 inducing release of the presequence, which then attaches to the Tim23 channel (step 3) (Chacinska et al., 2005; Mokranjac et al., 2005). The membrane potential (Δψ) activates the Tim23 channel and generates an electrophoretic force on the presequence. Tim21 also associates with the respiratory chain. The close association of the TIM23 complex with complexes III (bc1-complex) and IV (cytochrome c oxidase) supports the Δψ-dependent insertion of preproteins (step 4) (van der Laan et al., 2006). Now the import pathways separate. The translocation of inner membrane proteins, which carry a hydrophobic sorting signal, is arrested and the preproteins are laterally released into the inner membrane and integrated into the lipid phase (step 5a: sorting, stop transfer mechanism) (Glick et al., 1992; Chacinska et al., 2005; van der Laan et al., 2007). Most proteins, however, are imported completely into the matrix, an activity that depends on the import motor. The motor subunits assemble in a sequential manner. The J proteins, Pam16 and Pam18, associate with the TIM23 complex while it is still bound to the respiratory chain (Figure 3C, step 5b) (Wiedemann et al., 2007). Pam17 also assembles at an early stage (step 6) and is released when the preprotein emerges on the matrix side (step 7) (Popov-Celeketic et al., 2008a). The assembly of Tim44-mtHsp70 generates the active motor that binds to the polypeptide chain in transit and drives it into the matrix (step 7). MPP removes the presequence during or after translocation and the mature matrix protein achieves its active, folded form (step 8).
Thus, TIM23 interacts with different complexes during the import reaction cycle: the TOM complex, complexes III and IV of the respiratory chain, and the motor PAM. The model shown in Figure 3C depicts the main import steps for preproteins that are either laterally sorted or require motor activity. However, the two routes are not strictly separated, and an exchange may take place. This is particularly evident for preproteins, which are laterally sorted into the inner membrane (by the Tim21-containing complex) but also contain tightly folded domains that additionally require motor activity for unfolding. These preproteins depend on both activities, lateral sorting and the motor, and thus require cooperation of all TIM23-PAM subunits for efficient import. The formation of a single-entity TIM23-PAM machine during the import reaction cycle thus may be of particular importance for those preproteins.
The Carrier Pathway to the Inner Membrane
Arrival at the Outer membrane
It is generally assumed that precursor proteins synthesized on cytosolic ribosomes are guided by cytosolic targeting factors or chaperones to the mitochondrial surface. The role of cytosolic factors in mitochondrial protein import is best illustrated by the metabolite carrier proteins, which are imported into mitochondria and then inserted into the mitochondrial inner membrane. Molecular chaperones of the Hsp70 and Hsp90 families bind to the hydrophobic precursor carrier proteins in the cytosol and prevent their aggregation (Figure 4A, stage I). The precursorchaperone complexes are recognized by the surface receptor Tom70, which possesses binding sites for both the precursor proteins and the chaperones (Wiedemann et al., 2001; Young et al., 2003; Wu and Sha, 2006; Zara et al., 2009). The carrier precursor proteins contain multiple internal targeting signals and induce Tom70 molecules to form oligomers (Figure 4A, stage II). The binding of several Tom70 proteins is thought to prevent aggregation of the hydrophobic precursor. In an ATP-dependent step, the chaperones are released and the precursor is then transferred, in a loop formation, into the TOM channel (stage IIIa) (Wiedemann et al., 2001).
Figure 4. Intermembrane Space Chaperones: The Carrier Pathway, and Machinery for Import and Assembly.
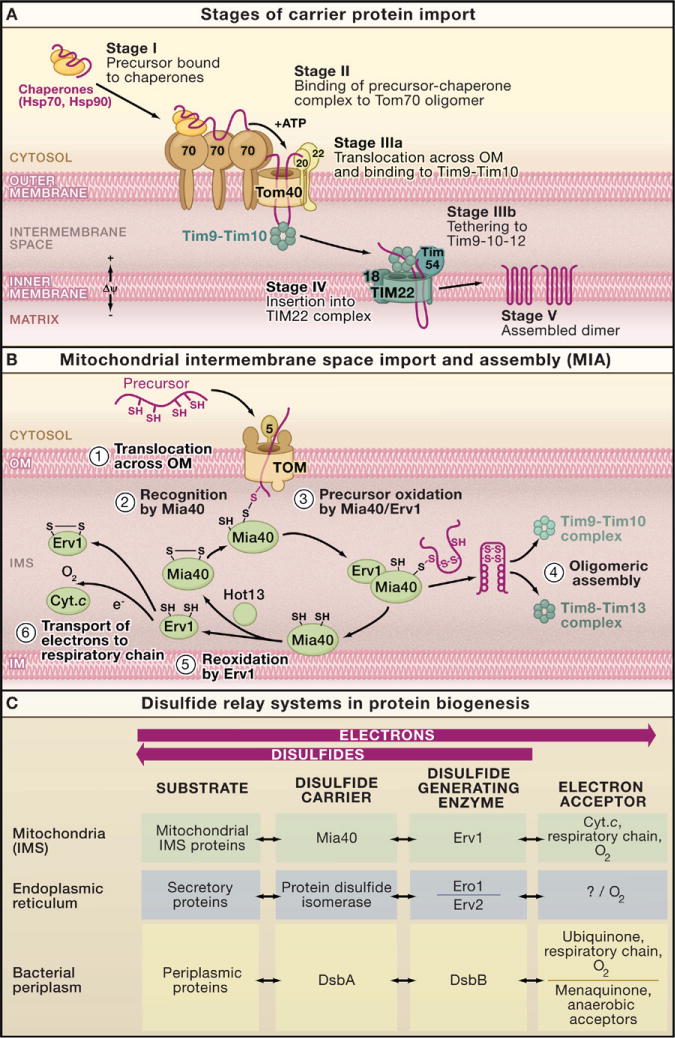
(A) Carrier pathway to the inner mitochondrial membrane. The noncleavable precursors of hydrophobic metabolite carriers of the inner membrane are imported in several stages. Cytosolic chaperones guide the precursor (stage I) to the receptor Tom70 (stage II). The precursor is transported through the translocase of the outer membrane (TOM complex) in a loop formation and interacts with the Tim9–Tim10 chaperone complex (stage IIIa). Tim9–Tim10 guides the precursor through the intermembrane space (IMS) to the carrier translocase of the inner membrane (TIM22 complex with bound Tim9-10-12) (stage IIIb). The membrane potential (Δψ) promotes insertion of the precursor into the inner membrane via the TIM22 complex (stage IV), followed by assembly into the mature form of the carrier protein (stage V).
(B) The machinery for import and assembly (MIA) of preproteins in the mitochondrial intermembrane space is required for small IMS proteins with cysteine motifs. The IMS receptor Mia40 binds to the precursor via a transient disulfide bond. The sulfhydryl oxidase Erv1 cooperates with Mia40 in the oxidation of the precursor protein. Erv1 reoxidizes Mia40 and transfers electrons to cytochrome c (Cyt. c). A third component, Hot13, assists in the oxidation of Mia40.
(C) Comparison of the disulfide relay systems of the mitochondrial intermembrane space, the endoplasmic reticulum, and the bacterial periplasm. In each system, disulfide bonds are introduced into substrate proteins and electrons are removed (oxidation of substrate proteins). A disulfide generating enzyme oxidizes a disulfide carrier that in turn oxidizes the substrate.
Journey through the Intermembrane Space
To complete translocation through the TOM channel, the carrier precursors have to bind to the Tim9–Tim10 complex of the intermembrane space (Figure 4A, stage IIIa). The hexameric Tim9–Tim10 complex forms a chaperone in the intermembrane space that guides the hydrophobic precursor proteins to the inner membrane (Curran et al., 2002a; Lu et al., 2004; Vasiljev et al., 2004; Webb et al., 2006). The Tim9–Tim10 complex not only acts as a chaperone for carrier precursor proteins, but also binds to precursors of other noncleavable inner membrane proteins, as well as to β-barrel proteins that are directed to the outer membrane.
A second hexameric chaperone complex Tim8–Tim13 is homologous to Tim9–Tim10 and functions in a related manner to guide hydrophobic precursors through the intermembrane space (Curran et al., 2002b; Davis et al., 2007). Analysis of Tim8 in humans revealed the first disease directly connected to mitochondrial protein import, Mohr-Tranebjaerg syndrome, also termed deafness dystonia syndrome (Roesch et al., 2002). Tim9–Tim10 is the major intermembrane space chaperone and both subunits are essential for cell viability; the Tim8–Tim13 subunits are dispensable for growth of yeast (Table 1).
Insertion into the Inner membrane
Tim9 and Tim10 are directly involved in the transfer of precursor proteins to the carrier translocase of the inner membrane (TIM22 complex). Tim9 and Tim10 assemble with an additional small Tim protein to form a membrane-bound Tim9-Tim10-Tim12 complex at the outer surface of the TIM22 complex (Figure 4A, stage IIIb) (Rehling et al., 2003; Neupert and Herrmann, 2007). A membraneintegrated subunit of the TIM22 complex, Tim54, exposes a large domain to the intermembrane space and may provide a binding site for the Tim9-Tim10-Tim12 complex (Kerscher et al., 1997; Wagner et al., 2008). Thus, the membrane protein substrate is chaperoned until the stage where it is inserted into the translocase channel formed by Tim22 (Rehling et al. 2003; Peixoto et al., 2007). The TIM22 complex contains two channels (twin-pore translocase) and derives external energy only from the membrane potential Δψ (stage IV) (Rehling et al., 2003). Inserted membrane protein precursors are then laterally released into the lipid phase by an as yet unknown mechanism, and are then assembled into their functional forms (stage V). The exact function of Tim18, a further membrane-embedded subunit of the TIM22 complex, is unknown; recent results suggest a role for Tim18 in the assembly of the translocase complex (Wagner et al., 2008).
Redox-Regulated Import
Mia40, an Intermembrane Space Receptor
It has been assumed that the mitochondrial intermembrane space freely exchanges small molecules with the cytosol and thus should have a similar milieu, including a reducing environment. It was thus a great surprise when a machinery was identified that formed disulfide bonds with incoming precursor proteins. The central component of this mitochondrial intermembrane space assembly machinery (MIA) is Mia40 (Figure 4B) (Chacinska et al., 2004; Naoé et al., 2004; Mesecke et al., 2005; Banci et al., 2009). It turns out that many intermembrane space proteins contain characteristic cysteine motifs and form intramolecular disulfide bonds (Curran et al., 2002a, 2002b; Lu et al., 2004; Webb et al., 2006; Müller et al., 2008). A great deal of biochemistry supported the presence of disulfide bonds in the Tim9–Tim10 complex; a high-resolution structure of the complex revealed two disulfide bonds within each subunit that are critical for the structural integrity of the complex (Webb et al., 2006). The majority of the cysteine-containing proteins in the intermembrane space are relatively small, typically smaller than 20 kDa, and it is likely that most are substrates of Mia40.
We only have limited knowledge about how the cysteine-containing intermembrane space precursors are translocated across the outer membrane. They likely use the TOM machinery as their import involves Tom5 (Kurz et al., 1999). Translocation occurs in the reduced form without disulfide bonds (Lu et al., 2004; Müller et al., 2008). Zinc ions seem to keep the precursors in the cytosol in a reduced, import-competent state (Morgan et al., 2009). Once the precursor emerges on the intermembrane space side, Mia40 binds to it via a hydrophobic cleft on its surface and the redox-active CPC motif (cysteine-proline-cysteine), forming a mixed disulfide bond with a cysteine residue of the precursor (Figure 4B) (Milenkovic et al., 2007, 2009; Sideris and Tokatlidis, 2007; Grumbt et al., 2007; Banci et al., 2009). Through this receptor function, Mia40 ensures that precursors are directed specifically into the intermembrane space.
A Redox-Driven Reaction Cycle
The precursor-Mia40 complex represents an intermediate stage that promotes the formation of disulfide bonds during the maturation of the precursors before they are released as folded proteins (Müller et al., 2008; Terziyska et al., 2009). However, Mia40 does not work alone but is part of a disulfide relay in the intermembrane space (Mesecke et al., 2005; Grumbt et al., 2007; Müller et al., 2008). The oxidation of precursor proteins by formation of disulfide bonds involves the removal of electrons from the precursors, leading to reduction of cysteine residues in Mia40 (Figures 4B and 4C). For the reoxidation of its cysteines, Mia40 cooperates with the sulfhydryl oxidase Erv1 that accepts electrons from Mia40 and transfers them via cytochrome c to the respiratory chain and hence to O2 (Farrell and Thorpe, 2005; Bihlmaier et al., 2007; Dabir et al., 2007). Alternatively, Erv1 can also utilize O2 directly, resulting in the production of hydrogen peroxide (Dabir et al., 2007). It is still an open question how the MIA system works under anaerobic conditions. An additional component of the MIA pathway is the cysteine-rich protein Hot13, which binds zinc ions. Hot13 may keep Mia40 in a zinc-free state and thus help to facilitate the oxidation of Mia40 by Erv1 (Curran et al., 2004; Mesecke et al., 2008).
Recently, there has been a further twist to the Mia40-Erv1 reaction cycle with the report of a ternary complex that contains all three partners, Erv1, Mia40, and the substrate (Stojanovski et al., 2008). This complex is thought to facilitate the transfer of multiple disulfide bonds from the sulfhydryl generating enzyme Erv1 to the precursor protein via Mia40. A physical connection of the partners may facilitate “disulfide channeling” that avoids multiple alternating contacts of Mia40 with Erv1 and substrate. Further studies are needed to elucidate which cysteine residues of Mia40 and Erv1 are involved in the channeling, and whether several copies of Mia40 and Erv1 work in such a ternary complex.
The MIA system not only drives the redox-regulated import of various intermembrane space proteins but also can import its own precursor proteins. The precursor of Erv1 uses the MIA pathway. For Mia40, two different scenarios have been observed. In lower eukaryotes, Mia40 is a large protein synthesized with a cleavable presequence that directs import and lateral sorting via the TIM23 complex (Chacinska et al., 2004; Naoé et al., 2004). In higher eukaryotes, Mia40 is small, consisting only of the conserved carboxy-terminal domain that contains the active cysteines, and is synthesized without a presequence (Hofmann et al., 2005; Grumbt et al., 2007; Chacinska et al., 2008; Banci et al., 2009). Like the typical MIA substrates, the small Mia40 protein is indeed imported via the MIA pathway (Chacinska et al., 2008). There are no obvious homologs of Mia40 in prokaryotes. We speculate that the first Mia40 molecules were imported by the classical presequence pathway (as is still the case in fungi). Then, after establishing an efficient MIA system, Mia40 itself could be imported via this pathway and the presequence could be dispensed with, as is seen in higher eukaryotes.
Disulfide Bond Formation in Different Compartments
The formation of disulfide bonds in newly synthesized proteins not only occurs in the mitochondrial intermembrane space, but also in the endoplasmic reticulum (ER) and the bacterial periplasm, which contain well characterized disulfide relay systems (Figure 4C) (Kadokura et al., 2004; Sevier et al., 2007). The basic principle involving a disulfide generating enzyme, a disulfide carrier, and substrate proteins is similar in all three systems. In each case, it is critical to have an electron-accepting system such as the respiratory chain and O2. Although Erv1 does not show homology to the main disulfide-generating enzymes of the ER (Ero1) and of bacteria (DsbB), a homolog exists in the ER, termed Erv2, which is able to oxidize protein disulfide isomerase (PDI) (Gerber et al., 2001; Gross et al., 2002). However, Mia40 does not show homology to the disulfide carriers of the ER (PDI) or bacteria (DsbA), and the current results suggest that important mechanistic differences exist. Mia40 forms stable intermediates with substrate proteins, and the ternary complex containing Erv1 permits the transfer of multiple disulfide bonds without intermittent release (Stojanovski et al., 2008). In the ER and in bacterial systems, the substrate intermediates formed with PDI and DsbA are typically short-lived (Kadokura et al. 2004). Although the environment in the mitochondrial intermembrane space is more oxidizing than that in the cytosol, it is significantly more reducing than the environment in the ER (Hu et al., 2008). The ternary complex of substrate-Mia40-Erv1 may prevent premature release of partially folded precursor intermediates that are prone to misfolding, thus ensuring that only the stably folded, fully oxidized forms are released.
Multiple Pathways to the Outer Membrane
The mitochondrial outer membrane contains two major classes of membrane proteins: β-barrel proteins that are inserted into the lipid phase by multiple transmembrane β strands, and α-helical proteins that are anchored in the membrane by one or more hydrophobic α-helical segments. All outer membrane proteins are synthesized in the cytosol and thus have to be imported into mitochondria.
The SAM Machinery for β-Barrel Sorting
The major representatives of mitochondrial β-barrel proteins are the abundant porin (VDAC) and the essential core of the TOM complex, Tom40. These two classes of proteins, although functionally distinct, have sequence similarities that suggest they were derived from a common ancestral mitochondrial porin (Pusnik et al., 2009). The precursors of β-barrel proteins are initially targeted to the TOM complex and are translocated through the Tom40 channel into the intermembrane space (Figure 5A). Thus, the precursor of Tom40 requires a pre-existing mature Tom40 for its entry into mitochondria. In the intermembrane space the Tim9–Tim10 and Tim8–Tim13 chaperone complexes bind to the precursors (Hoppins and Nargang, 2004; Wiedemann et al., 2004) and guide them to the sorting and assembly machinery for mitochondrial outer membrane proteins called the SAM complex (Paschen et al., 2003; Wiedemann et al., 2003). The β-barrel precursors are inserted into a hydrophilic environment within the SAM complex and are subsequently released into the lipid phase (Paschen et al., 2003; Wiedemann et al., 2003; Gentle et al., 2004; Kutik et al., 2008b). Porin molecules then associate to form homo-oligomers. Tom40 sequentially assembles with the other Tom proteins to form the mature TOM complex; the assembly starts with the smallest Tom protein, Tom5, followed by further small Tom proteins and the Tom receptors.
Figure 5. Protein Insertion into the Mitochondrial Outer Membrane.
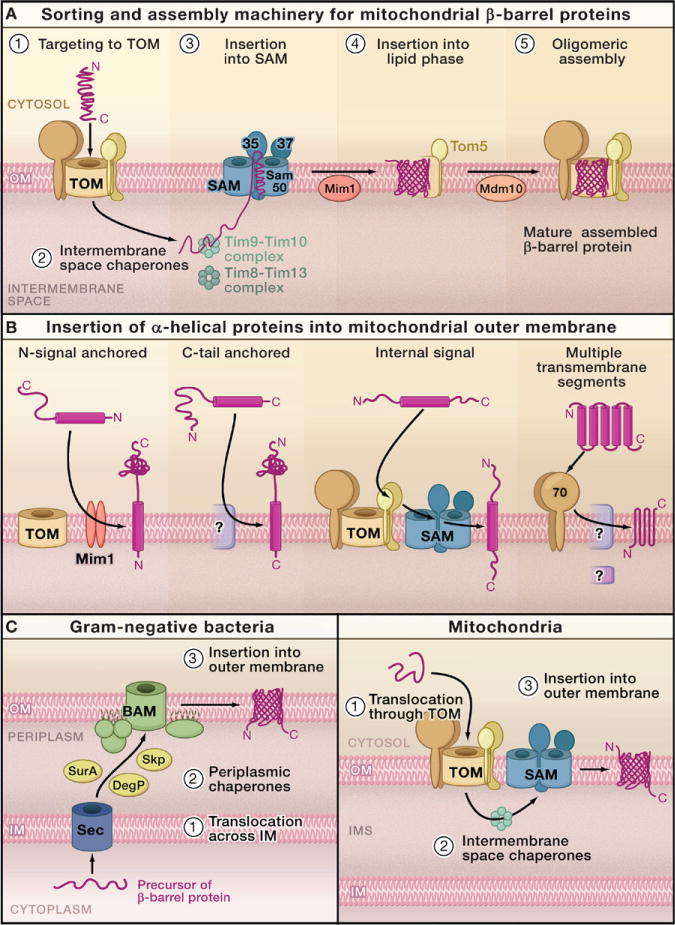
(A) Sorting of β-barrel proteins. The precursors of β-barrel proteins are initially transported via the translocase of the outer membrane (TOM complex) into the intermembrane space (IMS). IMS chaperone complexes transfer the precursors to the sorting and assembly machinery (SAM complex). The two essential subunits, Sam50 and Sam35, cooperate in insertion of the precursors into the outer membrane. Steps 1–3 show the general pathway for sorting of β-barrel proteins. Steps 4 and 5 depict specific additional steps of the assembly pathway of the β-barrel protein, Tom40. Mim1 supports integration of the Tom40 precursor into the outer membrane; Tom40 then associates with Tom5 and a further Tom40 molecule, followed by Mdm10-supported assembly of the oligomeric TOM complex.
(B) Insertion of α-helical proteins. Mitochondrial outer membrane proteins with α-helical transmembrane segments that typically function as sorting signals are imported by different pathways, depending on the location of the sorting signal. For different precursor proteins, a dependence on Mim1, Tom70, the TOM complex and/or SAM has been reported, but the mechanisms of membrane integration have not yet been defined.
(C) Evolutionary conservation of β-barrel sorting from Gram-negative bacteria to mitochondria. Bacteria synthesize the precursor proteins in the cytoplasm. The precursors are translocated through the Sec machinery to the periplasm and guided by chaperones to the β-barrel assembly machinery (BAM) of the outer membrane. For mitochondrial β-barrel proteins, the site of synthesis and the initial translocation through the TOM complex are different from bacteria, yet the subsequent steps of chaperone-guided transfer through the IMS and insertion into the outer membrane by the SAM complex share characteristics with the bacterial system. Sam50 is homologous to the central BAM component BamA (Omp85/YaeT). However, differences are also apparent as the other SAM and BAM subunits, as well as the chaperones of the IMS and periplasm, are not homologous to each other, and the lipid composition of the mitochondrial outer membrane is different from that of the bacterial outer membrane.
For a long time, Tom40 was the only known outer membrane protein involved in mitochondrial biogenesis that was essential for cell viability. The identification of the SAM complex revealed two more essential proteins, Sam50 and Sam35, emphasizing the crucial role of SAM for mitochondrial biogenesis (Kozjak et al., 2003; Paschen et al., 2003; Gentle et al., 2004; Ishikawa et al., 2004). The β-barrel protein Sam50 represents the core of the SAM complex (Figure 5A). Biophysical assays show that Sam50 forms a channel and that the channel activity is sensitive to the internal targeting signal (β signal, Figure 2B) of the β-barrel precursor proteins (Kutik et al., 2008b). However, the specific response to the β signal cannot be mediated by Sam50 alone but depends on the second essential SAM protein, Sam35, which binds to the signal in a receptor-like manner. The topology of Sam35 has not been fully determined. One possibility is that Sam35 is embedded in a hydrophilic environment between Sam50 molecules and thus binds to the β signal arriving in the SAM complex (Kutik et al., 2008b). The third subunit, Sam37, is involved in the release of precursor proteins from the SAM complex (Chan and Lithgow, 2008). Different models have been discussed regarding how the precursor proteins can be laterally released from a β-barrel channel. (1) As discussed for Tom40, the numerous hydrogen bonds that stabilize a β-barrel membrane protein make a lateral exit for precursor proteins from a monomeric channel unlikely, although it has not yet been excluded that such a mechanism could exist. (2) The SAM channel may be formed by several Sam50 molecules, and release of precursor proteins involves movement of the Sam50 molecules. The proposed topology of Sam35 as a signal-recognizing subunit located between several Sam50 molecules perhaps best fits this second model. (3) A third possibility is that the Sam50 barrel acts as a scaffold during insertion of β-barrel proteins into the outer membrane at the protein-lipid interface. In vitro studies of bacterial β-barrel protein insertion have found “molten disc” intermediates, which have a partial secondary structure with the β strands sitting flat on the membrane surface (Tamm et al., 2004). Sam50 might increase the kinetics of strand insertion by providing some local distortions in the lipid phase of the membrane, assisting formation of the barrel intermediates. A further clarification of this issue will require a structural analysis of the SAM complex.
Several other outer membrane proteins may promote the assembly pathway of the TOM complex (Figure 5A). Mdm10, originally identified for its role in maintaining mitochondrial morphology, associates with a fraction of SAM complexes to form a large complex that promotes the sequential assembly of Tom40 with receptor proteins such as Tom22 (Meisinger et al. 2004). Mdm10 is present in two different protein complexes, the SAM complex and a MDM/mitochore complex that contains two additional proteins, Mdm12 and Mmm1 (Boldogh et al., 2003). Mutants of each of these proteins affect β-barrel assembly (Meisinger et al., 2007). Future studies should address whether these morphology proteins interact directly with precursor proteins or are involved in providing the proper membrane environment for β-barrel assembly. The outer membrane protein Mim1, which promotes the insertion of some α-helical proteins into the outer membrane (Figure 5B), was also found to promote the assembly of β-barrel proteins (Ishikawa et al., 2004; Popov-Celeketic et al., 2008b). Mim1 associates transiently with the SAM complex and may modulate SAM function (Becker et al., 2008).
Pathways for α-Helical Proteins
The studies reported so far suggest a remarkable variety of mechanisms for the insertion of α-helical proteins into the outer membrane of mitochondria (Figure 5B). (1) Mim1 promotes the insertion of several so-called signal-anchored proteins (Becker et al., 2008; Hulett et al., 2008; Popov-Celeketic et al., 2008b). These proteins contain a single transmembrane segment at the amino terminus that functions as both sorting signal and membrane anchor. In the majority of cases, the insertion of signal-anchored proteins does not seem to require the TOM complex, though for some precursors an involvement of the core of the TOM complex (but not of Tom receptors) has been reported (Ahting et al., 2005). (2) For tail-anchored proteins, which contain a single transmembrane segment and sorting signal at the carboxy terminus, an insertase has not yet been defined. The available results suggest that neither the TOM complex nor Mim1 are required, but it does seem clear that the lipid composition of the membrane is important (Setoguchi et al., 2006; Kemper et al., 2008). (3) The precursor of the central receptor, Tom22, surprisingly requires both the TOM complex and SAM complex for its biogenesis. Tom22 contains several internal sequences required for its import, a hydrophobic segment in the middle of the precursor, and hydrophilic elements. Tom receptors are required for the recognition of the Tom22 precursor, and the SAM complex promotes the integration of the precursor into the outer membrane (Stojanovski et al., 2007). Thus, the function of the SAM complex is not restricted to the membrane insertion of β-barrel proteins but also includes some α-helical protein substrates. (4) Another variation of outer membrane insertion was identified for proteins with several transmembrane α-helical segments. These precursors require the receptor Tom70 but none of the other Tom proteins (Otera et al., 2007). Tom70 probably functions in a chaperone-like manner to transfer the precursors into the outer membrane. It remains to be shown whether insertion uses the protein-lipid interface or is mediated by an unidentified insertase. In addition, factors of the intermembrane space are required for the insertion of proteins that contain several transmembrane segments (Otera et al., 2007). The intermembrane space chaperones that guide the precursors of carrier proteins and β-barrel proteins are of course interesting candidates for this task.
We have only limited information on the components and mechanisms that direct α-helical proteins into the mitochondrial outer membrane. Given that many proteins with important functions in mitochondrial fusion and fission and programmed cell death are α-helical proteins, the analysis of their biogenesis will attract a lot of attention in the future.
Evolutionary Conservation of β-Barrel Protein Sorting
The principle of β-barrel protein assembly has been inherited from the bacterial ancestors of mitochondria. Sam50 is homologous to BamA, also termed Omp85 or YaeT, which forms the core of the β-barrel assembly machinery of the bacterial outer membrane (Kozjak et al., 2003; Paschen et al., 2003; Voulhoux et al., 2003; Gentle et al., 2004; Kim et al., 2007). In both bacteria and mitochondria, the β-barrel precursors are delivered to the BAM or SAM complex by chaperones in the topologically equivalent periplasm or intermembrane space, respectively (Figure 5C). What is clearly different in mitochondrial and bacterial systems is the site of synthesis of the precursors. Bacterial β-barrel proteins are synthesized within the cytoplasm and have to be exported by the Sec machinery to reach the periplasm. To reach the evolutionarily conserved insertion pathway from the intermembrane space side, mitochondrial β-barrel precursors first have to be imported via the TOM complex (Figure 5C). Walther et al. (2009) reported recently that bacterial proteins expressed in yeast were imported into mitochondria using the SAM pathway, thus demonstrating that the pathway of membrane insertion has been conserved.
The chloroplast protein import channel Toc75 is also homologous to BamA/Omp85 (Schleiff and Soll, 2005). Toc75 serves as a channel to import precursor proteins from the cytosol, thus providing evidence that this family of transporters can translocate precursors into a double-membraned structure. In early eukaryotic evolution, Sam50/BamA was already present as a β-barrel channel, whereas none of the Tom proteins show homology to bacterial proteins. Hence, we speculate that Sam50 may have been used as an early entry site for mitochondria until the Tom40 translocation channel was established as the main import site.
Conclusions and Perspectives
The first mitochondrial import components, belonging to the presequence pathway, were identified about 20 years ago. Today, four pathways have been characterized in detail, and it is evident that the hunt for new mitochondrial import components and pathways is not yet complete. Although the four defined import pathways all use the TOM complex (Figure 1B), the biogenesis of outer membrane proteins indicates that further pathways exist. These pathways either bypass the TOM complex completely, or do not use the Tom40 channel but rather use the protein-lipid interface at the TOM complex boundary. For preprotein import into the inner membrane, only the main classes of precursor proteins have been studied in detail: the presequence-carrying proteins and the noncleavable multispanning proteins like the metabolite carriers. It will be interesting to see whether smaller inner membrane proteins that contain only a single transmembrane segment will simply use the known TIM23 or TIM22 machineries, or whether new insertases in the inner membrane await discovery. Moreover, the export machinery for those few crucial proteins encoded by the mitochondrial genome shows a higher complexity than anticipated. In addition to the main component Oxa1, which is homologous to YidC of bacteria and Alb3 of chloroplasts, several more factors have now been identified (Table 1) (Preuss et al., 2005; Fiumera et al., 2007). The exact molecular functions of these export components are the subject of ongoing research.
High-resolution structures of a number of components of the import machinery have been solved, including receptors, chaperones, and the processing peptidase, providing important insights into the mechanisms of preprotein recognition, transfer, and processing. A major challenge, however, remains solving the structures of the membrane-embedded translocase complexes. Low-resolution electron microscopy analysis of purified translocases has provided first glimpses of the organization of the translocases and the presence of channels, but has not yet allowed functional conclusions to be drawn regarding the mechanism of preprotein translocation (Ahting et al., 2001; Paschen et al., 2003; Rehling et al., 2003; Model et al., 2008).
A field that has been largely neglected is the crucial role of lipids in protein insertion. The proper lipid environment seems to be important for the function of translocase complexes (van der Laan et al., 2007) and the insertion of precursor proteins (Kemper et al., 2008). The dimeric phospholipid cardiolipin, which is mainly present in the inner mitochondrial membrane, is of critical importance for the organization and function of many protein complexes in the membrane, including the presequence translocase (van der Laan et al., 2007; Claypool et al., 2008; Kutik et al., 2008a). Another intriguing question is the connection between the protein sorting machinery of the outer membrane and the machinery that maintains mitochondrial morphology as both SAM and MDM components influence protein biogenesis and mitochondrial morphology (Meisinger et al., 2004, 2007). Relevant to both the role of lipids and the regulation of mitochondrial membrane morphology is the connection between mitochondria and the ER, as components of the mitochondrial fusion machinery are involved in forming proteinaceous ER-mitochondria contact sites (de Brito and Scorrano, 2008). Such contact sites are involved in Ca2+ signaling but would also provide a means for lipid transfer between the two membrane systems. It is possible that so-called “morphology components” have several functions. They may provide structural elements for the organization of the membranes and also play roles in protein assembly and lipid biogenesis. A recent study showed that several morphology (MDM) proteins are involved in regulating mitochondrial phospholipids, although it is not yet known whether these proteins play a direct or indirect role in lipid metabolism (Osman et al., 2009). A close functional connection between protein biogenesis and lipid biosynthesis was demonstrated for the inner membrane protein Tam41; identified because of its influence on the organization of the TIM23-PAM machinery, Tam41 was subsequently shown to be required for the biosynthesis of cardiolipin (Kutik et al., 2008a). Elucidation of the functional network and interplay of components involved in protein and lipid biogenesis, mitochondrial morphology, and mitochondrial-ER connections will represent a major focus of future research.
Most studies on mitochondrial protein import have been performed in fungal model organisms. The great genetic and biochemical tractability of baker’s yeast has made it an ideal subject to define the components and molecular mechanisms of protein biogenesis. Studies in higher eukaryotes, particularly mammalian systems (Ryan and Hoogenraad, 2007), will be of enormous importance to address the unexplored field of how mitochondrial biogenesis is regulated. Proteomics studies have revealed numerous protein kinases and protein phosphatases, phosphorylated proteins, and GTP-binding proteins of mitochondria, with the functional importance unknown in most cases (Sickmann et al., 2003; Pagliarini et al., 2008). Similarly, the characterization of import machinery components that are involved in human mitochondrial diseases will attract increasing attention. Studies in yeast will be helpful in elucidating the molecular functions of human disease-related components. Typically, proteins that are defective in a human disease are not essential for cell viability in yeast; good examples include Tim8, which is involved in the deafness dystonia syndrome (Roesch et al., 2002), and a subunit of the m-AAA protease that is affected in hereditary spastic paraplegia where the proteolytic processing of a mitochondrial ribosomal protein is impaired (Nolden et al., 2005). Finally, the different views of the role of mitochondrial preprotein translocases in the targeting and assembly of proapoptotic and antiapoptotic factors in the outer membrane (Setoguchi et al., 2006) will stimulate further mechanistic analysis of the role of mitochondria in programmed cell death.
Acknowledgments
The authors are supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746, Excellence Initiative of the German Federal and State Governments (EXC 294), Gottfried Wilhelm Leibniz Program, and Fonds der Chemischen Industrie.
References
- Ahting U, Thieffry M, Engelhardt H, Hegerl R, Neupert W, Nussberger S. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J Cell Biol. 2001;153:1151–1160. doi: 10.1083/jcb.153.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahting U, Waizenegger T, Neupert W, Rapaport D. Signal-anchored proteins follow a unique insertion pathway into the outer membrane of mitochondria. J Biol Chem. 2005;280:48–53. doi: 10.1074/jbc.M410905200. [DOI] [PubMed] [Google Scholar]
- Alder NN, Jensen RE, Johnson AE. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell. 2008;134:439–450. doi: 10.1016/j.cell.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cefaro C, Ciofi-Baffoni S, Gallo A, Martinelli M, Sideris DP, Katrakili N, Tokatlidis K. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat Struct Mol Biol. 2009;16:198–206. doi: 10.1038/nsmb.1553. [DOI] [PubMed] [Google Scholar]
- Becker T, Pfannschmidt S, Guiard B, Stojanovski D, Milenkovic D, Kutik S, Pfanner N, Meisinger C, Wiedemann N. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J Biol Chem. 2008;283:120–127. doi: 10.1074/jbc.M706997200. [DOI] [PubMed] [Google Scholar]
- Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J Biol Chem. 2003;278:8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol. 2007;179:389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh IR, Nowakowski DW, Yang HC, Chung H, Karmon S, Royes P, Pon LA. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell. 2003;14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner K, Rehling P, Truscott KN. The carboxyl-terminal third of the dicarboxylate carrier is crucial for productive association with the inner membrane twin-pore translocase. J Biol Chem. 2005;280:6215–6221. doi: 10.1074/jbc.M412269200. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuán Szklarz LK, Schulze-Specking A, Truscott KN, Guiard B, Meisinger C, Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, et al. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Guiard B, Müller JM, Schulze-Specking A, Gabriel K, Kutik S, Pfanner N. Mitochondrial biogenesis, switching the sorting pathway of the intermembrane space receptor Mia40. J Biol Chem. 2008;283:29723–29729. doi: 10.1074/jbc.M805356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Lithgow T. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol Biol Cell. 2008;19:126–136. doi: 10.1091/mbc.E07-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauwin JF, Oster G, Glick BS. Strong precursor-pore interactions constrain models for mitochondrial protein import. Biophys J. 1998;74:1732–1743. doi: 10.1016/S0006-3495(98)77884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol. 2008;182:937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Leuenberger D, Oppliger W, Koehler CM. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 2002a;21:942–953. doi: 10.1093/emboj/21.5.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Leuenberger D, Schmidt E, Koehler CM. The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J Cell Biol. 2002b;158:1017–1027. doi: 10.1083/jcb.200205124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Leuenberger D, Leverich EP, Hwang DK, Beverly KN, Koehler CM. The role of Hot13p and redox chemistry in the mitochondrial TIM22 import pathway. J Biol Chem. 2004;279:43744–43751. doi: 10.1074/jbc.M404878200. [DOI] [PubMed] [Google Scholar]
- Dabir DV, Leverich EP, Kim SK, Tsai FD, Hirasawa M, Knaff DB, Koehler CM. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 2007;26:4801–4811. doi: 10.1038/sj.emboj.7601909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Alder NN, Jensen RE, Johnson AE. The Tim9p/10p and Tim8p/13p complexes bind to specific sites on Tim23p during mitochondrial protein import. Mol Biol Cell. 2007;18:475–486. doi: 10.1091/mbc.E06-06-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- Dienhart MK, Stuart RA. The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol Biol Cell. 2008;19:3934–3943. doi: 10.1091/mbc.E08-04-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmeier K, Hönlinger A, Bömer U, Dekker PJT, Eckerskorn C, Lottspeich F, Kübrich M, Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- D’Silva PR, Schilke B, Hayashi M, Craig EA. Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane. Mol Biol Cell. 2008;19:424–432. doi: 10.1091/mbc.E07-08-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Koppen M, Wagener J, Zick M, Israel L, Bernacchia A, Jagasia R, Rugarli EI, Imhof A, Neupert W, et al. OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA protease in mitochondria. Mol Biol Cell. 2007;18:3582–3590. doi: 10.1091/mbc.E07-02-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki M, Kanamori T, Nishikawa S, Shin I, Schultz PG, Endo T. Tom40 protein import channel binds to non-native proteins and prevents their aggregation. Nat Struct Biol. 2003;10:988–994. doi: 10.1038/nsb1008. [DOI] [PubMed] [Google Scholar]
- Esaki M, Shimizu H, Ono T, Yamamoto H, Kanamori T, Nishikawa S, Endo T. Mitochondrial protein import: requirement of presequence elements and TOM components for precursor binding to the TOM complex. J Biol Chem. 2004;279:45701–45707. doi: 10.1074/jbc.M404591200. [DOI] [PubMed] [Google Scholar]
- Farrell SR, Thorpe C. Augmenter of liver regeneration: a flavin-dependent sulfhydryl oxidase with cytochrome c reductase activity. Biochemistry. 2005;44:1532–1541. doi: 10.1021/bi0479555. [DOI] [PubMed] [Google Scholar]
- Fiumera HL, Broadley SA, Fox TD. Translocation of mitochondrially synthesized Cox2 domains from the matrix to the intermembrane space. Mol Cell Biol. 2007;27:4664–4673. doi: 10.1128/MCB.01955-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel K, Buchanan SK, Lithgow T. The alpha and the beta: protein translocation across mitochondrial and plastid outer membranes. Trends Biochem Sci. 2001;26:36–40. doi: 10.1016/s0968-0004(00)01684-4. [DOI] [PubMed] [Google Scholar]
- Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J, Mühlenhoff U, Hofhaus G, Lill R, Lisowsky T. Yeast Erv2p is the first microsomal FAD-linked sulfhydryl oxidase of the Erv1p/Alrp protein family. J Biol Chem. 2001;276:23486–23491. doi: 10.1074/jbc.M100134200. [DOI] [PubMed] [Google Scholar]
- Glick BS, Brandt A, Cunningham K, Müller S, Hallberg RL, Schatz G. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- Gross E, Sevier CS, Vala A, Kaiser CA, Fass D. A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p. Nat Struct Biol. 2002;9:61–67. doi: 10.1038/nsb740. [DOI] [PubMed] [Google Scholar]
- Grumbt B, Stroobant V, Terziyska N, Israel L, Hell K. Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J Biol Chem. 2007;282:37461–37470. doi: 10.1074/jbc.M707439200. [DOI] [PubMed] [Google Scholar]
- Hell K, Herrmann JM, Pratje E, Neupert W, Stuart RA. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc Natl Acad Sci USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Rothbauer U, Mühlenbein N, Baiker K, Hell K, Bauer MF. Functional and mutational characterization of human MIA40 acting during import into the mitochondrial intermembrane space. J Mol Biol. 2005;353:517–528. doi: 10.1016/j.jmb.2005.08.064. [DOI] [PubMed] [Google Scholar]
- Hoppins SC, Nargang FE. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J Biol Chem. 2004;279:12396–12405. doi: 10.1074/jbc.M313037200. [DOI] [PubMed] [Google Scholar]
- Hulett JM, Lueder F, Chan NC, Perry AJ, Wolynec P, Likic VA, Gooley PR, Lithgow T. The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J Mol Biol. 2008;376:694–704. doi: 10.1016/j.jmb.2007.12.021. [DOI] [PubMed] [Google Scholar]
- Hu J, Dong L, Outten CE. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J Biol Chem. 2008;283:29126–29134. doi: 10.1074/jbc.M803028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutu DP, Guiard B, Chacinska A, Becker D, Pfanner N, Rehling P, van der Laan M. Mitochondrial protein import motor: differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase. Mol Biol Cell. 2008;19:2642–2649. doi: 10.1091/mbc.E07-12-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J Cell Biol. 2004;166:621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan PS, Esser K, Pratje E, Michaelis G. Som1, a third component of the yeast mitochondrial inner membrane peptidase complex that contains Imp1 and Imp2. Mol Gen Genet. 2000;263:483–491. doi: 10.1007/s004380051192. [DOI] [PubMed] [Google Scholar]
- Kadokura H, Tian H, Zander T, Bardwell JC, Beckwith J. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science. 2004;303:534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- Kanamori T, Nishikawa S, Nakai M, Shin I, Schultz PG, Endo T. Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc Natl Acad Sci USA. 1999;96:3634–3639. doi: 10.1073/pnas.96.7.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniely S, Regev-Rudzki N, Pines O. The presequence of fumarase is exposed to the cytosol during import into mitochondria. J Mol Biol. 2006;358:396–405. doi: 10.1016/j.jmb.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Kemper C, Habib SJ, Engl G, Heckmeyer P, Dimmer KS, Rapaport D. Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J Cell Sci. 2008;121:1990–1998. doi: 10.1242/jcs.024034. [DOI] [PubMed] [Google Scholar]
- Kerscher O, Holder J, Srinivasan M, Leung RS, Jensen RE. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J Cell Biol. 1997;139:1663–1675. doi: 10.1083/jcb.139.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M, Keil P, Schneider H, van der Klei IJ, Pfanner N, Neupert W. The mitochondrial receptor complex: a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- Komiya T, Rospert S, Koehler C, Looser R, Schatz G, Mihara K. Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the ‘acid chain’ hypothesis. EMBO J. 1998;17:3886–3898. doi: 10.1093/emboj/17.14.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozany C, Mokranjac D, Sichting M, Neupert W, Hell K. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat Struct Mol Biol. 2004;11:234–241. doi: 10.1038/nsmb734. [DOI] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- Krayl M, Lim JH, Martin F, Guiard B, Voos W. A cooperative action of the ATP-dependent import motor complex and the inner membrane potential drives mitochondrial preprotein import. Mol Cell Biol. 2007;27:411–425. doi: 10.1128/MCB.01391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S, Rissler M, Guan XL, Guiard B, Shui G, Gebert N, Heacock PN, Rehling P, Dowhan W, Wenk MR, et al. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J Cell Biol. 2008a;183:1213–1221. doi: 10.1083/jcb.200806048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Krüger V, Prinz C, Meisinger C, Guiard B, Wagner R, et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell. 2008b;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- Kurz M, Martin H, Rassow J, Pfanner N, Ryan MT. Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol Biol Cell. 1999;10:2461–2474. doi: 10.1091/mbc.10.7.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dudek J, Guiard B, Pfanner N, Rehling P, Voos W. The presequence translocase-associated protein import motor of mitochondria: Pam16 functions in an antagonistic manner to Pam18. J Biol Chem. 2004;279:38047–38054. doi: 10.1074/jbc.M404319200. [DOI] [PubMed] [Google Scholar]
- Lill R, Mühlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- Liu Q, D’Silva P, Walter W, Marszalek J, Craig EA. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science. 2003;300:139–141. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
- Lu H, Allen S, Wardleworth L, Savory P, Tokatlidis K. Functional TIM10 chaperone assembly is redox-regulated in vivo. J Biol Chem. 2004;279:18952–18958. doi: 10.1074/jbc.M313045200. [DOI] [PubMed] [Google Scholar]
- Martinez-Caballero S, Grigoriev SM, Herrmann JM, Campo ML, Kinally KW. Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J Biol Chem. 2007;282:3584–3593. doi: 10.1074/jbc.M607551200. [DOI] [PubMed] [Google Scholar]
- Meinecke M, Wagner R, Kovermann P, Guiard B, Mick DU, Hutu DP, Voos W, Truscott KN, Chacinska A, Pfanner N, et al. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science. 2006;312:1523–1526. doi: 10.1126/science.1127628. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Rissler M, Chacinska A, Sanjuán Szklarz LK, Milenkovic D, Kozjak V, Schönfisch B, Lohaus C, Meyer HE, Yaffe MP, et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfannschmidt S, Rissler M, Milenkovic D, Becker T, Stojanovski D, Youngman MJ, Jensen RE, Chacinska A, Guiard B, et al. The morphology proteins Mdm12/Mmm1 function in the major β-barrel assembly pathway of mitochondria. EMBO J. 2007;26:2229–2239. doi: 10.1038/sj.emboj.7601673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Mesecke N, Bihlmaier K, Grumbt B, Longen S, Terziyska N, Hell K, Herrmann JM. The zinc-binding protein Hot13 promotes oxidation of the mitochondrial import receptor Mia40. EMBO Rep. 2008;9:1107–1113. doi: 10.1038/embor.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D, Gabriel K, Guiard B, Schulze-Specking A, Pfanner N, Chacinska A. Biogenesis of the essential Tim9–Tim10 chaperone complex of mitochondria: site-specific recognition of cysteine residues by the intermembrane space receptor Mia40. J Biol Chem. 2007;282:22472–22480. doi: 10.1074/jbc.M703294200. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Ramming T, Müller JM, Wenz LS, Gebert N, Schulze-Specking A, Stojanovski D, Rospert S, Chacinska A. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol Biol Cell. 2009;20:2530–2539. doi: 10.1091/mbc.E08-11-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg P, Nilsson S, Stahl A, Eriksson AC, Glaser E, Mäler L. NMR solution structure of the mitochondrial F1β presequence from Nicotiana plumbaginifolia. J Mol Biol. 2004;336:1129–1140. doi: 10.1016/j.jmb.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Model K, Meisinger C, Prinz T, Wiedemann N, Truscott KN, Pfanner N, Ryan MT. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat Struct Biol. 2001;8:361–370. doi: 10.1038/86253. [DOI] [PubMed] [Google Scholar]
- Model K, Meisinger C, Kühlbrandt W. Cryo-electron microscopy structure of a yeast mitochondrial preprotein translocase. J Mol Biol. 2008;383:1049–1057. doi: 10.1016/j.jmb.2008.07.087. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Popov-Celeketic D, Hell K, Neupert W. Role of Tim21 in mitochondrial translocation contact sites. J Biol Chem. 2005;280:23437–23440. doi: 10.1074/jbc.C500135200. [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Bourenkov G, Hell K, Neupert W, Groll M. Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J. 2006;25:4675–4685. doi: 10.1038/sj.emboj.7601334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Popov-Celeketic D, Mapa K, Gevorkyan-Airapetov L, Zohary K, Hell K, Azem A, Neupert W. Role of Tim50 in the transfer of precursor proteins from the outer to inner membrane of mitochondria. Mol Biol Cell. 2009;20:1400–1407. doi: 10.1091/mbc.E08-09-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan B, Ang SK, Yan G, Lu H. Zinc can play chaperone-like and inhibitor roles during import of mitochondrial small Tim proteins. J Biol Chem. 2009;284:6818–6825. doi: 10.1074/jbc.M808691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller JM, Milenkovic D, Guiard B, Pfanner N, Chacinska A. Precursor oxidation by Mia40 and Erv1 promotes vectorial transport of proteins into the mitochondrial intermembrane space. Mol Biol Cell. 2008;19:226–236. doi: 10.1091/mbc.E07-08-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoé M, Ohwa Y, Ishikawa D, Ohshima C, Nishikawa S, Yamamoto H, Endo T. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J Biol Chem. 2004;279:47815–47821. doi: 10.1074/jbc.M410272200. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Brinker A, Paschen SA, Moarefi I, Hayer-Hartl M, Neupert W, Brunner M. The protein import motor of mitochondria: a targeted molecular ratchet driving unfolding and translocation. EMBO J. 2002;21:3659–3671. doi: 10.1093/emboj/cdf358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Haag M, Potting C, Rodenfels J, Dip PV, Wieland FT, Brügger B, Westermann B, Langer T. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J Cell Biol. 2009;184:583–596. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Taira Y, Horie C, Suzuki Y, Suzuki H, Setoguchi K, Kato H, Oka T, Mihara K. A novel insertion pathway of mitochondrial outer membrane proteins with multiple transmembrane segments. J Cell Biol. 2007;179:1355–1363. doi: 10.1083/jcb.200702143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- Peixoto PM, Grana F, Roy TJ, Dunn CD, Flores M, Jensen RE, Campo ML. Awaking TIM22, a dynamic ligand-gated channel for protein insertion in the mitochondrial inner membrane. J Biol Chem. 2007;282:18694–18701. doi: 10.1074/jbc.M700775200. [DOI] [PubMed] [Google Scholar]
- Perocchi F, Jensen LJ, Gagneur J, Ahting U, von Mering C, Bork P, Prokisch H, Steinmetz LM. Assessing systems properties of yeast mitochondria through an interaction map of the organelle. PLoS Genet. 2006;2:e170. doi: 10.1371/journal.pgen.0020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov-Celeketic D, Mapa K, Neupert W, Mokranjac D. Active remodeling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 2008a;27:1469–1480. doi: 10.1038/emboj.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov-Celeketic J, Waizenegger T, Rapaport D. Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J Mol Biol. 2008b;376:671–680. doi: 10.1016/j.jmb.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Preuss M, Ott M, Funes S, Luirink J, Herrmann JM. Evolution of mitochondrial Oxa proteins from bacterial YidC: inherited and acquired functions of a conserved protein insertion machinery. J Biol Chem. 2005;280:13004–13011. doi: 10.1074/jbc.M414093200. [DOI] [PubMed] [Google Scholar]
- Pusnik M, Charrière F, Mäser P, Waller R, Dagley MJ, Lithgow T, Schneider A. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol Biol Evol. 2009;26:671–680. doi: 10.1093/molbev/msn288. [DOI] [PubMed] [Google Scholar]
- Rainey RN, Glavin JD, Chen HW, French SW, Teitell MA, Koehler CM. A new function in translocation for the mitochondrial i-AAA protease Yme1: import of polynucleotide phosphorylase into the intermembrane space. Mol Cell Biol. 2006;26:8488–8497. doi: 10.1128/MCB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Model K, Brandner K, Kovermann P, Sickmann A, Meyer HE, Kühlbrandt W, Wagner R, Truscott KN, Pfanner N. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science. 2003;299:1747–1751. doi: 10.1126/science.1080945. [DOI] [PubMed] [Google Scholar]
- Roesch K, Curran SP, Tranebjaerg L, Koehler CM. Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum Mol Genet. 2002;11:477–486. doi: 10.1093/hmg/11.5.477. [DOI] [PubMed] [Google Scholar]
- Ryan MT, Hoogenraad NJ. Mitochondrial-nuclear communications. Annu Rev Biochem. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Igura M, Obita T, Ose T, Kojima R, Maenaka K, Endo T, Kohda D. Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J. 2007;26:4777–4787. doi: 10.1038/sj.emboj.7601888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E, Soll J. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 2005;6:1023–1027. doi: 10.1038/sj.embor.7400563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell DJ, Hebert DN. Protein translocons: multifunctional mediators of protein translocation across membranes. Cell. 2003;112:491–505. doi: 10.1016/s0092-8674(03)00110-7. [DOI] [PubMed] [Google Scholar]
- Setoguchi K, Otera H, Mihara K. Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. EMBO J. 2006;25:5635–5647. doi: 10.1038/sj.emboj.7601438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevier CS, Qu H, Heldman N, Gross E, Fass D, Kaiser CA. Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell. 2007;129:333–344. doi: 10.1016/j.cell.2007.02.039. [DOI] [PubMed] [Google Scholar]
- Shariff K, Ghosal S, Matouschek A. The force exerted by the membrane potential during protein import into the mitochondrial matrix. Biophys J. 2004;86:3647–3652. doi: 10.1529/biophysj.104.040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Shönfisch B, Perschil I, Chacinska A, Guiard B, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris DP, Tokatlidis K. Oxidative folding of small Tims is mediated by site-specific docking onto Mia40 in the mitochondrial intermembrane space. Mol Microbiol. 2007;65:1360–1373. doi: 10.1111/j.1365-2958.2007.05880.x. [DOI] [PubMed] [Google Scholar]
- Slutsky-Leiderman O, Marom M, Iosefson O, Levy R, Maoz S, Azem A. The interplay between components of the mitochondrial protein translocation motor studied using purified components. J Biol Chem. 2007;282:33935–33942. doi: 10.1074/jbc.M704435200. [DOI] [PubMed] [Google Scholar]
- Stojanovski D, Guiard B, Kozjak-Pavlovic V, Pfanner N, Meisinger C. Alternative function for the mitochondrial SAM complex in biogenesis of α-helical TOM proteins. J Cell Biol. 2007;179:881–893. doi: 10.1083/jcb.200706043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Milenkovic D, Müller JM, Gabriel K, Schulze-Specking A, Baker MJ, Ryan MT, Guiard B, Pfanner N, Chacinska A. Mitochondrial protein import: precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase. J Cell Biol. 2008;183:195–202. doi: 10.1083/jcb.200804095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm LK, Hong H, Liang B. Folding and assembly of β-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Shiota T, Yamano K, Watanabe K, Yokota M, Yamamoto H, Sesaki H, Endo T. Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J Cell Biol. 2009;184:129–141. doi: 10.1083/jcb.200808068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AB, Smith BS, Kitada S, Kojima K, Miyaura H, Otwinowski Z, Ito A, Deisenhofer J. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure. 2001;9:615–625. doi: 10.1016/s0969-2126(01)00621-9. [DOI] [PubMed] [Google Scholar]
- Terziyska N, Grumbt B, Kozany C, Hell K. Structural and functional roles of the conserved cysteine residues of the redox-regulated import receptor Mia40 in the intermembrane space of mitochondria. J Biol Chem. 2009;284:1353–1363. doi: 10.1074/jbc.M805035200. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr Biol. 2006;16:2271–2276. doi: 10.1016/j.cub.2006.10.025. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Meinecke M, Dudek J, Hutu DP, Lind M, Perschil I, Guiard B, Wagner R, Pfanner N, Rehling P. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat Cell Biol. 2007;9:1152–1159. doi: 10.1038/ncb1635. [DOI] [PubMed] [Google Scholar]
- van Wilpe S, Ryan MT, Hill K, Maarse AC, Meisinger C, Brix J, Dekker PJT, Moczko M, Wagner R, Meijer M, et al. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- Vasiljev A, Ahting U, Nargang FE, Go NE, Habib SJ, Kozany C, Panneels V, Sinning I, Prokisch H, Neupert W, et al. Reconstituted TOM core complex and Tim9/Tim10 complex of mitochondria are sufficient for translocation of the ADP/ATP carrier across membranes. Mol Biol Cell. 2004;15:1445–1458. doi: 10.1091/mbc.E03-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Wagner K, Gebert N, Guiard B, Brandner K, Truscott KN, Wiedemann N, Pfanner N, Rehling P. The assembly pathway of the mitochondrial carrier translocase involves four preprotein translocases. Mol Cell Biol. 2008;28:4251–4260. doi: 10.1128/MCB.02216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Papic D, Bos MP, Tommassen J, Rapaport D. Signals in bacterial β-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc Natl Acad Sci USA. 2009;106:2531–2536. doi: 10.1073/pnas.0807830106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CT, Gorman MA, Lazarou M, Ryan MT, Gulbis JM. Crystal structure of the mitochondrial chaperone TIM9•10 reveals a six-bladed α-propeller. Mol Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N, Ryan MT. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001;20:951–960. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Kozjak V, Chacinska A, Schönfisch B, Rospert S, Ryan MT, Pfanner N, Meisinger C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Truscott KN, Pfannschmidt S, Guiard B, Meisinger C, Pfanner N. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J Biol Chem. 2004;279:18188–18194. doi: 10.1074/jbc.M400050200. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, van der Laan M, Hutu DP, Rehling P, Pfanner N. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J Cell Biol. 2007;179:1115–1122. doi: 10.1083/jcb.200709087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox AJ, Choy J, Bustamante C, Matouschek A. Effect of protein structure on mitochondrial import. Proc Natl Acad Sci USA. 2005;102:15435–15440. doi: 10.1073/pnas.0507324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sha B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat Struct Mol Biol. 2006;13:589–593. doi: 10.1038/nsmb1106. [DOI] [PubMed] [Google Scholar]
- Yamano K, Kuroyanagi-Hasegawa M, Esaki M, Yokota M, Endo T. Step-size analyses of the mitochondrial Hsp70 import motor reveal the Brownian ratchet in operation. J Biol Chem. 2008a;283:27325–27332. doi: 10.1074/jbc.M805249200. [DOI] [PubMed] [Google Scholar]
- Yamano K, Yatsukawa Y, Esaki M, Hobbs AE, Jensen RE, Endo T. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J Biol Chem. 2008b;283:3799–3807. doi: 10.1074/jbc.M708339200. [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- Zara V, Ferramosca A, Robitaille-Foucher P, Palmieri F, Young JC. Mitochondrial carrier protein biogenesis: role of the chaperones Hsc70 and Hsp90. Biochem J. 2009;419:369–375. doi: 10.1042/BJ20082270. [DOI] [PMC free article] [PubMed] [Google Scholar]


